Abstract
β-Barrel proteins of the Omp85 (Outer membrane protein, 85 kD) superfamily exist in the outer membranes of Gram-negative bacteria, mitochondria, and chloroplasts. Prominent Omp85 proteins in bacteria and mitochondria mediate biogenesis of other β-barrel proteins and are indispensable for viability. In Arabidopsis (Arabidopsis thaliana) chloroplasts, there are two distinct types of Omp85-related protein: Toc75 (Translocon at the outer envelope membrane of chloroplasts, 75 kD) and OEP80 (Outer Envelope Protein, 80 kD). Toc75 functions as a preprotein translocation channel during chloroplast import, but the role of OEP80 remains elusive. We characterized three T-DNA mutants of the Arabidopsis OEP80 (AtOEP80) gene. Selectable markers associated with the oep80-1 and oep80-2 insertions segregated abnormally, suggesting embryo lethality of the homozygous genotypes. Indeed, no homozygotes were identified among >100 individuals, and heterozygotes of both mutants produced approximately 25% aborted seeds upon self-pollination. Embryo arrest occurred at a relatively late stage (globular embryo proper) as revealed by analysis using Nomarski optics microscopy. This is substantially later than arrest caused by loss of the principal Toc75 isoform, atToc75-III (two-cell stage), suggesting a more specialized role for AtOEP80. Surprisingly, the oep80-3 T-DNA (located in exon 1 between the first and second ATG codons of the open reading frame) did not cause any detectable developmental defects or affect the size of the AtOEP80 protein in chloroplasts. This indicates that the N-terminal region of AtOEP80 is not essential for the targeting, biogenesis, or functionality of the protein, in contrast with atToc75-III, which requires a bipartite targeting sequence.
Chloroplasts and mitochondria evolved from bacteria through endosymbiosis. Recently, a family of β-barrel proteins related to Omp85 (Outer membrane protein, 85 kD) from Neisseria meningitidis was found in the outer membranes of Gram-negative bacteria, mitochondria, and chloroplasts (Yen et al., 2002; Gentle et al., 2005). Some bacteria contain multiple Omp85-related proteins, with the different homologs playing distinct roles in protein secretion (Jacob-Dubuisson et al., 2004) or the sorting of β-barrel proteins to the outer membrane (Voulhoux et al., 2003; Wu et al., 2005). By contrast, there appears to be just a single functional Omp85 homolog in mitochondria (Sam50 [Sorting and assembly machinery, 50 kD]; alternatively, Tob55 [Topogenesis of β-barrel proteins, 55 kD]), and this mediates β-barrel insertion into the membrane (Kozjak et al., 2003; Paschen et al., 2003; Gentle et al., 2004), as well as the insertion of other outer membrane proteins (Stojanovski et al., 2007).
Unlike mitochondria, chloroplasts contain at least two distinct types of Omp85 homolog, namely, Toc75 (Translocon at the outer envelope membrane of chloroplasts, 75 kD) and OEP80 (Outer Envelope Protein, 80 kD). Because homologs exist in extant cyanobacteria (one of which was shown to be essential for viability), these proteins are postulated to be derived from a common ancestor in the original endosymbiont (Bölter et al., 1998; Reumann et al., 1999, 2005). Detailed phylogenetic analyses suggested that Toc75 and OEP80 diverged early in the evolution of chloroplasts (Inoue and Potter, 2004).
Pea (Pisum sativum) Toc75 (psToc75) is one of the most abundant proteins in the chloroplast outer envelope membrane (Cline et al., 1981). It associates with precursor proteins in vitro (Perry and Keegstra, 1994; Schnell et al., 1994), and was reconstituted as a cation-selective ion channel in artificial liposomes (Hinnah et al., 1997), suggesting that it forms a major component of the preprotein translocation channel (Bédard and Jarvis, 2005; Kessler and Schnell, 2006; Smith, 2006). Unlike other outer membrane proteins, Toc75 is synthesized as a larger precursor with a bipartite targeting signal (Tranel et al., 1995; Tranel and Keegstra, 1996); the first part is a standard transit peptide for chloroplast import (Inoue et al., 2001) and the second part acts as an intraorganellar targeting signal that is cleaved by an envelope-bound type I signal peptidase (Inoue and Keegstra, 2003; Inoue et al., 2005; Baldwin and Inoue, 2006).
Arabidopsis (Arabidopsis thaliana) possesses four genomic sequences with homology to psToc75 on chromosomes I, III, IV, and V; these are termed atTOC75-I, -III, -IV, and -V, respectively (Jackson-Constan and Keegstra, 2001; Eckart et al., 2002). Among them, atTOC75-I, -III, and -IV are highly homologous to one another and to the pea gene, with predicted amino acid sequence identities ranging from 60% to 75%. Previous work demonstrated that atTOC75-I is a pseudogene and that atTOC75-IV encodes a truncated protein that lacks a transit peptide and is expressed at very low levels (Baldwin et al., 2005). By contrast, atToc75-III is synthesized as a larger precursor with a bipartite transit peptide, like psToc75, and is highly expressed (Inoue and Keegstra, 2003; Baldwin et al., 2005). Disruption of atTOC75-III with T-DNA insertions arrested embryo development at a very early stage (the two-cell stage; Baldwin et al., 2005; Hust and Gutensohn, 2006). These results indicate that atToc75-III is the most direct functional ortholog of psToc75.
The fourth Omp85 homolog of Arabidopsis chloroplasts, atToc75-V, is more distantly related to psToc75, sharing only 22% identity (Eckart et al., 2002). A putative pea ortholog of atToc75-V was not associated with the protein translocation machinery, suggesting that its role is not directly related to chloroplast protein import (Eckart et al., 2002). The pea protein appeared to be smaller (apparent size approximately 66 kD) than the conceptual translation of atTOC75-V (approximately 80 kD), whereas the latter was predicted to carry a transit peptide. Thus, it was proposed that atToc75-V has an 11-kD targeting sequence that is removed to yield a mature protein of 69 kD (Eckart et al., 2002). However, subsequent work suggested that atToc75-V (a polypeptide of 732 residues, as encoded by the originally annotated open reading frame) was targeted to isolated chloroplasts without undergoing any change in size (Inoue and Potter, 2004). Additionally, the imported protein was similar in size to a native chloroplast protein recognized by an atToc75-V antibody (approximately 80 kD) and was significantly larger than atToc75-III (approximately 75 kD). Based on these observations, and on the absence of evidence supporting a role in protein translocation, the protein was renamed with the more general designation, AtOEP80 (Inoue and Potter, 2004).
The Toc75 and OEP80 subfamilies are both widely distributed in different plant species (Inoue and Potter, 2004). However, in contrast with Toc75, the function of OEP80 remains elusive. Here, we demonstrate that AtOEP80 is essential for viability and reveal that the N-terminal part of the protein (corresponding to the region between the first and second AUG codons of the annotated open reading frame) is not required for its biogenesis or function.
