Abstract
The fragile X mental retardation protein (FMRP) is a RNA-binding protein proposed to post-transcriptionally regulate the expression of genes important for neuronal development and synaptic plasticity. We previously demonstrated that FMRP binds to its own FMR1 mRNA via a guanine-quartet (G-quartet) RNA motif. However, the functional effect of this binding on FMR1 expression was not established. In this work, we characterized the FMRP binding site (FBS) within the FMR1 mRNA by a site directed mutagenesis approach and we investigated its importance for FMR1 expression. We show that the FBS in the FMR1 mRNA adopts two alternative G-quartet structures to which FMRP can equally bind. While FMRP binding to mRNAs is generally proposed to induce translational regulation, we found that mutations in the FMR1 mRNA suppressing binding to FMRP do not affect its translation in cellular models. We show instead that the FBS is a potent exonic splicing enhancer in a minigene system. Furthermore, FMR1 alternative splicing is affected by the intracellular level of FMRP. These data suggest that the G-quartet motif present in the FMR1 mRNA can act as a control element of its alternative splicing in a negative autoregulatory loop.
INTRODUCTION
The most frequent cause of inherited mental retardation, fragile X syndrome, is caused by the absence of the RNA-binding protein Fragile X Mental Retardation (FMRP). In neurons, FMRP is associated with a limited subset of brain mRNAs together with other proteins within large ribonucleoparticles, the composition of which is still incompletely known (1–3). Within these mRNPs, FMRP is proposed to act as a regulator of translation or transport of specific target mRNAs. However, the molecular mechanisms of FMRP action on specific target mRNAs are still poorly understood. As a clue to the function of FMRP, the study of its mRNA targets appears an essential step. The guanine-quartet (G-quartet) structural motif was identified as a high affinity determinant of the interaction of FMRP with mRNAs (4,5). RNA G-quartet is not the only proposed target of FMRP since U-rich sequences (6), a kissing-loop motif (7) and the BC-1 RNA (9) were also found to mediate the interaction of FMRP with mRNAs. However, FMRP target mRNAs bearing the kissing-loop motif have not yet been identified and the interaction mediated via BC1 is under debate (8). Thus, at present, G-quartet still appears as a main consensus motif found in mRNAs of mammalian genes found associated with FMRP (10,11), and/or demonstrated to be affected by the absence of FMRP. Genes carrying potential or verified G-quartets include the microtubule associated protein 1B MAP1B (12), the post-synaptic density protein PSD95 (13,14), the catalytic subunit of protein phosphatase 2A (PP2Ac) (15), or the amyloid precursor protein APP (16), all important for neuronal development and synapse plasticity. However, the role of the FMRP/G-quartet interaction remains unclear as no direct evidence of its effect on post transcriptional control has been provided up to now and recent work indicated that the association of FMRP with polyribosomes (17) would not be mediated by G-quartets (7).
To address these questions, we analyzed in this work the interaction between FMRP and its own mRNA, FMR1, one of the best characterized targets of FMRP where the G-quartet motif had been identified (2). Because the interaction between FMRP and its own mRNA was suggestive of an autoregulatory loop, we tested whether FMRP could control its own expression. To determine the function of the FMR1 mRNA/FMRP interaction, we performed mutations within the G-quartet motif of the FMRP binding site (FBS) of FMR1, which abolished FMRP binding in vitro without changing the amino acid sequence of the protein and we examined at which level the FMRP/FBS interaction could play a role. Our data provide several lines of evidence for a role of the FBS and its binding to FMRP in alternative splicing regulation of the FMR1 gene.
MATERIALS AND METHODS
Plasmids and constructions
Plasmid pTL1 (18) was used to transiently or stably express FMR1 longest isoform 1 in the various cell lines described in text. Flag and cMyc tags were introduced in frame at N-terminus of FMR1 to give pTL1-Flag-FMR1 and pTL1 cMyc-FMR1. Mutations disrupting G-quartet within the FBS were introduced into pTL1-Flag-FMR1 using Quick Change Site Directed Mutagenesis kit (Stratagene, Cedar Creek, TX, USA). Primers used for mutagenesis are given in Supplementary Material available online. The SXN13 minigene constructions (19) were produced by inserting dsDNA fragments of FBS within exon 2 using SalI/BamHI sites. Plasmid pTAP–FMRP was constructed by inserting FMR1 Iso1 in frame at its N-terminus with TAP tag of pBS 1539 (20) into MluI site of pTRE2 vector (Clontech, Mountain View, CA, USA).
Cell culture and transfections
HeLa cells and FMR1−/− mouse embryonic immortalized fibroblasts (21) were cultured in DMEM supplemented with 10% fetal bovine serum, 100 µg/ml penicillin–streptomycin. PC12 Tet-On cells (Clontech) were grown in RPMI supplemented with 10% horse serum, 5% fetal bovine serum, 125 µg/ml hygromycin, 100 µg/ml of penicillin–streptomycin, in a 5% CO2 incubator at 37°C. PC12 Tet-On cells were stably transfected with pTAP–FMRP using Lipofectamine (Invitrogen, Carlsbad, CA, USA) according to manufacturer recommendations. The pHyg resistance vector was used in the cotransfection as a selection marker. Transfected cells were cultured in medium containing 125 µg/ml hygromycin and 1 µg/ml doxycyclin, and individual double stable selected cells were tested for the presence of the TAP–FMRP fusion protein by western blot using 1C3 anti-FMRP. PC12 Tet-On clone ‘1’ was selected for its tight regulation of TAP-FMRP expression. To induce exogenous human FMRP Iso1 expression in stably transfected cell lines, doxycyclin was added to cells to a final concentration of 250 ng/ml for 48 h.
For the determination of SXN minigene splicing efficiency, HeLa cells or FMR1−/− mouse embryonic immortalized fibroblasts at 40% confluency were transfected with 1.5 µg SXN vector using JetPEI (Polyplus) in 60 mm diameter plates. After 24 h, total RNA was extracted using Genelute mammalian total RNA kit (Sigma, Steinheim, Germany) and 5 µg was used for extension with the SXN primer described below.
