Abstract
The TNF locus on chromosome 6p21 encodes a family of proteins with key roles in the immune response whose dysregulation leads to severe disease. Transcriptional regulation is important, with cell type and stimulus-specific enhancer complexes involving the proximal TNF promoter. We show how quantitative chromatin profiling across a 34 kb region spanning the TNF locus has allowed us to identify a number of novel DNase hypersensitive sites and characterize more distant regulatory elements. We demonstrate DNase hypersensitive sites corresponding to the lymphotoxin alpha (LTA) and tumour necrosis factor (TNF) promoter regions, a CpG island in exon 4 of lymphotoxin beta (LTB), the 3′ end of nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 (NFKBIL1) and 3.4 kb upstream of LTA. These sites co-localize to highly conserved DNA sequences and show evidence of cell type specificity when lymphoblastoid, Jurkat, U937, HeLa and HEK293T cell lines are analysed using Southern blotting. For Jurkat T cells, we define histone modifications across the locus. Peaks of acetylated histone H3 and H4, together with tri-methyl K4 of histone H3, correspond to hypersensitive sites, notably in exon 4 of LTB. We provide evidence of a functional role for an intergenic DNase I hypersensitive site distal to LTA in Jurkat cells based on reporter gene analysis, with evidence of recruitment of upstream stimulatory factors (USF) transcription factors.
INTRODUCTION
The discovery of regulatory elements within DNA sequences remains a major priority in the post-genomic era (1). The sensitivity of DNA to digestion by the non-specific endonuclease DNase I has been highly informative as a tool to identify important regulatory sites including promoters, enhancers, locus control regions, silencers and insulators (2–4). DNase hypersensitivity results when the DNA is rendered more susceptible to enzymatic cleavage through loss or remodelling of one or more nucleosomes, events characteristically associated with active regulatory elements. The gold standard for detection of DNase I hypersensitive sites has been Southern blotting although a number of alternative approaches have been developed including cloning and sequencing (5,6), quantitative PCR (7) and the use of microarray platforms (8–10). For specific genomic regions, quantitative chromatin profiling using real time PCR to define hypersensitive sites is an attractive approach as it is reported to be highly sensitive and specific (7). Despite this, the sensitivity of the approach is yet to be independently replicated.
We sought to apply quantitative chromatin profiling to the TNF locus on chromosome 6p21 in order to systematically map DNase hypersensitive sites across the region. The locus contains genes encoding members of the TNF family, a group of proteins with key roles in immunity and inflammation which have been the focus of intensive basic science and translational research. These include lymphotoxin alpha (LTA), tumour necrosis factor (TNF) and lymphotoxin beta (LTB). The regulation of expression of these genes is critical to mounting an effective and coordinated immune response. Dysregulation can lead directly to severe disease as illustrated by studies showing the role of TNF in the pathophysiology of septic shock (11) and cerebral malaria (12) as well as Crohn's disease (13) and rheumatoid arthritis (14)—conditions in which biological therapies using antibodies and other inhibitors of TNF are dramatically impacting patient care.
A coordinated and specific level of TNF gene expression is required to respond to a diverse array of stimuli, ranging from antigen binding by B and T cells, to bacterial lipopolysaccharide, viruses, parasites, mitogens and cytokines. Intensive study of the transcriptional regulation of TNF has defined cell type and stimulus-specific enhancer complexes involving binding by Ets, Elks-1, ATF-2, c-jun, Egr-1, Sp1 and NFATp transcription factors to the highly conserved proximal TNF promoter, and recruitment of co-activator proteins including CREB binding protein and p300 (15–19). A number of other regulatory elements have been reported including within the third intron of TNF (20) and the 3′-UTR (21) together with more distal NFκB elements in the promoter (22,23) and downstream of TNF (24). The transcriptional regulation of other genes in the TNF locus is much less well characterized. The proximal LTA promoter is highly conserved with NFκB playing an important role in LTA inducibility in T cells and HTLV infected cell lines (25,26) while there is evidence that binding by HMGAIa (27) and NFAT (28) is important at more distal sites in the promoter. There is evidence of context specificity in regulation of LTA transcription with CD40 and IL4 responsive regions involving NFκB and STAT respectively (29). For LTB, the highly conserved proximal promoter contains important binding sites for transcriptional regulation including NFκB and Ets transcription factors, with the latter playing a stimulus-specific role (30,31).
It remains unclear whether more distant regulatory elements such as enhancers, locus control regions, silencers or insulators play a role in the specific or coordinated regulation of this cluster of genes. Polymorphism in the region has been associated with a range of autoimmune, inflammatory and infectious diseases including myocardial infarction (32), rheumatoid arthritis (33), cerebral malaria (34) and leprosy (35). The fine mapping of disease associations in this region has proved problematic due to the extent of linkage disequilibrium and difficulty of defining functionally important variants. The resolution of DNase hypersensitive sites in intergenic or other genomic regions would facilitate both our understanding of the regulation of this cluster of genes, and attempts to fine map and identify regulatory polymorphism. The data relating to DNase hypersensitivity is limited mainly to the TNF and LTA promoter regions (20,36–43). In human monocytes, monocyte-like cell lines and T cell lines, constitutive DNase hypersensitive sites have been identified localizing to the proximal TNF promoter (20,36–40). In Jurkat T cells, hypersensitive sites were also reported in the third intron of TNF and 3′-UTR, as well as the first intron of LTB; the same pattern was found for the monocytic cell line THP-1 except the TNF intronic and 3′-UTR hypersensitive sites were absent for this cell line (20,40). A DNase I hypersensitive site in the LTA promoter region was reported in THP-1 cells and primary human monocytes (38). DNase hypersensitive sites have also been identified in porcine peripheral blood mononuclear cells corresponding to the TNF promoter and third intron, together with the LTA promoter/5′-UTR (41); and in murine T cells involving the TNF and LTA promoter regions, as well as sites 5 kb upstream of LTA and 3 kb downstream of the TNF transcriptional start site (42).
In this study, we sought to systematically define and validate DNase I hypersensitivity across a 34 kb region spanning the TNF locus and flanking sequences. We were able to successfully establish the approach of quantitative chromatin profiling in our laboratory, to validate previously reported DNase hypersensitive sites in the locus and to demonstrate a number of novel sites that are candidate locations for regulatory elements. These sites were confirmed and further resolved by Southern blotting a panel of human cell lines including Jurkat T cells, the monocyte-like cell line U937, HeLa and HEK293T cells. Overall we report six novel DNase hypersensitive sites in the TNF locus which show cell type specificity and demonstrate striking co-localization to evolutionarily conserved sequence elements. We complement this work with chromatin profiling for specific histone modifications across the locus in Jurkat T cells, and provide evidence that one of the DNase hypersensitive sites in an intergenic region 3.4 kb upstream of LTA is associated with enhancer activity and recruitment of the upstream stimulatory factors (USF) family of transcription factor proteins in a human T cell line.
MATERIALS AND METHODS
Cell culture
Human lymphoblastoid cell lines (LCLs), BL41, Jurkat and U937 cells were grown in RPMI 1640 (Sigma–Aldrich, Gillingham, Dorset, UK) at 37°C in 5% CO2; HeLa and HEK293T cells were grown in DMEM (Sigma-Aldrich). Culture media for all lines was supplemented with 2 mM Glutamine (Sigma), 100 U/ml penicillin (Sigma), 0.1 mg/ml streptomycin (Sigma) and 10% FCS (Sigma). Cells were harvested in mid log phase. Mitogens used for cell stimulation where indicated were a combination of 125 nM ionomycin (Sigma) and 200 nM PMA (Sigma) (final concentration) applied to cells for 6 h unless otherwise stated. The following LCLs were used: GM12156 obtained from Coriell Institute for Medical Research (Camden, NJ, USA); PGF, European Collection of Cell Cultures (Salisbury, Wiltshire, UK); and QBL (Fred Hutchinson Cancer Research Center International Histocompatibility Working Group Cell and Gene Bank, Seattle, WA, USA).
DNase I digestions
We followed the methodology described by Dorschner et al. (7) with some modifications. Cells in suspension were pelleted at 500 g, washed twice with cold PBS and counted using a haemocytometer. Cells were resuspended in aliquots using 19 ml ice-cold buffer A (15 mM Tris–Cl pH 8, 15 mM NaCl, 60 mM KCl, 1 mM EDTA pH 8, 0.5 mM EGTA pH 8, 0.5 mM spermidine, 0.15 mM spermine) then 6 ml of 0.08% NP-40 in buffer A was added drop-wise to each of the cell aliquots and incubated on ice for 10 min. Lysis efficiency was confirmed by Trypan Blue staining. Nuclei were centrifuged at 1000 g for 3 min at 4°C then washed in 20 ml fresh buffer A before resuspension in buffer A to a concentration of 108 nuclei per ml. Serial dilutions of DNase (Roche, Burgess Hill, West Sussex, UK) in DNase buffer (10 mM Tris pH 8, 3 mM CaCl2, 75 mM NaCl) were prepared on ice. DNase digestions (20–160 U/ml) of 107 nuclei per DNase treatment were performed for 3 min at 37°C after which an equal volume of stop buffer (50 mM Tris–Cl pH 8, 100 mM NaCl, 100 mM EDTA pH 8) was added. ‘No DNase’ controls were used with sample incubation both on ice and at 37°C as per DNase digestions. Samples were then incubated at 37°C for 30 min with 6 μg RNase A (Roche) before incubating overnight at 55°C with proteinase K (25 μg/ml final concentration) and SDS (0.05%). DNA was purified by phenol–chloroform extraction and ethanol precipitation. DNA was further purified by precipitation with 2 M ammonium acetate pH 5.2 (Sigma) and ethanol, washed with 70% ethanol and resuspended in 500 μl 10 mM Tris pH 8. Samples were quantified using an ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA).
