Abstract
Fruit color-break is the visual manifestation of the developmentally regulated transition of chloroplasts to chromoplasts during fruit ripening and often involves biosynthesis of copious amounts of carotenoids concomitant with massive breakdown of chlorophyll. Regulation of chlorophyll breakdown at different physiological and developmental stages of the plant life cycle, particularly at fruit color-break, is still not well understood. Here, we present the dynamics of native chlorophyllase (Chlase) and chlorophyll breakdown in lemon (Citrus limon) fruit during ethylene-induced color-break. We show, using in situ immunofluorescence on ethylene-treated fruit peel (flavedo) tissue, that citrus Chlase is located in the plastid, in contrast to recent reports suggesting cytoplasmic localization of Arabidopsis (Arabidopsis thaliana) Chlases. At the intra-organellar level, Chlase signal was found to overlap mostly with chlorophyll fluorescence, suggesting association of most of the Chlase protein with the photosynthetic membranes. Confocal microscopy analysis showed that the kinetics of chlorophyll breakdown was not uniform in the flavedo cells. Chlorophyll quantity at the cellular level was negatively correlated with plastid Chlase accumulation; plastids with reduced chlorophyll content were found by in situ immunofluorescence to contain significant levels of Chlase, while plastids containing still-intact chlorophyll lacked any Chlase signal. Immunoblot and protein-mass spectrometry analyses were used to demonstrate that citrus Chlase initially accumulates as an approximately 35-kD precursor, which is subsequently N-terminally processed to approximately 33-kD mature forms by cleavage at either of three consecutive amino acid positions. Chlase plastid localization, expression kinetics, and the negative correlation with chlorophyll levels support the central role of the enzyme in chlorophyll breakdown during citrus fruit color-break.
Color-break and the enhanced biosynthesis of aroma compounds toward fruit ripening are strategies developed by plants to attract seed dispersal agents to fruits that contain fully mature seeds. In citrus, developmentally regulated fruit color-break involves accumulation of copious amounts of carotenoid pigments concomitant with chlorophyll breakdown (Masaya et al., 2004). Both metabolic processes occur within the plastids of the flavedo tissue (the pigmented outer part of the peel) and are the visual manifestation of the transformation of the flavedo photosynthetically active chloroplasts into chromoplasts (Thomson and Whatley, 1980; Goldschmidt, 1988; Kobayashi, 1991).
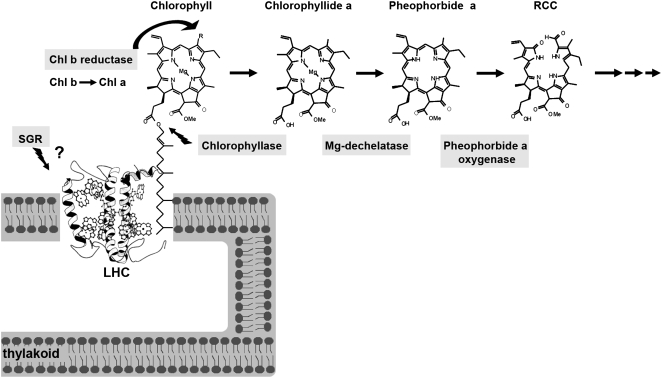
The chlorophyll molecule is a central player in harvesting light energy channeled for photosynthesis in leaves, yet it likely assumes other tissue-specific roles in the fruit peel, such as providing green pigmentation that camouflages the fruit until the seeds are mature. Indeed, the photosynthetic role in fruit appears to be limited, as photosynthetic levels of fruit peel chloroplasts have been documented to be quite low and account for only a few percent of the sugar accumulating in the fruit (Piechulla et al., 1987). Regulation of chlorophyll breakdown at the different physiological and developmental stages of the plant life cycle and particularly at fruit color-break is still not well understood. Tight regulation of the breakdown pathway would be expected to avoid accumulation of photodynamic chlorophyll intermediate metabolism products that are known to lead to oxidative damage upon exposure of the tissue to light (Hu et al., 1998; Mock et al., 1999; Molina et al., 1999; Ishikawa et al., 2001; Mach et al., 2001; Pružinská et al., 2003; Harpaz-Saad et al., 2007). The enzymatic breakdown of chlorophyll into phytol and the primary cleavage product of the porphyrin ring occurs in four consecutive steps catalyzed by chlorophyllase (Chlase), recently shown to catalyze the rate-limiting step in photosynthetic leaves (Harpaz-Saad et al., 2007), magnesium-dechelatase (not yet demonstrated to be an enzymatic step), pheophorbide a oxygenase (PaO), and red chlorophyll catabolite (RCC) reductase, respectively (Chlase: Jacob-Wilk et al., 1999; Tsuchiya et al., 1999; PaO: Pružinská et al., 2003; Alós et al., 2006; RCC reductase: Wüthrich et al., 2000; Fig. 1). Further downstream, the porphyrin breakdown products are modified and exported to the vacuole, where they are stored indefinitely (Hörtensteiner, 1999, 2004; Matile et al., 1999; Takamiya et al., 2000). Beyond the established enzymatic chlorophyll breakdown pathway, recent data from stay-green mutants suggests the involvement of an additional player in chlorophyll degradation: the SGR gene product, which likely functions upstream of the catabolic enzymes (Aubry et al., 2008) and has been suggested to function as a destabilizer of the light-harvesting complex (LHC; Park et al., 2007; Fig. 1).
Figure 1.
Graphic model of the proteins involved in chlorophyll breakdown in higher plants and chemical structures of chlorophyll and chlorophyll catabolites. The protein SGR, involved in stay-green mutations, is suggested to function upstream of the chlorophyll catabolic enzymes by destabilizing the LHC (Park et al., 2007). The enzyme chlorophyll b reductase is involved in maintaining the balance between chlorophylls by catalyzing the conversion of chlorophyll b to a. The catabolic steps of the chlorophyll breakdown pathway, which lead to loss of the typical green color, are outlined together with the structures of chlorophyll and the intermediate breakdown products as follows: (1) Chlase catalyzes the cleavage of the hydrophobic thylakoid-anchoring phytol chain from the chlorophyll porphyrin ring, resulting in the product chlorophyllide; (2) removal of the Mg ion from chlorophyllide, yielding pheophorbide a, has not yet been shown to be an enzymatic step (labeled as Mg-dechelatase); and (3) PaO catalyzes the cleavage of the porphyrin ring, resulting in the RCC, which is further metabolized by additional enzymes downstream, exported to the vacuole, nonenzymatically converted to nonfluorescent chlorophyll catabolites (NCCs), and stored indefinitely.
Citrus are classified as non-climacteric fruit; however, a role for ethylene during ripening, and in particular in chlorophyll breakdown during fruit color-break, is well established (Purvis and Barmore, 1981; Goldschmidt et al., 1993; Porat et al., 1999). Furthermore, the application of exogenous ethylene dramatically enhances the speed of color-break of detached mature green citrus fruit, which is a slow process for fruit left untreated on trees (Amir-Shapira et al., 1987; Iglesias et al., 2001). Thus, induction of fruit color-break by exogenous ethylene provides a defined time frame that enables the study of chlorophyll breakdown in citrus fruit at the molecular level. Indeed, previous work has demonstrated the correlation between chlorophyll breakdown and expression of the gene encoding Chlase in ethylene-treated fruit (Jacob-Wilk et al., 1999), suggesting that regulation of chlorophyll catabolism at the gene expression level occurs. However, Chlase gene expression is not increased during color-break of untreated fruits (Jacob-Wilk et al., 1999), raising the possibility that an additional posttranslational mode of regulation may be involved in regulating its function (Harpaz-Saad et al., 2007).