RESULTS AND DISCUSSION
The AtOEP80 Gene Is Expressed throughout Development
To gain initial insight into the in vivo role of AtOEP80, we examined its mRNA expression using publicly available microarray data and the Genevestigator V3 analysis tool (https://www.genevestigator.ethz.ch; Zimmermann et al., 2004; Grennan, 2006). For comparison, we also analyzed the other expressed Toc75-related Arabidopsis genes (atTOC75-III and atTOC75-IV), as well as the gene for a major component of the TIC (Translocon at the Inner envelope membrane of Chloroplasts) complex, atTIC110. A developmental time course revealed that all four genes are expressed throughout the life cycle (Supplemental Fig. S1A). The atTOC75-III and atTIC110 genes were expressed most strongly, whereas atTOC75-IV was expressed at very low levels. The AtOEP80 gene exhibited an intermediate expression level, equivalent to approximately 25% of that observed for atTOC75-III. Interestingly, developmental fluctuations in the expression of atTOC75-III and atTIC110 were paralleled in the AtOEP80 expression pattern, albeit with reduced amplitude, which is consistent with the hypothesis that the AtOEP80 function is related to plastid biogenesis (Inoue and Potter, 2004).
When expression of the four genes was considered on an anatomical basis (Supplemental Fig. S1B), a similar trend was observed: That is, in most cases, atTOC75-III and atTIC110 displayed the highest levels, AtOEP80 an intermediate level, and atTOC75-IV the lowest level. Interestingly, there were some notable exceptions to this rule, in pollen, mature embryos, and the endosperm. In embryos, AtOEP80 expression was approximately 40% higher than that of atTOC75-III. Given that atToc75-III, like atTic110, is essential during embryogenesis (Baldwin et al., 2005; Inaba et al., 2005; Kovacheva et al., 2005; Hust and Gutensohn, 2006), this observation suggests that AtOEP80 may also be important during seed development.
Inactivation of AtOEP80 Causes Seed Abortion
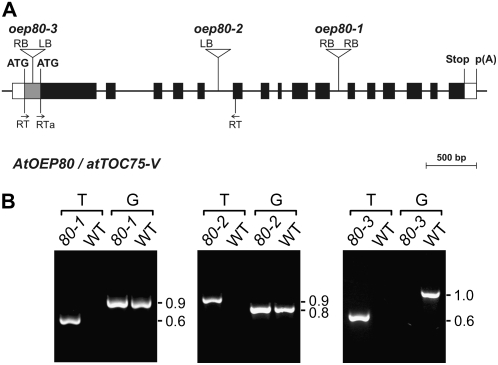
To assess the importance of OEP80 in plastids, we identified three different Arabidopsis lines with T-DNA insertions in the AtOEP80 gene (Fig. 1A). These mutants carry insertions in the tenth intron (oep80-1), the fifth intron (oep80-2), and the first exon (oep80-3) of the gene. With one exception, all T-DNA junction sequences were amplified and sequenced to obtain precise positional information (Fig. 1A); the 3′ side of the oep80-2 T-DNA insertion could not be amplified, presumably because it is incomplete and lacks a left border (LB) or a right border (RB) sequence. Interestingly, analysis of the T-DNA-associated selectable marker in segregating populations of oep80-1 and oep80-2 revealed significant deviations from standard Mendelian inheritance: Only two antibiotic-resistant plants were observed for every one antibiotic-sensitive plant (Supplemental Table S1). These data imply that the homozygous genotype is lethal in each case; they also indicate that both mutants carry just a single T-DNA insertion locus. Consistent with the first conclusion, when we analyzed 78 antibiotic-resistant oep80-1 plants and 41 antibiotic-resistant oep80-2 plants, by either progeny testing on selective medium or PCR analysis using gene- and T-DNA-specific primers, all 119 individuals were found to be heterozygous for the mutation. Examples of the PCR-based genotyping experiments we conducted are shown in Figure 1B: PCR reactions (T) utilizing one T-DNA border primer and one AtOEP80 primer gave a clear amplification product in both mutants, as did additional reactions (G) utilizing two AtOEP80 gene-specific primers flanking the insertion site.
Figure 1.
Basic characterization of the AtOEP80 T-DNA insertion mutants. A, Schematic showing the structure of the AtOEP80 gene and the location of each T-DNA insertion. Protein-coding exons are represented by black boxes and untranslated regions by white boxes; introns are represented by thin lines between the boxes. The gray area at the 5′ end of exon 1 represents a putative untranslated region, or alternatively encodes a nonessential cleavable peptide. Locations of primers used for RT-PCR analysis (RT; Fig. 4A) are shown. T-DNA insertion sites are indicated precisely, but the insertion sizes are not to scale. ATG, Potential translation initiation codons; Stop, translation termination codon; p(A), polyadenylation site. B, Analysis of mutant genotypes by PCR. Genomic DNA extracted from wild-type and mutant plants (oep80-1, 80-1; oep80-2, 80-2; and oep80-3, 80-3) was analyzed by PCR. Appropriate T-DNA- and AtOEP80-gene-specific primers were employed. Two different primer combinations were used: The first (T) comprised one T-DNA primer (RB for oep80-1; LB for oep80-2 and oep80-3) and one gene-specific primer (reverse in the case of oep80-1 and oep80-3; forward in the case of oep80-2); the second (G) comprised two gene-specific primers flanking the T-DNA insertion site. The results shown for oep80-1 and oep80-2 are representative of those obtained for all antibiotic-resistant plants tested; amplification using both T and G indicated the presence of both mutant and wild-type alleles, respectively, and demonstrated that the plants were heterozygous. Results shown for oep80-3 are representative of those obtained for all homozygotes tested; absence of amplification using the G primers indicated that the wild-type allele was not present. Sizes of the amplicons are indicated at right (in kb).
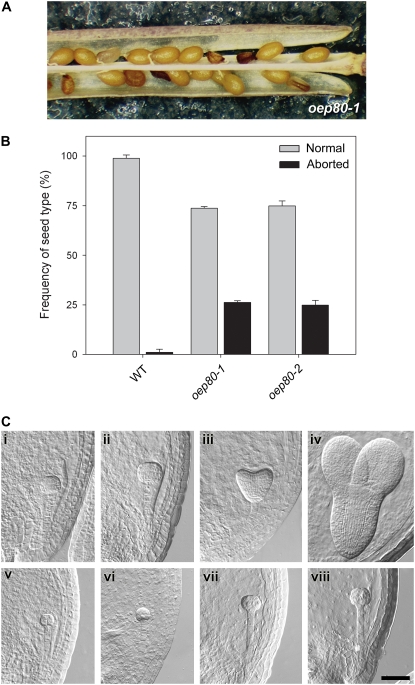
The absence of homozygous individuals from segregating populations suggested that the mutations might be lethal. To assess the possibility of embryo lethality, we inspected mature siliques of heterozygous oep80-1 and oep80-2 plants. In both cases, aborted seeds were observed and these occurred with a frequency of almost exactly 25% (Fig. 2, A and B), strongly supporting the notion that the homozygous genotypes were responsible for seed abortion. Identification of two independent mutant alleles that give rise to the same phenotype is widely accepted as proof of a causal relationship (Sjögren et al., 2004; Baldwin et al., 2005; Puyaubert et al., 2008). Thus, our data provide very strong evidence of an essential role for AtOEP80. Consistent with this conclusion, we observed that the transgenic overexpression of an AtOEP80 cDNA could efficiently complement the distorted segregation and seed abortion defects of oep80-1, enabling the identification of apparently normal homozygous mutants at Mendelian frequency (S.-C. Hsu, R. Patel, P. Jarvis, and K. Inoue, unpublished data).
Figure 2.
