Abstract
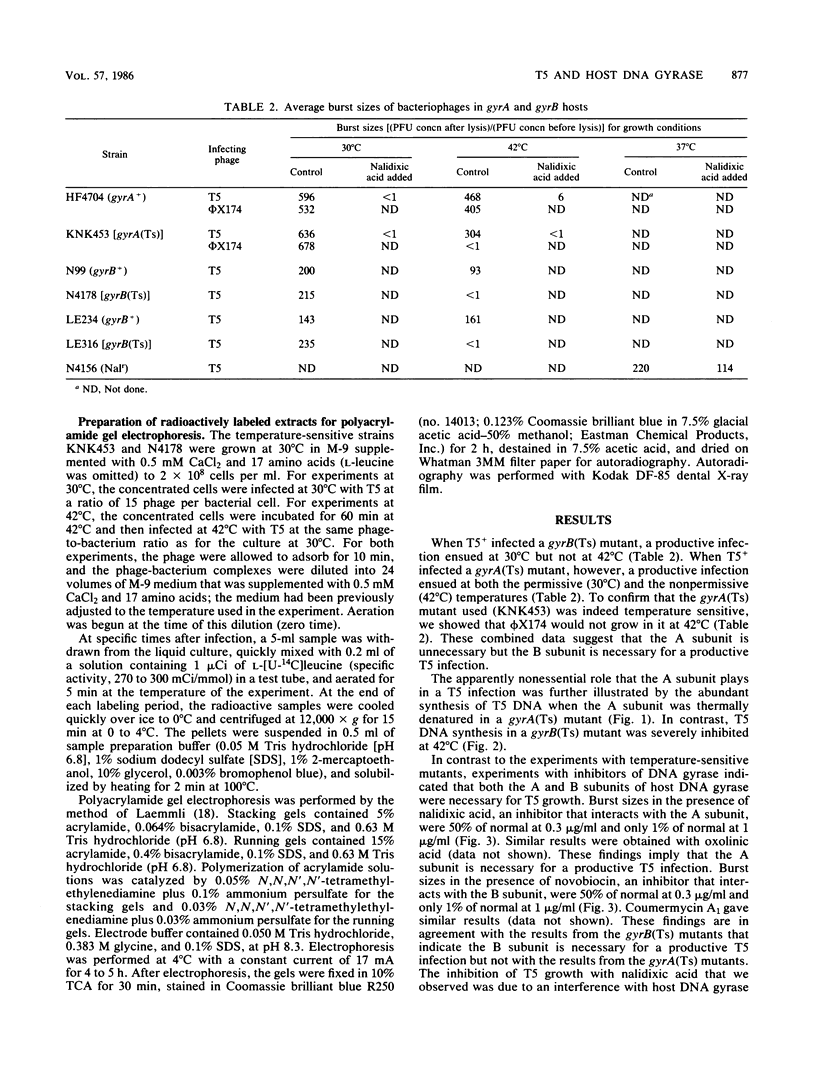
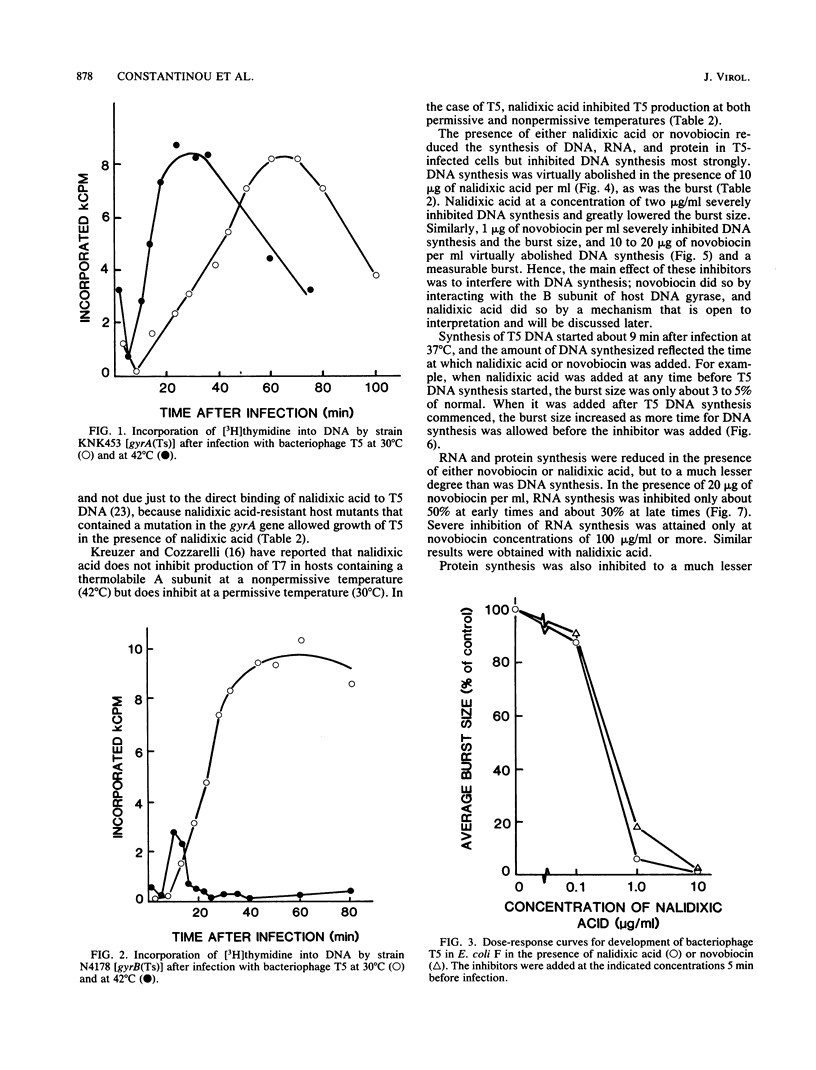
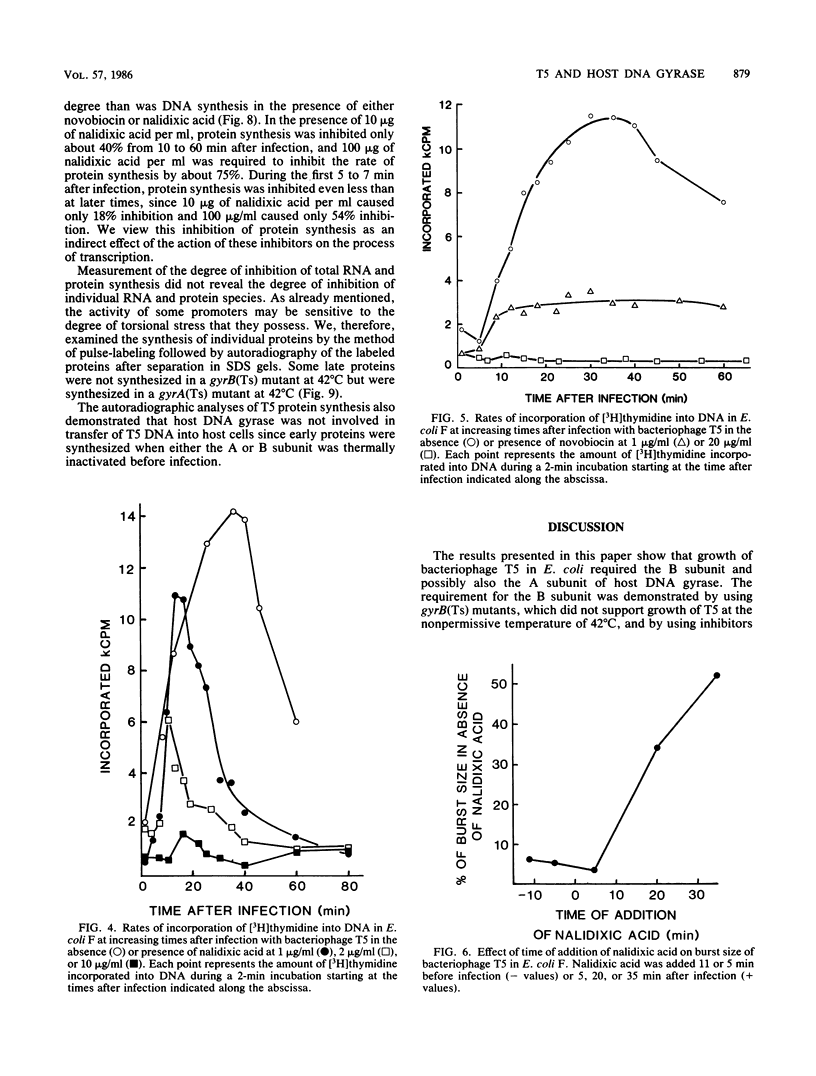
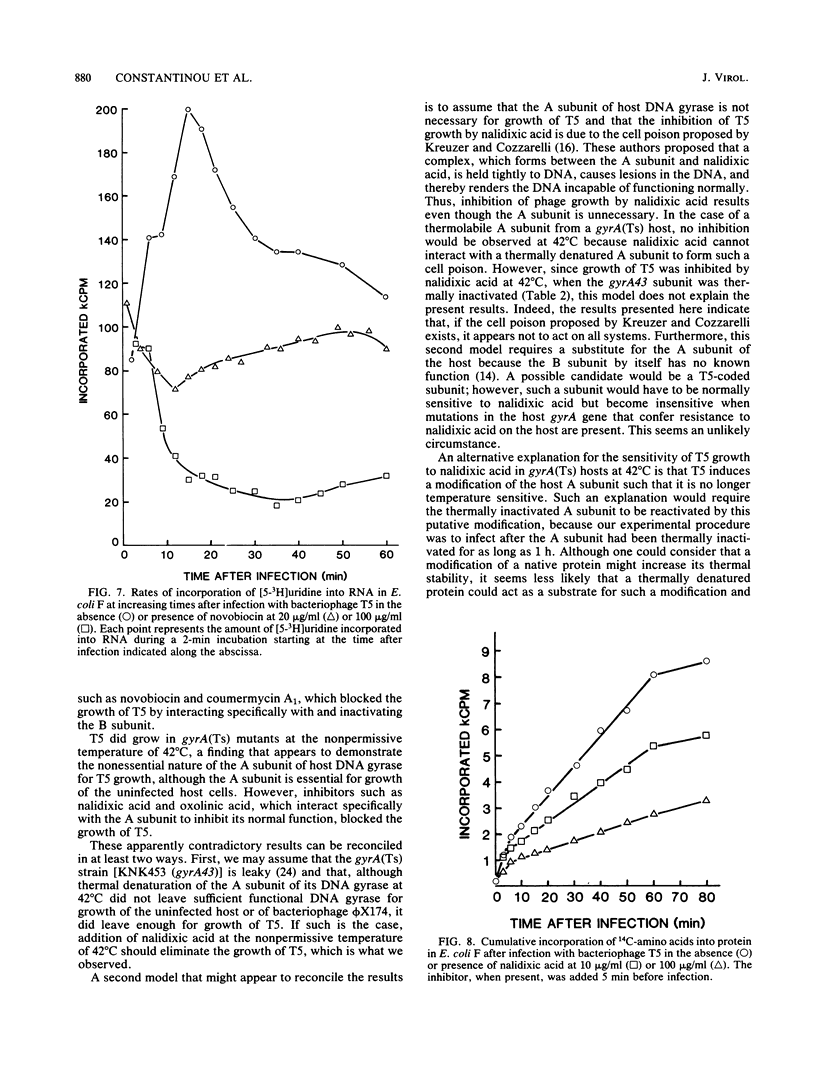
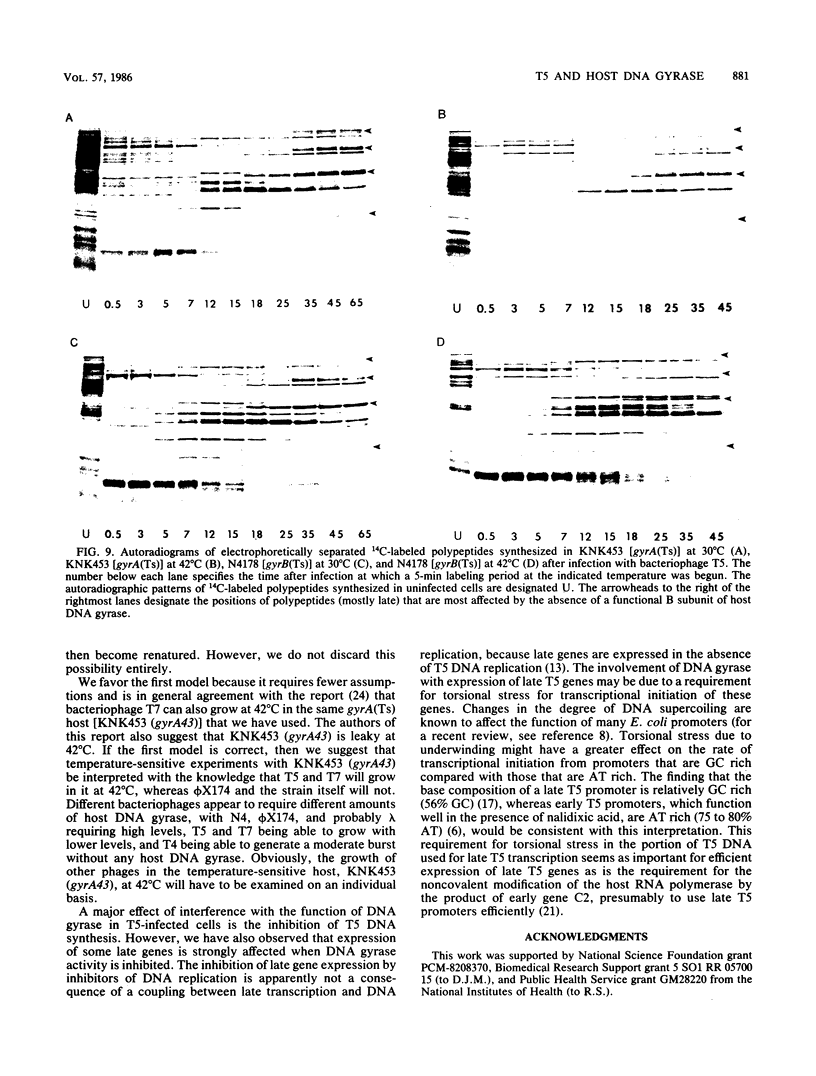
Bacteriophage T5 did not grow at the nonpermissive temperature of 42 degrees C in Escherichia coli carrying a temperature-sensitive mutation in gyrB [gyrB(Ts)], but it did grow in gyrA(Ts) mutants at 42 degrees C. These findings indicate that the A subunit of host DNA gyrase is unnecessary, whereas the B subunit is necessary for growth of T5. The necessity for the B subunit was confirmed by a strong inhibition of T5 growth by novobiocin and coumermycin A1, which interfere specifically with the function of the B subunit of host DNA gyrase. However, T5 growth was also strongly inhibited by nalidixic acid, which interferes specifically with the function of the A subunit. This inhibition was due to the interaction of nalidixic acid with the A subunit and not just to its binding to DNA, because appropriate mutations in the gyrA gene of the host conferred nalidixic acid resistance to the host and resistance to T5 growth in such a host. The inhibition by nalidixic acid was also not due to a cell poison formed between nalidixic acid and the A subunit (K. N. Kreuzer and N. R. Cozzarelli, J. Bacteriol. 