Intracellular localization is often one of the keys to understanding enzyme function and regulation in vivo and has been a controversial issue with regard to the enzyme Chlase. Krossing (1940) was first in demonstrating Chlase activity in a chloroplast fraction, followed by others (barley [Hordeum vulgare] leaves: Matile et al., 1997; citrus fruit: Trebitsh et al., 1993; citrus leaves: Garcia and Galindo, 1991; Ginkgo (Ginkgo biloba) leaves: Okazawa et al., 2006), some of which associated Chlase activity specifically with the plastid membrane fraction (Ardao and Vennesland, 1960; Terpstra, 1974, 1976; Tarasenko et al., 1986; Harpaz-Saad et al., 2007) or specifically with thylakoids (Okazawa et al., 2006). However, different attempts to establish the association of Chlase with specific thylakoid-located pigment-protein complexes have yielded ambiguous results. On the one hand, Chlase activity has been detected in components of the LHC of PSII (LHC II) that were separated electrophoretically (Tarasenko et al., 1986; Brandis et al., 1996), while on the other hand, attempts to separate solubilized complexes by density gradient centrifugation have failed to confirm an intimate association between Chlase and LHC II (Schellenberg and Matile, 1995; Brandis et al., 1996). Moreover, purified LHC II has been found to be devoid of Chlase activity (Schellenberg and Matile, 1995). Hirschfeld and Goldschmidt (1983) showed that Chlase activity is present in the thylakoid-free chromoplast membranes prepared from citrus fruit, suggesting that Chlase is present in plastid envelope membranes (the only membranes present in chromoplasts). More recent work (Matile et al., 1997) demonstrated association of Chlase activity with the plastid inner-envelope membrane from barley leaves, similar to the results obtained for PaO, the third enzyme of the chlorophyll catabolic pathway (Matile and Schellenberg, 1996).
The relatively recent cloning of genes encoding Chlase from several species has added to the confusion. Citrus and Chenopodium Chlase were suggested to be N-terminally processed based on comparison of the gene open reading frame to the N-terminal sequence of the purified protein. However, the apparently cleaved N-terminal sequences are not typical of N-terminal plastid transit peptides (Jacob-Wilk et al., 1999; Tsuchiya et al., 1999), leading to speculation that some Chlases may be targeted elsewhere in the cell (Takamiya et al., 2000). Recent work, showing that expression of the two known Arabidopsis (Arabidopsis thaliana) Chlase genes (AtCLH1 and AtCLH2) fused to GFP (C and N terminally) yielded cytoplasmic-localized proteins, questions the actual role of the known Chlase genes/enzymes in chlorophyll catabolism (Schenk et al., 2007).
In this work, we studied the dynamics of citrus Chlase in chlorophyll breakdown within its natural fruit flavedo tissue during ethylene-induced color break. We demonstrate, using in situ immunofluorescence, that Chlase is localized to plastids, and mostly associated with chlorophyll, in fruit flavedo. Levels of Chlase enzyme in the flavedo cells/plastids are negatively correlated with chlorophyll levels, further supporting the role of citrus Chlase in chlorophyll breakdown. Immunoblot and protein-mass spectrometry (MS) analyses demonstrate that Chlase initially accumulates as an approximately 35-kD precursor, which is subsequently N-terminally processed to approximately 33-kD mature forms by cleavage at one of three consecutive amino acids.
RESULTS
In Situ Immunofluorescence Establishes Localization of Chlase to the Plastid in Ethylene-Treated Citrus Fruit Peel
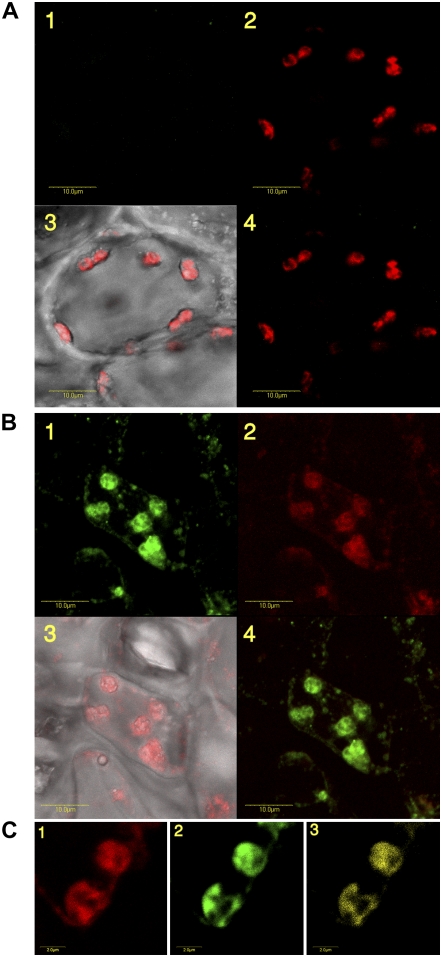
The intracellular location of Chlase has been subject to continuous controversy. We therefore pursued a study on the intracellular localization of citrus Chlase within the endogenous lemon (Citrus limon) fruit peel tissue during ethylene-induced fruit color-break. In situ immunofluorescence was chosen as the method to study localization in the endogenous citrus tissue, because it provides an unbiased picture of the distribution of any given protein within the cell. Mature green lemon fruits were treated with ethylene for 24 h to induce chlorophyll breakdown as well as Chlase expression. Flavedo (the outer pigmented part of the peel) of the ethylene-treated lemons was sectioned, fixed, and subjected to immunofluorescence analysis. To obtain fixed scanning conditions, the confocal parameters were calibrated on control cross-section samples. Emission levels, green (515–525 nm) and red (BA660IF), were fixed and used on all samples. Confocal microscopy images were recorded for bright-field, green fluorescence of the Alexa-488 secondary antibody and red autofluorescence of chlorophyll. Control sample sections dressed only with the secondary antibody (Alexa-488-conjugated anti-rabbit) do not show any fluorescence of the Alexa-488 fluorophore in any of the cells/chloroplasts (representative cell shown in Fig. 2A), while chlorophyll fluorescence of the chloroplasts is clearly visible (Fig. 2A, 2 and 3). Sections dressed with affinity-purified anti-citrus Chlase antibody followed by the secondary antibody showed fluorescence of the Alexa-488 fluorophore, corresponding to Chlase detection, in an estimated 50% of the flavedo cells. The Alexa-488 fluorescence (green) was limited almost exclusively to plastids (Fig. 2B, 1), overlapping with the red fluorescence of chlorophyll (Fig. 2B, 2 and 4). Low levels of fluorescence were also detected in small spherical structures distributed throughout the cells (Fig. 2B, 1).
Figure 2.