Southern blotting
About 15 μg DNase-treated or control DNA was digested using the restriction enzyme Sca I (NEB, Hitchin, Hertfordshire, UK) overnight at 37°C. DNA was precipitated on ice using 2 M ammonium acetate pH 5.2 and 2.5 volumes 100% ethanol combined with 1 μl glycogen (Roche). DNA was pelleted by centrifugation in a microfuge at maximum speed at 4°C for 10 min, washed in ice-cold 70% ethanol and air-dried before re-suspending in 21 μl 10 mM Tris pH 8. Samples were quantified by spectrophotometer and equal amounts loaded on a 0.8% 1 × TBE agarose gel which was run at 30 V overnight. The agarose gel was soaked for 20 min in freshly prepared denaturation solution (0.4 M NaOH, 1.5 M NaCl) then transferred onto Hybond N+ membrane (GE Healthcare Life Sciences, Amersham, Buckinghamshire, UK) by upward capillary transfer. After washing with 5 × SSC, the membrane was UV crosslinked using Stratalinker 2400 (Stratagene/Agilent Technologies, La Jolla CA, USA). Membranes were pre-hybridized with Church hybridization buffer [10 mg/ml BSA, 0.25 M phosphate buffer, 5% SDS, 1 mM EDTA, 0.25 M NaCl and 0.1 mg/ml sonicated herring sperm DNA (Roche)] at 65°C for at least 30 min. Probes for Southern blotting were generated by PCR amplification of 50 ng genomic DNA using Platinum Taq polymerase (Invitrogen, Paisley, Renfrewshire, UK) with initial denaturation at 96°C for 1 min; then 30 cycles of 94°C for 45 s, 60°C for 30 s, 72°C for 45 s then 72°C for 5 min. Primers were used at 0.2 μM; primer sequences are available on request. PCR products were gel-purified and 50 ng of probe was radiolabelled with α32P dCTP using random nonamers and Klenow following the manufacturer's instructions for the Megaprime DNA labelling system (GE Healthcare Life Sciences). Unincorporated nucleotides were removed using a ProbeQuant G-50 micro column (GE Healthcare Life Sciences). The probe was denatured at 95°C for 5 min, placed on ice, then added to fresh Church hybridization buffer and incubated with the membrane at 65°C overnight in a hybridization oven. Membranes were then washed in low stringency wash buffer (2 × SSC 1% SDS) followed by stringent wash buffer (0.5 × SSC 0.1% SDS) and exposed to an autorad.
Mapping DNase I hypersensitive sites using quantitative PCR
We followed the methodology described by Dorschner et al. (7) for quantitative chromatin profiling. Primers were designed to amplify 250 bp amplicons which were contiguous or minimally overlapping. Primer design followed the parameters described by Dorschner et al. (7) using Primer 3 (44) software. These included an optimal product size of 250 bp (±50); primer Tm (optimal 60°C ± 2),%GC (50% optimal, range 40–80) and length (optimal 24, range 19–27); and poly X (maximum 4). Primers were scanned for repetitive sequences using a human mispriming repeat library and verified for specificity using the standalone In-Silico PCR (Jim Kent, http://genome.ucsc.edu/cgi-bin/hgPcr). Primer sequences are shown in Supplementary Table S1A. Relative DNase I sensitivity of untreated and DNase-treated DNA was measured by real time quantitative PCR. Individual reactions comprised 0.9 μM forward and reverse primer, Power Sybr Green PCR master mix (Applied Biosystems, Warrington, Cheshire, UK) (containing SYBR® Green 1 Dye, AmpliTaq Gold® DNA Polymerase LD, dNTPs with dUTP/dTTP blend, Passive Reference 1 and optimized buffer components), and 20 ng template DNA made up to a 10 μl reaction volume with water. The samples and master mix were pipetted into 384 well reaction plates robotically using the Matrix Platemate Plus and plates sealed using a manual heat sealer and Abgene clear seal strong seals. Reactions were thermocycled on an ABI 7900HT (Applied Biosystems) with each reaction run in triplicate. To allow normalization of amplification efficiency, a dilution series for each sample analysed (40, 20 and 10 ng) was included on every individual 384 well plate with a primer pair amplifying a reference amplicon within an inactive and DNase-insensitive RHO (Rhodopsin) locus (3q21–q24) (7). Melt curve data for each primer pair used were recorded.
Analysis of quantitative PCR data
Data from primer pairs showing multiple peaks upon melting curve analysis were excluded from further analysis on the basis of potentially having multiple products. For each of four replicate DNase samples, at each primer a robust average of the triplicate raw values was generated, excluding any value >1 SD from the mean. In total, 20 primers were excluded on the basis of melting curve analysis and <0.5% of raw values were >1 SD from the raw mean. The amplification efficiency of test primers was normalized to that of the DNase-insensitive RHO locus primers. Using the RHO dilution series, present on each plate, a reference amplification curve was generated and amplification efficiency (E) was calculated as E = 10−1/slope (45). The amplification efficiency of all test primers was derived from the curve and normalized to the reference efficiency. A relative copy number ratio was generated using the 2−ΔΔ Ct equation (46), computed as 2−[(T− R)D − (T− R)U] where T is the test primer value, R is the RHO reference primer and D and U represent the DNase-treated and untreated samples, respectively. Statistical analysis of relative copy number ratios closely followed that of Dorschner et al. (7), where a baseline of DNase I sensitivity measurements across the region is calculated as a lowess-smoothed robust mean. The robust mean was calculated as a 20% trimmed mean and symmetric lowess smoothing was performed with a span of 0.2. Outliers were identified and the remaining data centred around the baseline. Signal-to-noise ratios (SNR) were computed as described in Dorschner et al. (7) where the SNR for each amplicon is the absolute deviation of it's trimmed mean from the baseline, divided by the median average deviation of the centred baseline. To identify putative DHS sites, locations were identified whose trimmed mean lay outside the lower 90% confidence limit of the baseline and whose SNR exceeded a threshold of 2.
RNA extraction, cDNA synthesis and quantitative PCR
Total RNA was prepared using the RNeasy Mini kit (Qiagen, Crawley, West Sussex, UK) following the manufacturer's instructions with inclusion of a DNase I digestion step on the RNeasy minicolumn. Following quantification, cDNA was prepared using SuperScript III (Invitrogen) using random primers and following the manufacturer's instructions which included digestion with RNase H (Invitrogen) following cDNA synthesis. Control RT reactions omitting the reverse transcriptase were included for each sample. Quantitative PCR was performed using SYBR Green Supermix (BioRad, Hemel Hempstead, Hertfordshire, UK) on an iQ Cycler (BioRad). PCR efficiency was determined and melt-curve analysis performed for gene-specific primer sets (sequences listed in Supplementary Table S1B). Relative gene transcript levels were determined by the ΔΔCt method.
Chromatin immunoprecipitation
About 100 × 106 cells in mid log phase were subjected to formaldehyde cross-linking (1% final concentration) for 15 min at room temperature, quenched by addition of glycine (0.125 M final concentration). Cells were lysed and nuclei isolated as described (47) with sonication in the presence of 212–300 micron glass beads (Sigma) using a Branson 450 Sonifier with a constant power amplitude of 40% for six pulses of 30 s on ice, cooling on ice for 1 min between pulses. Sonicated material was adjusted to 0.5% Sarkosyl and gently mixed for 10 min at room temperature before centrifugation at 10 000g at 4°C for 10 min to remove cell debris. Immunoprecipitation was performed using sheep anti-rabbit IgG coated Dynabeads (Invitrogen) pre-incubated with specific primary antibodies (histone H3 tri-methyl K4 rabbit ab 8580-50) (Abcam, Cambridge, Cambridgeshire, UK), anti-diacetylated histone H3 06-599 (Upstate/Millipore, Watford, Hertfordshire, UK), anti-tetracetylated histone H4 #06-866 (Upstate), anti-USF1 sc229 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-USF2 sc862 (Santa Cruz) and anti-actin #A-2066 (Sigma) in PBS with 5 mg/ml BSA. For histone IPs, chromatin was incubated with antibody-coated beads in 10 mM Tris pH 8, 1 mM EDTA, and 1% Triton X-100, 0.1% sodium deoxycholate, and 5 μg/ml pepstatin at 4°C overnight in a rotating platform. Beads were washed eight times in RIPA buffer (10 mM Tris pH 8, 1 mM EDTA, 1% NP-40, 0.7% sodium deoxycholate, 0.5 M LiCl) with 2-min incubation on ice between washes. For USF IPs, the IP buffer used was 17.5 mM Tris–HCl pH 8, 1.4 mM EDTA, 0.55% Triton X-100, 0.01% SDS, 16.7 mM NaCl; IPs were washed twice in a low-salt wash buffer (20 mM Tris HCl pH 8, 2 mM EDTA, 1% Triton X-100, 0.1% SDS, 150 mM NaCl), then twice in high-salt wash buffer (same as low salt wash buffer but containing 500 mM NaCl) and then twice in LiCl wash buffer (10 mM Tris HCl pH 8, 1 mM EDTA, 1% NP-40, 1% DOC, 250 mM LiCl), rotating for 5 min between each wash. 1× complete protease inhibitor cocktail (Roche) was used in all IP and wash buffers. Samples were washed once with TE and eluted in TE containing 1% SDS. Samples were vortexed briefly to resuspend the beads and incubated at 65°C for 10 min, centrifuged for 30 s at maximum speed and supernatant transferred to a new tube. Cross-links were reversed at 65°C overnight then digested with proteinase K for 2 h at 37°C. DNA fragments were purified by phenol–chloroform extraction and precipitated using 3 M NaAc (pH 5.2) in the presence of glycogen. DNA was treated with 0.5 μg RNase A and following spin column purification was eluted in 10 mM Tris pH 8. For quantitative chromatin immunoprecipitation analysis by real time PCR we followed the methodology described by De Gobbi and colleagues (48). Primer sets used for DHS profiling were used to amplify 250 bp amplicons at ∼1 kb intervals across the TNF locus with the amount of DNA immunoprecipitated by a specific histone antibody quantified relative to that of non-immunoprecipitated (input) DNA and normalized relative to a control sequence in the 18S ribosomal RNA gene.