140:424-435, 1979) because nalidixic acid inhibited growth of T5 in a gyrA(Ts) mutant (KNK453) at 42 degrees C. We suggest that T5 grows in KNK453 at 42 degrees C because its gyrA(Ts) mutation is leaky for T5. Inhibition of T5 growth due to inactivation of host DNA gyrase was caused mainly by inhibition of T5 DNA replication. In addition, however, late T5 genes were barely expressed when host DNA gyrase was inactivated.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alonso J. C., Sarachu A. N., Grau O. DNA gyrase inhibitors block development of Bacillus subtilis bacteriophage SP01. J Virol. 1981 Sep;39(3):855–860. doi: 10.1128/jvi.39.3.855-860.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird J. P., Bourguignon G. J., Sternglanz R. Effect of nalidixic acid on the growth of deoxyribonucleic acid bacteriophages. J Virol. 1972 Jan;9(1):17–21. doi: 10.1128/jvi.9.1.17-21.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman L. D., Hoffman M. S., McCorquodale D. J. Pre-early proteins of bacteriophage T5: structure and function. J Mol Biol. 1971 Dec 28;62(3):551–564. doi: 10.1016/0022-2836(71)90155-0. [DOI] [PubMed] [Google Scholar]
- Bourguignon G. J., Levitt M., Sternglanz R. Studies on the mechanism of action of nalidixic acid. Antimicrob Agents Chemother. 1973 Oct;4(4):479–486. doi: 10.1128/aac.4.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wyngaert M., Hinkle D. C. Involvement of DNA gyrase in replication and transcription of bacteriophage T7 DNA. J Virol. 1979 Feb;29(2):529–535. doi: 10.1128/jvi.29.2.529-535.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K. Biology of bacterial deoxyribonucleic acid topoisomerases. Microbiol Rev. 1984 Dec;48(4):273–289. doi: 10.1128/mr.48.4.273-289.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco S. C., Zivin R., Rothman-Denes L. B. Novel template requirements of N4 virion RNA polymerase. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3220–3224. doi: 10.1073/pnas.75.7.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Gellert M., Fisher L. M., O'Dea M. H. DNA gyrase: purification and catalytic properties of a fragment of gyrase B protein. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6289–6293. doi: 10.1073/pnas.76.12.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson H. E., McCorquodale D. J. Genetic and physiological studies of bacteriophage T5. 2. The relationship between phage DNA synthesis and protein synthesis in T5-infected cells. Biochem Biophys Res Commun. 1971 May 21;43(4):735–740. doi: 10.1016/0006-291x(71)90677-2. [DOI] [PubMed] [Google Scholar]