Subcellular localization of Chlase in ethylene-treated citrus fruit peel by in situ immunofluorescence. Mature green lemon fruit were treated with ethylene (20 μL L−1) at 25°C in the dark for 24 h in a 4-L sealed container. The container was ventilated once after 12 h, followed by injection of fresh ethylene, to maintain CO2 levels below 1%. The flavedo of the treated fruit was dissected and used for obtaining cross sections (70 μm thick) using a vibratome (see “Materials and Methods”). Tissue sections were fixed and dressed with Alexa-488-conjugated goat anti-rabbit secondary antibody as a control (A) or affinity-purified anti-citrus Chlase antibody (rabbit) followed by Alexa-488-conjugated goat anti-rabbit secondary antibody (B and C). Fluorescence was visualized using a laser scanning confocal microscope as described in “Materials and Methods.” Images of representative cells (A and B) are presented as follows: 1, green fluorescence of the Alexa-488 secondary antibody corresponds to detection by anti-Chlase antibody; 2, red fluorescence corresponds to chlorophyll autofluorescence; 3, confocal image recorded simultaneously in transmitted and red fluorescence mode (i.e. chlorophyll fluorescence superimposed on the bright-field image); and 4, confocal image recorded simultaneously for red and green fluorescence (i.e. immunofluorescence resulting from detection by anti-Chlase superimposed on the chlorophyll autofluorescence). Chloroplasts demonstrating green fluorescence (i.e. detection by anti-Chlase antibody) were further analyzed by the confocal microscope colocalization program analysis tool. Images of representative chloroplasts (C) are presented as follows: 1, red fluorescence corresponds to chlorophyll autofluorescence; 2, green fluorescence of the Alexa-488 secondary antibody corresponds to detection by anti-Chlase antibody; and 3, sectors of yellow color indicate colocalization of chlorophyll autofluorescence and immunofluorescence resulting from detection by anti-Chlase antibody. Colocalized sectors in the fluorescing chloroplasts account for 76% of the total fluorescence observed. Scale bar = 10 μm (A and B) and 2 μm (C).
The intra-organellar localization of Chlase has been no less controversial than the intracellular location, and data supporting plastid localization mostly point to localization in the inner envelope membrane of the plastid (Hirschfeld and Goldschmidt, 1983; Matile et al., 1997). However, these data specifically addressed potential inner envelope localization and were not designed to provide an unbiased picture of the intra-organellar distribution of Chlase. We therefore induced color-break in citrus fruit by ethylene and looked at magnifications of flavedo cell plastids in which Chlase was detected by in situ immunofluorescence. Although the confocal microscope resolution is limiting for analysis of intra-organellar compartmentalization, the high magnifications show colocalization of most of the Alexa-488 and the chlorophyll fluorescence within the plastid, suggesting that most of the Chlase protein is associated with the photosynthetic membranes. This observation was supported by the confocal microscope colocalization program analysis tool, where the red channel, corresponding to the emission of chlorophyll autofluorescence (Fig. 2C, 1), and the green channel, corresponding to Chlase immunofluorescence (Fig. 2C, 2), were merged, resulting in a colocalization signal shown in yellow (Fig. 2C, 3). The percentage of the yellow signal from total fluorescence observed was calculated to be approximately 74.6% (sd = 1.5; an average of three different images).
Expression of Chlase Is Correlated with Chlorophyll Breakdown at the Cellular Level
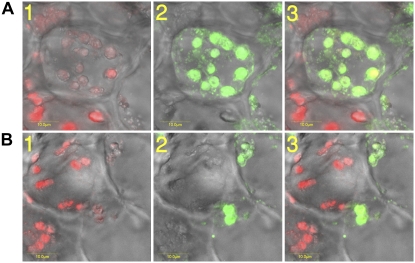
It was previously shown that treatment of maturing citrus fruit with exogenous ethylene dramatically enhances the speed of color break at the whole tissue level (Shimokawa et al., 1978; Purvis and Barmore, 1981). The current work, based on confocal microscopy, shows that ethylene-induced chlorophyll breakdown is not synchronous between cells of the flavedo tissue; while some cells display reduced chlorophyll levels after 24 h of ethylene treatment, in other cells the chlorophyll appears to be still intact (Fig. 3). Thus, it appears that the ethylene response kinetics is not uniform in citrus flavedo tissue at the cellular level. We therefore investigated the correlation between ethylene-induced chlorophyll breakdown and Chlase expression at the cellular level by in situ immunofluorescence (Fig. 3), monitored by laser scanning confocal microscopy. Immunofluorescence of two representative sample sections is presented (Fig. 3), showing a clear negative correlation between Chlase protein levels (green florescence; Fig. 3A, 1, and B, 1) and chlorophyll levels (red florescence; Fig. 3A, 2, and B, 2) also presented in a merged format (Fig. 3A, 3, and B, 3). Chloroplasts in cells displaying high levels of Chlase appear to have undergone massive chlorosis, while chloroplasts in cells lacking Chlase appear to contain mostly intact chlorophyll. We note that quenching of the chlorophyll fluorescence by the Alexa-488 fluorophore or vice versa has been ruled out (Alexa-488 excitation and emission = 494 and 519 nm, respectively; chlorophyll a/b excitation and emission = 425/460 and 650/670 nm, respectively), and thus the results reflect the actual levels of fluorescence of chlorophyll and Alexa-488.
Figure 3.
Citrus Chlase expression versus chlorophyll levels in ethylene-treated lemon fruit monitored by in situ immunofluorescence. Mature green lemon fruit were treated with ethylene (20 μL L−1) at 25°C in the dark for 24 h in a 4-L sealed container. The container was ventilated once after 12 h, followed by injection of fresh ethylene, to maintain CO2 levels below 1%. The flavedo of the treated fruit was dissected and used for obtaining cross sections (70 μm thick) using a vibratome. Tissue sections were fixed and dressed with affinity-purified anti-citrus Chlase antibody (rabbit) followed by Alexa-488-conjugated (goat anti-rabbit) secondary antibody. Fluorescence was visualized using a laser scanning confocal microscope as described in “Materials and Methods.” Images of representative cells (A and B) are presented as follows: 1, confocal image recorded simultaneously in transmitted and red fluorescence mode (i.e. chlorophyll fluorescence superimposed on the bright-field image); 2, confocal image recorded simultaneously in transmitted and green fluorescence mode (i.e. immunofluorescence resulting from the Alexa-488 secondary antibody, corresponding to detection by anti-Chlase antibody, superimposed on the bright-field image); 3, confocal image recorded simultaneously for transmitted, red, and green fluorescence (i.e. red fluorescence corresponds to chlorophyll autofluorescence, green immunofluorescence resulting from the Alexa-488 secondary antibody, corresponding to detection by anti-Chlase antibody, superimposed on the bright-field image). Scale bar = 10 μm (A and B).
Accumulation and Processing Kinetics of Chlase Protein in Response to Ethylene Induction
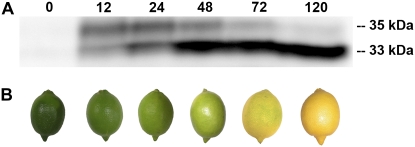
Purified citrus Chlase protein was previously shown to be de novo synthesized following ethylene treatment and was characterized, based on N-terminal sequencing, to be a processed protein lacking the N-terminal 21 amino acids encoded by the cDNA (Trebitsh et al., 1993; Jacob-Wilk et al., 1999). However, the precursor form of citrus Chlase, as well as putative precursors of Chlases from other plant species, were never observed in their respective endogenous plant systems, and it is unclear whether the precursor forms accumulate or are immediately processed (Trebitsh et al., 1993; Tsuchiya et al., 1999). To establish the kinetics of citrus Chlase protein expression and processing in response to ethylene during fruit color break, mature green lemon fruits were treated with (20 μL L−1) ethylene for up to 120 h. The flavedo tissue was harvested, and proteins were extracted and analyzed by immunoblot using citrus Chlase-specific antibodies (Fig. 4). Chlase protein was not detected in nontreated fruit flavedo (time point 0) but accumulated after 12 h of ethylene treatment (Fig. 4A), predominantly in the “precursor” form (approximately 35 kD). Levels of the precursor form were maintained for 24 h, after which they gradually decreased concomitantly with a dramatic increase in the levels of the “mature” Chlase form (approximately 33 kD). Fruit color-break was monitored and documented at the same time points used for protein extraction (Fig. 4B). Deep green mature lemons (0 h) became light green after the first 24 h, yellow-green after 48 h, and bright yellow after 120 h, demonstrating the correlation between Chlase protein accumulation, processing (maturation), and fruit peel color-break.