Reporter gene assays
The DNA fragment corresponding to the human LTA promoter and 5′ UTR (chr 6:31647635–31648498) was generated by PCR amplification of genomic DNA from the PGF cell line which provides the reference sequence of the human genome and is homozygous over the MHC (49). PCR products were cloned into BglII/NcoI sites upstream of the firefly luciferase gene in the pGL3-Basic vector (Promega, Southampton, Hampshire, UK). Using the same approach the DHS 44500 conserved region (313 bp) (31644374–31644686) was cloned into BamHI/SalI sites downstream of the luciferase gene. Alternative constructs were prepared also using the BamHI/SalI sites, inserting the conserved region in reverse orientation, or the flanking region telomeric (flank1, 319 bp) (31644049–31644367) or centromeric (flank2, 316 bp) (31644697–31645012) of DHS 44500. All constructs were verified by DNA sequencing. Jurkat cells were transiently transfected using Lipofectamine LTX and PLUS reagent (Invitrogen) according to the manufacturer's instructions, allowed to recover after transfection for 1 h, then where indicated were stimulated with 125 nM ionomycin and 200 nM PMA. Cells were harvested after 24 h and luciferase assays performed following the manufacturer's protocol. Firefly luciferase constructs were co-transfected with pRL-TK (Promega) encoding Renilla luciferase to allow normalization of transfection efficiencies. Two independent endotoxin-free preparations of all constructs were analysed in transfection experiments.
Solid phase DNase I footprinting
This was performed as previously described (50,51). Forward and reverse radiolabelled DNA probes spanning DHS 44500 (31644349–690 and 31644347–686, respectively) were generated by PCR using PGF genomic DNA as template with one biotinylated primer and one primer end labelled with γ32P using T4 polynucleotide kinase (primer sequences given in Supplementary Table S1C). Binding reactions comprised radiolabelled DNA probe adsorbed onto magnetic Dynabeads M-280 Streptavidin (Invitrogen) in binding reaction buffer either alone (naked DNA) or incubated with crude nuclear extract (NE) prepared from Jurkat cells. DNA-binding reactions were subjected to DNase I digestion (0.0075 − 0.25 U) for 30 s, washed and analysed on a 7% acrylamide urea gel (7 M). Areas of protection were localized with a Maxam–Gilbert sequencing ladder.
NEs, electrophoretic mobility shift assays
Nuclear extracts were prepared from Jurkat T cells as previously described (52). Oligonucleotide probes for the following sites were radiolabelled: ‘USF DHS 44500’ spanning the putative USF binding site in DHS 44500 (For: agctCACATGCACGTGACACTGAG, Rev: agctCTCAGTGTCACGTGCATGTG); ‘USF MLP’ spanning the USF binding site from the adenovirus major late promoter (For: agctGTAGGCCACGTGACCGGGT, Rev: agctACCCGGTCACGTGGCCTAC); ‘EGR’ spanning an EGR binding site (For: agctAAATCCCCGCCCCCGCGATGGA, Rev: agctTCCATCGCGGGGGCGGGGATTT). Electrophoretic mobility shift assays (EMSA) were performed as previously described (23); for supershift analysis, antibodies to USF1 (sc229), USF2 (sc862) and Oct1 (sc232) were used (Santa Cruz).
Resequencing
A total of 92 chromosomes were resequenced over the region 31644170–31644820 for unrelated individuals from each of the Caucasian (CEU) and African (YRI) cohorts of the International HapMap Project with 2× coverage using two amplicons (Agencourt Bioscience, Beverly MA, USA). One CEU sample failed quality control for one amplicon. Identifiers of specific individuals resequenced are available on request. All coordinates in the paper are from NCBI build 35.
RESULTS
DNase hypersensitive site mapping of the TNF locus and flanking regions by quantitative chromatin profiling
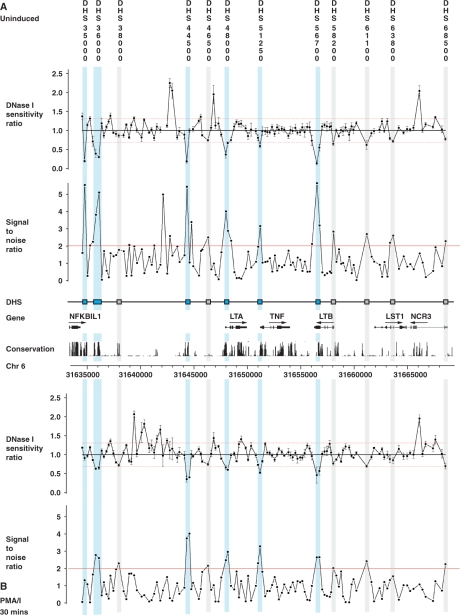
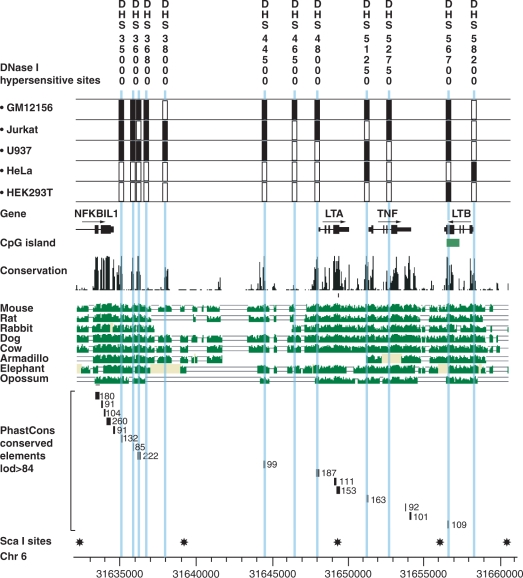
We aimed to define putative regulatory regions in the TNF locus and flanking genomic regions. DNase hypersensitivity assays to map sites of chromatin accessibility associated with transcriptionally active chromatin have been a powerful method of defining regulatory elements. This is particularly useful to screen intergenic regions which may contain enhancer and other regulatory elements acting at a distance from a gene. We first validated the approach of quantitative chromatin profiling using tiled PCR amplicons (7) for a known DNase hypersensitive site in the α-globin complex on chromosome 16p13.1 (53) using a lymphoblastoid B cell line GM12156 (Supplementary Figure S1A and B). We then proceeded to map DNase hypersensitive sites across the TNF locus by analysing a 34 kb genomic region centromeric to nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 (NFKBIL1), which included LTA, TNF, LTB, leukocyte specific transcript 1 (LST1) and natural cytotoxicity triggering receptor 3 (NCR3) genes using GM12156 (Figure 1). DNase hypersensitive sites were found in the LTA and TNF promoter and 5′-UTR, denoted ‘DHS 48000’ and ‘DHS 51250’, which localized within 0.1 kb of the transcriptional start sites of the two genes, respectively. Both hypersensitive sites were present in unstimulated and stimulated cells, being more pronounced and extensive (involving two rather than one PCR amplicons) in chromatin from stimulated cells. Two DNase hypersensitive sites were observed 3′ to NFKBIL1: ‘DHS 35000’ was found 0.3 kb downstream of exon 4 of NFKBIL1 and observed only in unstimulated cells; ‘DHS 36000’ was 1.6 kb 3′ to NFKBIL1 and present in chromatin from both unstimulated and stimulated cells. In the intergenic region between NFKBIL1 and LTA, a DNase hypersensitive site (denoted ‘DHS 44500’) was found 3.4 kb upstream of LTA. This was present in unstimulated and stimulated cells. A hypersensitive site (denoted ‘DHS 56700’) was also observed at the 3′ end of LTB, localizing to exon 4, and lying within the only significant CpG island in this 34 kb region. In unstimulated cells, a further five regions showed SNRs supporting the presence of a hypersensitive site, however the 90% confidence threshold of DNase I sensitivity ratios in these cases was not met in all but two instances. Interestingly, four of these sites displayed similar marginal evidence in the stimulated cells where an additional site was also suggested with evidence very near to thresholds (‘DHS 38000’).
Figure 1.
Quantitative chromatin profiling for a 34 kb region spanning the TNF locus and flanking genes in GM12156. This was carried out for four replicate DNase I digestions to give DNase I sensitivity measurements (ratios of copy number in treated compared to untreated samples) from 140 contiguous PCR amplicons designed to tile across the region (31634558–31668876). All samples were quantified in triplicate by Q-PCR. Relative DNase I sensitivity measurements are shown (DNase I-treated versus untreated) together with SNRs with genomic coordinates given along the x-axis. GM12156 was used, either resting (panel A) or after induction with 125 nM ionomycin plus 200 nM PMA for 30 min (panel B). Analysis revealed strong evidence for six hypersensitive sites (highlighted in light blue on figure): ‘DHS 35000’ 0.3 kb 3′ to exon 4 of NFKBIL1 in unstimulated cells (PCR amplicon coordinates 31634776–31635055); ‘DHS 36000’ 1.7 kb and 1.5 kb 3′ to exon 4 of NFKBIL1 in unstimulated cells (31636120–31636410) and stimulated cells (31635824–31636410) respectively; ‘DHS 44500’ 3.4 kb 5′ to LTA in unstimulated cells (31644348–31644622) and stimulated cells (31644348–31644807); ‘DHS 48000’ within 0.1 kb of the LTA transcriptional start site in unstimulated cells (31648007–31648254) extending up to 0.3 kb downstream in stimulated cells (31648007–31648393); ‘DHS 51250’ within 0.1 kb of the TNF transcriptional start site in stimulated cells (31651222–31651465) which was also present in unstimulated cells but to a lesser extent; ‘DHS 56700’ in exon 4 of LTB in unstimulated cells (31656528–31656800) and exon 4 and intron 3 of LTB in stimulated cells (31656528–31657053). In addition, six further sites are suggested with marginal evidence (shown highlighted in grey on figure).
