Abstract
During senescence and at times of stress, plants can mobilize needed nitrogen from chloroplasts in leaves to other organs. Much of the total leaf nitrogen is allocated to the most abundant plant protein, Rubisco. While bulk degradation of the cytosol and organelles in plants occurs by autophagy, the role of autophagy in the degradation of chloroplast proteins is still unclear. We have visualized the fate of Rubisco, stroma-targeted green fluorescent protein (GFP) and DsRed, and GFP-labeled Rubisco in order to investigate the involvement of autophagy in the mobilization of stromal proteins to the vacuole. Using immunoelectron microscopy, we previously demonstrated that Rubisco is released from the chloroplast into Rubisco-containing bodies (RCBs) in naturally senescent leaves. When leaves of transgenic Arabidopsis (Arabidopsis thaliana) plants expressing stroma-targeted fluorescent proteins were incubated with concanamycin A to inhibit vacuolar H+-ATPase activity, spherical bodies exhibiting GFP or DsRed fluorescence without chlorophyll fluorescence were observed in the vacuolar lumen. Double-labeled immunoelectron microscopy with anti-Rubisco and anti-GFP antibodies confirmed that the fluorescent bodies correspond to RCBs. RCBs could also be visualized using GFP-labeled Rubisco directly. RCBs were not observed in leaves of a T-DNA insertion mutant in ATG5, one of the essential genes for autophagy. Stroma-targeted DsRed and GFP-ATG8 fusion proteins were observed together in autophagic bodies in the vacuole. We conclude that Rubisco and stroma-targeted fluorescent proteins can be mobilized to the vacuole through an ATG gene-dependent autophagic process without prior chloroplast destruction.
In C3 plants, 75% to 80% of total leaf nitrogen is distributed to mesophyll chloroplasts, and most of this nitrogen is allocated into proteins (Makino and Osmond, 1991). The most abundant plant protein is Rubisco (EC 4.1.1.39), which catalyzes two competing reactions, photosynthetic CO2 fixation and photorespiratory carbon oxidation. Rubisco accounts for 12% to 30% of total leaf protein in C3 species (Evans, 1989). The degradation of Rubisco and most other stromal proteins begins at an early stage of senescence, and the released nitrogen can be remobilized to growing organs and finally stored in seeds (Friedrich and Huffaker, 1980; Mae et al., 1983). In addition, these proteins are also degraded under carbon-limited conditions, which are caused by darkness (Wittenbach, 1978), and their carbon is used mainly as substrates of respiration. Despite its important function to allow nutrient recycling, the degradation mechanism is not clearly understood yet (for review, see Krupinska, 2006; Feller et al., 2008).
Autophagy is known to be a major system for the bulk degradation of intracellular proteins, and the mechanism has been studied in depth in yeast and animals (for review, see Ohsumi, 2001; Levine and Klionsky, 2004). In those systems, the cytosol, including entire organelles, is engulfed in membrane-bound vesicles that are delivered to the vacuole (yeast) or the lysosome (animals). These vesicles and contents are then degraded by a variety of resident hydrolases. There are two types of autophagy: microautophagy and macroautophagy (Klionsky and Ohsumi, 1999). In microautophagy, the cytosol is engulfed by an invaginated vacuolar membrane. In macroautophagy, the cytosol is sequestered into a double-membrane vesicle called an autophagosome. The outer membrane of the autophagosome then fuses to the vacuolar membrane, thus delivering the inner membrane structure, the autophagic body, into the vacuolar lumen. Genetic analysis in yeast identified 16 autophagy genes (ATGs) essential for the formation of the autophagosome (Tsukada and Ohsumi, 1993; Thumm et al., 1994; Barth et al., 2001). In particular, it has been revealed that two ubiquitin-like conjugation pathways, ATG12 and ATG8 systems, are essential for autophagosome formation (Ohsumi, 2001).
The plant vacuole, an acidic compartment that is rich in lytic hydrolases, has also been implicated in autophagy (for review, see Thompson and Vierstra, 2005; Bassham et al., 2006). A recent genome-wide search revealed that Arabidopsis (Arabidopsis thaliana) has many genes homologous to ATGs in yeast, suggesting that the autophagic process is highly conserved in both organisms (Doelling et al., 2002; Hanaoka et al., 2002). Several knockout mutants of AtATG genes were isolated, and the roles of these genes were analyzed. Among AtATGs that are related to ubiquitin-like conjugation pathways, AtATG7 (Doelling et al., 2002), AtATG4s and AtATG8s (Yoshimoto et al., 2004), AtATG5 (Thompson et al., 2005), and AtATG10 and AtATG12 (Suzuki et al., 2005; Phillips et al., 2008) were characterized. The results indicated that those conjugation pathways also function in plants as they do in yeast. In addition, using transgenic Arabidopsis expressing a GFP-ATG8 fusion protein, a monitoring system for autophagy in plants was established (Yoshimoto et al., 2004; Contento et al., 2005; Thompson et al., 2005).
The atg mutants can complete their life cycles but show accelerated leaf senescence even under favorable plant growth conditions, suggesting that autophagy plays some role even in nutrient-rich conditions. The mutants are hypersensitive to either nitrogen or carbon starvation, both of which result in an accelerated loss of chlorophyll and some chloroplast proteins (Doelling et al., 2002; Hanaoka et al., 2002; Thompson et al., 2005; Phillips et al., 2008). The ATG-dependent autophagic process is not essential for the degradation of chloroplast proteins; other proteolytic systems must be able to degrade chloroplast contents when the ATG system is compromised (Levine and Klionsky, 2004; Bassham et al., 2006). However, prior experiments have not revealed whether the ATG-dependent autophagic degradation of chloroplast proteins occurs in wild-type plants.
By immunoelectron microscopy (IEM) of fixed tissues, we previously found that Rubisco is sometimes localized in small spherical bodies, named Rubisco-containing bodies (RCBs), that are located mostly in the cytoplasm and occasionally in the vacuole of naturally senescent leaves of wheat (Triticum aestivum; Chiba et al., 2003). RCBs are 0.4 to 1.2 μm in diameter and have a staining density similar to that of the stroma. They also contain another stromal protein, Gln synthetase (EC 6.3.1.2), but do not include thylakoid membranes. RCBs seem to be surrounded by isolation membranes, which are considered to be intermediate structures of autophagosomes, in the cytoplasm. These features indicate that only a part of the stroma is pinched off from chloroplasts and is mobilized to the vacuole for degradation, possibly by autophagy. This concept is consistent with the evidence that the chloroplast number remains constant until late in senescence but the stromal proteins are gradually degraded soon after leaf maturation (Hörtensteiner and Feller, 2002). Our observations clearly differ from the autophagy of whole chloroplasts, which has been reported by other researchers (Wittenbach et al., 1982; Minamikawa et al., 2001; Niwa et al., 2004).
In this study, we aimed to examine the involvement of the ATG-dependent autophagic process on the mobilization of the most abundant stromal protein, Rubisco, and stroma-targeted fluorescent proteins as a marker for the stromal components to the vacuole via RCBs. Using transgenic Arabidopsis expressing stroma-targeted fluorescent proteins, we first visualized RCBs in living wild-type cells. The atg5 mutation, which compromises the progression of autophagy, disrupted the accumulation of RCBs. In addition, stroma-targeted DsRed and GFP-ATG8 were colocalized in spherical bodies in the vacuole. RCBs could be visualized directly by a RBCS-GFP fusion whose expression was driven by its own promoter and that is integrated into a GFP-labeled Rubisco holoenzyme. Thus, Rubisco and likely other stroma-localized proteins can be mobilized to the vacuole by the ATG-dependent autophagic process without chloroplast destruction.
RESULTS
Visualization of RCBs by Stroma-Targeted GFP in Living Cells
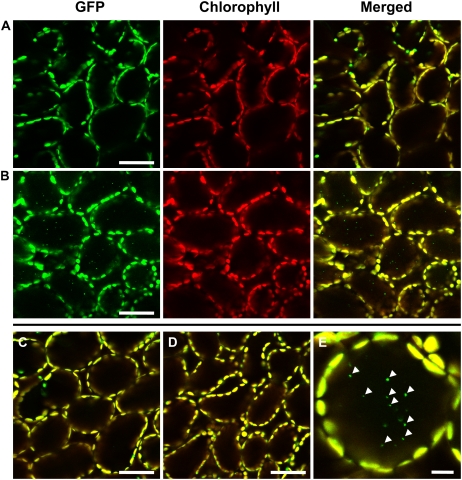
We examined transgenic Arabidopsis in which GFP, expressed in the nucleus under the control of a double 35S promoter (Kay et al., 1987), is targeted to the stroma of chloroplasts by a transit peptide of the Arabidopsis RECA protein (named CT-GFP; Köhler et al., 1997; Holzinger et al., 2007). As reported previously, GFP fluorescence was detected within chloroplasts in mesophyll cells when excised leaves of these plants were immediately observed by laser-scanning confocal microscopy (LSCM; Fig. 1A). We treated excised leaves with concanamycin A, an inhibitor of vacuolar H+-ATPase, because it had previously been reported that the interior pH of the vacuole increases in the presence of this drug, resulting in an accumulation of autophagic bodies in the lumen of the vacuole (Yoshimoto et al., 2004). When mature leaves, in which the degradation of Rubisco and major stromal proteins are already progressing, were excised from plants and then incubated in darkness in the presence of concanamycin A in a carbon- and nutrient-free medium, the GFP signal was also observed in small spherical bodies in the center of cells, in addition to broad GFP signals in the chloroplast stroma (Fig. 1, B and E). These bodies are around 1 μm in diameter and do not exhibit chlorophyll autofluorescence. These features are similar to those of the RCBs that were observed in leaf mesophyll cells of wheat fixed for electron microscopy (Chiba et al., 2003). The small GFP-labeled bodies exhibit the random motion characteristic of Brownian movement within a cell (Supplemental Movie S1). The movement of the GFP-labeled bodies is clearly different from that caused by cytoplasmic streaming, indicating that the GFP bodies are located in the vacuole. The vacuole is the largest compartment of mature plant cells, such as mesophyll cells, and may occupy as much as 80% to over 90% of the total cell volume (Matile, 1987).