Figure 4.
Expression and processing kinetics of citrus Chlase during ethylene-induced fruit color-break. Mature green lemon fruit were treated with ethylene (20 μL L−1) at 25°C in the dark for 0, 12, 24, 48, 72, or 120 h in a 600-L sealed container. The container was ventilated once every 24 h, followed by injection of fresh ethylene, to maintain CO2 levels below 1%. The flavedo of the treated and control fruit was peeled, frozen in liquid nitrogen, and ground to a powder. A, Total protein from each time point was acetone precipitated, extracted in USB protein extraction buffer, separated by SDS-PAGE (30 μg protein/lane), blotted, and dressed with anti-citrus Chlase antibodies. Protein bands bound by the Chlase-specific antibodies were visualized by chemiluminescence. Relative molecular weights of the bands detected by the antibody are denoted. B, Representative lemon fruit phenotype was documented for each time point of ethylene treatment.
Identification of Precursor and Mature Citrus Chlase Forms by Mass Spectrometry
The kinetics of citrus Chlase precursor expression and processing during fruit color-break presented above was inferred based on immunodetection using anti-citrus Chlase antibodies; however, further verification of Chlase forms and the putative processing site are required to reach established conclusions. Therefore, the putative precursor and mature Chlase forms were purified from ethylene-treated lemon peel (sample A after 12 h of treatment and sample B after 120 h of treatment) by immunoprecipitation followed by SDS-PAGE. Purified bands of approximately 35 kD and approximately 33 kD were subjected to proteolysis followed by identification of peptides by tandem MS (MS/MS) analysis.
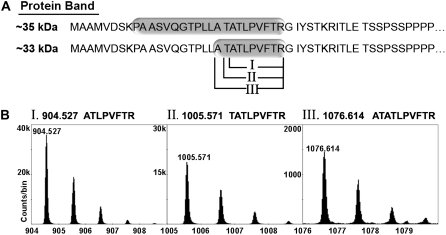
Both protein bands yielded peptides confirmed by MS/MS to be Chlase versions. MS analysis of peptides derived from the approximately 35-kD protein band resulted in 44% overall sequence coverage corresponding to citrus Chlase (data not shown). The tryptic polypeptide (K)PAASVQGTPLLATATLPVFTR, which starts at amino acid 9 of the Chlase open reading frame, was identified in the sample (Fig. 5), confirming that the approximately 35-kD protein band is the precursor Chlase version that includes the first 21 amino acids encoded by the open reading frame.
Figure 5.
MS-based identification of precursor and mature Chlase versions and the N-terminal processing sites. Mature green lemon fruit were treated with ethylene (20 μL L−1) at 25°C in the dark for 12 or 120 h in a 600-L sealed container. The container was ventilated once every 24 h, followed by injection of fresh ethylene, to maintain CO2 levels below 1%. The flavedo of the treated and control fruit was peeled, frozen in liquid nitrogen, and ground to a powder. Total protein from each time point was acetone precipitated, extracted in RIPA buffer, and Chlase versions were immunoprecipitated using affinity-purified anti-citrus Chlase antibodies. Putative precursor (approximately 35 kD) and mature (approximately 33 kD) Chlase versions were separated from the antibody by SDS-PAGE, visualized by colloidal Coomassie staining, and excised from the gel. Protein bands were digested with trypsin in-gel and eluted peptides were analyzed by MS/MS. A, The N-terminal tryptic and semi-tryptic peptides detected after trypsinization of the approximately 35-kD and 33-kD protein bands, respectively. Tryptic and semi-tryptic peptides are highlighted on the N-terminal amino acid sequence of Chlase (starting at Met-1). Roman numerals (I, II, III) designate the three different semi-tryptic peptides detected. B, Semi-tryptic peptides I, II, and III observed in the TOF mass spectrum of the HPLC fraction that eluted at 23 min. In each case, the multiple peaks correspond to the normal isotopic distribution (i.e. to the inclusion of 0, 1, 2, or 3 13C atoms in the peptide molecule). Mass of the monoisotopic peaks (zero 13Cs) appears above each peak and above each together with the sequences that were confirmed by MS/MS measurements (Table I). Note the different vertical scales in the three segments; all three peptides were observed in the same mass spectrum, collected at the same time under identical instrumental conditions, so the range of observed intensities of the monoisotopic peaks in the three peptide spectra (from approximately 1,500 counts/bin to almost 40,000 counts/bin) provides an estimation of the quantity of each of the detected peptides.
MS analysis of peptides derived from the approximately 33-kD protein band resulted in 74% overall sequence coverage corresponding to citrus Chlase (data not shown). To explore possible N-terminal processed peptides, arbitrary in silico sequence fragments of citrus Chlase were scanned against the observed mass spectra in search of relevant semi-tryptic peptides. Monoisotopic peaks were found for three semi-tryptic sequence candidates starting at locations 20 to 22 of the open reading frame: (1) ATLPVFTR @ m+H = 904.527; (2) TATLPVFTR @ m+H = 1,005.571; and (3) ATATLPVFTR @ m+H = 1,076.614 (Fig. 5). The retention times for all three sequences are in good agreement with the approximately 24-min value predicted by the peptide hydrophobicity calculator (Krokhin et al., 2006). The MS/MS measurement provides daughter ion pattern data that confirms their sequence identities (Table I) and demonstrates that the approximately 33-kD protein band contains the processed/mature Chlase versions. As all three peptides appeared in the same spectrum (i.e. they were acquired under identical instrument conditions), rough quantity estimation was performed based on their monoisotopic peak intensities. On this basis, the sample appears to contain approximately 63% ATLPVFTR (1, i.e. corresponding to processing after amino acid 21), approximately 33% TATLPVFTR (2, i.e. corresponding to processing after amino acid 20), and approximately 4% ATATLPVFTR (3, i.e. corresponding to processing after amino acid 19), demonstrating that Chlase processing occurs not only following amino acid 21 of the precursor, as previously known, but rather at three consecutive positions.
Table I.
Confirmation of N-terminal semi-tryptic peptides by MS/MS
Daughter ions (b ions) observed from MS/MS spectra of the three N-terminal semi-tryptic peptides (I, II, and III) derived from the 33-kD protein band (see Fig. 4). A full set of y-ions was also observed for peptides I and II, but not for III, probably due to the low parent ion intensity.