Primer extension
Primer extensions to detect G-quartet structure within RNAs were performed as described in (4) using primer 5′-TCCATCTGTTGTTCTCCTTT for FMR1 and 5′-AGAACCTCTGGGTCCAAGGGTAG for SXN minigene Exon 2.
RNA-binding assays
RNA-binding assays were performed using RNAs T7 in vitro transcribed labeled with [α-32P]ATP. Affinities were determined using competition gel shift assays with GST–FMRP as described previously (4). Briefly, 32P-labeled FMR1 mRNA fragment N19 encompassing the FBS was incubated with 0.1 pmol GST–FMRP in the presence of increasing concentrations of unlabeled N19 or mutant N19-ΔG4 competitor RNA.
Polysomes preparation
Polysomes were prepared from four 10 cm diameter confluent HeLa cell plates. Twenty minutes before harvest, 90 µg/ml cycloheximide was added to cultures. Cells were lyzed in 200 mM Tris–HCl pH 7.5, 5 mM MgCl2, 100 mM KCl, 10 U/ml RNasin (Promega, France), 1 mM DTE, 0.5% NP40 at 4°C. Supernatant of 10 min centrifugation at 13 000 r.p.m. was loaded onto 15–45% sucrose gradient run 2 h at 36 000 r.p.m. at 4°C. Polysomal fractions were precipitated with 0.1 M NaCl and 2.5 vol. ethanol and the mRNAs from these fractions were purified with GenElute Mammalian Total RNA kit (Sigma).
In situ hybridization
In situ hybridization were performed as described in (22) using oligonucleotide modified with fluorophore CY3 (GE Healthcare, France) and directed against the Flag sequence of exogenous FMR1 (5′-CTTGTCATCGTCGTCCTTGTAGTCCATGAATTCGCCCTATA).
Western and northern blots
Immunoblot analyses were performed with 1C3 antibody (1/2000), anti-Flag (1/1000 Sigma), anti-cMyc (1/500, Ozyme, France) and anti-βactin (1/1000) as described (18).
Northerns were performed according to (23). Radioactive probes were prepared using kit ‘dsDNA all-in-one-random-prime’ (Sigma) with FMR1-3′ UTR and 28S rRNA encoding cDNAs.
Real-time PCR
Total RNA (1 µg) prepared with Genelute mammalian total RNA kit (Sigma) was retrotranscribed with SuperscriptIII (Invitrogen) using random priming, and real-time PCR were performed using the Brilliant SYBR-Green QPCR Core Reagent Kit (Stratagene) on MX4000 apparatus (Stratagene). The following oligonucleotides were used for qRT–PCR.16Ra 5′-GTGGACGATTATCTGTTCGGGAA, R15/16 5′-CGTCGTTTCCTTTGAAGCC, P14/15F 5′-GATATACTTCAGGAACTAATTC, p14/15.1F 5′-GATATACTTCAGCTCCAACAG, p14/15.2F 5′-GATATACTTCAGAATCTGACC, 11/13F 5′-CAAAAGTCCAGAGGGGGATG, 5′UTR-FMR1.F 5′-GCGAGGAAGGACGAGAAGAT, 5′UTR-FMR1.R 5′-TGGTGGGAATCTCACATCATGG, R13/15 5′-CAGAATTAGTTCCTTTAAGTAG, R13/15.1 5′-GTGGTCAGATTCTTTAAGTAG, R13/15.2 5-CTGTTGGAGCTTTAAGTAG, F-GAPDH 5′-GGATGCAGGGATGATGTTC and R-GAPDH 5′-TGCACCACCAACTGCTTAG.
2D- PAGE
Protein extraction and first dimension: cells were harvested by centrifugation and resuspended in 10 mM Tris, 1 mM EDTA and 250 mM sucrose. Lysis was performed with rehydration buffer (7 M urea, 2 M thiourea, 4% CHAPS, 0.4% ampholytes, 20 mM DTT). DNA was eliminated by 3 min sonication. A total of 100 µg of proteins were diluted in 135 µl of rehydratation buffer, which were used to rehydrate Biorad ReadyStrip IPG pH 3–10 strips. Isoelectric focusing 30 min at 500 V and 250 Vh, 30 min at 1000 V and 500 Vh and 1 h at 4000 V and 8000 Vh using the MultiphorII system (GE Healthcare). Second dimension: strips were equilibrated for 20 min in 50 mM Tris–HCl pH 8.8, 6 M urea, 30% glycerol, 2% SDS, 50 mM DTT. Strips were placed on vertical 1.0 mm 10% SDS polyacrylamide gels and sealed with 0.5% agarose sealing solution. Electrophoresis was performed in standard running buffer at 150 V for 1 h.
RT–PCR
Total RNA was prepared from cortices of 10 days old wild-type (Wt) or FMRP−/− male mice using Trizol reagent (Invitrogen) followed by RNeasy purification (Qiagen, Hilden, Germany) and their synaptoneurosomal fractions were prepared according to (24). Total RNA (1 µg) was retrotranscribed with SuperscriptIII (Invitrogen) using random priming. One microliter of RT reaction (1/10) was used to perform PCR reactions in 25 µl reaction volume with the following primers: F13 5′-GTGGGAACAAAAGACAGCATCG, R15 5′-CCTCTGCGCAGGAAGCTC, R4 CACCAACAGCAAGGCTCTTT, F2-3 5′-TTGAAAACAACTGGCAACCA, F-GAPDH and R-GAPDH. Reactions were performed as follows, initial denaturation 3 min at 95°C, then 30 s at 95°C, 30 s at 60°C and 30 s at 72°C, with 40 cycles.