Embryo lethality of the oep80-1 and oep80-2 mutations. A, Appearance of aborted seeds within mature siliques of oep80-1 heterozygous plants. The aborted seeds are smaller in size than the normal seeds and have a darker, shriveled appearance. B, Frequency of aborted seeds within mature siliques of wild-type, oep80-1, and oep80-2 plants. The data shown are means (±sd) derived from analyses of three or four siliques from each of three to six independent plants per genotype. C, Analysis of embryo development in oep80-1 using Nomarski optics. Equivalent developmental series for normal (i–iv) and mutant (v–viii) embryos within immature oep80-1 heterozygous siliques. Normal embryos: i, early globular stage; ii, late globular stage; iii, heart stage; iv, torpedo stage. Corresponding mutant embryos from the same siliques: v, proembryo stage; vi, early globular stage; vii and viii, raspberry-like globular stage. Embryo cell stage names refer to the morphology of the embryo proper. All images are at the same magnification. Bar = 50 μm.
There are numerous precedents for embryo lethality caused by lesions in chloroplast proteins (Uwer et al., 1998; Apuya et al., 2001). In fact, in a large-scale screen for mutations affecting seed development, >25% of the identified loci were predicted to encode plastidic proteins (McElver et al., 2001). Most relevant among these previous studies are those pertaining to two major components of the chloroplast protein import machinery: atToc75-III (Baldwin et al., 2005; Hust and Gutensohn, 2006) and atTic110 (Inaba et al., 2005; Kovacheva et al., 2005). Interestingly, the aborted seeds in oep80 plants (Fig. 2A) appeared to be significantly larger than those observed previously in toc75-III mutants and were more similar in size to those in the tic110 mutant (Baldwin et al., 2005; Kovacheva et al., 2005). This suggested that the oep80 mutation, like tic110, may act at a later stage in embryogenesis than toc75-III.
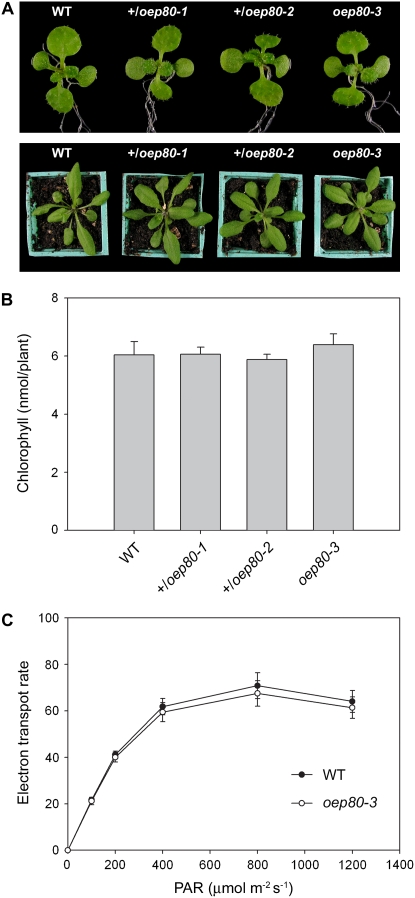
Whereas the toc75-III mutations appeared to be completely recessive (Baldwin et al., 2005), heterozygous tic110 plants were visibly pale and exhibited quantifiable defects in chloroplast biogenesis (Kovacheva et al., 2005). In this regard, the oep80 mutations are more similar to the toc75-III mutations, because heterozygotes could not be distinguished from the wild type, not only at a macroscopic level (Fig. 3A), but also in relation to chlorophyll content (Fig. 3B) and photosynthetic performance (Supplemental Table S2). This indicates that a single copy of the AtOEP80 gene is able to produce sufficient quantities of the protein for normal growth under standard, controlled conditions. The greater dosage dependency of tic110 may reflect a higher rate of turnover of the atTic110 protein or the absence of excess expression capacity for atTic110 in the wild type.
Figure 3.
Phenotypic analysis of the oep80 mutants. A, Plants of the indicated genotypes were grown on selective medium (except for the wild type) in vitro for 8 d, rescued to nonselective medium, and then photographed on day 14 (top). Additional similar plants were transferred to soil on day 14 and then allowed to grow for a further 10-d period prior to photography (bottom). Representative plants are shown in both cases. B, Chlorophyll concentrations in 14-d-old plants grown as described in A were determined photometrically. Values shown are means (±se) derived from 16 independent samples per genotype, each one containing six plants. Units are nmol chlorophyll a + b per plant. C, Analysis of photosynthesis in the oep80-3 mutant. Light response curves of photosynthetic electron transport rates in wild-type and mutant plants were determined by measuring chlorophyll fluorescence. Values were recorded at different irradiances of photosynthetically active radiation (PAR), ranging from 0 to 1,200 μmol photons m−2 s−1. Units for the data shown are μmol electrons m−2 s−1, assuming that 84% of the incident light is absorbed and that the transport of each electron utilizes two photons (Meyer et al., 1997; Aronsson et al., 2007). Measurements were done on fully grown leaves from 10 different 29-d-old plants per genotype grown under identical conditions. Values shown are means (±sd).
Homozygous oep80-1 Embryos Arrest at the Globular Stage
To determine more precisely the stage at which developmental arrest occurs, we conducted a thorough examination of developing embryos in wild-type and mutant plants using Nomarski optics. Figure 2C shows equivalent developmental series for normal (i–iv) and mutant (v–viii) embryos within immature siliques of self-pollinated oep80-1 heterozygotes. When normal embryos were at the globular stage (Fig. 2C, i and ii), mutant embryos (equivalent to 26% of the total number; Table I) were retarded at the proembryo stage (Fig. 2C, v and vi). As normal embryos progressed to the heart stage (Fig. 2C, iii), mutant embryos (equivalent to approximately 24% to 29% of the total number; Table I) developed to the globular stage (Fig. 2C, vii), but began to take on an abnormal, raspberry-like appearance, with protuberances on the surface of the embryo proper (Yadegari et al., 1994; Apuya et al., 2002). By the time the normal embryos had reached the torpedo stage (Fig. 2C, iv), the raspberry-like phenotype of the mutant embryos had become even more pronounced (Fig. 2C, viii). The mutant embryos (equivalent to approximately 22% of the total number; Table I) did not develop beyond the globular stage. In contrast with the situation in oep80-1 siliques, where two clear, distinct classes of embryos could be observed (normal and mutant), embryos within individual wild-type siliques rarely spanned more than two consecutive developmental stages (Table I).
Table I.
Distribution of embryo phenotypes in single siliques of oep80-1 heterozygotesa
| Genotype | Siliqueb | Proembryoc | Globular (Raspberry-Like) | Globular | Heart | Torpedo | N.D.d | Total Scored | Proportion Delayed or Abnormal |
|---|---|---|---|---|---|---|---|---|---|
| Wild type | 4 | 54 | – | – | – | – | 9 | 54 | – |
| 6 | 1 | – | 58 | – | – | 6 | 59 | – | |
| 8 | – | – | 14 | 46 | – | 1 | 60 | – | |
| 10 | – | – | 1 | 48 | 7 | 0 | 56 | – | |
| 12 | – | – | – | 9 | 48 | 10 | 57 | – | |
| oep80-1 | 4 | 54 | – | – | – | – | 6 | 54 | – |
| 6 | 17 | – | 49 | – | – | 1 | 66 | 0.26 | |
| 8 | – | 17 | 41 | 12 | – | 1 | 70 | 0.24 | |
| 10 | – | 16 | – | 39 | 1 | 1 | 56 | 0.29 | |
| 12 | – | 14 | – | 2 | 47 | 1 | 63 | 0.22 |
Embryo developmental stage names refer to the morphology of the embryo proper.