Figure 1.
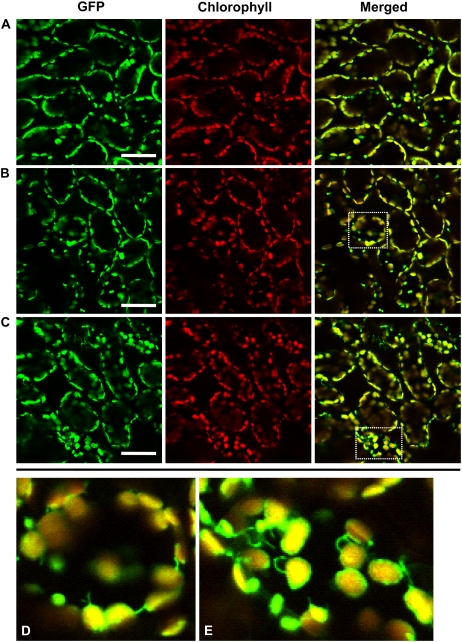
Visualization of stroma-targeted GFP in living leaf cells of Arabidopsis by LSCM. A, A fresh leaf excised from the plant was observed immediately. B and C, Excised leaves were incubated in 10 mm MES-NaOH (pH 5.5) with the addition of 1 μm concanamycin A (B) or 1% (v/v) dimethyl sulfoxide (C). D, Excised leaves were incubated in MS medium with 1 μm concanamycin A. Excised leaves were incubated at 23°C for 20 h in darkness. Stroma-targeted GFP appears green, and chlorophyll fluorescence appears red. In merged images, the overlap of GFP and chlorophyll fluorescence appears yellow. In C and D, merged images are shown. E, Magnification of a mesophyll cell of leaves incubated in the conditions described for B. Spherical bodies having GFP (arrowheads) were observed. The image in E was taken from Supplemental Movie S1. Bars = 50 μm (A–D) and 10 μm (E).
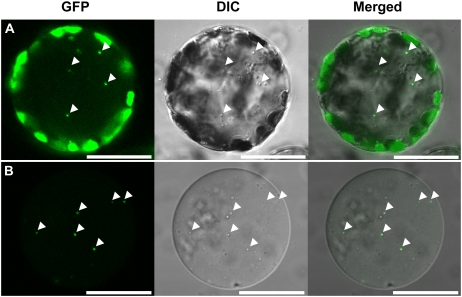
To further confirm the localization of the GFP bodies within cells, cells were released as protoplasts from leaves that were incubated in the same conditions as the leaves described in Figure 1B. GFP bodies were found in the center of a protoplast (Fig. 2A) and exhibited Brownian movement (data not shown), as seen previously in leaves (Supplemental Movie S1). GFP bodies were also found in vacuoles that were isolated from protoplasts (Fig. 2B). Differential interference contrast images clearly indicated that some of spherical bodies, which are localized in the vacuolar sap, are identical to the GFP bodies (Fig. 2). The GFP bodies were rarely seen when concanamycin A was absent (Fig. 1C). The GFP bodies are likely to be rapidly degraded in the vacuole when concanamycin A is absent. GFP bodies were not seen when leaves were incubated in Murashige and Skoog (MS) medium, which contains Suc, even in the presence of concanamycin A (Fig. 1D).
Figure 2.
Detection of the GFP-labeled spherical bodies in a protoplast (A) and a vacuole (B). Protoplasts were prepared from leaves that were incubated with 1 μm concanamycin A in 10 mm MES-NaOH (pH 5.5) at 23°C for 20 h in darkness. Vacuoles were released from the thermally lysed protoplasts and were fractionated by a Ficoll density gradient. Stroma-targeted GFP appears green, and images by differential interference contrast (DIC) are shown in gray. GFP bodies, indicated by arrowheads, are identical to spherical bodies that are localized in the vacuolar sap. Bars = 25 μm.
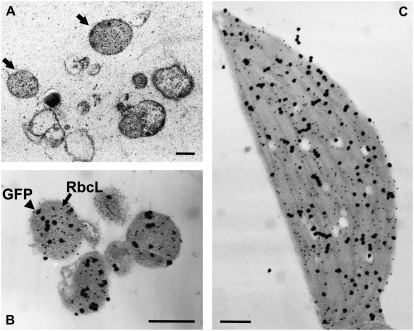
We analyzed the leaves of CT-GFP-containing plants incubated with concanamycin A by IEM with antibodies to RbcL, the large subunit of Rubisco. Spherical bodies containing RbcL, namely, RCBs, were found in the vacuolar compartment (Fig. 3A). In addition, double immunolabeling proved that stroma-targeted GFP was localized in RCBs as well as in chloroplasts (Fig. 3, B and C). When the leaves of CT-GFP-containing plants were analyzed by immunoblotting with anti-GFP antibodies following SDS-PAGE, a single band of 29 kD, which corresponds to the mature form of CT-GFP after cleavage of the transit sequence, was detected irrespective of incubation conditions (Supplemental Fig. S1). On the other hand, a band of 37 kD that would correspond to the premature form of GFP carrying the RECA transit peptide was not found even upon overexposure of the immunoblot. These data clearly indicate that GFP bodies found in the vacuole of living cells are RCBs and must be derived from chloroplasts in which processing of the CT-GFP transit peptide has occurred.
Figure 3.
Detection of Rubisco-containing bodies in concanamycin A-treated leaves of Arabidopsis expressing stroma-targeted GFP by IEM. Leaves were incubated with 1 μm concanamycin A in 10 mm MES-NaOH (pH 5.5) at 23°C for 20 h in darkness. A, Leaf sections were immunolabeled with anti-RbcL antibodies followed by 15-nm gold-conjugated goat anti-rabbit IgG. Spherical bodies containing RbcL (RCBs; arrowheads) were found in the vacuolar compartment. B and C, Leaf sections were double immunolabeled with anti-GFP and anti-RbcL antibodies. After immunolabeling with anti-GFP antibodies followed by 15-nm gold-conjugated goat anti-rabbit IgG, silver enhancement of gold particles was performed. By this treatment, 15-nm gold particles were covered by the shell of metallic silver and grown to around 100 nm. After that, sections were immunolabeled with anti-RbcL antibodies followed by 15-nm gold-conjugated goat anti-rabbit IgG. Black particles of about 100 nm reflect the existence of GFP, and smaller dots, the nonenhanced gold particles that are 15 nm in size, visualize RbcL. GFP colocalizes with RbcL in both Rubisco-containing bodies (B) and chloroplasts (C). Bars = 1 μm.
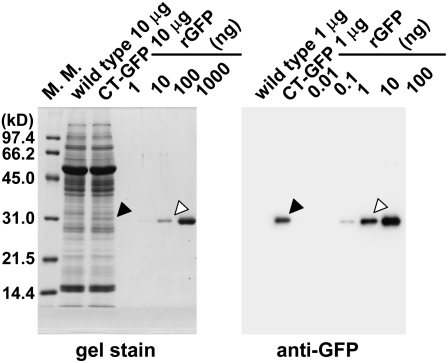
We analyzed the amount of CT-GFP expressed in order to verify that is does not accumulate at levels unusual for genuine chloroplast proteins (Fig. 4). Although the expression of CT-GFP is driven by a double 35S promoter, it accumulates only to approximately 1% of total soluble protein, which is much less than the amount of Rubisco, which accounts for about 50% of total soluble leaf protein. Taken together with previous observations of RCBs in senescing wild-type wheat leaves (Chiba et al., 2003), these results make it very unlikely that RCBs found in living cells of the transgenic plants could be artifactual products resulting from the expression of a fluorescent protein in chloroplasts.
Figure 4.