Identification of DNase hypersensitive sites by Southern blotting
The DNase hypersensitive sites DHS 48000 and DHS 51250 near the LTA and TNF transcriptional start sites are consistent with the well-documented importance of these regions in transcriptional initiation of these genes, and hypersensitive sites have been previously reported in the proximal TNF and LTA promoter regions (20,36–39). Our quantitative chromatin profiling data also identified several other DNase hypersensitive sites including within intergenic regions as well as the only CpG island in the TNF locus in exon 4 of LTB. We proceeded to try to confirm the presence of these hypersensitive sites by conventional Southern blotting of Sca I restriction fragments spanning a 28.4 kb region from NFKBIL1 to LTB which encompassed the hypersensitive sites defined by quantitative chromatin profiling.
Context specificity has been a hallmark of many studies analysing the transcriptional regulation of TNF and LTA, and we therefore also sought to determine whether these sites of DNase hypersensitivity were cell type specific.
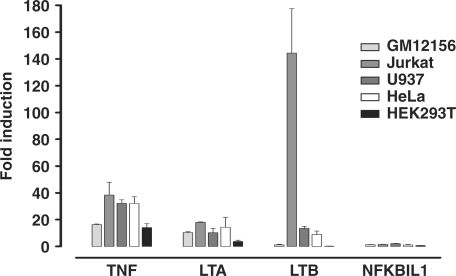
Chromatin was analysed from a panel of diverse human cell lines commonly used in gene expression studies including the Jurkat T cell leukaemia cell line; the U937 histiocytic lymphoma cell line which displays monocytic characteristics; the cervical epithelial carcinoma cell line HeLa; and the embryonic kidney cell line HEK293T. Quantitative real time PCR assays of relative transcript abundance were carried out to assess how expression of the genes of interest within the TNF locus varied between cell lines. All the cell types, including GM12156, showed inducible expression of LTA and TNF after 6 h of stimulation with a combination of PMA and ionomycin (Figure 2). Expression of LTB was highly inducible in Jurkat cells. However the levels of expression varied between cell types, being highest for LTA in the LCL and Jurkat cell lines, for TNF in the LCL and U937 cells, and for LTB in Jurkat cells (Supplementary Figure S2).
Figure 2.
Inducibility of genes in the TNF locus by mitogen stimulation. Inducibility of NFKBIL1, LTA, TNF and LTB is shown as the ratio of transcript abundance in unstimulated cells versus cells induced with a combination of PMA and ionomycin for 6 h. Transcript levels were assayed using gene-specific primers normalized to an ACTB control primer set by real time Q-PCR using the ΔΔCt method. Mean ± SD of three replicates are shown.
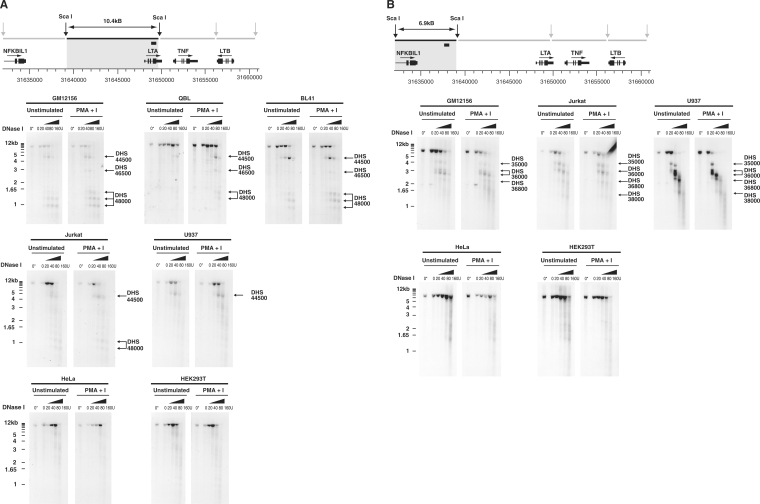
The most marked DNase sensitivity seen on quantitative chromatin profiling of GM12156 was at DHS 44500, a hypersensitive site within an intergenic region located 3.4 kb 5′ to LTA which has very recently been reported to be of regulatory significance in murine T cells (denoted HSS-9) (42). We analysed independent chromatin preparations from the same cell line by Southern blotting a 10.4 kb restriction fragment that spanned the region of interest 5′ to LTA. In the presence of higher concentrations of DNase, a clear faster migrating labelled band 4.8 kb in size was detected, demonstrating a DNase hypersensitive site at the position of DHS 44500 revealed by quantitative chromatin profiling (Figure 3A). This hypersensitive site is present constitutively and after induction with mitogen. A number of much smaller bands were also detected migrating at 1.4, 1.2 and 0.95 kb which localize to the LTA promoter and 5′-UTR (within 0.1 kb of the LTA transcriptional start site, and 0.2 kb and 0.4 kb downstream respectively) and refine the DNase hypersensitivity corresponding to DHS 48000 found on quantitative chromatin profiling. In mitogen stimulated cells, a weak 2.9 kb band was also observed, denoted DHS 46500, and located at 1.5 kb 5′ to LTA. This site was of borderline significance on earlier quantitative chromatin profiling but was subsequently found to be present in other B cell lines (Figure 3A) and to co-localize with a conserved region of DNA (see following section). We investigated whether the DHS 44500 hypersensitive site was present in a second EBV immortalized cell line, QBL. Southern blotting demonstrated the presence of the site in chromatin from both induced and uninduced cells (Figure 3A). We then investigated whether this site was restricted to EBV immortalized LCLs. In the non-EBV transformed B cell line BL41, we found that the hypersensitive site was also present constitutively. We found the same hypersensitive site in a T cell line (Jurkat) and the human monocyte-like cell line U937, however the site was not observed in HeLa cells or HEK293T cells (Figure 3A).
Figure 3.
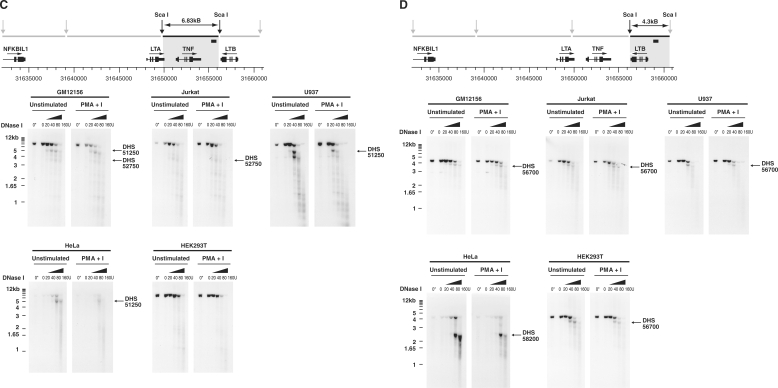
DNase hypersensitive sites in the TNF locus detected by Southern blotting for a range of cell types. Sca I restriction fragments were hybridized to radiolabelled probe and the pattern of cleavage analysed for nuclei incubated with increasing amounts of DNase I (20–160 U) at 37°C, or without exposure to DNase I incubated on ice (0*) or at 37°C (0). DNase hypersensitive sites are indicated by arrows with nomenclature following that used in analysis of quantitative chromatin profiling. LCLs (GM12156, QBL and a non-EBV transformed cell line BL41), Jurkat, U937, HeLa and HEK293T cells were analysed. (A) Southern blotting for region 5′ to LTA gene. A 10.4 kb Sca I restriction fragment (31639037–31649422) was hybridized to a labelled 247 bp probe (31649015–31649262). (B) Southern blotting for NFKBIL1 and 3′ region. A 6.92 kb Sca I restriction fragment (31632112–31639036) was hybridized to a labelled 266 bp probe (31637483–31637748). (C) Southern blotting in region of TNF. A 6.83 kb Sca I restriction fragment (31649420–31656247) hybridized to a labelled 245 bp probe (31655630–31655874). (D) Southern blotting in region of LTB. A 4.3 kb Sca I restriction fragment (31656244–31660544) was hybridized to a labelled 206 bp probe (31658807–31659012).
We investigated whether the novel DNase hypersensitive sites 3′ to NFKBIL1 found on quantitative chromatin profiling were present in GM12156 and other cell types (Figure 3B). Southern blotting a 6.9 kb restriction digest spanning the region resolved the hypersensitive site DHS 36000, noted on quantitative chromatin profiling, to a higher fidelity with two bands present of 3 and 2.7 kb corresponding to 1.3 and 1.6 kb 3′ to exon 4 of NFKBIL1. A number of weaker bands were also present in the LCLs which were more clearly demonstrated in the Jurkat and U937 cell lines as DNase hypersensitive sites, namely DHS 35000 seen as a discrete 4 kb band on Southern blotting corresponding to a region 0.3 kb 3′ to exon 4 of NFKBIL1; while 2.2 and 1.4 kb bands were also present (DHS 36800 and DHS 38000). No effect of cell stimulation with mitogen was observed. None of these sites were consistently demonstrated on Southern blots of DNase I digested chromatin from HeLa or HEK293T cell lines.
The DNase hypersensitive site DHS 51250 in the TNF promoter region identified on quantitative chromatin profiling in GM12156 was confirmed by Southern blotting (Figure 3C). A 5 kb fragment was observed in addition to the 6.83 kb full length Sca I digest in GM12156, U937 and HeLa cell lines but not Jurkat or HEK293T cells. In Jurkat and GM12156 cells, a faster migrating 3.5 kb fragment was observed localizing to intron 3 of TNF and denoted DHS 52750. Two DNase hypersensitive sites were found to be present constitutively in and around the LTB gene. DHS 56700, present within exon 4 of LTB on quantitative chromatin profiling, was clearly demonstrated on Southern blotting DNase digested chromatin from a range of cell types (Figure 3D). These included GM12156, Jurkat, U937 and HEK293T cells. Interestingly this DNase hypersensitive site was absent in HeLa cells where a 2.3 kb fragment (DHS 58200) was observed localizing within 0.1 kb 5′ to exon 1 of LTB. The results of our DNase I hypersensitive site mapping experiments across the TNF locus are summarized in Figure 4.
Figure 4.
Sequence conservation analysis and DNase I hypersensitivity map of TNF locus. Data from quantitative chromatin profiling and southern blotting for GM121565, Jurkat, U937, HeLa and HEK293T cells is summarized in the context of genomic structure and DNA sequence conservation analysis between species. The presence or absence of DNase hypersensitive sites in different cell types is indicated by filled and open boxes, respectively. Positions of Sca I restriction sites used in Southern blotting analysis are also shown with genomic coordinates from NCBI build 35.