Siliques were numbered consecutively from the top of the inflorescence, such that the oldest siliques have the highest numbers.
Proembryo stage includes one-cell to 16-cell stage embryos.
Not determined; seeds that were not classified for technical reasons.
A very similar raspberry-like, globular-stage arrest phenotype was reported for the atTic110 knockout mutant, tic110 (Kovacheva et al., 2005). The protuberances on the surface of the embryo proper, responsible for the raspberry-like appearance of the oep80 and tic110 mutant embryos, are in fact a characteristic feature of many mutants that arrest during this stage of embryogenesis. It is thought that they are caused by cellular maturation processes that normally occur during late embryogenesis and continue to proceed in the mutants in spite of the block in embryo growth and morphogenesis (Yadegari et al., 1994; Apuya et al., 2002). Unlike oep80, the tic110 mutation was also associated with abnormal, asymmetric, or unsynchronized cell divisions at earlier developmental stages in some embryos (Kovacheva et al., 2005). Because no such early defects were observed in the AtOEP80 knockout, the oep80 mutant phenotype can be considered somewhat less severe and later acting than tic110. Nevertheless, the oep80-mediated block in growth occurs considerably earlier than the stage during which photosynthetic establishment normally commences (heart stage; Apuya et al., 2001), and so the data indicate that the role of AtOEP80 is not directly associated with photosynthesis.
The late-acting effect of oep80 contrasts with the much earlier defect reported for the atToc75-III knockout mutation, toc75-III (Baldwin et al., 2005). In toc75-III, embryo arrest was observed to occur when the embryo proper was composed of just two cells. Because the two genes are expressed at comparable levels in embryos (Supplemental Fig. S1B), this difference in phenotype severity between toc75-III and oep80 may reflect differing roles of the proteins. Whereas atToc75-III is believed to be the channel responsible for the import of most proteins (Bédard and Jarvis, 2005; Kessler and Schnell, 2006; Smith, 2006), AtOEP80 has been proposed to play a more specialized role in the biogenesis of outer envelope β-barrel proteins, like certain bacterial and mitochondrial Omp85 proteins (Inoue and Potter, 2004).
Homozygous oep80-3 Plants Are Phenotypically Normal
In contrast with the situation for oep80-1 and oep80-2, the selectable marker associated with the oep80-3 T-DNA segregated normally, exhibiting standard Mendelian inheritance: three antibiotic-resistant plants were observed for every one antibiotic-sensitive plant (Supplemental Table S1). This implies that the homozygous oep80-3 genotype is not lethal, which is surprising given the location of the T-DNA in the first exon (Fig. 1A). Families containing only antibiotic-resistant individuals were identified and these were confirmed as oep80-3 homozygotes by PCR analysis (Fig. 1B). Remarkably, homozygous oep80-3 mutants were indistinguishable from wild type. The mutant was of a similar size and color to wild type throughout development (Fig. 3A) and contained normal levels of chlorophyll (Fig. 3B). Chlorophyll fluorescence measurements did not reveal any differences in photosynthetic performance between oep80-3 homozygotes and wild type (Fig. 3C; Supplemental Table S2), nor did assays of nonphotosynthetic development (root length, hypocotyl length in etiolated plants, and de-etiolation efficiency; Supplemental Table S2).
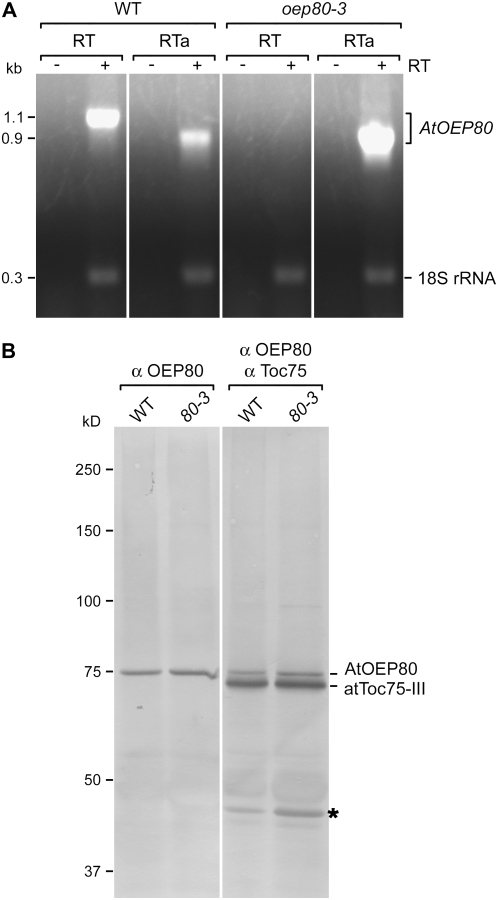
The aforementioned data provide strong evidence that there are no phenotypic consequences associated with the oep80-3 T-DNA insertion. This initially suggested that the mutant may encode a truncated form of AtOEP80 and that the missing N-terminal region is not essential. To investigate this possibility, we first of all analyzed AtOEP80 expression by reverse transcription (RT)-PCR. Using the RT pair of amplification primers (which flank the oep80-3 T-DNA insertion site; Fig. 1A), an amplicon of the expected size was obtained for wild type, but no expression was detected in oep80-3 (Fig. 4A). However, when the forward amplification primer was replaced with the RTa primer (located downstream of the T-DNA; Fig. 1A), we observed clear evidence of AtOEP80 mRNA expression in the mutant (Fig. 4A). In fact, the detected transcript was overexpressed in the mutant, relative to wild type, presumably as a consequence of cauliflower mosaic virus 35S enhancer sequences in the T-DNA construct. These data indicate that a truncated AtOEP80 message is indeed produced in the oep80-3 mutant.
Figure 4.
Analyses of mRNA and protein expression in the oep80-3 mutant. A, Analysis of AtOEP80 mRNA expression. Total RNA extracted from wild-type and homozygous oep80-3 mutant plants was analyzed by RT-PCR. Each reaction contained two primer pairs: The first specifically amplified a 1.1-kb fragment from the wild-type AtOEP80 transcript (locations of the RT primers used are indicated in Fig. 1A) or a 0.9-kb fragment from the wild-type and oep80-3 mutant transcripts (the forward RT primer was replaced with RTa in this case; see Fig. 1A); the second amplified a 315-bp fragment derived from 18S rRNA and served as a positive control. Reactions lacking reverse transcriptase (−RT) were included as negative controls. Images from different portions of the same gel are separated by vertical lines. Sizes of the amplicons are indicated at left (in kb). B, Analysis of AtOEP80 protein expression. Isolated chloroplast samples (equivalent to 20 μg [left] or 10 μg [right] chlorophyll) were separated by SDS-PAGE and then analyzed by immunoblotting using antiserum against AtOEP80 only (left), or a mixture of antisera against AtOEP80 and psToc75 (right). Protein bands corresponding to AtOEP80 and atToc75-III are indicated at right. Positions of molecular mass standards are indicated at left (sizes in kD); note that the 75-kD standard migrates more slowly than atToc75-III and at approximately the same speed as endogenous AtOEP80. A 40-kD protein band that was nonspecifically recognized by the psToc75 antiserum is indicated with an asterisk. Images from different portions of the same gel are separated by a vertical line.