| Daughters | Ion | m/z Observed | MH+ Calc. | Δ |
|---|---|---|---|---|
| mD | ||||
| (I) 904.527 ATLPVFTR | ||||
| ATLPVFTR | b8 | 904.529 | 904.526 | 3 |
| ATLPVFT | b7-18 | 712.416 | 712.404 | 12 |
| ATLPVF | b6 | 629.371 | 629.366 | 5 |
| ATLPV | b5 | 482.299 | 482.298 | 1 |
| ATLP | b4 | 383.228 | 383.230 | −2 |
| ATL | b3 | 286.177 | 286.177 | 0 |
| AT | b2 | 173.090 | 173.093 | −3 |
| (II) 1,005.571 TATLPVFTR | ||||
| TATLPVFTR | b9 | 1,005.580 | 1,005.573 | 7 |
| TATLPVFT | b8-18 | 813.453 | 813.451 | 2 |
| TATLPVF | b7-18 | 712.401 | 712.404 | −3 |
| TATLPV | b6-18 | 565.327 | 565.335 | −8 |
| TATLP | b5 | 484.283 | 484.277 | 6 |
| TATL | b4-18 | 369.215 | 369.214 | 1 |
| TAT | b3-18 | 256.129 | 256.130 | −1 |
| TA | b2 | 173.098 | 173.093 | 5 |
| (III) 1,076.614 ATATLPVFTR | ||||
| ATATLPVFTR | b10 | 1,076.616 | 1,076.610 | 6 |
| ATATLPVF | b8 | 801.449 | 801.451 | −2 |
| ATATLP | b6-18 | 537.312 | 537.304 | 8 |
| ATATL | b5-18 | 440.249 | 440.251 | −2 |
| ATAT | b4-18 | 327.175 | 327.167 | 8 |
| ATA | b3-18 | 226.123 | 226.120 | 3 |
| AT | b2 | 173.093 | 173.093 | 0 |
DISCUSSION
Due to continuous controversy in the literature, determining Chlase enzyme intracellular localization has become a key to understanding its role, mode of action, and regulation in the process of chlorophyll breakdown. In this work, we provide an unbiased localization study demonstrating that citrus Chlase expressed during ethylene-induced fruit color-break is a plastid-localized protein. Furthermore, Chlase protein levels are clearly negatively correlated with chlorophyll levels at the cellular level. Both findings are supportive of the role of Chlase in chlorophyll catabolism in the plastid during citrus fruit color break. This conclusion is further supported by previous work demonstrating that citrus Chlase transiently expressed in tobacco (Nicotiana tabacum) leaf protoplasts is localized to the chloroplast and leads to chlorophyll breakdown in vivo (Harpaz-Saad et al., 2007). We note that our results differ from recent data suggesting cytoplasmic localization of Arabidopsis Chlases based on transient expression of N- and C-terminal GFP fusions (Schenk et al., 2007). Similarly, our previous attempts to reveal the subcellular localization of citrus Chlase by transient expression of both precursor and mature Chlase fused to GFP (N-terminal, C-terminal, and internal fusions) in tobacco protoplasts resulted in cytoplasmic localization concomitant with the loss of in vivo enzyme function (Harpaz-Saad et al., 2007). We therefore speculate that GFP fusions may interfere with the natural localization of Chlases due to masking of amino- and carboxy-terminal targeting and modification signals (Tian et al., 2004). We also note that the N terminus of most Chlases does not conform to any known criteria for chloroplast transit peptides and recently provided evidence that the citrus Chlase N terminus does not function as a plastid-targeting transit peptide (Harpaz-Saad et al., 2007). Therefore, it is likely that a different domain of Chlase (i.e. not the N terminus) is responsible for plastid targeting, and perhaps it is not surprising that the amino-terminal 47 amino acids of Arabidopsis Chlase (AtCLH2) did not direct a fused GFP reporter to the plastid (Schenk et al., 2007). Based on our current results, generated on the endogenous enzyme in a natural system, we suggest that citrus Chlase and its homologs in other plant species are plastid localized.
Beyond the intracellular plastid localization that was established in this work, Chlase intra-organellar localization is still a major challenge, limiting our understanding of the regulation of Chlase function. Matile et al. (1997) demonstrated enrichment of Chlase activity in purified Brassica and barley inner envelope chloroplast membranes, suggesting that the enzyme is located in the envelope membrane and is spatially separated from the chlorophyll located in the photosynthetic membranes. The interaction between the enzyme and the substrate was speculated to be “latent” and to involve proteins that function in transporting chlorophyll molecules from the photosynthetic membranes to the proposed sites of breakdown in the inner envelope membrane. However, the results of Matile et al. (1997), suggesting inner envelope membrane localization, account for only 6% of the total Chlase activity, while the remaining 94% of Chlase enzyme may possibly be located elsewhere and warrant further investigation. While the resolution of the confocal microscope is insufficient to provide a complete picture of intra-organellar localization, our results showing colocalization of approximately 75% of the chlorophyll and Chlase signals suggests that the majority of Chlase enzyme is associated with the photosynthetic membranes. This finding is in line with recent results that suggest that Chlase and its substrate chlorophyll come into contact when the enzyme reaches intact chlorophyll located in the photosynthetic membranes (Harpaz-Saad et al., 2007). Yet, some of the Chlase enzyme is clearly located in the plastid envelope membrane both in citrus photosynthetic membrane-free chromoplasts and in barley and Brassica chloroplasts (Hirschfeld and Goldschmidt, 1983; Matile et al., 1997). Because recent results suggest contact between the envelope and photosynthetic membranes (Shimoni et al., 2005), perhaps the mobilization of envelope-located Chlase to the stromal lamellae is facilitated by membrane trafficking.
Accumulation of the Chlase precursor form in its natural tissue was not previously seen in any plant species and was made possible by modulating protein extraction procedures. In this work, we show that during ethylene-induced color-break, citrus Chlase initially accumulates primarily in the precursor form (approximately 35 kD), which is subsequently N-terminally processed to a mature enzyme form (approximately 33 kD), demonstrating that processing is not an immediate event. The role of Chlase N-terminal processing in chlorophyll breakdown during citrus fruit color-break remains unresolved. However, previous work involving expression of citrus Chlase in heterologous systems suggests that the cleaved N-terminal 21 amino acids are not necessary for targeting the protein to the plastid, and their removal results in a more active enzyme in vivo (Harpaz-Saad et al., 2007). Furthermore, detection of both precursor and mature citrus Chlase forms in chloroplasts, purified from tobacco protoplasts transiently expressing citrus Chlase, suggests that processing occurs within the plastid and thus may be involved in enzyme regulation.
The N-terminal processing site of citrus Chlase following amino acid 21 was previously inferred by comparing the N-terminal protein sequence of the mature protein obtained by Edman degradation (Trebitsh et al., 1993) to the open reading frame encoded by the cDNA (Jacob-Wilk et al., 1999). In the current work, more sensitive tools (MS) were used to show that processing occurs at three consecutive positions: following amino acids 19, 20, and 21 at a ratio of 4%:33%:63%, respectively. The current MS/MS results conform to the previous Edman degradation results (Trebitsh et al., 1993), because the most abundant form of mature Chlase (approximately 63%) is the version processed following amino acid 21. The functional significance of multiple processing sites is not clear, but a similar example has previously been described (Zybailov et al., 2008). One possibility is that the processing protease is not precise, resulting in multiple processing sites at different efficiencies. A second possibility is that a stepwise proteolysis is catalyzed by one or more proteolytic enzymes resulting in different lengths of the N terminus of the semi-tryptic peptide detected. Whether or not a stepwise processing has any regulatory significance is currently unclear and needs to be further investigated.
The N-terminal processing site has been experimentally determined up to now for only two Chlases: Citrus sinensis (Trebitsh et al., 1993; Jacob-Wilk et al., 1999; this work) and Chenopodium album (Tsuchiya et al., 1999). While the similarity of the N-terminal sequences between these Chlases is extremely low, processing occurs in both proteins at amino acids of similar nature, mostly before or after Thr in Citrus (this work) and before Ser in Chenopodium (Tsuchiya et al., 1999). It remains to be determined whether N-terminal processing in all plant Chlases occurs at Ser/Thr residues and what other features (i.e. sequence or structural features) direct processing site specificity.