SDS-PAGE and immunoblot analysis of chloroplast stroma-targeted GFP. Total soluble proteins (10 μg for gel stain, 1 μg for immunoblotting) extracted from fresh leaves expressing chloroplast stroma-targeted GFP (CT-GFP) and from fresh leaves of wild-type Columbia (wild type) as a control and a dilution series of a known amount of recombinant GFP (rGFP; amounts indicated in nanograms) were separated by SDS-PAGE and either stained with Coomassie Brilliant Blue R250 (gel stain) or analyzed by immunoblotting. Black arrowheads indicate the mature form of CT-GFP after cleavage of the transit peptide. White arrowheads indicate similar amounts of rGFP to CT-GFP on a gel or a membrane. The sizes of molecular mass markers (M. M.; kilodaltons) are indicated at left of the stained gel.
Involvement of an ATG-Dependent Autophagic Process in the Accumulation of RCBs
To clarify whether an ATG-dependent autophagic process is involved in the accumulation of RCBs, we first examined the effect of the atg5 mutation on the accumulation of RCBs in the vacuoles of living cells. We chose plants carrying the atg5 mutation because atg5-1 plants cannot form the ATG12-ATG5 conjugate that is essential for autophagy in yeast and animals and fail to accumulate ATG8-GFP-labeled autophagic bodies in the vacuolar lumen (Suzuki et al., 2005; Thompson et al., 2005; Phillips et al., 2008). Fluorescence of stroma-targeted GFP was obvious in atg5-1 (Fig. 5A) as well as in wild-type plants (Fig. 1A). Immunoblot analysis revealed that there is no difference in the expression of GFP between wild-type plants and atg5-1 (data not shown). When mature leaves of atg5-1 were incubated in the presence of concanamycin A, the accumulation of spherical bodies labeled with GFP was not observed (Fig. 5B). Many stromules (stroma-filled tubules that extend from the surface of plastids; for review, see Kwok and Hanson, 2004a; Natesan et al., 2005) appeared in mesophyll cells after incubation irrespective of the addition of concanamycin A (Fig. 5, B–E). Such stromules were not found in similar leaves of wild-type plants (Fig. 1, B and C). Stromules are much less common in leaf tissue than in tissue from other plant organs (Köhler and Hanson, 2000).
Figure 5.
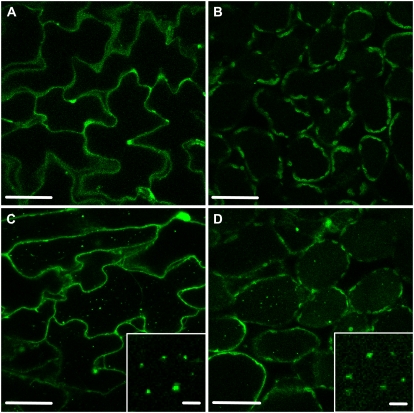
Effect of the atg5 mutation on the behavior of stroma-targeted GFP in mesophyll cells. A, A fresh leaf excised from an atg5 mutant (atg5-1) expressing stroma-targeted GFP was observed immediately. B and C, Excised leaves were incubated in 10 mm MES-NaOH (pH 5.5) with the addition of 1 μm concanamycin A (B) or 1% (v/v) dimethyl sulfoxide (C) at 23°C for 20 h in darkness. Stroma-targeted GFP appears green, and chlorophyll fluorescence appears red. In merged images, the overlap of GFP and chlorophyll fluorescence appears yellow. No spherical bodies having GFP were observed in the vacuole of concanamycin A-treated leaves (B). Many stromules were found after incubation irrespective of the addition of concanamycin A. D and E, Magnifications of stromule-rich cells indicated by the dashed-line areas in B and C, respectively. Bars = 50 μm.
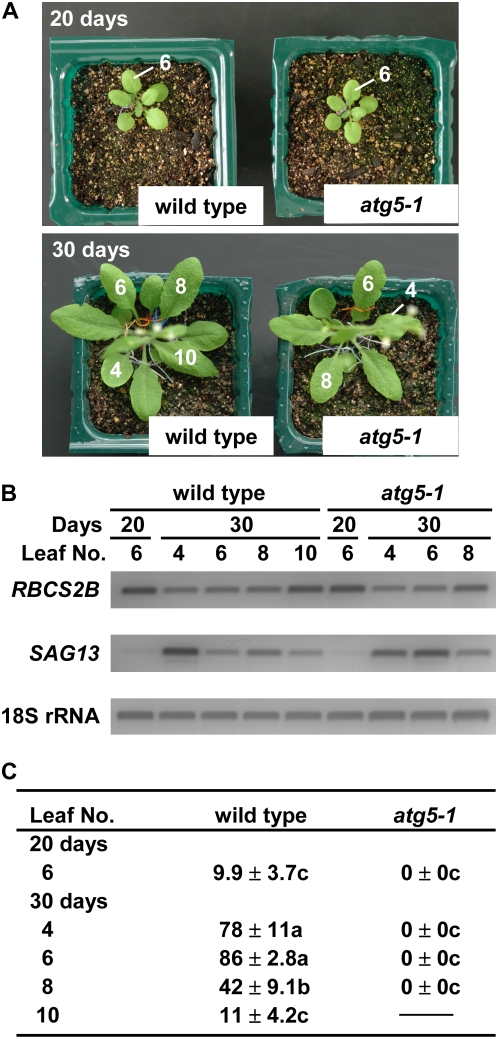
To elucidate the effect of the atg5 mutation and the leaf age on the appearance of RCBs more clearly, a statistical analysis was performed (Fig. 6). Under the growth conditions used, wild-type plants started to bolt at around 25 d after sowing and finally had 10 rosette leaves on average. The atg5-1 plants grew normally, as shown previously (Fig. 6A; Thompson et al., 2005), but they started to bolt around 2 d earlier than wild-type plants and finally had eight to nine rosette leaves under our growth conditions. In atg5-1 plants, the enhanced chlorosis of leaves was not observed until 30 d. We monitored the expression of two genes in order to verify that leaf senescence is progressing similarly in wild-type and atg5-1 plants in the leaves used for the examination of RCB formation. In lower leaves of atg5-1 plants at 30 d, the decline of RBCS2B transcripts, a marker for the young, photosynthetically active stage of leaf development (Hensel et al., 1993), and the increase of SAG13 transcripts, a marker for an early stage of leaf senescence (Weaver et al., 1998), were similar to those observed in wild-type plants (Fig. 6B).
Figure 6.
Effects of leaf age and the atg5 mutation on the appearance of Rubisco-containing bodies. A, Photographs of wild-type and atg5-1 plants expressing stroma-targeted GFP grown for 20 and 30 d after sowing. Rosette leaves were numbered in order of appearance. B, Expression of RBCS2B and SAG13 as marker genes for leaf age. Total RNA from leaves was isolated and subjected to semiquantitative reverse transcription-PCR using gene-specific primers. PCR products were electrophoresed, stained with SYBR Green I, and detected by a fluorescence image analyzer. 18S ribosomal RNA was used as an internal control. C, Percentage of mesophyll cells accumulating Rubisco-containing bodies. Excised leaves were incubated in 10 mm MES-NaOH (pH 5.5) with 1 μm concanamycin A at 23°C for 20 h in darkness. Four quadrangular regions (318 μm × 318 μm each, containing 120–470 mesophyll cells) per leaf were monitored by LSCM, and the number of cells accumulating Rubisco-containing bodies was counted. Values (percentages) are given as means ± se (n = 5). Statistical analysis was performed by ANOVA followed by Tukey's test. Values with the same letters are significantly different from each other at P < 0.05.
In wild-type plants, the appearance of RCBs was highly related to the leaf age (Fig. 6C). In mature and early senescent leaves (leaves 6 and 4 at 30 d), in which the level of RBCS2B has obviously declined compared with the uppermost leaves (leaf 10) or expanding leaves (leaf 6 at 20 d), most mesophyll cells accumulated RCBs, as shown in the image analysis (Fig. 1B). In contrast, RCBs were rarely seen in expanding (leaf 6 at 20 d) and young uppermost (leaf 10 at 30 d) leaves. In atg5-1 plants, RCBs were not seen at any stage of leaf development examined (Fig. 6C).
Autophagy in plants can be monitored with the use of transgenic Arabidopsis expressing a GFP-ATG8 fusion protein (Yoshimoto et al., 2004; Contento et al., 2005; Thompson et al., 2005). Spherical bodies labeled with GFP-ATG8, namely, autophagic bodies, accumulate in the vacuole when roots are incubated with concanamycin A (Yoshimoto et al., 2004). We examined leaf cells to determine whether autophagic bodies were also present in leaves in the same transgenic plants (Fig. 7). GFP-ATG8 was predominantly cytoplasmic in both epidermal and mesophyll cells of excised leaves (Fig. 7, A and B). When the leaves were incubated in the presence of concanamycin A, we could find many autophagic bodies in the vacuole in both epidermal and mesophyll cells (Fig. 7, C and D). Autophagic bodies found in leaves were around 1 μm in diameter and exhibited the random motion characteristic of Brownian movement (Supplemental Movie S2). These features are similar to those of RCBs found in this study and also to autophagic bodies previously found in roots (Yoshimoto et al., 2004), hypocotyls (Thompson et al., 2005), and suspension cell protoplasts (Contento et al., 2005). These results confirm that ATG-dependent autophagy occurs in leaves under the conditions used in our experiments. A faint signal of GFP was also observed in the vacuolar lumen (Fig. 7D), suggesting that some autophagic bodies are digested and the released GFP was not immediately degraded. It has been shown that vacuole-targeted GFP fluorescence is not observed because GFP is easily degraded by vacuolar papain-type Cys protease(s), which have an acidic optimum pH (Tamura et al., 2003). However, the GFP degradation can be abolished when the maintenance of interior acidic pH of the vacuole is affected by the inhibition of vacuolar H+-ATPase activity by concanamycin A (Tamura et al., 2003).