Sequence conservation and phylogenetic footprinting
DNA sequence of regulatory significance is typically subject to evolutionary selective pressure leading to DNA sequence conservation when genomic sequences from different species are compared (54). We proceeded to analyse the 34 kb region to identify sequence conservation and evidence of selection producing phylogenetic footprints. We used a conservative threshold to define conserved regions with a lod score of ≥99 by analysis of PhastCons Conserved Elements (17-way Vertebrate Multiz Alignment) (55) on the UCSC Genome Browser Database. This revealed 11 regions, which included exonic sequences and known promoter regions together with a number of intergenic regions. Strikingly, 6 out of the 11 highly conserved regions aligned exactly to the DNase hypersensitive sites we mapped by quantitative chromatin profiling and Southern blotting (Figure 4).
Analysis for PhastCons Conserved Elements (17-way Vertebrate Multiz Alignment) (55) on the UCSC Genome. One of these highly conserved regions (score 438, lod 99) spanned a 118bp region (from 31644420 to 31644538) within DNase hypersensitive site DHS 44500 located 3.4kB 5' to LTA. This was associated with a peak (between 31644420 and 31644700) in predicted regulatory potential based on comparison of seven species analysed by ESPERR (Evolutionary and Sequence Pattern Extraction through Reduced Representations) with a maximum score of 0.36 (mean 0.2, variance 0.009), a score above 0.01 indicating a very marked resemblance to alignment patterns typical of regulatory elements in the training set used (56). Very high sequence conservation was observed over the DNase hypersensitive sites found at the LTA and TNF promoter regions, DHS 48000 (score 510, lod 187) and DHS 51250 (score 494, lod 163) respectively. DHS 56700 was noted to align with a CpG island within LTB and show strong sequence conservation (score 449, lod 109). Similarly high levels of conservation were noted for DHS 35000 (score 471, lod 132) and DHS 36000 (score 529, lod 222) 3′ to NFKBIL1.
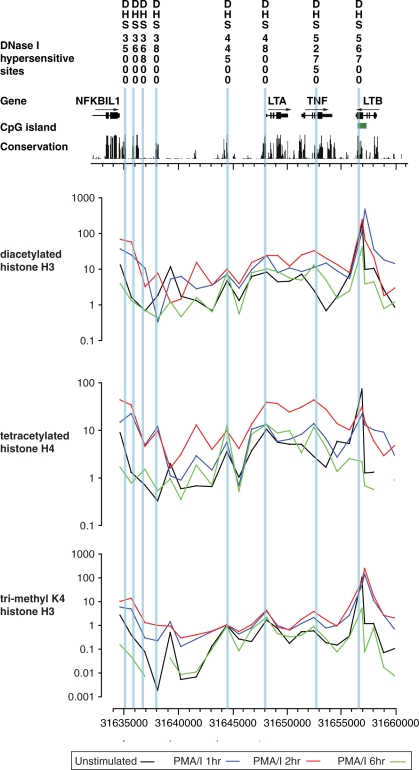
Histone modifications in Jurkat T cells across the TNF locus
We proceeded to analyse histone modifications over a 25.4 kb region spanning the TNF locus from NFKBIL1 to LTB. Jurkat T cells were chosen for this analysis as a representative cell type in which the majority of DNase hypersensitive sites were found and all the genes expressed, with significant inducibility for LTA, TNF and LTB. We performed chromatin immunoprecipitation experiments using specific histone antibodies to identify modifications associated with active chromatin (57). Quantitative real time PCR was used to map relative enrichment of chromatin modifications across the region at 1 kb intervals (Figure 5). Histone modifications were assessed across a time course following mitogen stimulation with peak levels of enrichment relative to input controls seen at 2 h after induction. We found a striking peak of histone acetylation (H3 and H4) at exon 4 of LTB which was associated with a peak in tri-methylated lysine 4 of histone H3. Elsewhere across the locus, peaks of histone acetylation and tri-methylation were found at the 3′ end of NFKBIL1, and the TNF and LTA promoter regions. Enrichment of tri-methylated lysine 4 was also found corresponding to the conserved DHS 44500 in the intergenic region 3.4 kb upstream of LTA; this was also associated with inducible acetylation of histone H3 and H4 (Figure 5).
Figure 5.
Tiling path analysis of histone modifications across the TNF locus. Histone modifications revealed by chromatin immunoprecipitation experiments using antibodies to diacetylated histone H3 (H3Ac), tetracetylated histone H4 (H4Ac) and tri-methylated histone K4 H3 (H3K4Me3) are shown for Jurkat T cells, either in the resting state or harvested 1, 2 or 6 h after induction with 125 nM ionomycin plus 200 nM PMA. The immunoprecipitated DNA was analysed using real time Q-PCR using 24 primer pairs at an average of 1.1 kb intervals across the region (31634558–31660002). On the y-axis, the fold difference in enrichment of each of the PCR amplicons tiled across the region is expressed relative to input DNA. Genomic location is shown on the x-axis.
Functional characterization of DHS 44500 5′ to LTA
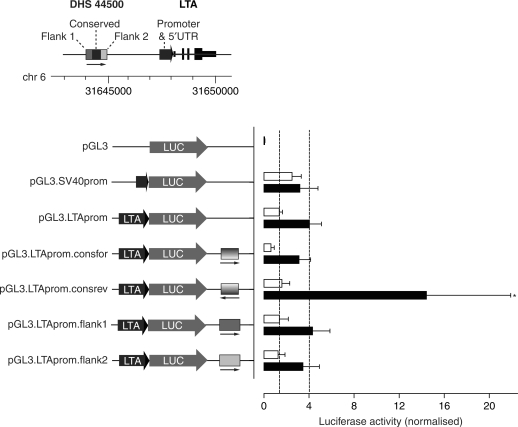
We proceeded to characterize further the regulatory significance of DHS 44500, a DNase hypersensitive site in the intergenic region upstream of LTA which showed sequence conservation and was associated with a peak of inducible histone modifications in Jurkat cells. We investigated first whether the segment of DNA spanning this site showed evidence of enhancer activity. We engineered a pGL3 LTA promoter construct in which the proximal LTA promoter region and 5′-UTR was placed immediately upstream of the luciferase reporter gene: the former encompasses the conserved proximal LTA promoter region previously reported as showing maximal activity on reporter gene assays (58,59). Transient transfection assays of Jurkat cells showed 3-fold induction of gene expression of this LTA promoter construct on stimulation of transfected cells with PMA and ionomycin. When in addition the 313 bp conserved region spanning DHS 44500 was inserted downstream of the luciferase gene, we found significant differences in level of expression following mitogen stimulation (Figure 6). This was dependent on the orientation of the DNA segment corresponding to DHS 44500, with induction after stimulation increased to 9.2-fold (P = 0.01 on paired t-testing versus pGL3 construct containing only the LTA promoter, two-tailed). In contrast, we found no difference in levels of gene expression when the flanking regions were inserted downstream of the luciferase gene.
Figure 6.
Effects of sequences spanning or flanking DHS 44500 on reporter gene expression in Jurkat T cells. Jurkat cells were transiently transfected with different pGL3 reporter constructs, either pGL3-Basic or driven by the LTA or SV40 promoters as indicated. The effects of inserting different sequences corresponding to the DHS 44500 or flanking regions were investigated. The mean ± SD of luciferase expression values are shown (normalized by pRL-TK) for five independent transfection experiments, each performed in duplicate. Open bars show expression for unstimulated cells, black bars values following mitogen stimulation (PMA/ionomycin). *P 0.01 on paired t-testing (two-tailed) versus pGL3.LTAprom.
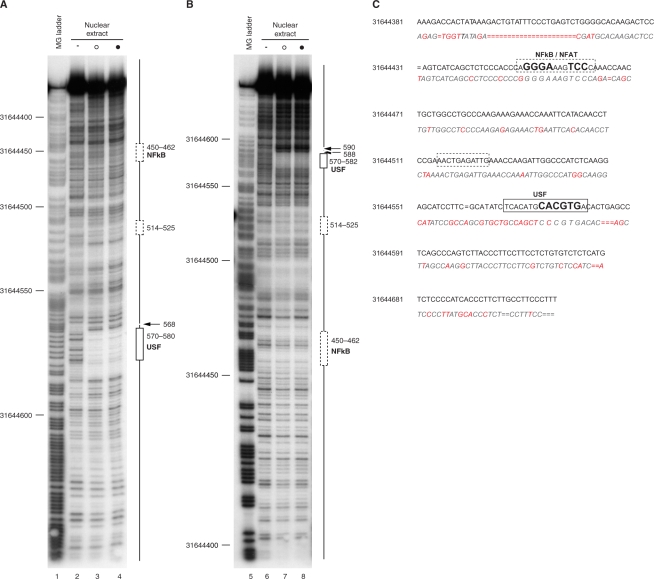
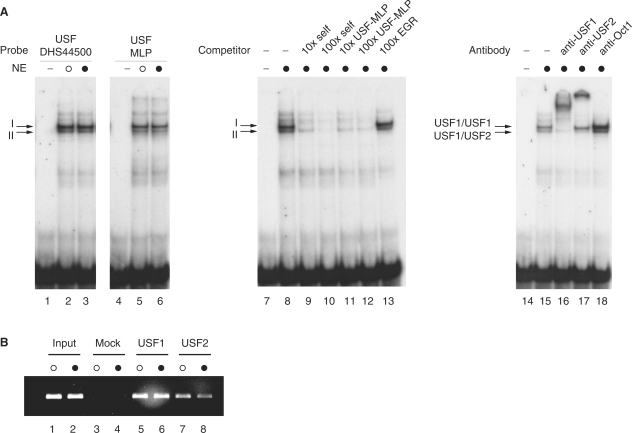
We then performed DNase I footprinting experiments in order to define the nature and extent of transcription factor binding over a 343 bp region spanning DHS 44500. Using NEs from both unstimulated and mitogen-induced Jurkat T cells, we observed a strong region of protection at 31644570–582, with flanking hypersensitivity to digestion, using probes radiolabelled on either the forward or reverse strands (Figure 7). This footprinted region spans a core E-box motif (CANNTG) with the sequence CACGTGAC reported to be bound with high affinity by members of the USF transcription factor family, in particular USF-1 (60). We further investigated recruitment of USF transcription factors at DHS 44500 using EMSA and chromatin immunoprecipitation experiments (Figure 8). Two major complexes (denoted I and II) were observed on EMSA using a radiolabelled oligoduplex probe corresponding to the region of protection seen on DNase I footprinting; these co-migrated with complexes seen with a probe matching a classical USF binding site from the adenovirus major late promoter (61). The complexes were found to be specific on competition assays, being effectively competed away by unlabelled self or the classical binding site but not by an unrelated probe. Supershift assays demonstrated complex I was a USF1 homodimer, and complex II a heterodimer of USF1/USF2. Chromatin immunoprecipitation experiments using the same cell line demonstrated that USF1 and USF2 were constitutively recruited in vivo (Figure 8). The mouse sequence corresponding to the human USF binding site in DHS 44500 shows a single nucleotide difference (CCCGTGAC versus CACGTGAC) but showed comparable in vitro binding of USF1/USF2 on EMSA (Supplementary Figure S3A). Our DNase I footprinting experiments also showed evidence of weak regions of protection in two other regions, 31644514–525 and 31644450–462 (Figure 7). The sequence within the latter (GGGAAAGTCC) corresponds to the consensus binding site for members of the Rel/NF-kB family of transcription factors p50/p65 (GGGRNYYYCC) (62,63) as well as members of the nuclear factor of activated T cells (NFAT) transcription factor family (64). EMSA experiments demonstrated binding of p50/p65 and p50/p50 to this region of DHS 44500 (Supplementary Figure S3B). Based on supershift experiments there may be other specific proteins recruited to both this site and the USF binding site. A specific complex was also observed binding to an oligonucleotide probe corresponding to the region 31644514–525 on EMSA; the specific DNA binding proteins involved remain to be identified (Supplementary Figure S3C).