RESULTS
The FBS contains two independent G-quartet structures stabilized by adenines
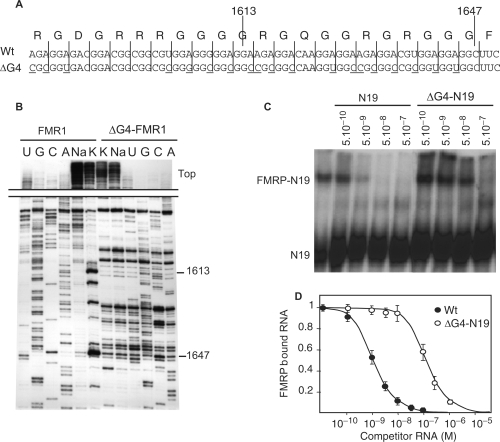
FMRP specifically binds to its own mRNA both in vitro (4,25,26) and in cells (27). The binding site of FMRP on its mRNA, here called the FBS, consists of a G-quartet motif present in the C-terminal coding region of FMR1 mRNA (4). The G-quartet motif is formed by the stacking of several guanine tetrad units. Adenines were also suspected to contribute to the structure in the FBS by forming intercalating adenine quartets. To investigate the function of the interaction between FMRP and its own mRNA, we constructed a series of mutants to inhibit FMRP/FBS interaction by disrupting the G-quartet structures. Previous work had suggested the presence of two distinct G-quartet structures (4). To test this hypothesis, two sets of mutations were constructed to disrupt either one or both potential structures, called ΔG1 and ΔG2 (Figure 1A). In a first step, mutations were essentially substitutions of As to Cs and Us at the wobble position of codons to preserve the encoded FMRP protein sequence and to test the contribution of adenines. The presence of G-quartets in the RNAs was indicated by the presence of potassium-dependent stops of reverse transcription as previously described (4). ΔG1 mutation, located around position 1613, suppressed the 1613 (G1) stop while the stop at position 1647 (G2) remained unchanged (Figure 1B). Conversely, ΔG2 mutations had the opposite effect, with the 1613 stop unchanged and the stop 1647 suppressed. These results indicate that two independent G-quartet structures exist in the FBS. Moreover, because the mutations left the guanine content of the FBS essentially unchanged while substituting several adenines, our results support a role for the adenines in stabilizing both FBS G-quartets. This stabilization effect can be explained by the formation of A-tetrads stacking within the G-quartet structure as previously proposed (4). When both sets of mutations were combined in mutant ΔG(1 + 2), the stop at position 1647 reappeared while the stop at position 1613 remained absent (Figure 1B). The reappearance of a G-quartet structure at G2 position within the ΔG(1 + 2) RNA despite the presence of mutations ΔG2 could be explained by the fact that the double mutant generated a different and more stable G-quartet structure because the G-content was essentially not affected by the mutations.
Figure 1.
Presence of two independent G-quartets in FBS. (A) Sequence of Wt and mutants (ΔG1, ΔG2, ΔG1 + 2) FBSs. The mutations are indicated in bold. The position of the two major stops of RT at 1613 and 1647 (+1 being A of start codon), in presence of 150 mM KCl, are shown. The circles above the sequences indicate the status of RT stop as determined in (B) (black circle, stop; open circle, absence of stop). (B) Autoradiograph of reverse transcriptions performed on full-length Wt or mutant FMR1 mRNA constructs and after separation on denaturing PAGE (see Material and methods section for details). The cation-dependent arrests at positions 1613 and 1647 reveal the 3′ edges of two distinct G-quartet structures. Lanes K and Na: extensions performed with 150 mM KCl and NaCl, respectively. The position and status of RT stops is shown with the circles as in (A). Sequencing lanes for mutants are shown. The full-length extension products seen on upper part of gel (top) reflects the strength of the different G-quartet structures.
FMRP binding to its own mRNA has no impact on FMR1 translation
We tested next the ability of these different mutant RNAs to interact with FMRP by gel shift assay as previously described (4). The mutant RNAs (ΔG1 and ΔG2) bound to FMRP with the same affinity as for the Wt FBS (data not shown). This indicated that FMRP equally binds one or the other structure. To completely disrupt G-quartet formation within FBS, a new set of mutations, consisting essentially of A to C substitutions at the wobble position of codons and favoring hairpin structures, was performed (Figure 2A). The mutations were inserted in full length FMR1 mRNA and the disruption of the G-quartet structure was confirmed by reverse transcriptase (RT) elongation test (Figure 2B). We have shown previously that a 425-long RNA fragment (N19) of FMR1 mRNA containing the FBS recapitulated a Wt-binding efficiency (4). To confirm the loss of interaction of FMRP on ΔG4-FBS, the mutations were inserted also in the N19 fragment (N19-ΔG4) and its interaction with FMRP was tested using gel shift assay (Figure 2C). The binding efficiency of N19-ΔG4 RNA was found to be decreased by more than a 100-fold compared with Wt N19 RNA (Figure 2D). This level of interaction, in the micromolar range, was assigned to nonspecific binding as previously determined (4).
Figure 2.
The disruption of G-quartets within FMR1 mRNA abolishes the interaction with FMRP. (A) Nucleotide sequences of the Wt or the G-quartet-less FMR1 mutant construction (ΔG4) with its amino acid sequence. Underlined nucleotides indicate the nucleotides mutated in ΔG4. (B) Cation-dependent arrest of reverse transcription showing the absence of G-quartet in ΔG4 ‘full-length’ FMR1 mRNA. (C) Competition experiments to compare the relative binding strength of FMRP for a 425-nt long RNA fragment encompassing the Wt FMRP binding site N19 and the mutant ΔG4-N19 by gel shift assay. Lane ‘–’ is control without competitor RNA. Position of free and FMRP-complexed 32P-labeled N19 RNA is shown. The molar concentration of unlabeled competitors is given at the top of figure. (D) Graph depicting the fraction of bound labeled N19 RNA plotted against competitor RNA concentrations as determined from C. Each point is the mean with standard deviation of at least three independent experiments.