Figure 7.
Visualization of autophagic bodies in concanamycin A-treated leaves of Arabidopsis expressing the GFP-ATG8 fusion protein. A and B, Epidermal (A) and mesophyll (B) cells of fresh leaves excised from the plant were observed immediately. C and D, Excised leaves were incubated in 10 mm MES-NaOH (pH 5.5) with the addition of 1 μm concanamycin A at 23°C for 20 h in darkness, and then epidermal (C) and mesophyll (D) cells of leaves were observed. Insets show magnifications of autophagic bodies. Bars = 50 μm in main images and 5 μm in the insets.
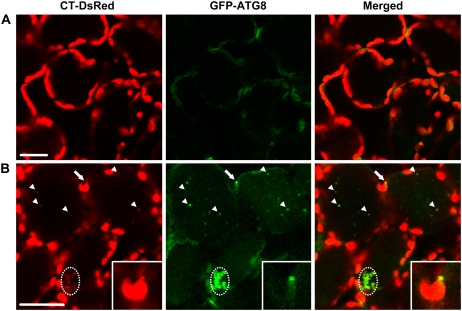
We next produced transgenic Arabidopsis expressing both stroma-targeted DsRed and GFP-ATG8 and observed the behavior of those proteins (Fig. 8; Supplemental Movie S3). DsRed fused to the C-terminal end of the RECA transit peptide correctly labeled the stroma of chloroplasts (Fig. 8A). When the leaves were incubated with concanamycin A, spherical bodies labeled with DsRed, namely, RCBs, were found in the vacuole (arrowheads in Fig. 8B). Most of the RCBs were also labeled with GFP-ATG8, but there were also some autophagic bodies that were not labeled with DsRed (Fig. 8B; Supplemental Movie S3). In addition, a strong signal of GFP-ATG8 was found on a chloroplast protrusion (arrows and insets in Fig. 8B). Some cells contained aggregated structures that were strongly labeled with GFP-ATG8 and DsRed (dashed-line circles in Fig. 8). The aggregated structures were moving slowly in a cell (Supplemental Movie S3), indicating that these are located in the vacuole. As the aggregated structures did not exhibit chlorophyll fluorescence, these were not whole chloroplasts. These structures are probably created by the aggregation of autophagic bodies after their transportation into the vacuole. Accumulation of an aggregated form of autophagic bodies in the vacuole has also been reported previously (Liu et al., 2005).
Figure 8.
Localization of stroma-targeted DsRed and the GFP-ATG8 fusion protein in living leaf cells. A, A fresh leaf excised from a plant expressing both stroma-targeted DsRed and GFP-ATG8 was observed immediately. B, Excised leaves were incubated in 10 mm MES-NaOH (pH 5.5) with the addition of 1 μm concanamycin A at 23°C for 20 h in darkness. Stroma-targeted DsRed appears red, and GFP-ATG8 appears green. In merged images, the overlap of DsRed and GFP-ATG8 appears yellow. Stroma-targeted DsRed and GFP-ATG8a were colocalized in a chloroplast protrusion (arrows), and free spherical bodies (arrowheads) and aggregated structures of spherical bodies were in the vacuole (dashed-line circles). Insets show magnifications of the chloroplast protuberance. The images in B are taken from Supplemental Movie S3. Bars = 25 μm.
RCBs Can Be Visualized Directly by GFP-Labeled Rubisco
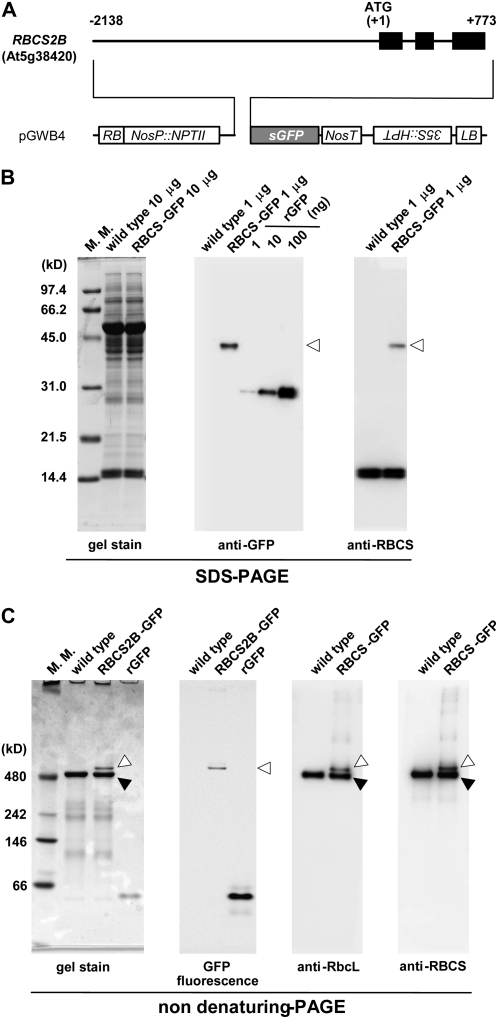
To monitor the mobilization of Rubisco to the vacuole in living cells directly, we created transgenic plants expressing a RBCS-GFP fusion under the control of its own promoter. A 2.9-kb PCR-amplified fragment containing the promoter and coding regions of the Arabidopsis RBCS2B gene was fused to the coding sequence of GFP in the pGWB4 vector (Fig. 9A; Nakagawa et al., 2007). In the transgenic plants, the presence of an RBCS2B-GFP fusion protein of 43 kD, which corresponds to the mature size after cleavage of the RCBS2B transit peptide, was confirmed by immunoblotting following SDS-PAGE. The amount of the fusion protein was approximately 1% of total soluble protein (Fig. 9B), much less than the amount of native RBCS protein that accumulates as a result of the expression of the four-member Arabidopsis RBCS gene family (Dedonder et al., 1993).
Figure 9.
Transgenic Arabidopsis expressing GFP-labeled Rubisco by the RBCS promoter. A, Organization of the RBCS2B-GFP fusion construct. Black bars represent exons. Numbers indicate nucleotide positions from the translation initiation codon (ATG). HPT, Hygromycin phosphotransferase; LB, left border; NosP and NosT, promoter and terminator sequences of nopaline synthase; NPTII, neomycin phosphotransferase; RB, right border; sGFP, synthetic GFP with the S65T mutation; 35S, cauliflower mosaic virus 35S promoter. B, SDS-PAGE analysis of the RBCS2B-GFP fusion. Total soluble proteins (10 μg for gel stain, 1 μg for immunoblotting) extracted from fresh leaves expressing the RBCS2B-GFP fusion (RBCS-GFP) and from fresh leaves of wild-type Columbia (wild type) as a control and a dilution series of a known amount of recombinant GFP (rGFP; amounts indicated in nanograms) were separated by SDS-PAGE and either stained with Coomassie Brilliant Blue R250 (gel stain) or analyzed by immunoblotting with anti-GFP or anti-RBCS antibodies. White arrowheads indicate the mature form of RBCS2B-GFP after cleavage of the RBCS2B transit peptide. C, Nondenaturing PAGE analysis of the RBCS2B-GFP fusion. Total soluble proteins (10 μg for gel stain and GFP fluorescence, 1 μg for immunoblotting) extracted from fresh leaves expressing the RBCS2B-GFP fusion (RBCS-GFP) and from fresh leaves of wild-type Columbia (wild type) as a control and 0.5 μg of recombinant GFP (rGFP) were separated by nondenaturing PAGE and stained with Coomassie Brilliant Blue R250 (gel stain), analyzed by an image analyzer (LAS-3000) to detect GFP fluorescence, or analyzed by immunoblotting with anti-RbcL or anti-RBCS antibody. Black arrowheads indicate native Rubisco holoenzyme, and white arrowheads indicate GFP-labeled Rubisco holoenzyme. The sizes of molecular mass markers (M. M.; kilodaltons) are indicated at left of the stained gels (B and C).
