Abstract
We developed a functional selection system based on randomized genetic elements (GE) to identify potential regulators of hepatitis C virus (HCV) RNA translation, a process initiated by an internal ribosomal entry site (IRES). A retroviral HCV GE library was introduced into HepG2 cells, stably expressing the Herpes simplex virus thymidine kinase (HSV-TK) under the control of the HCV IRES. Cells that expressed transduced GEs inhibiting HSV-TK were selected via their resistance to ganciclovir. Six major GEs were rescued by PCR on the selected cell DNA and identified as HCV elements. We validated our strategy by further studying the activity of one of them, GE4, encoding the 5′ end of the viral NS5A gene. GE4 inhibited HCV IRES-, but not cap-dependent, reporter translation in human hepatic cell lines and inhibited HCV infection at a post-entry step, decreasing by 85% the number of viral RNA copies. This method can be applied to the identification of gene expression regulators.
INTRODUCTION
In order to identify hepatitis C virus (HCV) genetic elements, either RNA sequences or protein domains, that may modulate the viral genome translation, we developed a functional selection procedure adapted from the genetic suppressor element (GSE) strategy, which acts by down-regulating a target gene through an inhibitory antisense RNA or a trans-dominant peptide (1).
HCV is a single-stranded RNA virus provoking a chronic liver inflammation, likely to evolve toward cirrhosis and hepatocellular carcinoma (2). HCV lifecycle as well as the functions of some HCV proteins are not yet fully elucidated (3). After HCV entry into target cells, the 9.6-kb viral RNA genome is translated as a single open reading frame into a precursor polyprotein cleaved by host and viral proteases to give rise to structural and non-structural viral proteins. The non-structural HCV proteins form a membrane-associated replication complex with the viral RNA and are sufficient for replication of the viral genome. They are not contained in the virus particles, and therefore HCV RNA replication totally depends upon efficient translation. The structural proteins assemble with the neo-synthesized viral RNA copies to form progeny virions. The HCV RNA genome is flanked on either side by untranslated regions (UTRs), crucial for viral gene expression. The 5′ UTR is folded in several stem-loop domains acting as an internal ribosome entry site (IRES) initiating translation by a cap-independent mechanism (4–6). The 5′ UTR also contains sequences essential for replication (7) and regulation of viral RNA abundance (8). Because of its peculiarity in promoting persistent infection, HCV has been suggested to modulate its own expression, in order to maintain the replication level below the threshold hampering host cell viability. Some viral proteins contribute to the balance between translation, replication and packaging steps of the viral RNA, as part of the virus strategy to control its lifecycle. Indeed, HCV core, NS4A, NS4B and NS5A proteins have been proposed to regulate HCV translation (9–14).
Our strategy uses a retroviral library of randomized HCV genetic elements (GE) for transducing recipient cells stably expressing the Herpes simplex virus thymidine kinase (HSV- TK) gene under the control of the HCV IRES. The TK activity works as a negative selectable marker: upon addition of ganciclovir (GCV), a nucleoside analogue, to the cultured cells, HSV-TK phosphorylates GCV, which turns into a genotoxic metabolite, leading to subsequent cell death. Transduced cells expressing a HCV library-derived GE able to inhibit the IRES-mediated synthesis of HSV-TK will survive the GCV treatment. In rescuing those elements integrated in GCV-resistant cells, we identified six major HCV sequences. We investigated the activity of one of them encoding the N-terminal domain of the NS5A protein, and showed that it inhibited HCV IRES-mediated translation and hence HCV replication in cultured cells.
MATERIALS AND METHODS
Random HCV fragment library
pGEM-HCV containing the full-length HCV H77 cDNA (C. Wychowski, unpublished data) was fragmented by DNAse I in the presence of Mn2+, according to (1). The fragment termini were rendered blunt-end by Klenow- (Invitrogen, Cergy-Pontoise, France) and Pfu- (Stratagene, Amsterdam-Zuidoost, Netherlands) DNA polymerases and ligated with a 50-fold excess of 5′–phosphorylated synthetic adaptors [5′AATCATCGATGGATGGATGG3′ (sense) 5′CCATCCATCCATCGATGATTAAA3′ (antisense)], in the presence of T4 DNA ligase (Invitrogen). These adaptors contained the Cla I site and ATG-initiating codons in the three different reading frames. After fractionation by agarose gel electrophoresis, the mixture of 100–300 bp DNA fragments was recovered from the gel, digested by Cla I (Roche, Meylan, France) and cloned into the Cla I site of the pLNCX retroviral vector (Clontech, Saint-Germain-en-Laye, France). Fifty thousand independent recombinant clones were pooled to generate the HCV-pLNCX library.
Generation of the reporter cell line
The pIF-TK-3′NC expression vector contains the sequences of the firefly luciferase (Fluc) gene, HCV 5′UTR (HCV H77, nt 1–371), HSV-TK gene and the HCV 3′UTR (HCV H77, nt 9376–9647), in the pcDNA3.1 Zeo vector (Invitrogen). Therefore, the HSV-TK coding sequence was obtained by PCR, using the pGT60-mcs plasmid (Invivogen, Toulouse, France) as template and the primers 5′AACCTCAAAGAAAACTGCAGATCTTGGCCTCGTACC3′ (forward) and 5′CTACACAGTGAGCGGCCGCGTCGACTCAATCTAGTC3′ (reverse). The PCR product was cloned between PstI and NotI sites into the pIRF expression vector [provided by A. Cahour, (11)], from which the Renilla luciferase (Rluc) gene has been excised, so that HSV-TK was in frame with the HCV IRES AUG341. Finally, the HCV 3′UTR obtained by PCR was inserted between NotI and XhoI sites.
HepG2 cells (grown in DMEM supplemented with 10% fetal calf serum, 2 mM l-glutamine and 1% non-essential amino acids, at 5% CO2) were transfected with pIF-TK-3′NC, using FuGENE (Roche), and selected with 0.5 mg/ml zeocin (Invitrogen) for 2–3 weeks. They were then cloned in the presence of 0.7 mg/ml zeocin and screened for Fluc activity, using the Luciferase Assay System (Promega, Charbonniéres-les-Bains, France), and GCV sensitivity. B1 was one of those clones. The Hep-TK cell line was obtained by pGT60-mcs tranfection in the same conditions as above, except that they were selected with hygromycin at 0.2 mg/ml (Invivogen). The transgene expression in both reporter cell lines was routinely checked by northern blots of HSV-TK mRNA, Fluc activity and GCV sensitivity.