The impact of the disruption of G-quartet structures within the FBS was then analyzed in various cell types [HeLa, Cos-7, and fibroblasts from FMR1−/− mice (21)] by transiently or stably expressing FMR1 bearing ΔG4 mutation. In these cells no difference in FMRP protein level could be detected between the cells expressing Wt or ΔG4 FMR1 (Figure 3A). Also, no difference could be detected between Wt and mutant FMR1 mRNA levels (Figure 3B). Furthermore, although mRNAs bearing G-quartets had been reported to be differently associated with polyribosomes in the absence of FMRP (10), we could not detect a change in the association of ΔG4-FMR1 mRNA with polysomes both in HeLa and in FMR1−/− mouse fibroblasts (Figure 3C). Finally, we did not observe any significant difference between Wt and ΔG4 FMR1 mRNAs localization in HeLa cells (Figure 3D). Thus, we concluded that the interaction between FMRP and the FBS had no detectable impact on FMR1 mRNA stability, translation and localization in the tested cells.
Figure 3.
FMRP binding on its own mRNA has no effect on FMR1 translational regulation. (A) Western blot analysis of pTL1 Flag-FMR1 and pTL1 Flag-ΔG4-FMR1 expression in HeLa cells. HeLa cells (6 × 106 cells) were transfected with the indicated amount plasmids (µg). Westerns blot on 15 µg total cell extracts using anti-cMyc, anti-Flag and anti-ßactin antibodies, revealed no difference between Wt and mutant ΔG4-FMR1 encoded protein levels (one of three independent experiments is presented, P < 0.05, similar results were obtained in Cos-7 and in FMR1−/−fibroblasts). (B) Northern blot analysis of FMR1 mRNA expression level with 15 µg of HeLa cell total RNA extracts using probes specific of pTL1 encoded FMR1 mRNAs and of 28S rRNA as internal control. The pBS is control lane without FMR1 encoded plasmid. No difference is observed between Wt and ΔG4 expression levels. (C) Localization of Wt and mutant ΔG4 FMR1 mRNAs in polyribosomes of HeLa cells. In the upper part is depicted a typical profile of polyribosomes separated on a 15–45% linear sucrose gradient registered at 254 nm optical density. The lower graphic represents the quantification by qRT–PCR of the FMR1 mRNA in the indicated pooled fractions using GAPDH mRNA as internal control. No significant difference was observed between Wt and ΔG4-FMR1 mRNAs in their localization in the different ribosome subsets. Similar results were obtained in the FMR1−/− cells. (D) Intracellular localization of Wt and ΔG4 mRNAs by fluorescence in situ hybridization in HeLa cells. Cy3 labeled anti-Flag oligo-deoxynucleotide probe (Flag) revealed a similar cytoplasmic and perinuclear localization for both mRNAs. DAPI staining of the nuclei is shown.
The FBS is a potent exonic splicing enhancer
A number of facts brought us to examine next a potential implication of the FBS in splicing. Firstly, the FBS is located nearby to alternatively spliced sites of FMR1 (Figure 4A). Secondly, because of its high purine content, the sequence of the FBS has analogies to an ESE consensus (28). Third, because of its shuttling activity, FMRP has been proposed to bind mRNAs already in the nucleus and therefore should be able to interact with pre-mRNAs (29,30).
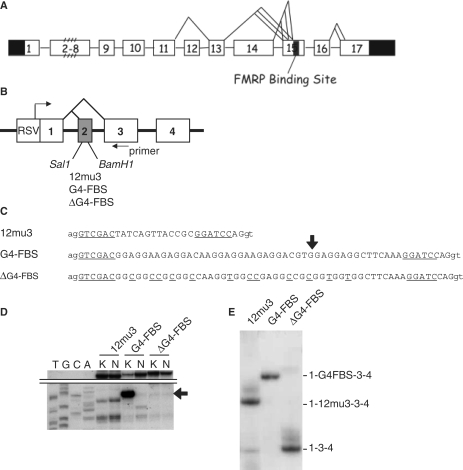
Figure 4.
The FBS has ESE properties. (A) Schematic structure of the human FMR1 gene with FBS localization. Untranslated regions are indicated in black and alternative splices are shown. (B) Schematic structure of the SXN13 minigene. RSV and Rous sarcoma virus promoter. (C) Sequence of the constructions inserted in the Sal1 and BamH1 sites of SXN13 minigene exon 2. The sequence 12mu3 is a moderate ESE in vitro selected (19). G4-FBS is a minimal fragment of FBS still able to form a G-quartet structure (see B hereafter). ΔG4-FBS is the corresponding mutant with genetic code preserved and G-quartet disrupted. The intron sequence is in lowercase and the exonic sequence is in uppercase. The SalI and BamHI sites are underlined. (D) In vitro analysis by reverse transcription of the presence of a G-quartet in SXN13 G4-FBS pre-mRNA but not in ΔG4-FBS. The RNAs were produced by T3 transcription and G-quartet formation was assayed by the RT elongation test as in Figure 1B. Lane K and N: extensions were performed with KCl and Na, respectively. The top of the gel with full-length extension products is shown. (E) Efficiency of splicing of SXN13 G4-FBS and ΔG4-FBS minigenes in HeLa cells. Primer SXN extensions were performed with RT on 5 µg total mRNA extracted from HeLa cells transfected with the different SXN13 minigene constructions. Results shown are visualized by autoradiography after migration on denaturing PAGE. The exonic content of the different transcripts is indicated.
The ability of the FBS to act as an ESE in vivo was tested by using the SXN13 minigene system (19) derived from the β-globin gene and composed of four exons, one of which (exon 2) being alternatively spliced (Figure 4B). The presence in exon 2 of a sequence with ESE properties (12MU3) induced exon 2 inclusion and resulted in a longer mRNA product (Figure 4C and E). A fragment of the FBS still able to form a G-quartet structure or its corresponding ΔG4 mutant was inserted within the second exon of the minigene to determine its ESE properties (Figure 4C and D). After transient transfection in HeLa cells of the plasmids bearing the different minigene constructions, RT elongation was directly performed on the total RNA extracted from the cells using a 5′ end 32P-labeled oligodeoxynucleotide priming within exon 3 of the minigene. The ESE properties of the FBS fragment were evaluated by measuring the ratio between the long RT product (bearing exon 2) and the short RT product (without exon 2) of the alternative splicing of the globin minigene. While the 12MU3 sequence was capable to specify exon 2 inclusion in about 80% of the splicing events (Figure 4E), the G4-FBS fragment induced a complete inclusion of the exon 2. Meanwhile, exon 2 was totally excised in ΔG4-FBS mutant. These data indicated that the FBS had potent exonic splicing enhancing properties on a minigene and these properties were linked to its ability to form a G-quartet structure.