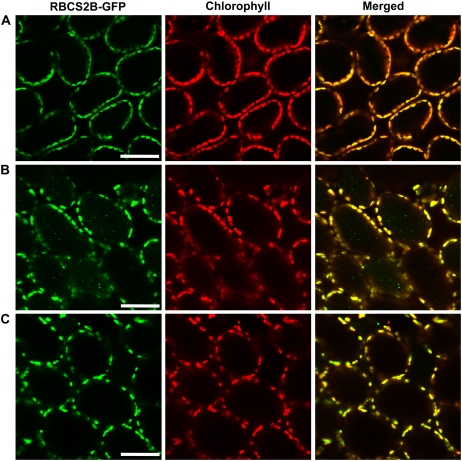
We investigated whether the RBCS2B-GFP fusion was integrated into the Rubisco enzyme, which is assembled from eight plastid-encoded RbcL and eight RBCS subunits, forming an L8S8 molecule of 550 kD in chloroplasts of higher plants (Knight et al., 1990). It has been shown previously that a pea (Pisum sativum) RBCS3A-cyan fluorescent protein fusion expressed under the control of a double 35S promoter can assemble into Rubisco holoenzyme and correctly label the chloroplast stroma and stromules in leaves of tobacco (Nicotiana tabacum; Kwok and Hanson, 2004b). Upon nondenaturing PAGE of leaf extracts of Arabidopsis RBCS-GFP transgenic plants, GFP fluorescence was observed in a single sharp band that was just above the native Rubisco holoenzyme; a band at the same location reacted with both anti-RbcL and anti-RBCS antibodies on immunoblots (Fig. 9C). As expected, no fluorescent bands were observed in wild-type plants. No fluorescent band was observed in the RBCS-GFP transgenic plants at the mobility expected for an unassembled GFP or the RBCS2B-GFP fusion protein. When the fluorescent band in the RBCS-GFP transgenic plants on nondenaturing PAGE (white arrowhead in Fig. 9C) was excised from a gel and then applied to SDS-PAGE, the band was separated into three bands, corresponding to RbcL of 53 kD, RBCS-GFP fusion of 43 kD, and RBCS of 15 kD (data not shown). These results indicated that all of the RBCS2B-GFP fusion proteins are integrated into Rubisco holoenzymes in leaves. In the transgenic plants, the GFP fluorescence was specifically localized in chloroplasts in mesophyll cells (Fig. 10A). There was no obvious GFP fluorescence in nongreen plastids of leaf epidermis and root (data not shown). These results confirmed that the RBCS2B-GFP fusion is correctly expressed under the control of the RBCS2B promoter.
Figure 10.
Visualization of Rubisco-containing bodies by GFP-labeled Rubisco. A, A fresh leaf excised from an RBCS2B-GFP transgenic plant (wild-type background) was observed immediately. B and C, Excised leaves from the RBCS2B-GFP transgenic plants of wild-type background (B) or atg5-1 background (C) were incubated in 10 mm MES-NaOH (pH 5.5) with the addition of 1 μm concanamycin A at 23°C for 20 h in darkness. GFP appears green, and chlorophyll fluorescence appears red. In merged images, the overlap of GFP and chlorophyll fluorescence appears yellow. Bars = 50 μm.
When mature leaves of RBCS-GFP transgenic plants were incubated in the presence of concanamycin A, accumulation of small spherical bodies having GFP fluorescence (i.e. similar to the RCBs seen in CT-GFP leaves; Fig. 1B) was observed (Fig. 10B). In contrast, the accumulation of GFP bodies was not observed in leaves of atg5-1 plants expressing the RBCS2B-GFP fusion (Fig. 10C). These results further support the mobilization of Rubisco to the vacuole via RCBs by ATG-dependent autophagy.
DISCUSSION
In this study, we succeeded in visualizing RCBs in the vacuoles of living cells. The atg5 mutation, which compromises the progression of autophagy (Suzuki et al., 2005; Thompson et al., 2005; Phillips et al., 2008), disrupted the accumulation of RCBs. GFP-ATG8 was colocalized with stroma-targeted DsRed in spherical bodies in the vacuole. Taken together, these data clearly indicate that Rubisco and stroma-targeted fluorescent proteins are partially mobilized to the vacuole in vivo and that this process requires ATG-dependent autophagy.
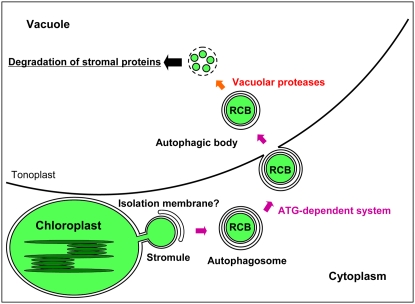
Based on the experiments reported in this study as well as results from our previous electron microscopic study (Chiba et al., 2003) and a number of related reports concerning the autophagic process, a proposed mechanism of the mobilization of Rubisco and possibly other stromal proteins to the vacuole for degradation is presented (Fig. 11). The electron microscopic study showed that RCBs in the cytoplasm are surrounded by membrane structures, which are morphologically considered to be isolation membranes characteristic of macroautophagy (Chiba et al., 2003). RCBs in the cytoplasm contain stromal proteins, RbcL, RBCS, and Gln synthetase, but do not contain thylakoid proteins, light-harvesting chlorophyll a/b protein of PSII, α,β-subunits of coupling factor 1 of ATPase, and cytochrome f (Chiba et al., 2003), suggesting that only the stromal portion of chloroplasts is incorporated into RCBs. The mobilization of RCBs by an ATG-dependent system to the vacuole is supported by our finding that GFP-ATG8, which is a marker for isolation membranes, autophagosomes, and autophagic bodies, was colocalized with stroma-targeted DsRed in the autophagic body (Fig. 8) and that the atg5 mutation disrupted the accumulation of RCBs (Figs. 5, 6, and 10).
Figure 11.
Proposed model of the degradation of Rubisco and possibly other stromal proteins by ATG-dependent autophagy via RCBs. Under the starvation conditions that induce autophagy, a stromule protruding from a chloroplast is sequestered by an isolation membrane. The RCB, namely, an autophagosome containing stromal contents from the chloroplast, is formed and transported into the vacuole. The RCB is degraded by vacuolar proteases.
The RCBs accumulated in the vacuole only when the vacuolar lytic activity was suppressed by the addition of concanamycin A (Fig. 1). In the ATG-dependent system, the outer membrane of an autophagosome fuses with the vacuolar membrane and the inner membrane structure, the autophagic body, is released to the vacuolar lumen and is rapidly disintegrated (Ohsumi, 2001). Therefore, under the usual cellular status in which the vacuolar lytic activity functions normally, RCBs should be rapidly destroyed and the stromal protein contents will be released and degraded by vacuolar proteases, as shown previously (Thayer and Huffaker, 1984; Bhalla and Dalling, 1986; Yoshida and Minamikawa, 1996).
In addition to its localization on autophagic bodies found in the vacuole, GFP-ATG8 fluorescence was observed on a chloroplast protrusion (Fig. 8), which might be an incipient stromule, such as those occasionally seen in wild-type mesophyll cells. We propose that a chloroplast protrusion or stromule might be sequestered from the main chloroplast body by an isolation membrane and thus generate an RCB, namely, an autophagosome containing a stromal portion of chloroplasts. The RCBs could then be transported into the vacuole and degraded by various vacuolar proteases. At present, the origin and the organization of isolation membranes are not fully understood even in yeast (Ohsumi, 2006; Suzuki and Ohsumi, 2007).
The release of stromal portions from plastids for degradation may possibly be one of several physiological functions of stromules. In cinephotomicrographic and video microscopic studies, chloroplast protuberances have been observed to segment into vesicular structures (Wildman et al., 1962; Gunning, 2005). The release of bodies approximately 1.5 μm in diameter from the ends of stromules has been observed (Gunning, 2005). In transgenic plants expressing stroma-targeted GFP, GFP bodies of similar size, which are presumably separated from plastids or stromules, have also been observed by LSCM (Arimura et al., 2001; Waters et al., 2004). Like RCBs, stromules in leaf cells do not contain detectable chlorophyll, and thylakoid membranes have not been seen within stromules visualized by electron microscopy (Köhler et al., 1997; Holzinger et al., 2007). Cyan fluorescent protein-labeled Rubisco is present in plastid stromules and can traffic between plastids (Kwok and Hanson, 2004b). Interestingly, stromules were frequently found in mesophyll chloroplasts in atg5-1 but not in wild-type plants after incubation in starvation conditions (Figs. 1 and 5). In general, stromules are abundant on chlorophyll-free plastids, such as those in roots, petals, and suspension-cultured cells, and are rarely seen on mesophyll chloroplasts (Köhler et al., 1997; Köhler and Hanson, 2000). Perhaps chloroplast protrusions and stromules cannot be sequestered by isolation membranes in atg5-1 cells, in which the ATG-dependent autophagy is compromised, consequently leading to an increase in stromule length and frequency. Alternatively, perhaps some stress caused by the disruption of the autophagic pathway results in an increase in the formation of stromules.
In yeast, the nucleus has been observed to be partially engulfed by the vacuole, a phenomenon termed “piecemeal microautophagy” of the nucleus (Roberts et al., 2003). Recently, partial engulfment of the endoplasmic reticulum by autophagosomes was shown in yeast (Hamasaki et al., 2005). However, organelles such as mitochondria and peroxisomes in yeast are entirely engulfed by the vacuole in microautophagy or entirely sequestered into autophagosomes, which then fuse with the vacuole in macroautophagy (Takeshige et al., 1992; Tuttle and Dunn, 1995). In cotyledons of germinated Vigna mungo seeds, microautophagy of a whole starch granule and macroautophagy of a whole mitochondrion were observed (Toyooka et al., 2001). It was reported that abnormal plastids can be removed, possibly by autophagy, in cotyledon cells of Arabidopsis mutants lacking AtTIC40 (Niwa et al., 2004).