Finally, much of the published data regarding the regulation of the chlorophyll breakdown pathway, and specifically regarding Chlase, was obtained in senescing leaves, involving the chloroplast-gerontoplast developmental transition (Matile et al., 1997; Fang et al., 1998; Kariola et al., 2005; Ben-Yaakov et al., 2006; Schenk et al., 2007) or in response to induction by mediators of biotic stress (Tsuchiya et al., 1999; Benedetti and Arruda, 2002). Here, we investigated the role of Chlase in fruit color-break, reflecting the developmentally regulated transition of chloroplasts to chromoplasts. We note that chromoplasts differ markedly from gerontoplasts, which are the plastid structures of senescing tissue (Sitte et al., 1980; Bouvier and Camara, 2007; Krupinska, 2007). Indeed, the transition to chromoplasts involves both catabolic and anabolic processes, while the transition to gerontoplasts during senescence is strictly catabolic. In this context, we note that recent work questions the role of Arabidopsis Chlases (AtCLH1 and AtCLH2) in chlorophyll breakdown during senescence, because double mutant plants do not display a significant delay in senescence-associated chlorophyll breakdown (Schenk et al., 2007). In contrast, data regarding the Chlase of citrus fruit in the native and the heterologous systems suggests that it plays a major role in chlorophyll catabolism in vital tissue (Harpaz-Saad et al., 2007; this work).We further note that AtCLH1 was demonstrated to be involved in chlorophyll turnover in healthy plants or in breakdown following treatment with methyl-jasmonate or in response to high light (Kariola et al., 2005). These results demonstrate a role for AtCLH1 in chlorophyll breakdown under natural physiological conditions in vital tissues, such as during natural turnover, in response to wounding or photobleaching. Thus, it is possible that regulation of chlorophyll breakdown may differ between vital physiological developmental stages (i.e. photosynthetic leaves and ripening fruit) versus senescence, which is a metabolite salvaging step in aging tissue in which diverse catabolic machineries are induced. Further work will be needed to investigate whether chlorophyll breakdown during these distinct plastid transitions is indeed regulated differently.
MATERIALS AND METHODS
Plant Material and Treatment
Mature green lemon (Citrus limon) fruit were treated with ethylene (20 μL L−1) at 25°C in the dark for 0, 12, 24, 48, 72, or 120 h in a 600-L (for protein extraction procedures) or 4-L (for immuno in situ procedure) sealed container. The container was ventilated once every 24 h or 12 h, respectively, followed by injection of fresh ethylene to maintain CO2 levels below 1%.
Extraction and Analysis of Protein from Citrus Flavedo
Flavedo Harvesting
The flavedo (the outer, pigmented part of the fruit peel) was peeled, frozen in liquid nitrogen, and ground to a powder. Ground powder samples were acetone precipitated by addition of 5 volumes (w/v) of cold acetone (−20°C), followed by overnight incubation at −20°C. Samples were than centrifuged for 10 min at 15,000g, 4°C.
Protein Extraction
Acetone-precipitated protein samples (45 mg) were homogenized and incubated in 450 μL of USB buffer (20 mm Tris-HCl, pH 7.5, 8 m urea, 4.5% SDS, and 1 m β-mercaptoethanol) for 1 h at 25°C. Extracts were centrifuged for 15 min at 15,000g at 25°C, the supernatant was transferred to a new tube, and the centrifugation step was repeated two additional times to obtain a clear soluble protein solution. Equal amounts of total protein from each sample were separated by SDS-PAGE (12%) and subjected to protein gel-blot analysis as described below.
Western-Blot Analysis
Proteins separated by SDS-PAGE were transferred onto nitrocellulose membranes (Schleicher and Schuell) as described (Page and Thorpe, 1996). Membranes were blocked and incubated overnight with the following antibodies: mouse anti-citrus Chlase polyclonal antibodies diluted 1:1,000 (Harpaz-Saad et al., 2007); and rabbit anti-LHC II polyclonal antibodies diluted 1:40,000 (Agrisera). Immunoblots were developed using the SuperSignal West Dura substrate (Pierce) after incubation for 1 h with goat anti-mouse horseradish peroxidase-conjugated secondary antibody diluted 1:10,000 (Jackson ImmunoResearch Laboratories). Dressed immunoblots were visualized in a FluorChem 8800 imaging system (Alpha Innotech). Between application of different antibodies, membranes were stripped using Restore protein gel-blot stripping buffer (Pierce).
In Situ Immunofluorescence Staining
Mature green lemon fruit were treated with ethylene as previously described. The flavedo was sliced for cross sections (70 μm thick) using a vibratome (model 100plus sectioning system; Technical Products International). Tissue cross sections were fixed for 1.5 h at 25°C in PBS (0.1 m sodium phosphate buffer, pH 7.4) containing 3% paraformaldehyde and 1% glutaraldehyde, followed by three 30-min washes with PBS. Membrane permeability was obtained by incubation in PBS buffer contain 1% Triton X-100 for 2 h at 25°C, followed by three 30-min washes with PBS. Samples were then incubated with the primary antibody (affinity-purified rabbit anti-Chlase polyclonal antibody at a concentration of 19 ng antibody/mL buffer) in PBS containing 0.05% Triton X-100 and 1% bovine serum albumin (BSA) for 1.5 h at 25°C, followed by three 30-min washes with PBS. Control samples were incubated in PBS containing 0.05% Triton X-100 and 1% BSA without antibody.
From this stage on, all procedures were carried out in the dark. Samples were incubated with the secondary antibody (Alexa-488-conjugated goat anti-rabbit; Jackson ImmunoResearch Laboratories) in PBS containing 0.05% Triton X-100 and 1% BSA for 1.5 h at 25°C, followed by three 30-min washes with PBS. The tissue samples were then mounted with Elvanol (DuPont) under a coverslip.
Confocal Microscopy and Image Analysis
Images were acquired using a IX81 fully automated Olympus microscope equipped with a 488-nm argon laser for Alexa-488 excitation and a narrow-band emission filter (515–525 nm) to filter Alexa-488 from chlorophyll green autofluorescence, a BA 660 IF emission filter for chlorophyll red autofluorescence, and a PLAPO60XWLSM (numerical aperture 1.00) objective. Transmitted light images were acquired using Nomarski differential interference contrast. Image analysis was carried out using the flow view software (Olympus).
Antibody Affinity Purification
Polyclonal anti-citrus Chlase antibodies were raised in rabbit (immunological services, Weizmann Institute of Science, Rehovot, Israel) against a recombinant StrepTag-thioredoxin-ChlaseΔN fusion protein expressed in bacteria as previously described (Harpaz-Saad et al., 2007). Anti-Chlase antibody affinity purification was performed by affinity binding to recombinant Chlase protein cross-linked to cyanogen bromide-activated Sepharose 4B as described by the manufacturer (GE Healthcare Bio-Sciences AB). Eluted antibody concentration was determined using the BCA Protein Assay kit as described by the manufacturer (Pierce).
Immunoprecipitation and Gel Purification of Chlase Versions
Acetone powder samples (3 g) of ethylene-treated (12 and 120 h) lemon fruit peel were homogenized and incubated in 30 mL of radioimmunoprecipitation assay (RIPA) buffer (20 mm Tris-HCl, pH 7.2, 100 mm NaCl, 0.1% SDS, 1% NP-40, 1% sodium deoxycholate) containing complete EDTA-free protease inhibitor cocktail (Roche) in a rotator at 4°C for 2 h. Extracts were centrifuged for 15 min at 15,000g at 4°C, the supernatant was transferred to a new tube, and the centrifugation step was repeated two additional times to obtain a clear soluble protein solution. The protein extract was then incubated with (40 μL) anti-Chlase antibodies (raised in mice; Harpaz-Saad et al., 2007) in a rotator at 4°C for 12 to 15 h.
Protein-A agarose beads (10 μL), prepared as described by the manufacturer (Sigma), were added and rotational incubation was continued for an additional 4 h. The immunoprecipitated complex was collected by centrifugation at 420g, 4°C for 2 min, and the supernatant was aspirated. The agarose beads were washed three times with ice-cold RIPA buffer. Immunoprecipitated protein was eluted in 30 μL of USB buffer (20 mm Tris-HCl, pH 7.5, 8 m urea, 4.5% SDS, and 1 m β-mercaptoethanol) by incubation for 15 min at 25°C, followed by centrifugation for 5 min at 420g, 20°C. The protein containing supernatant was transferred to a new tube.