RETROVIRAL LIBRARY PRODUCTION AND TRANSDUCTION IN TARGET CELLS
HEK 293T cells (3 × 106 per 6 cm diameter tissue culture dish) were transfected with 3.5 μg of HCV-pLNCX library (or insert-free pLNCX for the control), 5 μg of pVPack-GagPol (Stratagene) and 3 μg of pVPack-vesicular stomatitis virus G protein (Stratagene), using the calcium phosphate procedure. Two days after transfection, the retroviral supernatant was harvested and filtered through a 0.45 µm filter. B1 cells (8 × 104 per 10 cm diameter culture dish) were infected with 3 ml of either HCV library or insert-free retroviral supernatant, mixed with 10 μg/ml DEAE-Dextran (Sigma, Saint-Quentin Fallavier, France). The efficiency of retroviral vector transduction on B1 cells was monitored using a pLNCX-DsRed vector, producing the red fluorescent protein Ds-Red (Clontech, Saint-Germain-en-Laye, France), and was found to be higher than 50% in these conditions. The transduced cells were grown in a non-selective culture medium for a week, and then treated by 140 μM GCV ([9-(1,3-dihydroxy-2-propoxy)methyl]guanine; Invivogen) for one month, changing the medium every 2–3 days. The surviving cells were expanded and 8 × 104 cells per 10 cm diameter culture dish were exposed again to 140 μM GCV for a month.
Rescue of the HCV fragments from the selected cells and sub-cloning
Isolated B1 colonies surviving GCV treatment were picked and grown individually. HCV fragments were rescued by PCR from the DNA of the selected cells as in (1), using the primers 5′GCCCCAAGCTTGTTAACAACGATGGATG3′ (forward) and 5′ATGGCGTTACTTAAGCTAGCTCGCCAAACCTAC3′ (reverse). The forward primer was designed to eliminate the ClaI site and to provide a HindIII site. The PCR products were cloned into the TOPO-TA vector (Invitrogen) and sequenced using the reverse primer mentioned above. The inserted HCV fragments were identified as sequences inserted between the adapter sequences and verified to belong to HCV H77. The PCR products were digested by HindIII and ClaI (Roche) and ligated into the corresponding sites of the pLHCX retroviral vector (Clontech), carrying the hygromycine phosphotransferase gene. Retroviral transduction of each selected fragment was done as above in HepG2 or Huh7 cells, except that the transduced cells were further enriched by hygromycin selection (0.3 mg/ml; Invivogen) for 10 days. The expression of the fragment in the transduced cells was assessed by RT–PCR, using the same primers as above.
Cell viability assay
Cells were grown in a 96 well microplate, starting at 2000 cells per well. Cell viability was assessed using the CellTiter96 AQueous One Solution Reagent (Promega, Charbonniéres-les-Bains, France) and a spectrophotometric microplate reader (Bio-Tek, Winooski, VT, USA).
In vitro transcription of RNA
The pIRF-derived pIRF-3′NC vector, containing the HCV H77 3′UTR inserted between the NotI and XhoI sites, was linearized by XhoI and transcribed using the AmplicapTM T7 High Yield transcription kit (TEBU, Le Perray en Yvelines, France). The plasmids pFK-I389 luc/NS3-3/5.1, containing the subgenomic replicon HCV 1b sequence and the Fluc gene, and pFK-I389luc/NS3-3′/GND carrying the unactivating GND mutations in the NS5B gene [provided by R. Bartenschlager (15)] were linearized with ScaI and transcribed using the MEGAscript transcription kit (Ambion, Courtaboeuf, France). Capped RNA of the GEs were obtained by transcription of PCR products amplified from the corresponding pLHCX vector with the primers 5′ TAATACGACTCACTATAGGGATCAACGATGGATGGATGG3′(T7-forward, initiating codons in bold) and 5′CCATCCATCCATCGATGATTAAA3′ (reverse). The initiating AUG were mutated stepwise by successive PCRs, using T7-forward-mut primers, in which the first ATG (mut1), the 1st and 2nd (mut2) or the 3 ATG (mut3) were mutated in CTG. Finally, the first AUG was rescued using PCR product (mut3) as a template and a T7-forward-mut primer in which the first initiating codon was ATG and the 2nd and 3rd CTG (ATG1). At each step, the PCR products were purified (Qiagen, Courtaboeuf, France) and transcribed using the AmplicapTM T7 transcription kit (TEBU). The synthesized RNAs were treated by RQ1 DNAse, purified on RNeasy columns (Qiagen) or by acidic phenol:chloroform 5 : 1 extraction, and quantified by UV-absorbance at 260 nm. Their quality was verified by agarose gel electrophoresis and ethidium bromide staining.
RNA transient transfection
Huh7 cells (5 x 104 per well) were seeded into a 24-well plate and grown for 24 h in DMEM supplemented with 10% fetal calf serum, 2 mM l-glutamine and 1% non-essential amino acids. pIRF-3′NC or replicon RNA (150 femtomoles) was mixed to 200 μl of serum-free DMEM containing 2 μl DMRIE-C transfection reagent (Invitrogen) and immediately added to the cell layers, that were incubated at 37°C. At 18 h post-transfection, the cells were lysed in 100 µl ice-cold lysis buffer, and the luciferase activities were measured using the Dual Luciferase Assay (Promega) and normalized for total protein content, which was measured by the Bradford assay (Bio-Rad, Marnes-la-Coquette, France).