The N Terminus of “Full-Length” AtOEP80 Is Dispensable
To determine precisely the nature of the oep80-3 transcript, we amplified its 5′ end by RACE-PCR and sequenced the resulting product. The mutant mRNA was found to comprise approximately 86 to 90 nucleotides encoded by the T-DNA LB fused to an AtOEP80-encoded sequence at the expected position based on the previously determined T-DNA gene junction sequence (Fig. 1A; Supplemental Fig. S2). This transcript lacks the first AUG codon (AUG1) of the wild-type message and so is predicted to encode a truncated, approximately 74-kD protein of 680 residues starting from the second, in-frame AUG (AUG2); an approximately 80-kD polypeptide of 732 residues is encoded by initiation at AUG1. To test for the presence of this smaller protein, we analyzed isolated chloroplasts from wild-type and oep80-3 plants by immunoblotting. Surprisingly, the mutant chloroplasts contained an AtOEP80 protein of the same size as that in wild type and this migrated at a position just above atToc75-III (Fig. 4B; see also Supplemental Fig. S3A); the atToc75-III protein has a calculated molecular mass of approximately 75 kD and yet runs significantly faster than the 75-kD standard on a 7.5% SDS-PAGE gel. Assuming that AtOEP80 is approximately 74 kD in size, its slower migration than atToc75-III may be due to posttranslational modification of one of the proteins because there are no obvious differences in amino acid composition.
In an attempt to explain the above data, we used SDS-PAGE and immunoblotting to compare the sizes of different, in vitro-translated AtOEP80 proteins, imported into chloroplasts, with that of the endogenous protein. We previously took a similar approach to show that the full-length protein of 732 residues, following import, migrates in similar fashion to the endogenous protein as recognized by an AtOEP80-specific antibody (Inoue and Potter, 2004). At that time, we had been focusing on determining whether AtOEP80 is synthesized with a cleavable, approximately 11-kD transit peptide, and so might not have detected more subtle mobility differences between the proteins. Here, we used an affinity-purified sample of the antibody (Supplemental Fig. S3B) to improve sensitivity and clarity, and extended the gel electrophoresis time to thoroughly scrutinize any small mobility differences.
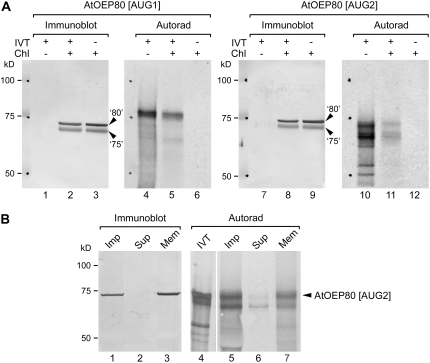
We prepared two translation reactions: one utilizing the full-length AtOEP80 cDNA as template (AtOEP80 [AUG1]) and another utilizing a truncated cDNA lacking the first 156 nucleotides of the coding sequence and starting from the second AUG codon (AtOEP80 [AUG2]) as template (Fig. 5A, lanes 4 and 10). Whereas the former reaction contained a single, major product of the expected size (approximately 80 kD), the latter contained a number of smaller proteins (presumably corresponding to initiation at downstream AUG codons; predicted sizes 71, 70, 59, 54, and 48 kD) in addition to the expected product of 74 kD. The prominence of alternative initiation products in the second translation reaction most likely reflects the suboptimal context of AUG2 in the translation system used (see Supplemental Appendix S1). In import experiments conducted in vitro, both proteins were recovered in chloroplasts (Fig. 5A, lanes 5 and 11). Proper membrane integration of the 680-residue protein was confirmed in a high-pH wash experiment: The longest translation product was recovered almost exclusively in the membrane fraction following alkaline treatment, whereas the most abundant shorter product was substantially released to the supernatant (Fig. 5B, compare lanes 6 and 7). These data confirm that the first 52 residues of the full-length 732-residue protein are not essential for import or membrane integration. Interestingly, whereas AtOEP80 translated from AUG1 (the 732-residue protein) migrated more slowly than endogenous AtOEP80, that translated from AUG2 (the 680-residue protein) migrated in a very similar position to the endogenous protein (Fig. 5A; compare lanes 5 and 2 and lanes 11 and 8; Fig. 5B, compare lanes 5 and 1 and lanes 7 and 3).
Figure 5.
Electrophoretic mobility comparisons between proteins imported into Arabidopsis chloroplasts in vitro and endogenous AtOEP80. A, Radiolabeled long (AtOEP80 [AUG1]; 732 residues) and short (AtOEP80 [AUG2]; 680 residues) forms of the AtOEP80 protein were generated by in vitro translation using different cDNA templates. These were incubated with Arabidopsis chloroplasts under import conditions and then the chloroplasts were recovered. In vitro translation products equivalent to 10% of the amount added to each import assay (IVT+/Chl−), Arabidopsis chloroplasts containing imported, radiolabeled proteins (IVT+/Chl+), and equivalent chloroplast samples lacking imported, radiolabeled protein (IVT−/Chl+) were resolved side-by-side using SDS-PAGE, blotted onto the same membrane, and then analyzed either by probing with AtOEP80 and psToc75 antisera (Immunoblot) or by autoradiography (Autorad). The positions of endogenous AtOEP80 and atToc75-III proteins are indicated at right (‘80’ and ‘75’, respectively). Positions of molecular mass standards are indicated at left (sizes in kD). Under the conditions used, the endogenous AtOEP80 protein migrated slower than atToc75-III, whereas both proteins ran faster than the 75-kD marker protein. B, In vitro translated, radiolabeled AtOEP80 (AtOEP80 [AUG2]; 680 residues) was incubated with Arabidopsis chloroplasts under import conditions. One-half of the recovered chloroplast sample was subjected to alkaline extraction using 0.1 m Na2CO3 and separated into soluble and membrane fractions as described (Inoue and Potter, 2004). In vitro translation products equivalent to 5% of the amount used for the import assay (IVT), unfractionated Arabidopsis chloroplasts containing imported, radiolabeled protein (Imp), and the supernatant (Sup) and membrane (Mem) fractions obtained after alkaline extraction were resolved side-by-side using SDS-PAGE, blotted onto the same membrane, and then analyzed either by probing with AtOEP80 antiserum (Immunoblot) or by autoradiography (Autorad). The position of imported AtOEP80 is indicated at right. Positions of molecular mass standards are indicated at left (sizes in kD); note that the 75-kD marker runs significantly slower than atToc75-III on a 7.5% SDS-PAGE gel.
These results may be explained in two different ways. One possibility is that the AtOEP80 protein is normally translated from a noncanonical, downstream initiation codon, even in wild type, with AUG2 being one candidate (see Supplemental Appendix S1; Supplemental Fig. S4). This hypothesis is supported by the fact that the oep80-3 mutant expresses a protein of the same size as that in wild type (Fig. 4; Supplemental Fig. S3) and by the comigration of the 680-residue protein translated from AUG2 with the endogenous protein in chloroplasts (Fig. 5). It is also noteworthy that the two most similar sequences present in the protein databases (OsI_006101 [EAY84868] and OsJ_005573 [EAZ22090], both from rice [Oryza sativa]) align with AtOEP80 only at positions downstream of the second Met. Such noncanonical initiation might have developmental or regulatory significance.