At the leaf stage used in this study, autophagy of whole chloroplasts (i.e. mobilization of them to the vacuole), which is monitored by differential interference contrast imaging and chlorophyll autofluorescence, was not observed even in the presence of concanamycin A. Why would chloroplasts be partially rather than fully degraded by autophagy? One possible explanation is that the autophagic machinery induced by starvation does not have sufficient size capacity to sequester whole chloroplasts. Chloroplasts are normally above 5 μm in size and are much larger than mitochondria or peroxisomes. Plant autophagic bodies found in roots were approximately 1.5 μm in size and contained mitochondria, endoplasmic reticulum, and Golgi bodies (Yoshimoto et al., 2004). Autophagic bodies found in leaf cells were also this size (Fig. 7, C and D) and, therefore, not large enough to engulf a 5-μm chloroplast. It could also be advantageous for a starving plant to reutilize materials from chloroplast proteins without destroying whole chloroplasts. If environmental conditions improve, then a chloroplast that has undergone only loss of some protein content could be rejuvenated and resume normal function. Furthermore, when only a portion of the content of a chloroplast is removed by autophagy, some basal functions of the chloroplast could be maintained under starvation conditions.
In the atg5 mutants, a gradual loss of Rubisco subunits occurred during the extended dark treatment (Thompson et al., 2005). We found that Rubisco also decreases during natural senescence in the atg mutants (H. Ishida, S. Wada, M. Izumi, and A. Makino, unpublished data). These results indicate that chloroplast proteins can be degraded by other mechanisms, such as plastid-localized proteases (Sakamoto, 2006), even if autophagy is disrupted. We observed that selective attenuation of the Rubisco degradation occurs in senescent leaves of the RBCS antisense rice (Oryza sativa; Ishizuka et al., 2004). In addition, we observed specific degradation of Rubisco by reactive oxygen under chilling-light conditions (Nakano et al., 2006). These previous findings as well as our current results indicate that chloroplast protein degradation in vivo is a complex and highly regulated phenomenon that merits further investigation.
RCBs were originally identified in naturally senescing wheat leaves by IEM (Chiba et al., 2003). To visualize RCBs accumulated in the vacuole, we used a 20-h dark treatment of detached leaves with concanamycin A in the absence of sugar. The accumulation of RCBs in the vacuole was mainly found in mature and senescent leaves and was rarely observed in young or expanding leaves under these conditions (Fig. 6). These observations are in agreement with prior findings that the degradation of Rubisco occurs mainly in mature and senescent leaves (Mae et al., 1983). RCBs started to accumulate in the vacuole after around 8 h in this condition (data not shown), while at night, leaves attached with seedlings are often in the dark for 10 h in the growth conditions used here. Therefore, our studies are also suggestive of a role of RCBs in the natural senescence of Arabidopsis, but further careful experimentation will be needed to understand the physiology of chloroplast stromal recycling by autophagy during the normal life cycle of the plant.
Even when mature leaves were incubated with concanamycin A, few RCBs were detected in the presence of Suc-containing MS medium (Fig. 1D). It was previously shown that the rate of the Rubisco degradation is accelerated under conditions of carbon or nutrient deficiency (Wardley et al., 1984; Ferreira and Teixeira, 1992). Similarly, plant autophagy is also stimulated under carbon- or nutrient-deprived conditions (Aubert et al., 1996; Moriyasu and Ohsumi, 1996; Yoshimoto et al., 2004; Thompson et al., 2005). In fact, several Arabidopsis mutants with disruptions in the autophagic pathway have shown accelerated leaf senescence and could not survive under severe starvation conditions (Doelling et al., 2002; Hanaoka et al., 2002; Thompson et al., 2005; Xiong et al., 2005; Phillips et al., 2008). It was reported that atg5 mutants die after 6 d of darkness, which is tolerated by wild-type Arabidopsis (Thompson et al., 2005). These results suggest that the degradation of stromal proteins by the ATG-dependent autophagic pathway via RCBs makes a contribution both to the recycling of nutrients for growing tissues and to the retention of chloroplast function under such stress conditions (i.e. carbon or nutrient deficiency rather than natural senescence). A reutilization of nutrients from chloroplast proteins without destroying whole chloroplasts is probably essential for light- and nutrient-limited plants. Examining the effect of nitrogen limitation and other stresses on whole plants with regard to RCB formation and mobilization will be valuable to understand the relative importance of the degradation pathway we have described.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Transgenic Arabidopsis (Arabidopsis thaliana) ecotype Columbia expressing stroma-targeted GFP (CT-GFP) was created as described previously (Köhler et al., 1997; Holzinger et al., 2007). The construct consists of the transit sequence from the Arabidopsis RECA gene fused to the S65TmGFP4 coding region, driven by a double cauliflower mosaic virus 35S promoter. The plasmid for generating transgenic Arabidopsis expressing the stroma-targeted DsRed was constructed in a similar manner. The DsRed2 coding region from pGDR (Goodin et al., 2002) was fused in-frame with the RECA transit peptide sequence and cloned into the XbaI and SstI sites of the pBI121 binary vector. The construct was introduced into Arabidopsis ecotype Wassilewskija by the floral-dip method for Agrobacterium tumefaciens-mediated transformation in planta (Clough and Bent, 1998). Seeds of a T-DNA knockout mutant of ATG5 (atg5-1; SAIL_129_B07.v1) were obtained from the Nottingham Arabidopsis Resource Centre. Transgenic Arabidopsis Wassilewskija plants expressing a GFP-ATG8a fusion protein were described previously (Yoshimoto et al., 2004). Transgenic Arabidopsis with atg5-1 background expressing stroma-targeted GFP and transgenic Arabidopsis ecotype Wassilewskija expressing both the stroma-targeted DsRed and the GFP-ATG8 fusion protein were obtained by sexual crosses. Arabidopsis transgenic plants expressing RBCS2B-GFP fusion were created as follows. A DNA fragment that starts from the 5′ region 2,138 bp upstream of the translation initiation site and terminates just before the stop codon of RBCS2B was amplified from the Arabidopsis genomic DNA by PCR using the primers 5′-CACCATTCTTGCCTTTGGTTCTCTAATGAAAG-3′ and 5′-AGCTTCGGTGAAGCTTGGG-3′ and cloned into pENTR/D/TOPO vector (Invitrogen). The coding region was transferred to the pGWB4 vector (Nakagawa et al., 2007) by an LR recombination reaction. The construct was introduced into Arabidopsis (wild-type Columbia and atg5-1 background) as described above. All plants were grown on soil (Metro-Mix 350; Sun Gro Horticulture) at 23°C on a 14/10-h photoperiod with fluorescent lamps in a growth room for 3 to 5 weeks.
Concanamycin A Treatment
To visualize RCBs and autophagic bodies, mature leaves were cut from plants, washed with 70% (v/v) ethanol and then with sterile water, infiltrated by a syringe briefly in solutions consisting of 10 mm MES-NaOH (pH 5.5) or a standard MS medium containing Gamborg's vitamins with 2% (v/v) Suc, either in the presence or the absence of 1 μm concanamycin A (Wako), and incubated at 23°C for 20 h in darkness. A stock solution of concanamycin A was made to 100 μm in dimethyl sulfoxide. Protoplasts were prepared from the concanamycin A-treated leaves by the procedure of Asai et al. (2000), except that 1 μm concanamycin A was added to each preparation medium. Vacuoles were prepared from the protoplasts by the procedure of Robert et al. (2007).
Image Analysis with LSCM
LSCM was performed with a Nikon C1si system equipped with a CFI Plan Apo VC60× water-immersion objective (numerical aperture = 1.20; Nikon). Leaves were mounted on slides with coverglasses. Both GFP and chlorophyll were excited with the 488-nm line of a multi-argon ion laser, and emission of GFP and chlorophyll was detected between 500 and 530 nm and over 650 nm by a multichannel detector with filters, respectively. There is no fluorescence bleedthrough between GFP and chlorophyll channels at the range of a laser power and gain used for image analysis (Supplemental Fig. S2). For observation of leaves expressing both the GFP-ATG8 fusion protein and stroma-targeted DsRed, GFP was excited with the 488-nm line of a multi-argon ion laser and DsRed was excited with the combination of the 488-nm line of a multi-argon ion laser and the 543-nm line of a helium-neon laser. The fluorescence spectra between 500 and 650 nm were then obtained at 5-nm resolution by a spectral detector, and separated signals of GFP and DsRed were obtained by an unmixing program using Nikon C1si software. Each experiment was performed at least three times using independent samples, and representative data are shown.