HCV production and infection
The plasmid pJFH1 containing the full-length cDNA of JFH1 isolate [provided by T. Wakita (16)] has been used to generate genomic HCV RNA, which was delivered to Huh7 cells by electroporation, as described previously (17). Viral stocks of about 107 JFH1 RNA copies/ml were obtained by amplification on Huh-7 cells. 5 × 104 Huh7 cells per well were plated into 24-well plates and infected 24 h later with JFH1 stocks. For HCV immunodetection, the cells were fixed with 3% paraformaldehyde, permeabilized with 0.1% Triton X-100, and labeled with anti-E2, as described (17).
Quantitative RT–PCR
Total RNA was extracted using Trizol reagent (Invitrogen). qRT-PCR amplifications were conducted on 50 ng of total RNA using the MyiQ TM real-time system and the iScriptTM One-Step RT-PCR with SYBR Green kit (both from, Bio-Rad). HSV-TK [5′GTACCCGAGCCGATGACTTACT3′ (forward), 5′CCCGGCCGATATCTCA3′ (reverse)] and GAPDH [5′GTACCCGAGCCGATGACTTACT3′ (forward), 5′CCCGGCCGATATCTCA3′ (reverse)] primers were used at 0.3 μM. Reactions were performed in 25 μl under the following conditions: step 1, 10 min at 50°C; step 2, 5 min at 95°C; step 3, 40 cycles with one cycle consisting of 10 s at 95°C, 30 s at 62°C and 30 s at 72°C. HSV-TK and GAPDH copy numbers were determined by comparison to serially diluted transcripts included in the RT–PCR analysis. JFH1 RNA was quantified under the same conditions as above, except that primers in the NS3 region were used [5′GGTCATCACGGTCCTGACTCC3′ (forward) 5′TGTCTCAACGGGGATGAAAT3′ (reverse)] and a 30 s at 84°C step was added at each step 3 cycle.
In vitro translation assay
RNA (500 ng) encoding either GE1 or GE4 were translated for increasing periods of time at 30°C in a final volume of 20 µl containing 1 µl amino acid mixture and 14 µl nuclease-treated rabbit reticulocyte lysate (RRL) (Promega), thus providing RRL1. The reaction were started by amino acid addition and stopped on ice. In a second step referred to as RRL2, 100 ng pIRF-3′NC RNA were translated for 15 min at 30°C in the presence of 1 µl amino acid mixture, 13 µl RRL and 2 µl RRL1. The reaction was started by amino acid addition and stopped by RNAse A (5 µg/ml final). Fluc and Rluc activities in RRL2 were measured using the Dual Luciferase Assay (Promega).
Peptide synthesis
The peptides used in this study were purchased at NeoMPS (Strasbourg, France), and solubilized in sterile water at 10 mM, except W20L which was soluble in 10% DMSO at 3 mM.
RESULTS
We generated an HCV-derived GE library by random HCV cDNA fragmentation. A 100–300 bp size limit of the fragments was chosen first because 30–100 amino acids could be in the range of the size of HCV proteins domains (3) and second, because nucleic acids of that length hybridize strongly enough to be efficient antisense GEs. Longer fragments might cover the whole IRES or some VHC proteins of small size, likely to provide efficient but not novel GEs. Fragments were inserted into a retroviral expression vector under the control of a robust promoter, allowing not only their clonal distribution but also their stable expression in mammalian cells, either as sense or antisense RNAs. In addition, these fragments could be translated into peptides from one of the start codons in all three open reading frames. Beside the random library, another premise for the GE selection is a stable reporter system in recipient cells. To this end, the bicistronic vector pLuc-IRES-HSVTK-3′UTR carrying the cap-dependent Fluc and the HCV IRES-dependent HSV-TK was transfected into human hepatic HepG2 cells and several stable transfectants were obtained, among which the B1 clone was chosen for its high luciferase activity and sensitivity to GCV.
Selection of HCV fragments inhibiting HCV IRES activity
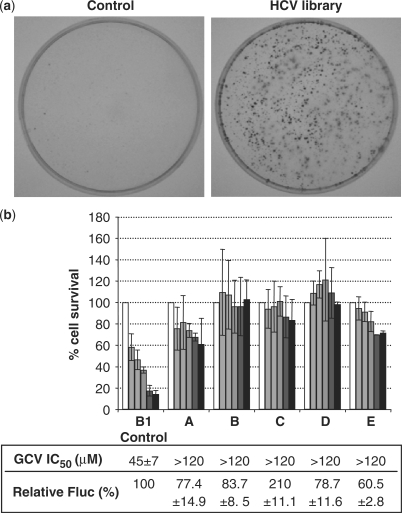
B1 cells were infected with pLNCX-derived retroviruses carrying the HCV GE library and subjected to GCV selection at 140 μM, corresponding to the IC90 for non-infected parent B1 cells. A control selection was performed in parallel on B1 cells infected with insert-free pLNCX-derived retroviruses. After two passages in the selective medium, many HCV library-transduced B1 cells survived to GCV selection, in contrast to control cells, all of which died (Figure 1a). Fifty surviving B1 colonies were picked and analysed individually for their GCV-resistant phenotype and Fluc activity. Most of them were resistant to high GCV concentrations, whereas their Fluc activity was found to be at least 60% that of control cells: Figure 1b shows the phenotype of five B1 clones surviving the selection, as an example. The detection of Fluc activity concomitantly to the loss of sensitivity to GCV ruled out the possibility of either the loss or the repression of the bicistronic transgene to explain GCV-resistance of the selected B1 colonies.
Figure 1.
Selection of HCV genetic elements conferring resistance to GCV in B1 cells. (a) Typical plates of control B1 cells transduced with insert-free retroviral vector (left) or B1 cells transduced with the retroviral HCV fragment library (right), after the second round of selection. In each selection round, transduced B1 cells were treated by 140 μM GCV for 1 month. Colonies were stained with crystal violet. (b) GCV resistance of individual selected B1 colonies. Each B1 colony picked from the selected HCV library-transduced cells was expanded, plated in a 96-well plate and treated for 2 weeks with increasing GCV doses, from 0 (white bars), 40, 60, 80, 100–120 μM (black bars). The results are expressed as the percentage of living cells (measured by the cell viability assay; see ‘Materials and Methods’ section) grown in the absence of GCV (mean ± SEM of duplicates). Control B1 cells and five representative colonies annotated A to E are shown. GCV IC50 deduced from these results, as well as the Fluc activity relative to that of control B1 cells, measured on 106 exponentially growing cells, are indicated.