However, in two different in vitro translation systems (wheat germ, Fig. 5A; rabbit reticulocyte, Fig. 5B), initiation at AUG2 appeared to be rather inefficient. Moreover, a recent proteomic study indicated that translation from AUG1 can occur in vivo; of 89 AtOEP80 peptides identified by Dunkley et al. (2006), one (FSSSSIR; positions 10–16 relative to Met-1/AUG1) was found to correspond to the sequence between AUG1 and AUG2 (K. Lilley, personal communication). Thus, an alternative to the noncanonical initiation possibility outlined above is that translation from AUG1 leads to the formation of a 732-residue precursor protein, which is processed to a lower molecular mass form during targeting or membrane insertion. Because the targeting, accumulation, and functionality of AtOEP80 were not detectably altered in the oep80-3 mutant (Figs. 3 and 4), one may conclude that any cleavable, N-terminal targeting sequence that is present is dispensable. It is noteworthy that neither of the AtOEP80 translation products changed in size upon import (Fig. 5). However, this may simply indicate that proteolytic processing is inefficient in the context of an in vitro protein import assay, as has been observed previously for Tic22 and Toc75 (Kouranov et al., 1999; Inoue and Keegstra, 2003).
CONCLUSION
Our aim was to assess the importance of the plastidic protein, AtOEP80, for plant growth and development. As a first step, we analyzed the expression of AtOEP80 using publicly available microarray data relative to well-known components of the protein translocation machinery of the plastid envelope. Expression levels of AtOEP80 paralleled those of atTOC75-III and atTIC110 throughout development, but at substantially lower levels (approximately 25% of the level of atTOC75-III). Interestingly, a different trend was observed in embryos, with AtOEP80 expression being approximately 40% higher than atTOC75-III expression, hinting at an important role for AtOEP80 during embryogenesis. Consistent with this notion, the knockout mutations oep80-1 and oep80-2 were embryo lethal in the homozygous state, demonstrating that AtOEP80 plays an essential role during early stages of plastid development. Developmental arrest occurred at a relatively late stage in oep80 (globular stage embryo proper), which contrasts with the early defect (two-cell stage) caused by loss of atToc75-III (Baldwin et al., 2005). This difference in phenotypic severity may reflect differences in the roles of the proteins: atToc75-III playing a wide-ranging role in the import of many proteins and AtOEP80 a more specialized role, perhaps in the biogenesis of a relatively small subset of proteins. Further experimentation will be required to determine the exact function of AtOEP80. Surprisingly, a third AtOEP80 mutant, oep80-3, which carries a T-DNA insertion in the first exon, was found to be viable and indistinguishable from wild type in the homozygous state. In spite of the fact that oep80-3 expresses a truncated transcript lacking the first AUG codon, the mutant was found to express an AtOEP80 protein of the same size as that in wild type. Together with data from in vitro translation, import, and immunoblotting experiments, this observation led to the conclusion that the N-terminal region of the putative full-length AtOEP80 protein of 732 residues is not required for targeting, membrane insertion, or functionality. This contrasts with the situation for atToc75-III, which requires a bipartite targeting sequence for proper biogenesis (Tranel and Keegstra, 1996; Inoue and Keegstra, 2003). Thus, there are two essential, Omp85-related β-barrel proteins in the outer envelope membrane of chloroplasts, but it would appear that these proteins have quite different requirements for membrane insertion.
MATERIALS AND METHODS
Plant Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants were of the Columbia-0 ecotype. For in vitro growth, seeds were surface sterilized, sown on Murashige and Skoog agar medium in petri plates, cold treated at 4°C, and thereafter kept in a growth chamber, as described previously (Aronsson and Jarvis, 2002). To select for the presence of T-DNA insertions, the following antibiotics were added to the medium: hygromycin B, 15 to 30 μg/mL (oep80-1); and sulfadiazine, 11.25 μg/mL (oep80-2 and oep80-3). All plants were grown under a long-day cycle (16 h light, 8 h dark).
Root length measurements were conducted as described previously (Constan et al., 2004), using plants grown on vertically oriented Murashige and Skoog agar plates under standard conditions for 10 d. Hypocotyl length measurements were done using plants germinated on soil and grown in sealed propagators in the dark for 5 d using a published method (Salter et al., 2003). De-etiolation experiments were conducted according to a previous report (Baldwin et al., 2005) and as described below, using plants grown on Murashige and Skoog medium lacking Suc. Following cold treatment, plates were exposed to standard light for 4 h to promote germination and then kept in darkness for 6 d; then, plates were transferred to continuous light for a further period of 2 d prior to scoring.
Identification of the oep80 Mutants
The T-DNA insertion lines were obtained from the following sources: oep80-1 was from the Csaba Koncz laboratory (pool 894, line 89350; Ríos et al., 2002); and oep80-2 and oep80-3 were from Genomanalyse im Biologischen System Pflanze-Kölner Arabidopsis T-DNA (GABI-Kat; lines 429H12 and 430F02, respectively; Rosso et al., 2003).
Mutant genotypes were assessed by PCR (Fig. 1B). Genomic DNA was extracted from Arabidopsis plants using a published protocol (Edwards et al., 1991) and PCR was conducted using standard procedures. The primers used were as follows: oep80-1 forward, 5′-CATGGATTGAAGGAGATGACAAGAG-3′; oep80-1 reverse, 5′-GAAACGAGCTGGTCCAATGTGTATG-3′; oep80-1 T-DNA RB, 5′-CAGTCATAGCCGAATAGCCTCTCCA-3′; oep80-2 forward, 5′-AGTAAGAACGAAAGATGGTGAGGA-3′; oep80-2 reverse, 5′-TCACTTTCCCTACACAGCTTGA-3′; oep80-2 and oep80-3 T-DNA LB, 5′-CCCATTTGGACGTGAATGTAGACAC-3′; oep80-3 forward, 5′-TCCTTGTTTGTTCATGTGTGTGA-3′; oep80-3 reverse, 5′-TCCTCACCATCTTTCGTTCTTACT-3′; and oep80-3 T-DNA RB, 5′-GCAAGTGGATTGATGTGATATCTCCAC-3′. The amplification products were analyzed by agarose gel electrophoresis and stained with ethidium bromide. The location of each T-DNA insertion was determined precisely by the sequencing of PCR products spanning both junctions (except in the case of oep80-2, where only one junction was identified).
Chlorophyll Quantification and Photosynthetic Measurements
Chlorophyll was extracted from 14-d-old plants grown in vitro and determined photometrically as described previously (Porra et al., 1989; Aronsson et al., 2003). Photosynthetic electron transport rates (Fig. 3C), as well as the photochemical efficiency of PSII (Fv/Fm) and photosynthetic performance index (Supplemental Table S2), were determined by measuring chlorophyll fluorescence using a continuous excitation fluorimeter (Handy PEA; Hansatech Instruments), according to the manufacturer's instructions and as described previously (Meyer et al., 1997; Strasser et al., 2004). The plants used for these chlorophyll fluorescence assays were grown on selective medium for 8 d (where appropriate), rescued to nonselective medium, and then transferred to soil after 2 weeks.