Protein Analysis
Leaves were homogenized in HEPES-NaOH (pH 7.5) containing 14 mm 2-mercaptoethanol and protease inhibitor cocktail (Complete Mini; Roche) in a chilled mortar and pestle. Total soluble proteins in extracts were quantified by Bradford assay (Bio-Rad) and mixed with an equal volume of SDS sample buffer consisting of 200 mm Tris-HCl (pH 8.5), 2% (w/v) SDS, 0.7 m 2-mercaptoethanol, and 20% (v/v) glycerol, boiled for 3 min, and then subjected to SDS-PAGE. Nondenaturing PAGE was performed as described previously (Ishida et al., 1999), except that a 4% to 12% gradient gel (Invitrogen) was used. GFP fluorescence on nondenaturing gels was detected by LAS-3000 (Fujifilm) equipped with a blue-light-emitting diode (460nmEPI) and a band-pass filter for GFP (510DF10). Immunoblotting was performed as described previously (Xu et al. 2006) with a mouse anti-GFP monoclonal antibody (Nacalai) and anti-RbcL and anti-RBCS antibodies prepared from rabbit anti-rice (Oryza sativa) Rubisco antiserum by affinity purification using RbcL and RBCS purified from wheat (Triticum aestivum) leaves as ligands, as described previously (Ishida et al., 1997). Signals were developed with horseradish peroxidase-conjugated secondary antibodies and chemiluminescent reagent (Pierce) and detected by ImageQuant400 (GE Healthcare).
IEM
IEM was performed with a transmission electron microscope (H-8100; Hitachi) operated at 75 kV as described in detail previously (Chiba et al., 2003) with the anti-RbcL antibodies described above and rabbit anti-GFP antibodies (Invitrogen). Our affinity-purified anti-RbcL antibodies are highly specific for RbcL (Ishida et al., 1997; Kokubun et al., 2002; Nakano et al., 2006). Double immunolabeling was performed using a silver-enhancing kit (BB International) according to the manufacturer's instructions. Using this silver-enhancing technique, since the shell of metallic silver covers both the first gold particles and the absorbed protein or IgG and protects the first immunoreagent from cross-reacting with the second one, two different primary antibodies raised in the same animal and the same secondary antibody can be used (Manigley and Roth, 1985).
Statistical Analysis
To examine the effects of leaf age and the atg5 mutation on the accumulation of RCBs, corresponding leaves in order of appearance from five independent wild-type and atg5-1 plants expressing stroma-targeted GFP at 20 and 30 d after sowing were collected and treated with concanamycin A as described above. After the treatment, each leaf was divided into four parts and a quadrangular region at the center of each part (318 μm × 318 μm each), which covers 30 to 120 mesophyll cells depending on leaf age, was monitored by LSCM with a CFI Plan Fluor 40× objective (numerical aperture = 0.75; Nikon) for 1 min (69 scans) and time-lapse images were obtained. The number of total cells and the number of cells containing RCBs were counted on the images. Statistical analysis was performed by ANOVA followed by Tukey's test using JMP6 (SAS Institute).
Semiquantitative Reverse Transcription-PCR Analysis
Total RNA was isolated from leaves using the RNeasy plant mini kit (Qiagen). Isolated RNA was treated with DNase (DNA-free; Ambion) prior to the synthesis of first-strand cDNA by the SuperScript III first-strand synthesis system for reverse transcription-PCR with oligo(dT)20 primer (Invitrogen). cDNA from 0.3 μg of total RNA was used as template for each 100-μL PCR. Gene-specific primers used for PCR were 5′-ACCTTCTCCGCAACAAGTGG-3′ and 5′-GAAGCTTGGTGGCTTGTAGG-3′ for RBCS2B (Acevedo-Hernández et al., 2005), 5′-CAGCTTGCCCACCCATTGTTA-3′ and 5′-GTCGTACGCACCGCTTCTTTCTTA-3′ for SAG13 (Barth et al., 2004), and QuantumRNA 18S internal standards for 18S ribosomal RNA (Ambion). PCR was terminated after 12 cycles for RBCS2B, 18 cycles for 18S ribosomal RNA, and 26 cycles for SAG13.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. SDS-PAGE and immunoblot analysis of chloroplast stroma-targeted GFP.
Supplemental Figure S2. Separation of GFP fluorescence and chlorophyll autofluorescence by LSCM.
Supplemental Movie S1. Movement of GFP-labeled spherical bodies in living cells of leaves of CT-GFP plants treated with concanamycin A and incubated in nutrient-free medium for 20 h in darkness.
Supplemental Movie S2. Movement of autophagic bodies in living cells of concanamycin A-treated leaves from transgenic plants expressing GFP-ATG.
Supplemental Movie S3. Visualization of stroma-targeted DsRed and GFP-ATG8 in living cells of concanamycin A-treated leaves.
Supplementary Material
Acknowledgments
We thank Dr. Jian Hua and her coworkers for technical advice about Arabidopsis transformation, Dr. Tsuyoshi Nakagawa for the gift of pGWB vector, Dr. Andrew O. Jackson for the gift of pGDR vector, the Nottingham Arabidopsis Resource Centre for providing seeds of an Arabidopsis T-DNA insertion mutant, Dr. Takafumi Uchida for the use of the LAS-3000, Dr. Tomoyuki Yamaya for the use of the ImageQuant400, Dr. Ikuko Hara-Nishimura and Dr. Yuki Fujiki for valuable suggestions, and Dr. Jin Su for technical advice about vector construction.
This work was supported by KAKENHI (grant nos. 18780042, 19039004, and 20780044 to H.I. and grant no. 17051002 to T.M.) and by the Energy Biosciences program of the U.S. Department of Energy (grant no. DE–FG02–89ER14030 to M.R.H.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hiroyuki Ishida (hiroyuki@biochem.tohoku.ac.jp).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Acevedo-Hernández GJ, Leon P, Herrera-Estrella LR (2005) Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. Plant J 43 506–519 [DOI] [PubMed] [Google Scholar]
- Arimura S, Hirai A, Tsutsumi N (2001) Numerous and highly developed tubular projections from plastids observed in tobacco epidermal cells. Plant Sci 160 449–454 [DOI] [PubMed] [Google Scholar]
- Asai T, Stone JM, Heard JE, Kovtun Y, Yorgey P, Sheen J, Ausubel FM (2000) Fumonisin B1-induced cell death in Arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell 12 1823–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert S, Gout E, Bligny R, Marty-Mazars D, Barrieu F, Alabouvette J, Marty F, Douce R (1996) Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: control by the supply of mitochondria with respiratory substrates. J Cell Biol 133 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth C, Moeder W, Klessig DF, Conklin PL (2004) The timing of senescence and response to pathogens is altered in the ascorbate-deficient Arabidopsis mutant vitamin c-1. Plant Physiol 134 1784–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth H, Meiling-Wesse K, Epple UD, Thumm M (2001) Autophagy and cytoplasm to vacuole targeting pathway both require Aut10p. FEBS Lett 508 23–28 [DOI] [PubMed] [Google Scholar]
- Bassham DC, Laporte M, Marty F, Moriyasu Y, Ohsumi Y, Olsen LJ, Yoshimoto K (2006) Autophagy in development and stress responses of plants. Autophagy 2 2–11 [DOI] [PubMed] [Google Scholar]
- Bhalla PL, Dalling MJ (1986) Endopeptidase and carboxypeptidase enzymes of vacuoles prepared from mesophyll protoplasts of primary leaf of wheat seedlings. J Plant Physiol 122 289–302 [Google Scholar]
- Chiba A, Ishida H, Nishizawa NK, Makino A, Mae T (2003) Exclusion of ribulose-1,5-bisphosphate carboxylase/oxygenase from chloroplasts by specific bodies in naturally-senescing leaves of wheat. Plant Cell Physiol 44 914–921 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Contento AL, Xiong Y, Bassham DC (2005) Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J 42 598–608 [DOI] [PubMed] [Google Scholar]
- Dedonder A, Rethy R, Fredericq H, Van Montagu M, Krebbers E (1993) Arabidopsis rbcS genes are differentially regulated by light. Plant Physiol 101 801–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277 33105–33114 [DOI] [PubMed] [Google Scholar]
- Evans JR (1989) Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia 78 9–19 [DOI] [PubMed] [Google Scholar]
- Feller U, Anders I, Mae T (2008) Rubiscolytics: fate of Rubisco after its enzymatic function in a cell is terminated. J Exp Bot 59 1615–1624 [DOI] [PubMed] [Google Scholar]
- Ferreira RM, Teixeira AR (1992) Sulfur starvation in Lemna leads to degradation of ribulose-bisphosphate carboxylase without plant death. J Biol Chem 267 7253–7257 [PubMed] [Google Scholar]
- Friedrich JW, Huffaker RC (1980) Photosynthesis, leaf resistance and ribulose-1,5-bisphosphate carboxylase degradation in senescing barley leaves. Plant Physiol 65 1103–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO (2002) pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J 31 375–383 [DOI] [PubMed] [Google Scholar]
- Gunning BE (2005) Plastid stromules: video microscopy of their outgrowth, retraction, tensioning, anchoring, branching, bridging, and tip-shedding. Protoplasma 225 33–42 [DOI] [PubMed] [Google Scholar]
- Hamasaki M, Noda T, Baba M, Ohsumi Y (2005) Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic 6 56–65 [DOI] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel LL, Grbić V, Baumgarten DA, Bleecker AB (1993) Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell 5 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzinger A, Buchner O, Lutz C, Hanson MR (2007) Temperature-sensitive formation of chloroplast protrusions and stromules in mesophyll cells of Arabidopsis thaliana. Protoplasma 230 23–30 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S, Feller U (2002) Nitrogen metabolism and remobilization during senescence. J Exp Bot 53 927–937 [DOI] [PubMed] [Google Scholar]