Characterization of the selected HCV elements
PCR analysis of the integrated proviral inserts revealed that each selected B1 colony contained one to three fragments of the starting HCV library, the size of which ranged from 50 to 195 bp. The PCR products were cloned in the same orientation as in the parental B1 colony. Six major HCV GE, homologous to five different regions of the HCV genome, were obtained out of the 50 clones, with a frequency of 15–50%. Five of them were sense-oriented, i.e. corresponded to the HCV RNA + strand, and in frame of the adaptor first AUG, suggesting that they are likely to act by encoding peptides derived from C, NS3, NS4B, NS5A or NS5B genes (Table 1), rather than antisense RNAs. GE6, the unique antisense-oriented GE, was identified in the NS5B region. GE4, 195 nucleotides long, was actually a composite element of fragments from the NS5A gene on the 5′ side of the sequence and from the core gene on the 3′ side; 15 nt in the 34 nt overlap between the 3′ end of the NS5A fragment (HCV H77 nt 6359–6392) and the 5′ end of the core fragment (nt 635–669) were conserved in both sequences. Surprisingly the core gene derived sequence in GE4 constituted the GE1 element (Table 1). Moreover, we observed that GE4 and GE1 on the one hand, and GE5 and GE6 on the other hand, were always selected together.
Table 1.
Characteristics of the selected HCV genetic elements (GE)
| GE | Frequencya (%) | Size (nt) | Regionb | HCV gened | Orientation | GCV IC50c (μM) |
|---|---|---|---|---|---|---|
| 1 | 50 | 77 | 635–712 | Core | Sense | 33.5 ± 8.4 |
| 2 | 25 | 64 | 4758–4822 | NS3 | Sense | 74 ± 30 |
| 3 | 15 | 61 | 6039–6100 | NS4B | Sense | 104.6 ± 26 |
| 4 | 25 | 195 | 6246–6392 635–712 | NS5A Core | Sense Sense | >120 |
| 5 | 40 | 66 | 7623–7689 | NS5B | Sense | >120 |
| 6 | 50 | 56 | 9023–9079 | NS5B | Anti sense | 42.8 ± 12.7 |
anumber of B1 colonies containing the GE relative to the total number of selected colonies (each selected B1 colony contained 1–3 GE).
bRegion of the HCV H77 strain genome corresponding to the GE.
cValues deduced from a cell viability assay realized as in Fig. 1b.
dViral gene from which the GE was derived.
The ability of the selected GEs to individually confer GCV resistance was evaluated by the cell viability assay after individual retroviral transduction into B1 cells. With a GCV IC50 over 100 μM, GE3, GE4 and GE5 promoted GCV resistance in B1 cells (Table 1). GEs 1, 2 or 6, the GCV IC50 of which were close to that of parental B1 cells (45 μM), were considered inactive. We noticed no perturbation of cell growth in any GE-transduced cell line, as compared to either parental or control B1 cell lines (data not shown).
GE4 effect on HCV RNA expression models
We chose GE4 for further investigation, because it individually reproduced the phenotype it was selected for, i.e. GCV resistance of B1 cells, suggesting that it might be a potential inhibitor of HCV IRES-dependent translation. We verified that Fluc activity was unchanged in GE4-transfected as compared to control B1 cells (107.1% ± 4.4 of control cells, n = 4), as well as the amount of HSV-TK mRNA relative to GAPDH mRNA (2.77 ± 1.31 for GE4 versus 1.15 ± 0.34 for control, n = 4). These results exclude an effect of GE4 on transcription of the bicistronic reporter transgene in B1 cells. Interestingly, GE4 did not alter GCV sensitivity of HepG2 cells expressing the HSV-TK gene in a cap-dependent manner (19 ± 6 μM for GE4, versus 17.5 ± 8.1 μM for control, n = 3), corroborating its lack of effect on cap-dependent translation on the one hand, and excluding a possible effect on TK enzymatic activity, GCV uptake, metabolism or cell death signaling, on the other hand. All together, these results are consistent with a GE4-induced inhibition of HCV IRES-mediated HSV-TK translation.
In order to validate its activity on other recipient cells and HCV IRES-dependent reporters, GE4 was introduced into Huh7 cells by retroviral transduction, and control cells were transduced with the empty retroviral vector. In contrast to HepG2 cells, the human hepatic Huh7 cells are currently considered to be the most permissive for HCV RNA expression models, including subgenomic replicons (15) and production of infectious virions (16). GE4 effect on RNA translation was first determined by transient transfection of pIRF-3′NC RNA, in which Fluc and Rluc (Renilla luciferase) ORFs were translated by cap- and HCV IRES-dependent mechanisms, respectively. As shown in Table 2, HCV IRES-dependent Rluc, but not cap-dependent Fluc, was markedly inhibited in GE4–Huh7 as compared to control cells. Next, GE4 effect was assessed using the HCV replicon, which carries the Fluc mRNA under the HCV IRES control. During the 18 h period post- transfection, the replicon RNA was poorly replicated, but mainly translated, as indicated by quite similar luciferase values obtained with replication-competent or defective replicons (data not shown). As shown in Table 2, normalized replicon Fluc activity was decreased by 80% in GE4-expressing cells as compared to control cells, whether the replicon was replication-competent or defective, indicating that GE4 greatly impaired HCV replicon translation.
Table 2.
Effect of GE4 on HCV IRES-dependent translation, using transient transfection of reporter RNA into Huh7 cells
| Luciferase activity in GE4-Huh7 cells (% control) | |
|---|---|
| Tranfection of pIRF-3′NC RNA | |
| Cap-dependent Fluc | 97.3 ± 8.4 |
| IRES-dependent Rluc | 54.8 ± 0.9 |
| Transfection of HCV replicon RNA | |
| Replication competent replicon | 25.4 ± 4.0 |
| Replication defective replicon | 19.6 ± 4.1 |
Control (transduced with empty pLHCX) or GE4-expressing Huh7 cells (transduced with GE4-pLHCX) were grown in 24-well plates before being transfected with 150 femtomoles/well of reporter RNA: either pIRF-3′NC RNA with cap-dependent Fluc and HCV IRES-dependent RLuc (a), or HCV Luc-replicon RNA with HCV IRES-dependent Fluc (b). Fluc or Rluc activities were measured 18 h post-transfection and normalized for total protein content. The results are expressed as percent of normalized luciferase in control cells (mean ± s.e.m of three independent experiments).