Isolation of RNA and RT-PCR
Total RNA was isolated from Arabidopsis seedlings with an RNeasy plant mini kit (Qiagen), and used to prepare cDNA with Superscript III and random primers (Invitrogen). Using the resultant cDNA as a template, PCR amplifications were performed using the following AtOEP80 gene-specific primers (Figs. 1A and 4A): RT forward, 5′-ATGCATTGTCACAACGATGA-3′; RTa forward, 5′-ATGCTCCAGTCGCTAAAGAATC-3′; and RT reverse, 5′-TCTACATCCCTCTTCCCTTGAA-3′. Control amplification of a sequence derived from 18S rRNA was performed according to the manufacturer's instructions (Ambion).
Chloroplast Isolation, Import, and Immunoblotting
Isolation of chloroplasts from plate-grown Arabidopsis seedlings and in vitro chloroplast protein import assays were performed essentially as described previously (Fitzpatrick and Keegstra, 2001; Inoue and Potter, 2004). For the preparation of radiolabeled precursor proteins, TNT coupled systems containing wheat germ extract (Fig. 5A) or rabbit reticulocyte lysate (Fig. 5B) were used (Promega). The cDNA construct encoding the short form of the AtOEP80 protein (AtOEP80 [AUG2]; 680 residues) was prepared by subcloning a PCR product (forward primer, 5′-ATGCTCCAGTCGCTAAAGAATC-3′; reverse primer, 5′-CTCGAGTTAGTTCCGCAGACCAAC-3′), amplified using pGEMT-AtOEP80 (Inoue and Potter, 2004) as template, into the pGEM-T Easy vector (Promega). Samples were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Bio-Rad). Imported proteins were detected by autoradiography. For immunodetection, membranes were incubated with antisera raised against psToc75 or AtOEP80 (see below). Immunoreactive proteins were detected using a secondary antibody conjugated with alkaline phosphatase and a 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium substrate mixture (Bio-Rad).
A previously described antiserum against residues 325 to 337 of AtOEP80 was employed (Inoue and Potter, 2004). Its specificity was confirmed in a competition experiment, and then the crude serum was purified by affinity chromatography for use in Figures 4 and 5 (Supplemental Fig. S3). Briefly, a 200-μg sample of the antigen peptide (Inoue and Potter, 2004) was coupled to 100 μL of UltraLink iodoacetyl gel (MicroLink peptide coupling kit; Pierce) in the provided minicolumn according to the manufacturer's instructions. The crude serum (300 μL) was applied to the column and incubated at room temperature for 2 h. The unbound fraction was collected, and then the column was washed 15 times with 300 μL of wash buffer (0.7 m NaCl, 0.05% [v/v] Tween 20) and a further three times with 300 μL of phosphate-buffered saline (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4, pH 7.4). Bound antibodies were eluted with 0.1 m Gly, pH 2.8, immediately neutralized with a 1/20th volume of 1 m Tris-HCl, pH 9, and stored at 4°C until further use.
Embryo Analysis by Light Microscopy
The analysis of cleared wild-type and oep80-1 mutant embryos using Nomarski optics (Fig. 2C) was performed as described previously (Goubet et al., 2003; Baldwin et al., 2005; Kovacheva et al., 2005). A microscope (model BHS; Olympus) equipped for differential interference contrast (model BH2-NIC; Olympus) was employed for these studies.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_568378, CAB51191, NP_192647, NP_172176, Q43715, EAY84868, and EAZ22090.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression patterns of the AtOEP80 gene relative to genes for other envelope proteins.
Supplemental Figure S2. Annotated T-DNA LB junction sequence for the oep80-3 mutant, illustrating the structure of the 5′ end of the expressed transcript.
Supplemental Figure S3. Specificity confirmation and affinity purification of the AtOEP80 antibody used in the immunoblotting experiments shown in Figures 4 and 5.
Supplemental Figure S4. In silico analysis of the 5′ region of the AtOEP80 mRNA.
Supplemental Table S1. Segregation of the T-DNA-associated selectable marker in each of the oep80 mutants.
Supplemental Table S2. Phenotypic analysis of the oep80 mutants, including photosynthetic measurements as well as studies on nonphotosynthetic growth.
Supplemental Appendix S1. Analysis of the 5′ region of the AtOEP80 mRNA in silico.
Supplementary Material
Acknowledgments
T-DNA lines were identified with help from Gabino Ríos and Csaba Koncz (oep80-1) and GABI-Kat (oep80-2 and oep80-3). We are grateful to Kathryn Lilley for providing AtOEP80 mass spectrometry peptide data, and to Michael Zuker for assistance with use of the mfold program and interpretation of the results. We thank Weihua Huang and Rebecca Shipman for insightful comments on the manuscript, and members of both laboratories for helpful discussions.
This work was supported by a University of California, Davis, Pomology Graduate Student Researcher Fellowship (to S.-C.H.), by a Jastro-Shields Fellowship (to S.-C.H.), by a University of California, Davis, Grant to Promote Extra-Mural Funding (to K.I.), by the Royal Society Rosenheim Research Fellowship (to P.J.), and by Biotechnology and Biological Sciences Research Council grants BBS/B/03629, BB/C006348/1, and BBS/D016541/1 (to P.J.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Kentaro Inoue (kinoue@ucdavis.edu) and Paul Jarvis (rpj3@le.ac.uk).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Apuya NR, Yadegari R, Fischer RL, Harada JJ, Goldberg RB (2002) RASPBERRY3 gene encodes a novel protein important for embryo development. Plant Physiol 129 691–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apuya NR, Yadegari R, Fischer RL, Harada JJ, Zimmerman JL, Goldberg RB (2001) The Arabidopsis embryo mutant schlepperless has a defect in the chaperonin-60α gene. Plant Physiol 126 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronsson H, Boij P, Patel R, Wardle A, Töpel M, Jarvis P (2007) Toc64/OEP64 is not essential for the efficient import of proteins into chloroplasts in Arabidopsis thaliana. Plant J 52 53–68 [DOI] [PubMed] [Google Scholar]
- Aronsson H, Combe J, Jarvis P (2003) Unusual nucleotide-binding properties of the chloroplast protein import receptor, atToc33. FEBS Lett 544 79–85 [DOI] [PubMed] [Google Scholar]
- Aronsson H, Jarvis P (2002) A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett 529 215–220 [DOI] [PubMed] [Google Scholar]
- Baldwin A, Wardle A, Patel R, Dudley P, Park SK, Twell D, Inoue K, Jarvis P (2005) A molecular-genetic study of the Arabidopsis Toc75 gene family. Plant Physiol 138 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin AJ, Inoue K (2006) The most C-terminal tri-glycine segment within the polyglycine stretch of the pea Toc75 transit peptide plays a critical role for targeting the protein to the chloroplast outer envelope membrane. FEBS J 273 1547–1555 [DOI] [PubMed] [Google Scholar]
- Bédard J, Jarvis P (2005) Recognition and envelope translocation of chloroplast preproteins. J Exp Bot 56 2287–2320 [DOI] [PubMed] [Google Scholar]
- Bölter B, Soll J, Schulz A, Hinnah S, Wagner R (1998) Origin of a chloroplast protein importer. Proc Natl Acad Sci USA 95 15831–15836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Andrews J, Mersey B, Newcomb EH, Keegstra K (1981) Separation and characterization of inner and outer envelope membranes of pea chloroplasts. Proc Natl Acad Sci USA 78 3595–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constan D, Froehlich JE, Rangarajan S, Keegstra K (2004) A stromal Hsp100 protein is required for normal chloroplast development and function in Arabidopsis. Plant Physiol 136 3605–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley TP, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA 103 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckart K, Eichacker L, Sohrt K, Schleiff E, Heins L, Soll J (2002) A Toc75-like protein import channel is abundant in chloroplasts. EMBO Rep 3 557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick LM, Keegstra K (2001) A method for isolating a high yield of Arabidopsis chloroplasts capable of efficient import of precursor proteins. Plant J 27 59–65 [DOI] [PubMed] [Google Scholar]