- Ishida H, Makino A, Mae T (1999) Fragmentation of the large subunit of ribulose-1,5-bisphosphate carboxylase by reactive oxygen species occurs near Gly-329. J Biol Chem 274 5222–5226 [DOI] [PubMed] [Google Scholar]
- Ishida H, Nishimori Y, Sugisawa M, Makino A, Mae T (1997) The large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase is fragmented into 37-kDa and 16-kDa polypeptides by active oxygen in the lysates of chloroplasts from primary leaves of wheat. Plant Cell Physiol 38 471–479 [DOI] [PubMed] [Google Scholar]
- Ishizuka M, Makino A, Suzuki Y, Mae T (2004) Amount of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) protein and levels of mRNAs of rbcS and rbcL in the leaves at different positions in transgenic rice plants with decreased content of Rubisco. Soil Sci Plant Nutr 50 233–239 [Google Scholar]
- Kay R, Chan A, Daly M, McPherson J (1987) Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 236 1299–1302 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Ohsumi Y (1999) Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol 15 1–32 [DOI] [PubMed] [Google Scholar]
- Knight S, Andersson I, Brändén CI (1990) Crystallographic analysis of ribulose 1,5-bisphosphate carboxylase from spinach at 2.4 A resolution. Subunit interactions and active site. J Mol Biol 215 113–160 [DOI] [PubMed] [Google Scholar]
- Köhler RH, Cao J, Zipfel WR, Webb WW, Hanson MR (1997) Exchange of protein molecules through connections between higher plant plastids. Science 276 2039–2042 [DOI] [PubMed] [Google Scholar]
- Köhler RH, Hanson MR (2000) Plastid tubules of higher plants are tissue-specific and developmentally regulated. J Cell Sci 113 81–89 [DOI] [PubMed] [Google Scholar]
- Kokubun N, Ishida H, Makino A, Mae T (2002) The degradation of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase into the 44-kDa fragment in the lysates of chloroplasts incubated in darkness. Plant Cell Physiol 43 1390–1395 [DOI] [PubMed] [Google Scholar]
- Krupinska K (2006) Fate and activities of plastids during leaf senescence. In RR Wise, JK Hoober, eds, The Structure and Function of Plastids. Springer, Dordrecht, The Netherlands, pp 433–449
- Kwok EY, Hanson MR (2004. a) Stromules and the dynamic nature of plastid morphology. J Microsc 214 124–137 [DOI] [PubMed] [Google Scholar]
- Kwok EY, Hanson MR (2004. b) GFP-labelled Rubisco and aspartate aminotransferase are present in plastid stromules and traffic between plastids. J Exp Bot 55 595–604 [DOI] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 6 463–477 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP (2005) Autophagy regulates programmed cell death during the plant innate immune response. Cell 121 567–577 [DOI] [PubMed] [Google Scholar]
- Mae T, Makino A, Ohira K (1983) Changes in the amounts of ribulose bisphosphate carboxylase synthesized and degraded during the life span of rice leaf (Oryza sativa L.). Plant Cell Physiol 24 1079–1086 [Google Scholar]
- Makino A, Osmond B (1991) Effects of nitrogen nutrition on nitrogen partitioning between chloroplasts and mitochondria in pea and wheat. Plant Physiol 96 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manigley C, Roth J (1985) Applications of immunocolloids in light microscopy. IV. Use of photochemical silver staining in a simple and efficient double-staining technique. J Histochem Cytochem 33 1247–1251 [DOI] [PubMed] [Google Scholar]
- Matile P (1987) The sap of plant cells. New Phytol 105 1–26 [DOI] [PubMed] [Google Scholar]
- Minamikawa T, Toyooka K, Okamoto T, Hara-Nishimura I, Nishimura M (2001) Degradation of ribulose-bisphosphate carboxylase by vacuolar enzymes of senescing French bean leaves: immunocytochemical and ultrastructural observations. Protoplasma 218 144–153 [DOI] [PubMed] [Google Scholar]
- Moriyasu Y, Ohsumi Y (1996) Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol 111 1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104 34–41 [DOI] [PubMed] [Google Scholar]
- Nakano R, Ishida H, Makino A, Mae T (2006) In vivo fragmentation of the large subunit of ribulose-1,5-bisphosphate carboxylase by reactive oxygen species in an intact leaf of cucumber under chilling-light conditions. Plant Cell Physiol 47 270–276 [DOI] [PubMed] [Google Scholar]
- Natesan SK, Sullivan JA, Gray JC (2005) Stromules: a characteristic cell-specific feature of plastid morphology. J Exp Bot 56 787–797 [DOI] [PubMed] [Google Scholar]
- Niwa Y, Kato T, Tabata S, Seki M, Kobayashi M, Shinozaki K, Moriyasu Y (2004) Disposal of chloroplasts with abnormal function into the vacuole in Arabidopsis thaliana cotyledon cells. Protoplasma 223 229–232 [DOI] [PubMed] [Google Scholar]
- Ohsumi Y (2001) Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2 211–216 [DOI] [PubMed] [Google Scholar]
- Ohsumi Y (2006) Protein turnover. IUBMB Life 58 363–369 [DOI] [PubMed] [Google Scholar]
- Phillips AR, Suttangkakul A, Vierstra RD (2008) The ATG12 conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S, Zouhar J, Carter C, Raikhel N (2007) Isolation of intact vacuoles from Arabidopsis rosette leaf-derived protoplasts. Nat Protocols 2 259–262 [DOI] [PubMed] [Google Scholar]
- Roberts P, Moshitch-Moshkovitz S, Kvam E, O'Toole E, Winey M, Goldfarb DS (2003) Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell 14 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W (2006) Protein degradation machineries in plastids. Annu Rev Plant Biol 57 599–621 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Ohsumi Y (2007) Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett 581 2156–2161 [DOI] [PubMed] [Google Scholar]
- Suzuki NN, Yoshimoto K, Fujioka Y, Ohsumi Y, Inagaki F (2005) The crystal structure of plant ATG12 and its biological implication in autophagy. Autophagy 1 119–126 [DOI] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y (1992) Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol 119 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Shimada T, Ono E, Tanaka Y, Nagatani A, Higashi SI, Watanabe M, Nishimura M, Hara-Nishimura I (2003) Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J 35 545–555 [DOI] [PubMed] [Google Scholar]
- Thayer SS, Huffaker RC (1984) Vacuolar localization of endoproteinases EP(1) and EP(2) in barley mesophyll cells. Plant Physiol 75 70–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AR, Vierstra RD (2005) Autophagic recycling: lessons from yeast help define the process in plants. Curr Opin Plant Biol 8 165–173 [DOI] [PubMed] [Google Scholar]
- Thumm M, Egner R, Kock B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH (1994) Isolation of autophagocytosis mutants of Saccaharomyces cerevisiae. FEBS Lett 349 275–280 [DOI] [PubMed] [Google Scholar]
- Toyooka K, Okamoto T, Minamikawa T (2001) Cotyledon cells of Vigna mungo seedlings use at least two distinct autophagic machineries for degradation of starch granules and cellular components. J Cell Biol 154 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333 169–174 [DOI] [PubMed] [Google Scholar]
- Tuttle DL, Dunn WA Jr (1995) Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J Cell Sci 108 25–35 [DOI] [PubMed] [Google Scholar]
- Wardley TA, Bhalla PL, Dalling MJ (1984) Changes in the number and composition of chloroplasts during senescence of mesophyll cells of attached and detached leaves of wheat (Triticum aestivum L.). Plant Physiol 75 421–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Fray RG, Pyke KA (2004) Stromule formation is dependent upon plastid size, plastid differentiation status and the density of plastids within the cell. Plant J 39 655–667 [DOI] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM (1998) A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol 37 455–469 [DOI] [PubMed] [Google Scholar]
- Wildman SG, Hongladarom T, Honda SI (1962) Chloroplasts and mitochondria in living plant cells: cinephotomicrographic studies. Science 138 434–436 [DOI] [PubMed] [Google Scholar]
- Wittenbach VA (1978) Breakdown of ribulose bisphosphate carboxylase and change in proteolytic activity during dark-induced senescence of wheat seedlings. Plant Physiol 62 604–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenbach VA, Lin W, Herbert RR (1982) Vacuolar localization of proteases and degradation of chloroplasts in mesophyll protoplasts from senescing primary wheat leaves. Plant Physiol 69 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Bassham DC (2005) AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J 42 535–546 [DOI] [PubMed] [Google Scholar]
- Xu Y, Ishida H, Reisen D, Hanson MR (2006) Upregulation of a tonoplast-localized cytochrome P450 during petal senescence in Petunia inflata. BMC Plant Biol 6 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Minamikawa T (1996) Successive amino-terminal proteolysis of the large subunit of ribulose 1,5-biphosphate carboxylase/oxygenase by vacuolar enzymes from French bean leaves. Eur J Biochem 238 317–324 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.