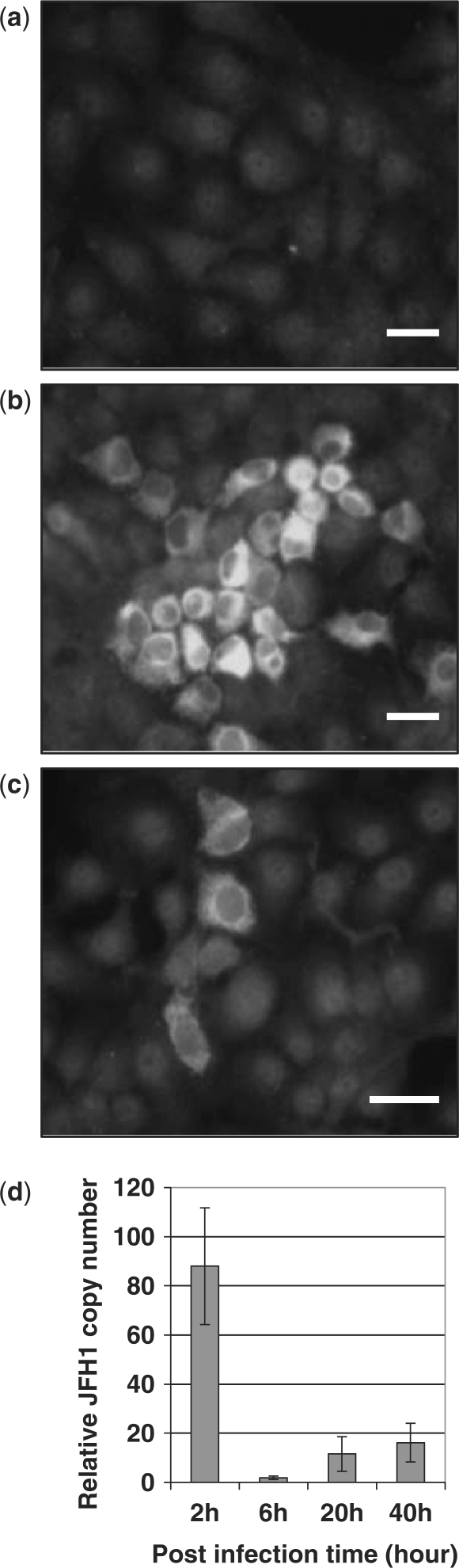
Last, we evaluated the GE4 effect on an HCV infection model in Huh7 cells from the HCV JFH1 strain (16). Both control and GE4-Huh7 cells were incubated with infectious HCV JFH-1 supernatant and analyzed for JFH1 E2 protein and genomic RNA 40 h later, during which the virus fulfilled its lifecycle. JFH1-infected cells were revealed by immunofluorescent staining of HCV E2 protein. Whereas no fluorescent cells were visible in non-infected cells (Figure 2a), infected cells are grouped in foci of 15–20 fluorescent cells in control Huh7 cell layer (Figure 2b). The number of fluorescent cells in GE4-expressing Huh7 cultures was noticeably reduced, as shown in Figure 2c as a representative field, suggesting that GE4 impaired JFH-1 viral lifecycle in those cells. This antiviral effect was measured using quantitative RT-PCR of JFH1 RNA, the replication of which depends on its translation. The amounts of viral RNA extracted from GE4-expressing cells were similar to that of control cells at 2 h post-infection, but were severely reduced thereafter, reaching 16.2 ± 7.9% of that of control cells at 40 h post-infection (Figure 2d). These results indicate that GE4 inhibited the replication cycle of HCV likely by interfering with a post-entry step.
Figure 2.
Antiviral effect of GE4. (a–c). Immunodetection of HCV E2 protein in HCV-infected Huh7 cells. Control (b) or GE4-transfected (c) Huh7 cells were grown on glass coverslips and incubated with 50-fold diluted JFH1 stock for 2 days. Non-infected cells (a) were used as negative control (X bar = 25 µm). (d) Post-infection kinetics of HCV RNA copy number in GE4-transfected Huh7 cells. Control or GE4-transfected Huh7 cells were incubated with undiluted JFH1 stock for 2 h at 37°C. After extensive washes, the cells were incubated further for 4, 18 and 38 h at 37°C. At the indicated post-infection time, total RNA was extracted and JFH1 RNA copies were measured by qRT–PCR and normalized to total RNA. The results are expressed as the percentage of normalized JFH1 copy number of control cells (mean ± SEM of two independent experiments).