- Higgins N. P., Peebles C. L., Sugino A., Cozzarelli N. R. Purification of subunits of Escherichia coli DNA gyrase and reconstitution of enzymatic activity. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1773–1777. doi: 10.1073/pnas.75.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Tomizawa J. I. Involvement of DNA gyrase in bacteriophage T7 DNA replication. Nature. 1977 Nov 3;270(5632):78–80. doi: 10.1038/270078a0. [DOI] [PubMed] [Google Scholar]
- Kreuzer K. N., Cozzarelli N. R. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J Bacteriol. 1979 Nov;140(2):424–435. doi: 10.1128/jb.140.2.424-435.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksenzenko V. N., Kamynina T. P., Pustoshilova N. M., Kryukov V. M., Bayev A. A. Cloning of bacteriophage T5 promoters. Mol Gen Genet. 1982;185(3):520–522. doi: 10.1007/BF00334154. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Liu C. C., Alberts B. M. T4 DNA topoisomerase: a new ATP-dependent enzyme essential for initiation of T4 bacteriophage DNA replication. Nature. 1979 Oct 11;281(5731):456–461. doi: 10.1038/281456a0. [DOI] [PubMed] [Google Scholar]
- McCarthy D. Gyrase-dependent initiation of bacteriophage T4 DNA replication: interactions of Escherichia coli gyrase with novobiocin, coumermycin and phage DNA-delay gene products. J Mol Biol. 1979 Jan 25;127(3):265–283. doi: 10.1016/0022-2836(79)90329-2. [DOI] [PubMed] [Google Scholar]
- McCorquodale D. J. The T-odd bacteriophages. CRC Crit Rev Microbiol. 1975 Dec;4(2):101–159. doi: 10.3109/10408417509111574. [DOI] [PubMed] [Google Scholar]
- Menzel R., Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983 Aug;34(1):105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- Shen L. L., Pernet A. G. Mechanism of inhibition of DNA gyrase by analogues of nalidixic acid: the target of the drugs is DNA. Proc Natl Acad Sci U S A. 1985 Jan;82(2):307–311. doi: 10.1073/pnas.82.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steck T. R., Drlica K. Involvement of DNA gyrase in bacteriophage T7 growth. J Virol. 1985 Jan;53(1):296–298. doi: 10.1128/jvi.53.1.296-298.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetler G. L., King G. J., Huang W. M. T4 DNA-delay proteins, required for specific DNA replication, form a complex that has ATP-dependent DNA topoisomerase activity. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3737–3741. doi: 10.1073/pnas.76.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]