Figure 7.
DNase I footprinting analysis of DHS 44500. DNA probes spanning DHS 44500 radiolabelled on either the forward (panel A) or reverse (panel B) strands were digested with DNase I either alone or in the presence of NEs from Jurkat T cells. Maxam Gilbert sequencing ladder (lanes 1 and 5); naked DNA (0.0075 U DNase I) (lanes 2 and 6); NEs from unstimulated cells (0.25 U DNase I) (open circle) (lanes 3 and 7) or cells induced with PMA and ionomycin for 2 h (0.25 U DNase I) (filled circle) (lanes 4 and 8). (C) Schematic showing sites of protection in context of human and murine sequences.
Figure 8.
USF1 and USF2 bind within DHS 44500. (A) EMSA experiments using radiolabelled probes corresponding to the putative USF binding site within DHS 44500 (lanes 1–3, 7–18) or to a USF binding site from the adenovirus major late promoter (lanes 4–6) were subjected to electrophoresis without NE (lanes 1, 4, 7 and 14), with NE from unstimulated cells (open circle) (lanes 2, 5) or cells induced with PMA and ionomycin for 2 h (filled circle) (lanes 3, 6, 8–13, 15–18). Competition experiments using 10× or 100× molar excess of unlabelled probe are shown (lanes 9–13); supershift experiments using antibodies as indicated (lanes 16–18). (B) Chromatin immunoprecipitation experiment using Jurkat T cells either unstimulated (open circle) or after stimulation with PMA and ionomycin (filled circle). Results of PCR amplification for primer pair at DHS 44500 (31644348–622) are shown for input controls (lanes 1 and 2); mock antibody immunoprecipitated controls (lanes 3 and 4); USF1 (lanes 5 and 6) and USF2 (lanes 7 and 8) immunoprecipitated DNA.
Finally, we sought to determine the extent of nucleotide diversity over a 650 bp region spanning DHS 44500 (31644170–031644820). A total of 92 chromosomes were resequenced for unrelated individuals from each of the CEU and YRI cohorts of the International HapMap Project (Supplementary Figure S4A–C). This analysis revealed two common SNPs were located near DHS 44500 but outside the conserved region, rs2844483 and rs2844484, of which rs2844484 had been previously validated but was of unknown allele frequency in dbSNP. We found the SNPs were in significant linkage disequilibrium with minor allele frequencies of 0.378 and 0.370 in the CEU population, and 0.25 and 0.261 in the YRI population respectively. These SNPs form part of an 8 kb haplotypic block extending into the 3′ region of NFKBIL1 which includes DHS 36000. In addition a novel SNP (ss99307025) was identified on one chromosome (GM12155) at 31644594 in the CEU but not YRI population.
DISCUSSION
We have presented data providing a comprehensive and systematic analysis of DNase hypersensitivity across the TNF locus. This genomic region encompasses a cluster of genes of critical importance in the immune response whose regulation remains incompletely understood. Our analysis of chromatin accessibility should facilitate further efforts to understand the transcriptional regulation of the locus and has highlighted a number of novel regions of potential functional significance. Our data demonstrates the successful application of quantitative chromatin profiling to interrogate a genomic locus spanning 34 kb. Quantitative chromatin profiling offers an attractive approach to allow screening of specific genomic regions with high specificity and sensitivity (7). Our data represents the first report to our knowledge of the successful application of this technique outside of the laboratory where it was developed and used as part of the ENCODE project (1). The approach remains relatively costly due to the number of real time PCR reactions required but our data shows that the number of replicate digestions can be reduced while maintaining high sensitivity due to the low variance and high SNR observed at hypersensitive sites. We made extensive use of Southern blotting analysis in this study to screen different cell types for DNase I hypersensitive sites: this proved to have higher resolution than quantitative chromatin profiling and was highly reproducible. Although more labour intensive, Southern blotting remains the gold standard in detection of hypersensitive sites and has previously been used for screening large genomic regions (65,66).
The TNF locus has been of significant research interest in terms of gene regulation with most work relating to transcriptional regulation of TNF. We confirmed the previously reported DNase hypersensitivity in the region of the human TNF promoter (20,36–39) and within intron 3 in Jurkat T cells (20). Cell type specificity was found for the hypersensitive sites we describe within the TNF locus, consistent with previous reports of specific regulatory mechanisms operating dependent on the stimulus and cell type (67). We have shown here that there are a number of novel hypersensitive sites in or near flanking genes of putative regulatory significance. Strikingly, the sites found 3′ to NFKBIL1, in an intergenic region between NKFBIL1 and LTA, and within exon 4 of LTB, co-localize to highly conserved regions of DNA. Our analysis of sequence conservation and phylogenetic footprinting concurs with previous reported analyses of human–mouse conservation which include this locus and we were able to define a number of novel conserved regions (42,68,69).
The transcriptional regulation of NFKBIL1 (also referred to as IKBL) is currently unknown. The encoded protein is thought to be a member of the I kappa B family as it contains ankyrin repeats which allow for interaction and regulation of NFκB/Rel proteins and, together with a bipartite protein targeting motif, determine localization to nuclear speckles (70,71). DNA sequence polymorphism 5′ to NFKBIL1 has been associated with susceptibility to rheumatoid arthritis (72) while resequencing of ancestral MHC haplotypes identified a 3′ untranslated region nucleotide substitution on the HLA B44 haplotype, an extended haplotype associated with susceptibility to autoimmune disease and cancer (73). Further characterization of the 3′ DNase I hypersensitive sites we have reported here is likely to be highly informative in terms of the regulation of NFKBIL1 and resolution of functional polymorphisms responsible for haplotypic associations with this locus. The haplotypic analysis presented here highlights how SNPs flanking DHS 44500 may be of potential relevance to such studies given the 8 kb haplotypic block extending into the 3′ region of NFKBIL1 which includes rs2844483 and rs2844484.
The novel DNase I site in exon 4 of LTB (DHS 56700) present across a range of cell types may be of particular significance given its co-localization to highly conserved sequence elements and the only CpG island in the region. Striking levels of histone H3 and H4 acetylation, together with tri-methylated lysine 4 of histone H3 were found at this site in Jurkat T cells. To date, functional analysis of LTB has been restricted to the proximal promoter region where Ets and NFkB binding sites have been shown to be important in PMA inducibility in Jurkat T cells (30,31). We did not find evidence of DNase hypersensitivity in the promoter region in Jurkat cells or a number of other cell types except HeLa, which may indicate cell context specificity in the regulation of LTB and highlights the need for further research into its regulation which includes regions outside of the classical promoter.
For the distal hypersensitive site (DHS 44500) localizing 3.4 kb upstream of LTA, we have presented data showing the DNase hypersensitivity is cell type specific, being present only in cell lines of haematological lineage, and is localized within a region showing chromatin modifications associated with transcriptionally active euchromatin in Jurkat T cells. Using the same cell line, we found that the region has enhancer activity in a reporter gene assay with evidence of recruitment of the USF family of basic helix-loop-helix leucine zipper transcription factors. This family of transcription factors are involved in regulation of many genes involved in the stress and immune responses, acting to modulate transcriptional activation, interact with coactivators and the pre-initiation complex, and recruit chromatin remodelling enzymes (60). A DNase hypersensitive site corresponding to DHS 44500 was reported in murine T cells during revision of this manuscript denoted HSS-9 (42). This study demonstrated a functional role for the region involving modulation of expression of TNF via an intrachromosomal looping mechanism (42). Further work is required to assess whether a similar mechanism may be operating in human T cells but our reporter gene data suggest the human sequence spanning DHS 44500 has the potential to modulate expression of LTA: this may be orientation-dependent and show human–murine differences. Our findings that DHS 44500 contained a binding site capable of binding p50 and p65 are consistent with published analysis of the murine sequence conserved at this site (denoted NFAT-8,394 in HSS-9) where both NFATp and p50/p65 were reported to bind in vitro; binding by NFATp was also demonstrated by ChIP assays in murine T cells (42). There is also some evidence that DHS 44500 may be present in porcine peripheral blood mononuclear cells as a site was noted approximately 4.4 kb upstream of LTA which lay outside the cloned TNF locus and was not analysed (41). We identified two common SNPs flanking DHS 44500 present in both Caucasian and African populations; their functional significance in terms of gene expression remains to be determined.