Analysis of GE4 mechanism of action on translation
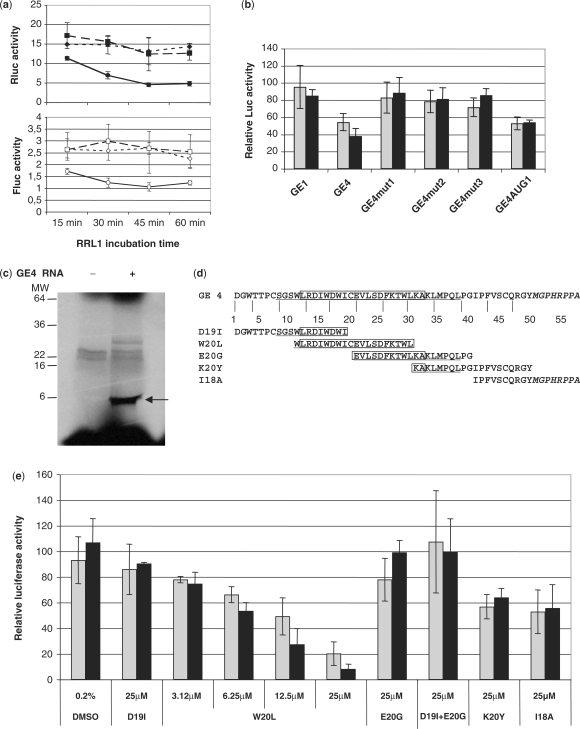
We used a two-step cell-free translation assay to determine if GE4 acted as RNA or peptide, and, if it acted as a peptide, from which reading frame it was translated. Indeed the retroviral vector constructs were designed in such a way that they could be translated in any of the three reading frames. GE4 RNA was first translated for increasing periods of time in RRL, thus providing RRL1 mixture. Identical RRL1 volumes, i.e. those containing the possible GE4 translation product, withdrawn at increasing GE4 RNA translation times, were then added to a second translation mixture (RRL2) containing pIRF-3′NC RNA to determine the effect on cap- and HCV IRES-dependent-translation. As shown in Figure 3a, both Fluc and Rluc activity in RRL2 were decreased in the function of GE4 translation time in RRL1, i.e. of increasing amounts of GE4 translation product. Unlike GE4, GE1, concomitantly selected with GE4 but individually inactive in B1 cells (Table 1), did not affect Fluc or Rluc much more than the control RRL1 incubated without RNA. We used this two-step translation assay to determine the activity of GE4-mut, in which the reading frames were successively mutated: neither GE4-mut1, GE4-mut2 or GE4-mut3 exerted an inhibition as strong as wild type GE4 (Figure 3b). Rescuing the first AUG in GE4-mut3 RNA restored a 50% inhibiting activity on both Fluc and Rluc. These results show that the GE4 translation product starting at the first AUG, rather than GE4 RNA, was responsible of the inhibition of pIRF-3′NC RNA translation. They also revealed that the activity of GE4 in the cell-free assay was not limited to HCV IRES-dependent translation, but also affected the cap-dependent one, contrary to what we observed in cells.
Figure 3.
Analysis of GE4 mechanism of action on translation. (a) Result of a typical two-step translation assay using RNA encoding GE1(squares), GE4 (circles) or no RNA (diamonds), incubated in RRL1 for the indicated period of time: cap-dependent Fluc (open symbols, lower figure) and HCV IRES-dependent Rluc (closed symbols, upper figure) activities in RLL2 are expressed as absolute RLU (× 106) values per assay (mean ± SEM of triplicate). (b) Effect of mutations in GE4 RNA on cap-dependent Fluc (light bars) and HCV IRES-dependent Rluc (dark bars) activities. GE4 RNA, either wild-type (GE4) or mutated at the level of the first AUG (mut1), the 1st and the 2nd AUGs (mut2), the 3AUGs (mut3) or the 2nd and 3rd AUGs (AUG1), was translated in RRL1 as in Figure 3a. The results are expressed as the percentage of RRL2 luciferase activity without RNA, obtained with a 45-min RRL1 (mean ± SEM of at least four experiments). (c) SDS–PAGE of the translation product of GE4 RNA (right lane), indicated by an arrow. GE4 RNA has been translated for 45 min in RRL1 mix, in the presence of S35-methionine, as recommended by the manufacturer. In the left lane, no RNA has been added. (d) Amino acid sequence of GE4 and GE4-derived synthetic peptides. The NS5A N-terminal membrane anchor domain is underlined. The amino acids L12–A32 form an amphipatic α-helix (boxed). (e) The synthetic peptides and the W20L solvent (0.2% DMSO) were tested for their effect on pIRF-3′NC RNA translation in the RRL2 translation assay. The luciferase activities are expressed as the percentage of activity in the absence of peptide (mean ± SEM of three independent experiments).
GE4 RRL1 was found to give rise to a 6 kDa peptide (Figure 3c), in agreement with the expected 58 amino acid sequences starting at the first AUG codon (Figure 3d). The 2nd and 3rd start codons would have led to 10 and 23 amino acid peptides, respectively, which could not be seen on the Figure 3c gel. This hypothetical 58 mer sequence contains at the N-terminus 7 amino acids belonging to NS4B, the N-terminal NS5A region (amino acids 8–50), followed by a sequence (amino acids 51–58) resulting from the translation of GE1 in a frame different from that of the HCV core protein, giving rise to a stop codon. Interestingly, the GE4 peptide sequence includes the whole N-terminal NS5A membrane anchor, which is organized in an amphipathic α-helix (27). We then synthesized a series of five overlapping peptides spanning the GE4 sequence (Figure 3d), and tested the activity of each of them in the RRL translation assay, using pIRF-3′NC reporter RNA. This screening revealed the strong, dose-dependent inhibition induced by W20L of both FLuc and RLuc activities, as compared to the other peptides (Figure 3e). W20L was more efficient at inhibiting RLuc than Fluc, so that the RLuc/FLuc ratio was 0.46 ± 0.08% of 25 μM of W20L. The combination of D19I and E20G, each containing half of W20L, could not restore the effect of W20L. Taken together, these results indicate that the activity of GE4 on translation may be related to a sequence spanning the NS5A amphipathic α-helix. The fact that GE4 and W20L affected both HCV IRES- and cap-dependent translation suggests that they may directly interact with a factor of the translation machinery.
DISCUSSION
In order to identify regulators of HCV translation in cultured cells, we developed a combinatorial approach inspired from the genetic suppressor element strategy (1,18–23), generating an original GE library derived from the viral genome, as well as recipient cells expressing a selectable reporter gene under the control of the targeted HCV IRES. Without using high-throughput sequencing facilities, we isolated six HCV genome derived-elements, which were present in at least 15% of the selected cell clones. All but one were sense-oriented and likely to act as peptides. None of the HCV fragments rescued in our study were antisense to the HCV RNA IRES or 3′UTR actually present in our reporter system, suggesting that antisense GEs had no selective advantage in our experimental conditions. The greater efficiency of phenotype suppression by peptides rather than by antisense constructs has been observed in other applications of the genetic suppressor element strategy (18,19). However, during our selection, the active GEs clearly did not act as negative trans-dominant peptides of the viral proteins they are derived from, because these are absent in our recipient system.