The overexpression of one FMRP isoform alters FMR1 alternative splicing pattern in PC12 cells
The fact that the FBS had a potent ESE activity in a minigene suggested that FMRP could regulate its own splicing by binding to FBS. To verify this hypothesis, we first tested whether the splicing efficiency of a globin minigene bearing the FBS fragment could be influenced by FMRP. Splicing of SXN13-G4-FBS minigene was analyzed in FMR1−/− mouse fibroblasts (21). In these cells, the expression of either FMRP major cytoplasmic isoform 7 or nuclear isoform 6 (18) by transient or stable transfection, had no detectable influence on SXN13-G4-FBS expression (data not shown). An absence of effect of FMRP on the minigene system could be due to the fact that the FBS was out of its natural context or had a too strong ESE effect on minigene splicing.
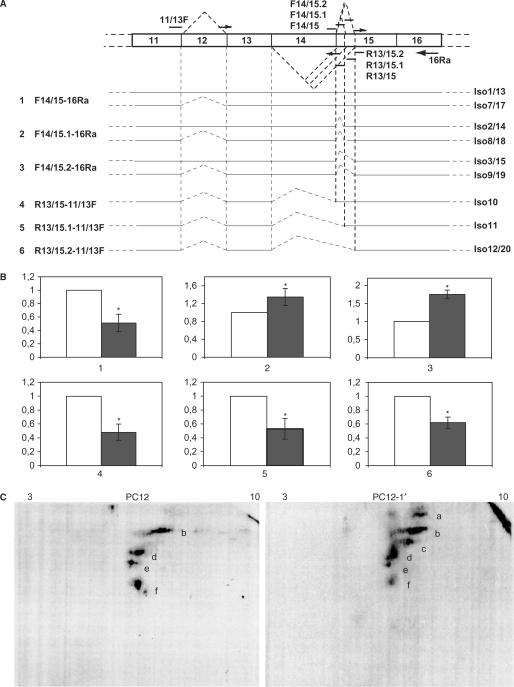
We then tested the influence of an overexpression of FMRP on FMR1 pre-mRNA splicing. The FBS is located close to two alternative splice sites within exon 15 of FMR1. The 3′ side of the FBS is located 110, 74 or 35 nucleotides downstream of the three different acceptor 5′ ends of exon 15 (4,31). The alternative splicing occurring at these three sites leads to six types of exon 15 variants, depending whether exon 14 is skipped or not. These three acceptor sites are used in different proportions in various tissues (32) suggesting the possibility of regulation at this level. Because the FBS is highly conserved (4) we tested the possible impact of FMRP/FBS interaction in rat cells. Rat phaeochromocytoma PC12 cells stably transfected with a tagged human isoform 1 FMRP (iso-1h) under the control of the inducible promoter Tet-On were used (PC12-1′ cells). We tested in these cells, the effect of iso-1h FMRP increase on endogenous (rat) FMR1 mRNA alternate splicing. Upon induction of iso-1h expression (Figure 5A) by doxycyclin treatment of the cells, the total amount of FMR1 mRNAs (rat + human) was found increased over 30-fold compared to its basal level in PC12 cells (Figure 5B left). Meanwhile, the global level of endogenous FMR1 mRNAs (rat) was not affected (Figure 5B right). The splicing events taking place around the FBS site within the endogenous FMR1 were analyzed by qRT–PCR using rat specific primer sets (Figure 6A). Our data showed that the products of exon 15 first acceptor site usage (including the longest isoform 1, the most frequent isoform 7 and isoforms 13 and 17) were decreased over 2-fold (Figure 6B). This decrease was concomitant with an increase in minor isoforms, products of exon 15 second and third acceptor site usage (1.4- and 1.8-fold respectively), including the minor isoforms 2, 3, 8, 9, 14, 15, 18 and 19. Thus, the overexpression of the full-length FMRP isoform alters FMR1 splicing events around the FBS in a manner that indicates a displacement of the equilibrium between major and minor isoforms. These data are in agreement with the hypothesis that FMRP binding to the FBS plays a role in regulating FMR1 splicing.
Figure 5.
The overexpression of exogenous FMR1 isoform 1 within PC12 cells does not affect the total level of endogenous FMR1 mRNAs. (A) Right (1C3), western blot analysis of FMRP expression in normal PC12 cells (PC12) and in PC12 expressing a tagged human FMRP isoform (PC12-1′) after doxycyclin induction. Left (Protein A), control western blot with TAG specific antibody to reveal the absence of degradation or abortive product in PC12-1′ extracts. (B) qRT–PCR data comparing the level of total (endogenous rat + exogenous human) and endogenous (rat) FMR1 mRNAs in the PC12 (white bar) and PC12-1′ cells (black bar). Data are the means from qRT–PCR triplicates using at least two independent RNA preparations. PC12 values were arbitrarily set to 1. Normalization was performed using the internal standard GAPDH.
Figure 6.
The overexpression of exogenous FMR1 isoform 1 within PC12 cells alters FMR1 splicing pattern. (A) Schematic structure of FMR1 region subjected to alternative splicing with the list of rat specific sets of primers used for the analysis. The primer localization with the isoforms they enable to measure is given. (B) Quantification by qRT–PCR of different FMR1 isoforms ratio between PC12 (white bars) and PC12-1′ (black bars) cells using the primer sets presented in (A). Data are means from qRT–PCR triplicates, normalized with GAPDH and using at least two independent RNA preparations. Values from PC12 were arbitrarily set to 1. *Student test P < 0.05. (C) Western blot with anti-FMRP (1C3) antibodies on PC12 and PC12-1′ cell extracts separated on 2D PAGE. Spots described in the article are identified from a to f. pH ranges are indicated at the top of the 2D PAGE.