The detailed map of DNase I hypersensitive sites we have presented here should facilitate our understanding of potentially distant regulatory elements controlling transcription of this important cluster of genes, and of the consequences of genetic diversity within such regions for gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the Wellcome Trust [grant number 074318/Z/04/Z to J.C.K]. J.C.K. is a Wellcome Trust Senior Research Fellow in Clinical Science. Funding to pay Open Access publication charges for this article was provided by the Wellcome Trust.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Narelle Maugeri for technical assistance together with other colleagues in the Knight lab for helpful advice and discussion; we are also grateful to Marco de Gobbi for technical advice.
Conflict of interest statement. None declared.
REFERENCES
- 1.The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elgin SC. The formation and function of DNase I hypersensitive sites in the process of gene activation. J. Biol. Chem. 1988;263:19259–19262. [PubMed] [Google Scholar]
- 3.Weintraub H, Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976;193:848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- 4.Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 5.Crawford GE, Holt IE, Mullikin JC, Tai D, Blakesley R, Bouffard G, Young A, Masiello C, Green ED, Wolfsberg TG, et al. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc. Natl Acad. Sci. USA. 2004;101:992–997. doi: 10.1073/pnas.0307540100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabo PJ, Humbert R, Hawrylycz M, Wallace JC, Dorschner MO, McArthur M, Stamatoyannopoulos JA. Genome-wide identification of DNaseI hypersensitive sites using active chromatin sequence libraries. Proc. Natl Acad. Sci. USA. 2004;101:4537–4542. doi: 10.1073/pnas.0400678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorschner MO, Hawrylycz M, Humbert R, Wallace JC, Shafer A, Kawamoto J, Mack J, Hall R, Goldy J, Sabo PJ, et al. High-throughput localization of functional elements by quantitative chromatin profiling. Nat. Methods. 2004;1:219–225. doi: 10.1038/nmeth721. [DOI] [PubMed] [Google Scholar]
- 8.Crawford GE, Davis S, Scacheri PC, Renaud G, Halawi MJ, Erdos MR, Green R, Meltzer PS, Wolfsberg TG, Collins FS. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat. Methods. 2006;3:503–509. doi: 10.1038/NMETH888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Follows GA, Dhami P, Gottgens B, Bruce AW, Campbell PJ, Dillon SC, Smith AM, Koch C, Donaldson IJ, Scott MA, et al. Identifying gene regulatory elements by genomic microarray mapping of DNase I hypersensitive sites. Genome Res. 2006;16:1310–1319. doi: 10.1101/gr.5373606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Meth. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- 11.Tracey KJ, Cerami A. Tumor necrosis factor, other cytokines and disease. Annu. Rev. Cell Biol. 1993;9:317–343. doi: 10.1146/annurev.cb.09.110193.001533. [DOI] [PubMed] [Google Scholar]
- 12.Kwiatkowski D, Hill AVS, Sambou I, Twumasi P, Castracane J, Manogue KR, Cerami A, Brewster DR, Greenwood BM. TNF concentration in fatal cerebral, non-fatal cerebral, and uncomplicated Plasmodium falciparum malaria. Lancet. 1990;336:1201–1204. doi: 10.1016/0140-6736(90)92827-5. [DOI] [PubMed] [Google Scholar]
- 13.Neurath MF, Fuss I, Pasparakis M, Alexopoulou L, Haralambous S, Meyer zum Buschenfelde KH, Strober W, Kollias G. Predominant pathogenic role of tumor necrosis factor in experimental colitis in mice. Eur. J. Immunol. 1997;27:1743–1750. doi: 10.1002/eji.1830270722. [DOI] [PubMed] [Google Scholar]
- 14.Brennan FM, Chantry D, Jackson A, Maini R, Feldmann M. Inhibitory effect of TNF alpha antibodies on synovial cell interleukin-1 production in rheumatoid arthritis. Lancet. 1989;2:244–247. doi: 10.1016/s0140-6736(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 15.Tsytsykova AV, Goldfeld AE. Inducer-specific enhanceosome formation controls tumor necrosis factor alpha gene expression in T lymphocytes. Mol. Cell Biol. 2002;22:2620–2631. doi: 10.1128/MCB.22.8.2620-2631.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falvo JV, Uglialoro AM, Brinkman BM, Merika M, Parekh BS, Tsai EY, King HC, Morielli AD, Peralta EG, Maniatis T, et al. Stimulus-specific assembly of enhancer complexes on the tumor necrosis factor alpha gene promoter. Mol. Cell Biol. 2000;20:2239–2247. doi: 10.1128/mcb.20.6.2239-2247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai EY, Falvo JV, Tsytsykova AV, Barczak AK, Reimold AM, Glimcher LH, Fenton MJ, Gordon DC, Dunn IF, Goldfeld AE. A lipopolysaccharide-specific enhancer complex involving Ets, Elk-1, Sp1, and CREB binding protein and p300 is recruited to the tumor necrosis factor alpha promoter in vivo. Mol. Cell Biol. 2000;20:6084–6094. doi: 10.1128/mcb.20.16.6084-6094.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barthel R, Tsytsykova AV, Barczak AK, Tsai EY, Dascher CC, Brenner MB, Goldfeld AE. Regulation of tumor necrosis factor alpha gene expression by mycobacteria involves the assembly of a unique enhanceosome dependent on the coactivator proteins CBP/p300. Mol. Cell Biol. 2003;23:526–533. doi: 10.1128/MCB.23.2.526-533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falvo JV, Brinkman BM, Tsytsykova AV, Tsai EY, Yao TP, Kung AL, Goldfeld AE. A stimulus-specific role for CREB-binding protein (CBP) in T cell receptor-activated tumor necrosis factor alpha gene expression. Proc. Natl Acad. Sci. USA. 2000;97:3925–3929. doi: 10.1073/pnas.97.8.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barthel R, Goldfeld AE. T cell-specific expression of the human TNF-alpha gene involves a functional and highly conserved chromatin signature in intron 3. J. Immunol. 2003;171:3612–3619. doi: 10.4049/jimmunol.171.7.3612. [DOI] [PubMed] [Google Scholar]
- 21.Seiler-Tuyns A, Dufour N, Spertini F. Human tumor necrosis factor-alpha gene 3′ untranslated region confers inducible toxin responsiveness to homologous promoter in monocytic THP-1 cells. J. Biol. Chem. 1999;274:21714–21718. doi: 10.1074/jbc.274.31.21714. [DOI] [PubMed] [Google Scholar]
- 22.Shakhov AN, Collart MA, Vassalli P, Nedospasov SA, Jongeneel CV. Kappa B-type enhancers are involved in lipopolysaccharide-mediated transcriptional activation of the tumor necrosis factor alpha gene in primary macrophages. J. Exp. Med. 1990;171:35–47. doi: 10.1084/jem.171.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Udalova IA, Knight JC, Vidal V, Nedospasov SA, Kwiatkowski D. Complex NF-kappaB interactions at the distal tumor necrosis factor promoter region in human monocytes. J. Biol. Chem. 1998;273:21178–21186. doi: 10.1074/jbc.273.33.21178. [DOI] [PubMed] [Google Scholar]
- 24.Kuprash DV, Udalova IA, Turetskaya RL, Rice NR, Nedospasov SA. Conserved kappa B element located downstream of the tumor necrosis factor alpha gene: distinct NF-kappa B binding pattern and enhancer activity in LPS activated murine macrophages. Oncogene. 1995;11:97–106. [PubMed] [Google Scholar]
- 25.Messer G, Weiss EH, Baeuerle PA. Tumor necrosis factor beta (TNF-beta) induces binding of the NF-kappa B transcription factor to a high-affinity kappa B element in the TNF-beta promoter. Cytokine. 1990;2:389–397. doi: 10.1016/1043-4666(90)90046-v. [DOI] [PubMed] [Google Scholar]
- 26.Paul NL, Lenardo MJ, Novak KD, Sarr T, Tang WL, Ruddle NH. Lymphotoxin activation by human T-cell leukemia virus type I-infected cell lines: role for NF-kappa B. J. Virol. 1990;64:5412–5419. doi: 10.1128/jvi.64.11.5412-5419.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fashena SJ, Reeves R, Ruddle NH. A poly(dA-dT) upstream activating sequence binds high-mobility group I protein and contributes to lymphotoxin (tumor necrosis factor-beta) gene regulation. Mol. Cell Biol. 1992;12:894–903. doi: 10.1128/mcb.12.2.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuprash DV, Boitchenko VE, Yarovinsky FO, Rice NR, Nordheim A, Ruhlmann A, Nedospasov SA. Cyclosporin A blocks the expression of lymphotoxin alpha, but not lymphotoxin beta, in human peripheral blood mononuclear cells. Blood. 2002;100:1721–1727. [PubMed] [Google Scholar]
- 29.Worm MM, Tsytsykova A, Geha RS. CD40 ligation and IL-4 use different mechanisms of transcriptional activation of the human lymphotoxin alpha promoter in B cells. Eur. J. Immunol. 1998;28:901–906. doi: 10.1002/(SICI)1521-4141(199803)28:03<901::AID-IMMU901>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 30.Kuprash DV, Osipovich OA, Pokholok DK, Alimzhanov MB, Biragyn A, Turetskaya RL, Nedospasov SA. Functional analysis of the lymphotoxin-beta promoter. Sequence requirements for PMA activation. J. Immunol. 1996;156:2465–2472. [PubMed] [Google Scholar]
- 31.Voon DC, Subrata LS, Karimi M, Ulgiati D, Abraham LJ. TNF and phorbol esters induce lymphotoxin-beta expression through distinct pathways involving Ets and NF-kappa B family members. J. Immunol. 2004;172:4332–4341. doi: 10.4049/jimmunol.172.7.4332. [DOI] [PubMed] [Google Scholar]