- Gentle I, Gabriel K, Beech P, Waller R, Lithgow T (2004) The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol 164 19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle IE, Burri L, Lithgow T (2005) Molecular architecture and function of the Omp85 family of proteins. Mol Microbiol 58 1216–1225 [DOI] [PubMed] [Google Scholar]
- Goubet F, Misrahi A, Park SK, Zhang Z, Twell D, Dupree P (2003) AtCSLA7, a cellulose synthase-like putative glycosyltransferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol 131 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grennan AK (2006) Genevestigator. Facilitating Web-based gene-expression analysis. Plant Physiol 141 1164–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnah SC, Hill K, Wagner R, Schlicher T, Soll J (1997) Reconstitution of a chloroplast protein import channel. EMBO J 16 7351–7360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hust B, Gutensohn M (2006) Deletion of core components of the plastid protein import machinery causes differential arrest of embryo development in Arabidopsis thaliana. Plant Biol (Stuttg) 8 18–30 [DOI] [PubMed] [Google Scholar]
- Inaba T, Alvarez-Huerta M, Li M, Bauer J, Ewers C, Kessler F, Schnell DJ (2005) Arabidopsis Tic110 is essential for the assembly and function of the protein import machinery of plastids. Plant Cell 17 1482–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Baldwin AJ, Shipman RL, Matsui K, Theg SM, Ohme-Takagi M (2005) Complete maturation of the plastid protein translocation channel requires a type I signal peptidase. J Cell Biol 171 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Demel R, de Kruijff B, Keegstra K (2001) The N-terminal portion of the preToc75 transit peptide interacts with membrane lipids and inhibits binding and import of precursor proteins into isolated chloroplasts. Eur J Biochem 268 4036–4043 [DOI] [PubMed] [Google Scholar]
- Inoue K, Keegstra K (2003) A polyglycine stretch is necessary for proper targeting of the protein translocation channel precursor to the outer envelope membrane of chloroplasts. Plant J 34 661–669 [DOI] [PubMed] [Google Scholar]
- Inoue K, Potter D (2004) The chloroplastic protein translocation channel Toc75 and its paralog OEP80 represent two distinct protein families and are targeted to the chloroplastic outer envelope by different mechanisms. Plant J 39 354–365 [DOI] [PubMed] [Google Scholar]
- Jackson-Constan D, Keegstra K (2001) Arabidopsis genes encoding components of the chloroplastic protein import apparatus. Plant Physiol 125 1567–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Fernandez R, Coutte L (2004) Protein secretion through autotransporter and two-partner pathways. Biochim Biophys Acta 1694 235–257 [DOI] [PubMed] [Google Scholar]
- Kessler F, Schnell DJ (2006) The function and diversity of plastid protein import pathways: a multilane GTPase highway into plastids. Traffic 7 248–257 [DOI] [PubMed] [Google Scholar]
- Kouranov A, Wang H, Schnell DJ (1999) Tic22 is targeted to the intermembrane space of chloroplasts by a novel pathway. J Biol Chem 274 25181–25186 [DOI] [PubMed] [Google Scholar]
- Kovacheva S, Bédard J, Patel R, Dudley P, Twell D, Ríos G, Koncz C, Jarvis P (2005) In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J 41 412–428 [DOI] [PubMed] [Google Scholar]
- Kozjak V, Wiedemann N, Milenkovic D, Lohaus C, Meyer HE, Guiard B, Meisinger C, Pfanner N (2003) An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J Biol Chem 278 48520–48523 [DOI] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer U, Köllner B, Willenbrink J, Krause GHM (1997) Physiological changes on agricultural crops induced by different ambient ozone exposure regimes. I. Effects on photosynthesis and assimilate allocation in spring wheat. New Phytol 136 645–652 [DOI] [PubMed] [Google Scholar]
- Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W (2003) Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature 426 862–866 [DOI] [PubMed] [Google Scholar]
- Perry SE, Keegstra K (1994) Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell 6 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975 384–394 [Google Scholar]
- Puyaubert J, Denis L, Alban C (2008) Dual targeting of Arabidopsis holocarboxylase synthetase1: a small upstream open reading frame regulates translation initiation and protein targeting. Plant Physiol 146 478–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Davila-Aponte J, Keegstra K (1999) The evolutionary origin of the protein-translocating channel of chloroplastic envelope membranes: identification of a cyanobacterial homolog. Proc Natl Acad Sci USA 96 784–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Inoue K, Keegstra K (2005) Evolution of the general protein import pathway of plastids. Mol Membr Biol 22 73–86 [DOI] [PubMed] [Google Scholar]
- Ríos G, Lossow A, Hertel B, Breuer F, Schaefer S, Broich M, Kleinow T, Jasik J, Winter J, Ferrando A, et al (2002) Rapid identification of Arabidopsis insertion mutants by non-radioactive detection of T-DNA tagged genes. Plant J 32 243–253 [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53 247–259 [DOI] [PubMed] [Google Scholar]
- Salter MG, Franklin KA, Whitelam GC (2003) Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426 680–683 [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Kessler F, Blobel G (1994) Isolation of components of the chloroplast protein import machinery. Science 266 1007–1012 [DOI] [PubMed] [Google Scholar]
- Sjögren LL, MacDonald TM, Sutinen S, Clarke AK (2004) Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol 136 4114–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD (2006) Protein import into chloroplasts: an ever-evolving story. Can J Bot 84 531–542 [Google Scholar]
- Stojanovski D, Guiard B, Kozjak-Pavlovic V, Pfanner N, Meisinger C (2007) Alternative function for the mitochondrial SAM complex in biogenesis of {alpha}-helical TOM proteins. J Cell Biol 179 881–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser RJ, Srivastava A, Tsimilli-Michael M (2004) Analysis of the chlorophyll a fluorescence transient. In G Papageorgiou, Govindjee, eds, Advances in Photosynthesis and Respiration: Chlorophyll Fluorescence, a Signature of Photosynthesis, Vol 19. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 321–362
- Tranel PJ, Froehlich J, Goyal A, Keegstra K (1995) A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J 14 2436–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranel PJ, Keegstra K (1996) A novel, bipartite transit peptide targets OEP75 to the outer membrane of the chloroplastic envelope. Plant Cell 8 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwer U, Willmitzer L, Altmann T (1998) Inactivation of a glycyl-tRNA synthetase leads to an arrest in plant embryo development. Plant Cell 10 1277–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J (2003) Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299 262–265 [DOI] [PubMed] [Google Scholar]
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D (2005) Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121 235–245 [DOI] [PubMed] [Google Scholar]
- Yadegari R, Paiva G, Laux T, Koltunow AM, Apuya N, Zimmerman JL, Fischer RL, Harada JJ, Goldberg RB (1994) Cell differentiation and morphogenesis are uncoupled in Arabidopsis raspberry embryos. Plant Cell 6 1713–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen MR, Peabody CR, Partovi SM, Zhai Y, Tseng YH, Saier MH (2002) Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim Biophys Acta 1562 6–31 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.