We examined also the splicing events leading to exon 14 skipping. Upon overexpression of the full-length FMRP, all transcripts lacking exon 14 were found decreased by 2-fold. Although the splicing events leading to exon 14 skipping are likely in relationship with those occurring between exons 14 and 15, they are quite rare events compared with the latter [(26) and our data not shown]. To confirm the alterations of FMR1 expression seen at the RNA level upon iso-1h overexpression, we analyzed FMRP isoform expression by western blotting after 2D PAGE. The use of monoclonal anti-FMRP 1C3 antibody (recognizing an N-terminal epitope) indeed revealed significant differences in FMRP isoforms upon iso-1h overexpression (Figure 6C). The identification of each protein spot is however extremely difficult due to the complexity of splice products. The highest product visible only in iso-1h expressing cells (Figure 6C, right, spot a) could correspond to the exogenous iso-1h transferred inefficiently due to its higher molecular weight. Spots b and d, which showed a broadening in the PC12-1′ cells compared to PC12, likely contained several isoform species of similar molecular weight. The origin of spot c, which appeared in PC12-1′ cells was unknown. Most remarkable is the decrease of spot f, which could correspond to the shortest FMRP isoforms 10 and 11 (48 and 47 kDa respectively) that were found decreased at mRNA level. Altogether, these data showed that an overexpression of one FMRP isoform was able to alter FMR1 alternative splicing pattern both at RNA and protein level.
The splicing pattern of FMR1 exon 15 is altered in the cortex of FMR1−/− mice
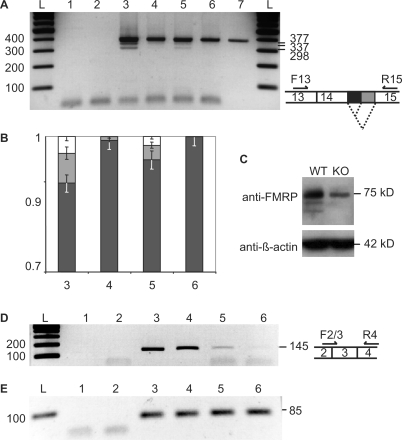
Following the observation that an overexpression of FMRP in cells leads to an alteration of its splicing at exon 15, we tested next whether the absence of FMRP could lead to similar defects. Thus, we analyzed the splicing pattern of FMR1 mRNAs in Wt and FMR1−/− mice where FMRP protein is absent but FMR1 mRNA is still expressed. FMR1−/− mice have been produced by the insertion of a neomycin cassette within exon 5 (33). The exon 15 splicing events were analyzed by RT–PCR (Figure 7A) on total RNA extracted from cortices of 10-days old Wt and FMR1−/− mice as well as in the synaptoneurosomal fractions (SN) of these extracts where FMRP function is considered to be prominent (34). In parallel, RT-PCRs were also performed on another part of FMR1 mRNA (exon 3, Figure 7D) and on GAPDH mRNA (Figure 7E) for normalization. As previously reported, while FMRP protein expression was abolished in FMR1−/− (Figure 7C), FMR1 mRNA remained expressed, although reduced to about 65% of Wt level (Figure 7A and 7D, compare lanes 3 to 4 and 5 to 6), possibly due to NMD events. RT–PCR performed across exon 15 (Figure 7A) revealed three bands corresponding to the three isoforms produced by the alternative branching of exon 14 and exon 15. Not surprisingly the isoforms lacking exon 14 are not detected in these PCR conditions because they are much less frequent events (26). Comparison of Wt and FMR1−/− exon 15 RT-PCR products (Figure 7B) revealed a marked difference concerning the smaller products that correspond to minor spliced mRNAs variants. Thus, these exon 15 minor splices disappeared in FMR1−/−, both in total cortical extracts and in SN fractions (Figure 7A lanes 4 and 6). These data indicate that FMRP absence alters the splicing pattern of FMR1 exon 15 in the cortex. While an overexpression of FMRP led to an increase of exon 15 minor splices, the absence of FMRP had the opposite effect. Altogether these data support a role for FMRP in the control of its own splicing at exon 15.
Figure 7.
The absence of FMRP alters exon 15 FMR1 mRNA splicing in mouse cortical extracts. (A) RT–PCR analysis of splicing events in exon 15 using primers F13 and R15, hybridizing in exons 13 and 15, respectively. The ethidium bromide stained PCR products separated on a 1.8 % agarose gel is shown (negative image). 1, no Taq polymerase control; 2, no RT control with Wt total cortical RNA; 3, Wt total cortical RNA; 4, KO total cortical RNA; 5, Wt total RNA from cortical synaptoneurosomes; 6, KO total RNA from cortical synaptoneurosomes; 7, control pTL1 plasmid; L, DNA ladder. Expected size of each PCR product is indicated on the right side of the gel. (B) densitometric analysis of RT–PCR products shown in (A) and expressed as the ratio of exon 15 isoforms for lanes 3, 4, 5 and 6, with the same color code as for splice scheme in (A). Error bars are standard deviations with n = 3. (C) Western blot analysis of FMRP expression in the cortical extracts. The band seen with anti-FMRP antibody in FMR1−/− (KO) corresponds to cross-reactivity with FXRs. (D) RT-PCR using primers F2-3 and R4, hybridizing over exons 2, 3 and 4, respectively. Samples tested are the same as in (A). (E) RT–PCR using primers F-GAPDH and R-GAPDH hybridizing in GAPDH with the same samples as in (A).