- 32.Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, Tsunoda T, Sato H, Hori M, Nakamura Y, Tanaka T. Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction. Nat. Genet. 2002;32:650–654. doi: 10.1038/ng1047. [DOI] [PubMed] [Google Scholar]
- 33.Ota M, Katsuyama Y, Kimura A, Tsuchiya K, Kondo M, Naruse T, Mizuki N, Itoh K, Sasazuki T, Inoko H. A second susceptibility gene for developing rheumatoid arthritis in the human MHC is localized within a 70-kb interval telomeric of the TNF genes in the HLA class III region. Genomics. 2001;71:263–270. doi: 10.1006/geno.2000.6371. [DOI] [PubMed] [Google Scholar]
- 34.McGuire W, Hill AV, Allsopp CE, Greenwood BM, Kwiatkowski D. Variation in the TNF-alpha promoter region associated with susceptibility to cerebral malaria. Nature. 1994;371:508–510. doi: 10.1038/371508a0. [DOI] [PubMed] [Google Scholar]
- 35.Alcais A, Alter A, Antoni G, Orlova M, Nguyen VT, Singh M, Vanderborght PR, Katoch K, Mira MT, Vu HT, et al. Stepwise replication identifies a low-producing lymphotoxin-alpha allele as a major risk factor for early-onset leprosy. Nat. Genet. 2007;39:517–522. doi: 10.1038/ng2000. [DOI] [PubMed] [Google Scholar]
- 36.Sariban E, Imamura K, Luebbers R, Kufe D. Transcriptional and posttranscriptional regulation of tumor necrosis factor gene expression in human monocytes. J. Clin. Invest. 1988;81:1506–1510. doi: 10.1172/JCI113482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sung SJ, Walters JA, Hudson J, Gimble JM. Tumor necrosis factor-alpha mRNA accumulation in human myelomonocytic cell lines. Role of transcriptional regulation by DNA sequence motifs and mRNA stabilization. J. Immunol. 1991;147:2047–2054. [PubMed] [Google Scholar]
- 38.Espel E, Garcia-Sanz JA, Aubert V, Menoud V, Sperisen P, Fernandez N, Spertini F. Transcriptional and translational control of TNF-alpha gene expression in human monocytes by major histocompatibility complex class II ligands. Eur. J. Immunol. 1996;26:2417–2424. doi: 10.1002/eji.1830261023. [DOI] [PubMed] [Google Scholar]
- 39.Skoog T, Hamsten A, Eriksson P. Allele-specific chromatin remodeling of the tumor necrosis factor-alpha promoter. Biochem. Biophys. Res. Commun. 2006;351:777–783. doi: 10.1016/j.bbrc.2006.10.114. [DOI] [PubMed] [Google Scholar]
- 40.Ranjbar S, Rajsbaum R, Goldfeld AE. Transactivator of transcription from HIV type 1 subtype E selectively inhibits TNF gene expression via interference with chromatin remodeling of the TNF locus. J. Immunol. 2006;176:4182–4190. doi: 10.4049/jimmunol.176.7.4182. [DOI] [PubMed] [Google Scholar]
- 41.Kuhnert P, Peterhans E, Pauli U. Chromatin structure and DNase I hypersensitivity in the transcriptionally active and inactive porcine tumor necrosis factor gene locus. Nucleic Acids Res. 1992;20:1943–1948. doi: 10.1093/nar/20.8.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsytsykova AV, Rajsbaum R, Falvo JV, Ligeiro F, Neely SR, Goldfeld AE. Activation-dependent intrachromosomal interactions formed by the TNF gene promoter and two distal enhancers. Proc. Natl Acad. Sci. USA. 2007;104:16850–16855. doi: 10.1073/pnas.0708210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tumanov AV, Nedospasov SA, Turetskaia RL. Chromatin organization in the tumor necrosis factor/lymphotoxin gene locus: correlation with tissue-specific expression. Mol. Biol. (Mosk) 1998;32:98–102. [PubMed] [Google Scholar]
- 44.Rozen S, Skaletsky HJ. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S, Misener S, editors. Totowa, NJ: Humana Press; 2000. pp. 365–386. [Google Scholar]
- 45.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 46.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 48.De Gobbi M, Viprakasit V, Hughes JR, Fisher C, Buckle VJ, Ayyub H, Gibbons RJ, Vernimmen D, Yoshinaga Y, de Jong P, et al. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science. 2006;312:1215–1217. doi: 10.1126/science.1126431. [DOI] [PubMed] [Google Scholar]
- 49.Stewart CA, Horton R, Allcock RJ, Ashurst JL, Atrazhev AM, Coggill P, Dunham I, Forbes S, Halls K, Howson JM, et al. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res. 2004;14:1176–1187. doi: 10.1101/gr.2188104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knight JC, Udalova I, Hill AV, Greenwood BM, Peshu N, Marsh K, Kwiatkowski D. A polymorphism that affects OCT-1 binding to the TNF promoter region is associated with severe malaria. Nat. Genet. 1999;22:145–150. doi: 10.1038/9649. [DOI] [PubMed] [Google Scholar]
- 51.Sandaltzopoulos R, Becker PB. Solid phase DNase I footprinting: quick and versatile. Nucleic Acids Res. 1994;22:1511–1512. doi: 10.1093/nar/22.8.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vyas P, Vickers MA, Simmons DL, Ayyub H, Craddock CF, Higgs DR. Cis-acting sequences regulating expression of the human alpha-globin cluster lie within constitutively open chromatin. Cell. 1992;69:781–793. doi: 10.1016/0092-8674(92)90290-s. [DOI] [PubMed] [Google Scholar]
- 54.Visel A, Bristow J, Pennacchio LA. Enhancer identification through comparative genomics. Semin. Cell Dev. Biol. 2007;18:140–152. doi: 10.1016/j.semcdb.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor J, Tyekucheva S, King DC, Hardison RC, Miller W, Chiaromonte F. ESPERR: Learning strong and weak signals in genomic sequence alignments to identify functional elements. Genome Res. 2006;16:1596–1604. doi: 10.1101/gr.4537706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 58.Jongeneel CV. In: Tumor Necrosis Factors: The Molecules and Their Emerging Role in Medicine. Beutler B, editor. New York: Raven Press Ltd; 1992. pp. 539–559. [Google Scholar]
- 59.Kuprash D, Turetskaya R, Boldin M, Udalova I, Kistanova E, Smirnova J, Nedospasov S. In: Tumour Necrosis Factor: Molecular and Cellular Biology and Clinical Relevance. Fiers W, Buurman WA, editors. Basel: Karger; 1993. pp. 19–26. [Google Scholar]
- 60.Corre S, Galibert MD. Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res. 2005;18:337–348. doi: 10.1111/j.1600-0749.2005.00262.x. [DOI] [PubMed] [Google Scholar]
- 61.Read ML, Clark AR, Docherty K. The helix-loop-helix transcription factor USF (upstream stimulating factor) binds to a regulatory sequence of the human insulin gene enhancer. Biochem. J. 1993;295((Pt 1)):233–237. doi: 10.1042/bj2950233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen FE, Ghosh G. Regulation of DNA binding by Rel/NF-kappaB transcription factors: structural views. Oncogene. 1999;18:6845–6852. doi: 10.1038/sj.onc.1203224. [DOI] [PubMed] [Google Scholar]
- 63.Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature. 1998;391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- 64.Macian F. NFAT proteins: key regulators of T-cell development and function. Nat. Rev. Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 65.Lee DU, Rao A. Molecular analysis of a locus control region in the T helper 2 cytokine gene cluster: a target for STAT6 but not GATA3. Proc. Natl Acad. Sci. USA. 2004;101:16010–16015. doi: 10.1073/pnas.0407031101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Monticelli S, Lee DU, Nardone J, Bolton DL, Rao A. Chromatin-based regulation of cytokine transcription in Th2 cells and mast cells. Int. Immunol. 2005;17:1513–1524. doi: 10.1093/intimm/dxh329. [DOI] [PubMed] [Google Scholar]
- 67.Goldfeld AE, Doyle C, Maniatis T. Human tumor necrosis factor alpha gene regulation by virus and lipopolysaccharide. Proc. Natl Acad. Sci. USA. 1990;87:9769–9773. doi: 10.1073/pnas.87.24.9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xie T, Rowen L, Aguado B, Ahearn ME, Madan A, Qin S, Campbell RD, Hood L. Analysis of the gene-dense major histocompatibility complex class III region and its comparison to mouse. Genome Res. 2003;13:2621–2636. doi: 10.1101/gr.1736803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuprash DV, Udalova IA, Turetskaya RL, Kwiatkowski D, Rice NR, Nedospasov SA. Similarities and differences between human and murine TNF promoters in their response to lipopolysaccharide. J. Immunol. 1999;162:4045–4052. [PubMed] [Google Scholar]
- 70.Albertella MR, Campbell RD. Characterization of a novel gene in the human major histocompatibility complex that encodes a potential new member of the I kappa B family of proteins. Hum. Mol. Genet. 1994;3:793–799. doi: 10.1093/hmg/3.5.793. [DOI] [PubMed] [Google Scholar]
- 71.Semple JI, Brown SE, Sanderson CM, Campbell RD. A distinct bipartite motif is required for the localization of inhibitory kappaB-like (IkappaBL) protein to nuclear speckles. Biochem. J. 2002;361:489–496. doi: 10.1042/0264-6021:3610489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Okamoto K, Makino S, Yoshikawa Y, Takaki A, Nagatsuka Y, Ota M, Tamiya G, Kimura A, Bahram S, Inoko H. Identification of I kappa BL as the second major histocompatibility complex-linked susceptibility locus for rheumatoid arthritis. Am. J. Hum. Genet. 2003;72:303–312. doi: 10.1086/346067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allcock RJ, Christiansen FT, Price P. The central MHC gene IKBL carries a structural polymorphism that is associated with HLA-A3,B7,DR15. Immunogenetics. 1999;49:660–665. doi: 10.1007/s002510050662. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.