To separate and isolate Chalse protein versions, immunoprecipitated protein samples were separated by 12.5% SDS-PAGE as described (Page and Thorpe, 1996). Gels were then stained in Colloidal Coomassie under gentle shaking at 25°C as follows. Gels were initially fixed by incubation for 1 h in fixation solution (50% ethanol; 12% acetic acid). Gels were then transferred to freshly made staining solution (10% [w/v] ammonium sulfate; 10% phosphoric acid; 0.12% colloidal Coomassie Brilliant Blue G-250 [Fluka]; 20% methanol, in HPLC grade distilled, deionized water). Gels were stained for 16 to 20 h, washed twice for 20 min in destaining solution (20% ethanol), followed by washing with distilled, deionized water for 20 min. Protein gel bands of the Chlase precursor and mature versions were carefully excised and submitted to analysis by MS as detailed below.
Mass Spectrometric Analyses of Protein Bands
Protein gel slices was first subjected in situ to a full tryptic digest overnight. In preliminary examinations, the digest was measured directly (without prior reversed-phase HPLC fractionation) in a matrix-assisted laser desorption/ionization tandem quadrupole/time-of-flight (MALDI-QqTOF) mass spectrometer (Loboda et al., 2000). For more detailed analyses, the proteolyzed fragments were separated into 40 1-min fractions by reversed-phase HPLC and deposited onto a MALDI target by a robot previously constructed in the Manitoba laboratory (Krokhin et al., 2004). The mass-to-charge spectrum for each spot was then acquired in the same TOF instrument. The candidate protein was initially identified using SMART, a software tool for retention time-based peptide mass fingerprinting developed at the University of Manitoba (Krokhin et al., 2006). We note that some protein samples were identified as a service in the Smoler Proteomics Center in Israel.
To explore possible N-terminal peptides, we generated in silico arbitrary sequence fragments from the citrus Chlase-deduced amino acid sequence and scanned them against the observed mass spectra, allowing up to 10 ppm variation in mass and ±5 min in retention time.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AF160869.
Acknowledgments
We thank Ms. Hanita Zemach for advice and assistance in preparing samples for in situ immunofluorescence and Dr. Einat Sadot for generous contributions of antibodies. We thank Dr. Hinanit Koltai for use of the Vibratome for tissue sectioning, and Dr. Edna Pesis, Dr. Amnon Lichter, Dr. Amnon Lers, Mr. Oleg Feygenberg, and Mr. Yohanan Zutahy for advice and assistance with ethylene treatments. We thank Arten Hadarim (R-10 Agriculture, Nir Zvi, Israel) for generous contributions of lemons.
This work was supported by the Israel Science Foundation (grant no. 1291/04) and by a Levi Eshkol Ph.D. scholarship (to T.A.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yoram Eyal (eyalab@volcani.agri.gov.il).
Open Access articles can be viewed online without a subscription.
References
- Alós E, Cercós M, Rodrigo MJ, Zacarías L, Talón M (2006) Regulation of color break in citrus fruits: changes in pigment profiling and gene expression induced by gibberellins and nitrate, two ripening retardants. J Agric Food Chem 54 4888–4895 [DOI] [PubMed] [Google Scholar]
- Amir-Shapira D, Goldschmidt EE, Altman A (1987) Chlorophyll catabolism in senescing plant tissues: in vivo breakdown intermediates suggest different degradative pathways for citrus fruit and parsley leaves. Proc Natl Acad Sci USA 84 1901–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardao C, Vennesland B (1960) Chlorophyllase activity of spinach chloroplastin. Plant Physiol 35 368–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry S, Mani J, Hörtensteiner S (2008) Stay-green protein, defective in Mendel's green cotyledon mutant, acts independent and upstream of pheophorbide a oxygenase in the chlorophyll catabolic pathway. Plant Mol Biol 67 243–256 [DOI] [PubMed] [Google Scholar]
- Ben-Yaakov E, Harpaz-Saad S, Galili D, Eyal Y, Goldschmidt E (2006) The relationship between chlorophyllase activity and chlorophyll degradation during the course of leaf senescence in various plant species. Isr J Plant Sci 54 129–136 [Google Scholar]
- Benedetti CE, Arruda P (2002) Altering the expression of the chlorophyllase gene ATHCOR1 in transgenic Arabidopsis caused changes in the chlorophyll-to-chlorophyllide ratio. Plant Physiol 128 1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier F, Camara B (2007) The Role of Plastids in Ripening Fruits. In Wise RR, Hoober JK, eds, The Structure and Function of Plastids. Springer, Dordrecht, The Netherlands, pp 419–432
- Brandis A, Vainstein A, Goldschmidt EE (1996) Distribution of chlorophyllase among components of chloroplast membranes in orange (Citrus sinensis) leaves. Plant Physiol Biochem 34 49–54 [Google Scholar]
- Fang Z, Bouwkamp JC, Solomos T (1998) Chlorophyllase activities and chlorophyll degradation during leaf senescence in non-yellowing mutant and wild type of Phaseolus vulgaris L. J Exp Bot 49 503–510 [Google Scholar]
- Garcia AL, Galindo L (1991) Chlorophyllase in citrus leaves: localization and partial purification of the enzyme. Photosynthetica 25 105–111 [Google Scholar]
- Goldschmidt EE (1988) Regulatory aspects of the chloro-chromoplast interconversions in senescing citrus fruit peel. Isr J Bot 37 123–130 [Google Scholar]
- Goldschmidt EE, Huberman M, Goren R (1993) Probing the role of endogenous ethylene in the degreening of citrus fruit with ethylene antagonists. Plant Growth Regul 12 325–329 [Google Scholar]
- Harpaz-Saad S, Azoulay T, Arazi T, Ben-Yaakov E, Mett A, Shiboleth YM, Hörtensteiner S, Gidoni D, Gal-On A, Goldschmidt EE, et al (2007) Chlorophyllase is a rate-limiting enzyme in chlorophyll catabolism and is posttranslationally regulated. Plant Cell 19 1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschfeld KR, Goldschmidt EE (1983) Chlorophyllase activity in chlorophyll-free citrus chromoplasts. Plant Cell Rep 2 117–118 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S (1999) Chlorophyll breakdown in higher plants and algae. Cell Mol Life Sci 56 330–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S (2004) The loss of green color during chlorophyll degradation: a prerequisite to prevent cell death? Planta 219 191–194 [DOI] [PubMed] [Google Scholar]
- Hu G, Yalpani N, Briggs SP, Johal GS (1998) A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant in maize. Plant Cell 10 1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias DJ, Tadeo FR, Legaz F, Primo-Millo E, Talon M (2001) In vivo sucrose stimulation of colour change in citrus fruits epicarps: interactions between nutritional and hormonal signals. Physiol Plant 112 244–250 [DOI] [PubMed] [Google Scholar]
- Ishikawa A, Okamoto H, Iwasaki Y, Asahi T (2001) A deficiency of coproporphyrinogen III oxydase causes lesion formation in Arabidopsis. Plant J 27 89–99 [DOI] [PubMed] [Google Scholar]
- Jacob-Wilk D, Holland D, Goldschmidt EE, Riov J, Eyal Y (1999) Chlorophyll breakdown by chlorophyllase: isolation and functional expression of the Chlase1 gene from ethylene-treated citrus fruit and its regulation during development. Plant J 20 653–661 [DOI] [PubMed] [Google Scholar]