DISCUSSION
In this work, we analyzed the functional impact of the interaction between FMRP, the protein absent in the fragile X syndrome and the binding site identified in its own mRNA (4). We previously demonstrated that the FBS is located within the region encoding the RGG domain of FMRP. One main structural feature of this site is its ability to adopt a guanine quadruplex or G-quartet motif. We showed here that the structure of the FBS was more complex than initially thought. Thus, we identified two independent G-quartet structures in the FBS. Mutations that abolished either one or the other structure (mutant ΔG1 and ΔG2) had no impact on FMRP binding efficiency in the context of a 425-nt long fragment (N19), indicating that FMRP can indistinctly bind to either one or the other structure. Furthermore, we showed that several adenines of the FBS play a role in the differential stability of the G-quartet structures, supporting the initial hypothesis that the structure involves intercalating adenine quartets (4) and as already observed for other G-quartet structures (35). Substitution of these adenines by pyrimidines does not however prevent formation of a G-quartet structure within the FBS and does not affect binding to FMRP in vitro. The elimination of both structures (mutant ΔG4), while keeping the encoded protein sequence unchanged, dramatically reduced FMRP binding to a non specific level and confirmed the absolute requirement for a G-quartet for efficient binding. We then tested the impact of mutation ΔG4 within the context of full FMR1 mRNA in cells. Surprisingly, no effect of G-quartet absence could be detected neither on mRNA translation and localization nor on polyribosomes association in HeLa cells. Thus, these observations do not support a role in a translationally controlled autoregulatory loop of the binding of FMRP to its own mRNA as initially proposed (4). The fact that the FBS site is purine-rich and localizes close to alternative splicing sites was suggestive of its potential function as a splicing regulator of FMR1. Indeed, mammalian ESEs were identified initially as purine-rich sequences that associate with specific SR-family proteins and promote the utilization of adjacent splice sites (28). When a fragment of the FBS that retained its ability to form a G-quartet was tested in a minigene system, a strong exonic splicing enhancer activity was observed. This activity was completely abolished in a mutant that had lost its ability to form the G-quartet although it kept a G-rich sequence (ΔG4). Thus, our data indicate that the FBS is a potent ESE and interestingly, the ESE activity of FBS seems to rely on its ability to adopt a G-quartet structure. These data suggest that the FBS may be a control element of FMR1 alternative splicing and the binding of FMRP could play a role in the control. Indeed, we showed that the equilibrium between short and long FMRP isoforms produced by exon 15 alternative splicing is altered by manipulating the level of FMRP protein (either by overexpression of the longest isoform 1 or in FMR1 KO cells where the FMR1 mRNA is still expressed). This supports the idea that FMRP binding to the FBS site controls the ratio between the different isoforms in an autoregulatory loop. The binding of FMRP longest isoform 1 on the FBS could counteract or modulate its ESE function (for instance by interfering with SR proteins) such as to favor the minor site inclusion. The two alternative G-quartet structures are equidistant (39 and 36 nt respectively) from the two alternative splicing sites in exon 15, suggesting that they could act as a molecular switch for controlling exon 15 alternative splicing. However, one cannot exclude at present that the observed effect of FMRP on the alternative splicing of its own mRNA may be indirect, involving for instance the translational control by FMRP of splicing factors.
The biological significance of a modulation of FMR1 alternative splicing is presently unclear in particular because it is not known whether the different isoforms of FMRP, some of which being present in very low amount, have different functions. Still a variation in their ratio is likely to have implication for the function of FMRP. For instance, the isoforms lacking the 5′ end of exon 15 produced by the alternative splicing at second and third acceptor sites both lack serine 499, the major known phosphorylation site of FMRP (36,37). This phosphorylation site was shown to modulate FMRP association to mRNAs in drosophila (37) and to affect translation in mammalian cells (36). Based on our observations, increased FMRP binding to FBS would result in a decrease in the synthesis of FMRP major isoforms (carrying a complete exon 15) together with an increase of minor isoforms (lacking serine 499) downregulating FMRP function in a negative autoregulatory loop.
In conclusion, while we could not show a translational effect of FMRP binding to its own FMR1 mRNA, our data support the implication of the FMRP/G-quartet interaction on the regulation of FMRP alternative splicing around exon 15. The fact that perturbations of the intracellular level of FMRP leads to modulation of exon 15 isoforms expression in a way susceptible to alter their RNA-binding properties suggests the existence of a possible autoregulatory loop. Our data suggest also that FMRP might be involved in splicing regulation of other genes containing G-quartet motifs in their protein coding sequence. This should be particularly prominent in neurons where FMRP is expressed at its highest level and even locally in dendrites where FMRP is present and splicing has been proposed to occur (38).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We would like to thank Barbara Bardoni, Bernard and Chantal Ehresmann, J. Tazzi, Nicolas Charlet and Cyril Bourgeois for helpful discussions, M. Beaulande and Eric Flatter for technical assistance, Enzo Lalli, Solange Pannetier, Edouard Bertrand and Tom Cooper for suggestions and material. This work was supported by National Institutes of Health (R01 HD40612-01), Agence Nationale de la Recheche (ANR-06-NEURO-015-01), Fondation Jérôme Lejeune to H.M., Fondation pour la Recherche Médicale to X.T., Association pour la Recherche sur le Cancer to M.C.D. Funding to pay the Open Access publication charges for this article was provided by GIE CERBM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zalfa F, Achsel T, Bagni C. mRNPs, polysomes or granules: FMRP in neuronal protein synthesis. Curr. Opin. Neurobiol. 2006;16:265–269. doi: 10.1016/j.conb.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Schaeffer C, Beaulande M, Ehresmann C, Ehresmann B, Moine H. The RNA binding protein FMRP: new connections and missing links. Biol. Cell. 2003;95:221–228. doi: 10.1016/s0248-4900(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 3.Garber K, Smith KT, Reines D, Warren ST. Transcription, translation and fragile X syndrome. Curr. Opin. Genet. Dev. 2006;16:270–275. doi: 10.1016/j.gde.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO. J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Yun SW, Seto J, Liu W, Toth M. The fragile X mental retardation protein binds and regulates a novel class of mRNAs containing U rich target sequences. Neuroscience. 2003;120:1005–1017. doi: 10.1016/s0306-4522(03)00406-8. [DOI] [PubMed] [Google Scholar]