Three of the six selected GEs were individually active, namely GE3, 4 and 5, but their activity appeared unrelated to the frequency of their presence in the B1 clones, as shown, for example, by GE1, individually inactive despite being the most frequent (Table 1). The reason for this discrepancy is unclear. It could reveal that the GCV selection was not strong enough or uncompleted, still leaving false positives, as previously mentioned (1). Alternatively, the coincidence of couples of GEs in the same B1 clone, like GE1 and GE4 for instance, may not be fortuitous: whether the properties of some of the selected GEs are functionally additive or synergistic would have to be examined (24).
The well-studied HCV genome and protein structures allowed association of the active, selected GEs to known functional domains. GE3 encodes a sequence located next to the fourth transmembrane segment of the HCV non-structural protein NS4B (25). GE5 is a region in the N-terminal ‘fingers’ of the HCV RNA-dependent RNA polymerase (26). GE4 encodes the N-terminal NS5A region including the totality of the membrane anchor domain, which is structured in an amphipathic α-helix (27). We studied the GE4 activity on translation to validate our selection. GE4 inhibited HCV IRES-dependent translation independently of recipient cells, reporter gene and HCV strain, without apparently affecting cap-dependent translation, since stable GE4 transfectants could be selected either in HepG2 or Huh7 cells. Accordingly, this sequence was able to inhibit HCV infection. A cell-free assay containing the cell components necessary for protein synthesis allowed us to specify that GE4 was functioning as a peptide rather than RNA, and that its active part was related to the NS5A-derived peptide likely to fold as an amphipathic α-helix. The main function of NS5A N-terminal domain is to anchor this non-structural protein, mandatory to HCV replication, to intracellular membrane through an in-plane amphipathic α-helix structure interacting with the cytosolic membrane leaflet (28). The current hypothesis is that multiple NS5A dimers could form a two-dimensional array on intracellular membranes, thereby creating a ‘railway’ that would allow RNA sliding (3). Interestingly, mutations affecting the positioning of fully conserved residues located at the cytosolic surface of the NS5A amphipathic α-helix impaired HCV RNA replication without interfering with NS5A membrane association, suggesting that the N-terminal helix performed some additional functions other than membrane anchoring (27). NS5A also interacts with a number of host cell factors, including the RNA-activated protein kinase, PKR (29). Few and controversial data about the effect of NS5A on HCV IRES-mediated translation have been reported (14,30); the central domain of NS5A rather than its N-terminal region was suggested to be responsible for all these effects. Thus, our approach led us to unveil an unsuspected function of the N-terminal NS5A sequence in the regulation of HCV IRES-dependent RNA translation. Whether this regulation functions in the context of the intact protein still remains to be investigated.
It was intriguing that the cell-free assay revealed an effect of GE4 peptide on cap-dependent translation, whereas this was not apparent in growing cells. Assuming that the GE4 peptide directly interacted with some factors involved in the translation machinery, as suggested by our results of cell-free assays, one hypothesis is that it could be related to limited rate of active eIF2 or eIF3, the only eukaryotic translation initiation factors required by HCV IRES (4–6). Indeed, it was reported that HCV IRES could be substantially inhibited by partial depletion of eIF2γ and eIF2Bγ, the guanine nucleotide-exchange factor responsible for recycling eIF2 to its active GTP-bound state, whereas general protein translation and cell growth remained unaffected, suggesting that HCV IRES activity may depend on high levels of active eIF2, in contrast to the cap (31). If GE4 peptide limited somehow the rate of active eIF2 (or eIF3), this would impair both HCV IRES-and cap-dependent translation in a cell-free system, but become less sensitive for cap-dependent translation in living cells, which may be able to regenerate these translation factors at a threshold sufficient for the cap, but not for the IRES.
In conclusion, this selection led to the discovery of functional domains of the HCV genome. This strategy is particularly adapted to viruses, which fulfill a lot of functions with a limited number of protein and genetic elements. This method is of general interest and might prove useful for the identification of regulatory elements in either viral or host cell genomes.
ACKNOWLEDGEMENTS
This work has been supported by grants from the INSERM (Grant # A02272GS), the ANRS (Grant # 2005-196), the Conseil Régional d’Aquitaine (Grant# 20020301117N) and the GEFLUC association. L.J. is a recipient of a Conseil Régional d’Aquitaine fellowship. Funding to pay the Open Access publication charges for this article was provided by INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Roninson IB, Gudkov AV. Genetic suppressor elements in the characterization and identification of tumor suppressor genes. Methods Mol. Biol. 2003;222:413–436. doi: 10.1385/1-59259-328-3:413. [DOI] [PubMed] [Google Scholar]
- 2.Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21–S29. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- 3.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat. Rev. Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 4.Honda M, Beard MR, Ping LH, Lemon SM. A phylogenetically conserved stem-loop structure at the 5' border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rijnbrand RC, Lemon SM. Internal ribosome entry site-mediated translation in hepatitis C virus replication. Curr. Top. Microbiol. Immunol. 2000;242:85–116. doi: 10.1007/978-3-642-59605-6_5. [DOI] [PubMed] [Google Scholar]