- Kariola T, Brader G, Li J, Palva ET (2005) Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell 17 282–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H (1991) Differentiation of amyloplasts and chromoplasts. In L Bogorad, IK Vasil, eds, The Photosynthetic Apparatus: Molecular Biology and Operation. Cell Culture and Somatic Cell Genetics of Plants, Vol 7B. Academic Press, San Diego, pp 395–415
- Krokhin OV, Craig R, Spicer V, Ens W, Standing KG, Beavis RC, Wilkins JA (2004) An improved model for prediction of retention times of tryptic peptides in ion pair reversed-phase HPLC: its application to protein peptide mapping by off-line HPLC-MALDI MS. Mol Cell Proteomics 3 908–919 [DOI] [PubMed] [Google Scholar]
- Krokhin OV, Spicer V, Standing KG, Wilkins JA, Ens W (2006) “SMART” (search by mass and retention time): a new tool for HPLC-MALDI MS peptide mass fingerprinting. In Proceedings of the 54th ASMS Conference on Mass Spectrometry and Allied Topics. American Society for Mass Spectrometry, Santa Fe, NM, abstract wp27/533
- Krossing G (1940) Versuche zur Lokalisation einiger Fermente in den verschiedenen Zellbestandteilen der Spinatblätter. Biochem Z 305 359–373 [Google Scholar]
- Krupinska K (2007) Fate and activities of plastids during leaf senescence. In RR Wise, JK Hoober, eds, The Structure and Function of Plastids. Springer, Dordrecht, The Netherlands, pp 433–449
- Loboda AV, Krutchinsky AN, Bromirski M, Ens W, Standing KG (2000) A tandem quadrupole/time-of-flight mass spectrometer with a matrix-assisted laser desorption/ionization source: design and performance. Rapid Commun Mass Spectrom 14 1047–1057 [DOI] [PubMed] [Google Scholar]
- Mach JM, Castillo AR, Hoogstraten R, Greenberg JT (2001) The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc Natl Acad Sci USA 98 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaya K, Yoshinori I, Hikaru M, Minoru S, Hiroshi H, Masamichi Y (2004) Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiol 134 824–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matile P, Hörtensteiner S, Thomas H (1999) Chlorophyll degradation. Annu Rev Plant Physiol Plant Mol Biol 50 67–95 [DOI] [PubMed] [Google Scholar]
- Matile P, Schellenberg M (1996) The cleavage of phaeophorbide a is located in the envelope of barley gerontoplasts. Plant Physiol Biochem 34 55–59 [Google Scholar]
- Matile P, Schellenberg M, Vicentini F (1997) Localization of chlorophyllase in the chloroplast envelope. Planta 201 96–99 [Google Scholar]
- Mock HP, Heller W, Molina A, Neubohn B, Sandermann H, Grimm B (1999) Expression of uroporphyrinogen decarboxylase or coproporphyrinogen oxidase RNA in tobacco induces pathogen defense responses conferring increased resistance to tobacco mosaic virus. J Biol Chem 274 4231–4238 [DOI] [PubMed] [Google Scholar]
- Molina A, Volrath S, Guyer D, Maleck K, Ryals J, Ward E (1999) Inhibition of protoporphyrinogen oxidase expression in Arabidopsis causes a lesion-mimic phenotype that induces systemic acquired resistance. Plant J 17 667–678 [DOI] [PubMed] [Google Scholar]
- Okazawa A, Tango L, Itoh Y, Fukusaki E, Kobayashi A (2006) Characterization and subcellular localization of chlorophyllase from Ginkgo biloba. Z Naturforsch 61(C) 111–117 [DOI] [PubMed] [Google Scholar]
- Page M, Thorpe R (1996) Protein blotting by electroblotting. In JM Walker, ed, The Protein Protocols Handbook. Humana Press, Totowa, NJ, pp 317–319
- Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, Lee SK, Jeong SW, Seo HS, Koh HJ, et al (2007) The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19 1649–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechulla B, Glick RE, Bahl H, Melis A, Gruissem W (1987) Changes in photosynthetic capacity and photosynthetic protein pattern during tomato fruit ripening. Plant Physiol 84 911–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porat R, Weiss B, Chen L, Daus A, Goren R, Droby S (1999) Effects of ethylene and 1-methylcyclopropene on the post-harvest qualities of ‘Shamuti’ oranges. Postharvest Biol Technol 15 155–163 [Google Scholar]
- Pružinská A, Tanner G, Anders I, Roca M, Hörtensteiner S (2003) Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc Natl Acad Sci USA 100 15259–15264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis AC, Barmore CR (1981) Involvement of ethylene in chlorophyll degradation in peel of citrus fruits. Plant Physiol 68 854–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg M, Matile P (1995) Association of components of the chlorophyll catabolic system with pigment-protein complexes from solubilized chloroplast membranes. J Plant Physiol 146 604–608 [Google Scholar]
- Schenk N, Schelbert S, Kanwischer M, Goldschmidt EE, Dörmann P, Hörtensteiner S (2007) The chlorophyllases AtCLH1 and AtCLH2 are not essential for senescence-related chlorophyll breakdown in Arabidopsis thaliana. FEBS Lett 27 5517–5525 [DOI] [PubMed] [Google Scholar]
- Shimokawa K, Sakanoshita A, Yaeo K (1978) Ethylene-enhanced chlorophyllase activity during degreening of Citrus unshiu. Sci Hortic (Amsterdam) 8 129–135 [Google Scholar]
- Shimoni E, Rav-Hon O, Ohad I, Brumfeld V, Reich Z (2005) Three-dimensional organization of higher-plant chloroplast thylakoid membranes revealed by electron tomography. Plant Cell 17 2580–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitte P, Falk H, Liedvogel B (1980) Chromoplasts. In FGC Czygan, G Fischer Verl, eds, Pigments in Plants. Stuttgart, New York, pp 117–148
- Takamiya KI, Tsuchiya T, Ohta H (2000) Degradation pathway(s) of chlorophyll: what has gene cloning revealed? Trends Plant Sci 5 426–431 [DOI] [PubMed] [Google Scholar]
- Tarasenko LG, Khodasevich EV, Orlovskaya KI (1986) Location of chlorophyllase in chloroplast membranes. Photobiochem Photobiophys 12 119–121 [Google Scholar]
- Terpstra W (1974) Properties of chloroplasts and chloroplast fragments as deduced from internal chlorophyll-chlorophyllide conversion. Z Planzenphysiol 71 129–143 [Google Scholar]
- Terpstra W (1976) Chlorophyllase and lemellar structure in Phaeodactylum triconutum. Situation of chlorophyllase in pigment membranes. Z Planzenphysiol 80 177–188 [Google Scholar]
- Thomson WW, Whatley JM (1980) Development of nongreen plastids. Annu Rev Plant Physiol 31 375–394 [Google Scholar]
- Tian GW, Mohanty A, Chary SN, Li S, Paap B, Drakakaki G, Kopec CD, Li J, Ehrhardt D, Jackson D, et al (2004) High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol 135 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh T, Goldschmidt EE, Riov J (1993) Ethylene induces de novo synthesis of chlorophyllase, a chlorophyll degrading enzyme, in citrus fruit peel. Proc Natl Acad Sci USA 90 9441–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T, Ohta H, Okawa K, Iwamatsu A, Shimaba H, Masuda T, Takamiya KI (1999) Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: finding of a lipase motif and the induction by methyl jasmonate. Proc Natl Acad Sci USA 96 15362–15367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich KL, Bovet L, Hunziker PE, Donnison IS, Hörtensteiner S (2000) Molecular cloning, functional expression and characterization of RCC reductase involved in chlorophyll catabolism. Plant J 21 189–198 [DOI] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van-Wijk KJ (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3 e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]