- 7.Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iacoangeli A, Rozhdestvensky TS, Dolzhanskaya N, Tournier B, Schutt J, Brosius J, Denman RB, Khandjian EW, Kindler S, Tiedge H. On BC1 RNA and the fragile X mental retardation protein. Proc. Natl Acad. Sci. USA. 2008;105:734–739. doi: 10.1073/pnas.0710991105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 10.Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 11.Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, Eberwine J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 12.Lu R, Wang H, Liang Z, Ku L, O’Donnell WT, Li W, Warren ST, Feng Y. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc. Natl Acad. Sci. USA. 2004;101:15201–15206. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Todd PK, Mack KJ, Malter JS. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc. Natl Acad. Sci. USA. 2003;100:14374–14378. doi: 10.1073/pnas.2336265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, di Penta A, Tabolacci E, Chiurazzi P, Neri G, Grant SG, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat. Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castets M, Schaeffer C, Bechara E, Schenck A, Khandjian EW, Luche S, Moine H, Rabilloud T, Mandel JL, Bardoni B. FMRP interferes with the Rac1 pathway and controls actin cytoskeleton dynamics in murine fibroblasts. Hum. Mol. Genet. 2005;14:835–844. doi: 10.1093/hmg/ddi077. [DOI] [PubMed] [Google Scholar]
- 16.Westmark CJ, Malter JS. FMRP mediates mGluR5-dependent translation of amyloid precursor protein. PLoS Biol. 2007;5:e52. doi: 10.1371/journal.pbio.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khandjian EW, Huot ME, Tremblay S, Davidovic L, Mazroui R, Bardoni B. Biochemical evidence for the association of fragile X mental retardation protein with brain polyribosomal ribonucleoparticles. Proc. Natl Acad. Sci. USA. 2004;101:13357–13362. doi: 10.1073/pnas.0405398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sittler A, Devys D, Weber C, Mandel JL. Alternative splicing of exon 14 determines nuclear or cytoplasmic localisation of fmr1 protein isoforms. Hum. Mol. Genet. 1996;5:95–102. doi: 10.1093/hmg/5.1.95. [DOI] [PubMed] [Google Scholar]
- 19.Coulter LR, Landree MA, Cooper TA. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol. Cell. Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 21.Mazroui R, Huot ME, Tremblay S, Filion C, Labelle Y, Khandjian EW. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 2002;11:3007–3017. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- 22.Verheggen C, Lafontaine DL, Samarsky D, Mouaikel J, Blanchard JM, Bordonne R, Bertrand E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO. J. 2002;21:2736–2745. doi: 10.1093/emboj/21.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sagerstrom CG, Sive HL. RNA blot analysis. In: Krieg PA, editor. A Laboratory Guide to RNA: Isolation, Analysis, and Synthesis. New York: Wiley-Liss; 1996. pp. 83–103. [Google Scholar]
- 24.Witzmann FA, Arnold RJ, Bai F, Hrncirova P, Kimpel MW, Mechref YS, McBride WJ, Novotny MV, Pedrick NM, Ringham HN, et al. A proteomic survey of rat cerebral cortical synaptosomes. Proteomics. 2005;5:2177–2201. doi: 10.1002/pmic.200401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sung YJ, Conti J, Currie JR, Brown WT, Denman RB. RNAs that interact with the fragile X syndrome RNA binding protein FMRP. Biochem. Biophys. Res. Commun. 2000;275:973–980. doi: 10.1006/bbrc.2000.3405. [DOI] [PubMed] [Google Scholar]
- 26.Ashley CT, Jr, Wilkinson KD, Reines D, Warren ST. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 27.Ceman S, Brown V, Warren ST. Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X-related proteins as components of the complex. Mol. Cell. Biol. 1999;19:7925–7932. doi: 10.1128/mcb.19.12.7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blencowe BJ. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 29.Eberhart DE, Malter HE, Feng Y, Warren ST. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum. Mol. Genet. 1996;5:1083–1091. doi: 10.1093/hmg/5.8.1083. [DOI] [PubMed] [Google Scholar]
- 30.Bardoni B, Sittler A, Shen Y, Mandel JL. Analysis of domains affecting intracellular localization of the FMRP protein. Neurobiol. Dis. 1997;4:329–336. doi: 10.1006/nbdi.1997.0142. [DOI] [PubMed] [Google Scholar]
- 31.Eichler EE, Richards S, Gibbs RA, Nelson DL. Fine structure of the human FMR1 gene. Hum. Mol. Genet. 1993;2:1147–1153. doi: 10.1093/hmg/2.8.1147. [DOI] [PubMed] [Google Scholar]
- 32.Verkerk AJ, de Graaff E, De Boulle K, Eichler EE, Konecki DS, Reyniers E, Manca A, Poustka A, Willems PJ, Nelson DL, et al. Alternative splicing in the fragile X gene FMR1. Hum. Mol. Genet. 1993;2:399–404. doi: 10.1093/hmg/2.4.399. [DOI] [PubMed] [Google Scholar]
- 33.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 34.Weiler IJ, Spangler CC, Klintsova AY, Grossman AW, Kim SH, Bertaina-Anglade V, Khaliq H, de Vries FE, Lambers FA, Hatia F, et al. Fragile X mental retardation protein is necessary for neurotransmitter-activated protein translation at synapses. Proc. Natl Acad. Sci. USA. 2004;101:17504–17509. doi: 10.1073/pnas.0407533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan B, Xiong Y, Shi K, Deng J, Sundaralingam M. Crystal structure of an RNA purine-rich tetraplex containing adenine tetrads: implications for specific binding in RNA tetraplexes. Structure. 2003;11:815–823. doi: 10.1016/s0969-2126(03)00107-2. [DOI] [PubMed] [Google Scholar]
- 36.Ceman S, O’Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum. Mol. Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- 37.Siomi MC, Higashijima K, Ishizuka A, Siomi H. Casein kinase II phosphorylates the fragile X mental retardation protein and modulates its biological properties. Mol. Cell. Biol. 2002;22:8438–8447. doi: 10.1128/MCB.22.24.8438-8447.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glanzer J, Miyashiro KY, Sul JY, Barrett L, Belt B, Haydon P, Eberwine J. RNA splicing capability of live neuronal dendrites. Proc. Natl Acad. Sci. USA. 2005;102:16859–16864. doi: 10.1073/pnas.0503783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.