- 6.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 7.Friebe P, Lohmann V, Krieger N, Bartenschlager R. Sequences in the 5' nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 2001;75:12047–12057. doi: 10.1128/JVI.75.24.12047-12057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of Hepatitis C Virus RNA Abundance by a Liver-Specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 9.Shimoike T, Mimori S, Tani H, Matsuura Y, Miyamura T. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J. Virol. 1999;73:9718–9725. doi: 10.1128/jvi.73.12.9718-9725.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Yamada O, Yoshida H, Iwai T, Araki H. Autogenous translational inhibition of core protein: implication for switch from translation to RNA replication in hepatitis C virus. Virology. 2002;293:141–150. doi: 10.1006/viro.2001.1270. [DOI] [PubMed] [Google Scholar]
- 11.Boni S, Lavergne JP, Boulant S, Cahour A. Hepatitis C virus core protein acts as a trans-modulating factor on internal translation initiation of the viral RNA. J. Biol. Chem. 2005;280:17737–17748. doi: 10.1074/jbc.M501826200. [DOI] [PubMed] [Google Scholar]
- 12.Li D, Takyar ST, Lott WB, Gowans EJ. Amino acids 1-20 of the hepatitis C virus (HCV) core protein specifically inhibit HCV IRES-dependent translation in HepG2 cells, and inhibit both HCV IRES- and cap-dependent translation in HuH7 and CV-1 cells. J. Gen. Virol. 2003;84:815–825. doi: 10.1099/vir.0.18697-0. [DOI] [PubMed] [Google Scholar]
- 13.Kato J, Kato N, Yoshida H, Ono-Nita SK, Shiratori Y, Omata M. Hepatitis C virus NS4A and NS4B proteins suppress translation in vivo. J. Med. Virol. 2002;66:187–199. doi: 10.1002/jmv.2129. [DOI] [PubMed] [Google Scholar]
- 14.He Y, Yan W, Coito C, Li Y, Gale M, Jr, Katze MG. The regulation of hepatitis C virus (HCV) internal ribosome-entry site-mediated translation by HCV replicons and nonstructural proteins. J. Gen. Virol. 2003;84:535–543. doi: 10.1099/vir.0.18658-0. [DOI] [PubMed] [Google Scholar]
- 15.Krieger N, Lohmann V, Bartenschlager R. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 2001;75:4614–4624. doi: 10.1128/JVI.75.10.4614-4624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rouille Y, Helle F, Delgrange D, Roingeard P, Voisset C, Blanchard E, Belouzard S, McKeating J, Patel AH, Maertens G, et al. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J. Virol. 2006;80:2832–2841. doi: 10.1128/JVI.80.6.2832-2841.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holzmayer TA, Pestov DG, Roninson IB. Isolation of dominant negative mutants and inhibitory antisense RNA sequences by expression selection of random DNA fragments. Nucleic Acids Res. 1992;20:711–717. doi: 10.1093/nar/20.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ossovskaya VS, Mazo IA, Chernov MV, Chernova OB, Strezoska Z, Kondratov R, Stark GR, Chumakov PM, Gudkov AV. Use of genetic suppressor elements to dissect distinct biological effects of separate p53 domains. Proc. Natl Acad. Sci. USA. 1996;93:10309–10314. doi: 10.1073/pnas.93.19.10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garkavtsev I, Riabowol K. Extension of the replicative life span of human diploid fibroblasts by inhibition of the p33ING1 candidate tumor suppressor. Mol. Cell Biol. 1997;17:2014–2019. doi: 10.1128/mcb.17.4.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gudkov AV, Kazarov AR, Thimmapaya R, Axenovich SA, Mazo IA, Roninson IB. Cloning mammalian genes by expression selection of genetic suppressor elements: association of kinesin with drug resistance and cell immortalization. Proc. Natl Acad. Sci. USA. 1994;91:3744–3748. doi: 10.1073/pnas.91.9.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gros L, Delaporte C, Frey S, Decesse J, de Saint-Vincent BR, Cavarec L, Dubart A, Gudkov AV, Jacquemin-Sablon A. Identification of new drug sensitivity genes using genetic suppressor elements: protein arginine N-methyltransferase mediates cell sensitivity to DNA-damaging agents. Cancer Res. 2003;63:164–171. [PubMed] [Google Scholar]
- 23.Dunn SJ, Khan IH, Chan UA, Scearce RL, Melara CL, Paul AM, Sharma V, Bih FY, Holzmayer TA, Luciw PA, Abo A. Identification of cell surface targets for HIV-1 therapeutics using genetic screens. Virology. 2004;321:260–273. doi: 10.1016/j.virol.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Levenson VV, Lausch E, Kirschling DJ, Broude EV, Davidovich IA, Libants S, Fedosova V, Roninson IB. A combination of genetic suppressor elements produces resistance to drugs inhibiting DNA replication. Somat Cell Mol. Genet. 1999;25:9–26. doi: 10.1023/b:scam.0000007136.49230.b3. [DOI] [PubMed] [Google Scholar]
- 25.Hugle T, Fehrmann F, Bieck E, Kohara M, Krausslich HG, Rice CM, Blum HE, Moradpour D. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology. 2001;284:70–81. doi: 10.1006/viro.2001.0873. [DOI] [PubMed] [Google Scholar]
- 26.Bressanelli S, Tomei L, Roussel A, Incitti I, Vitale RL, Mathieu M, De Francesco R, Rey FA. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl Acad. Sci. USA. 1999;96:13034–13039. doi: 10.1073/pnas.96.23.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penin F, Brass V, Appel N, Ramboarina S, Montserret R, Ficheux D, Blum HE, Bartenschlager R, Moradpour D. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 2004;279:40835–40843. doi: 10.1074/jbc.M404761200. [DOI] [PubMed] [Google Scholar]
- 28.Brass V, Bieck E, Montserret R, Wolk B, Hellings JA, Blum HE, Penin F, Moradpour D. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 2002;277:8130–8139. doi: 10.1074/jbc.M111289200. [DOI] [PubMed] [Google Scholar]
- 29.Thompson SR, Sarnow P. Regulation of host cell translation by viruses and effects on cell function. Curr. Opin. Microbiol. 2000;3:366–370. doi: 10.1016/s1369-5274(00)00106-5. [DOI] [PubMed] [Google Scholar]
- 30.Kalliampakou KI, Kalamvoki M, Mavromara P. Hepatitis C virus (HCV) NS5A protein downregulates HCV IRES-dependent translation. J. Gen. Virol. 2005;86:1015–1025. doi: 10.1099/vir.0.80728-0. [DOI] [PubMed] [Google Scholar]
- 31.Kruger M, Beger C, Li QX, Welch PJ, Tritz R, Leavitt M, Barber JR, Wong-Staal F. Identification of eIF2Bgamma and eIF2gamma as cofactors of hepatitis C virus internal ribosome entry site-mediated translation using a functional genomics approach. Proc. Natl Acad. Sci. USA. 2000;97:8566–8571. doi: 10.1073/pnas.97.15.8566. [DOI] [PMC free article] [PubMed] [Google Scholar]