Abstract
The mechanisms of influenza A virus mRNA intracellular transport are still not clearly understood. Here, we visualized the distribution and transport of influenza A virus mRNA in living cells using molecular beacon (MB) technology. Confocal-FRAP measurements determined that the transport of influenza A virus intronless mRNA, in both nucleus and cytoplasm, was energy dependent, being similar to that of Poly(A)+ RNA. Drug inhibition studies in living cells revealed that the export of influenza A virus mRNA is independent of the CRM1 pathway, while the function of RNA polymerase II (RNAP-II) may be needed. In addition, viral NS1 protein and cellular TAP protein were found associated with influenza A virus mRNA in the cell nucleus. These findings characterize influenza A virus mRNA transport in living cells and suggest that influenza A virus mRNA may be exported from the nucleus by the cellular TAP/p15 pathway with NS1 protein and RNAP-II participation.
INTRODUCTION
Influenza virus is one of the few RNA viruses to synthesize its mRNA in the nucleus of infected cells (1). The virus mRNAs are potential substrates for the cellular splicing machinery and need to be exported from the nucleus to enable the viral proteins to be synthesized (2). Uncovering the mechanisms of influenza virus mRNA export is of great importance to truly understand the replication and pathogenicity of the virus. The nuclear export of cellular mRNA is mediated by several proteins that bind to mRNA and to pre-mRNA precursors (3). However, unlike cellular intron-containing mRNAs, most influenza virus mRNAs are intronless. Hence, the export mechanisms of viral intronless mRNAs might be different from those of cellulr mRNAs. Moreover, because there are three different types of influenza virus mRNA, more than one mechanism of nuclear export might operate in virus-infected cells (1). The first type of influenza virus mRNA includes intronless mRNAs, such as PA, PB1, PB2, HA, NA and NP mRNA. The second type of viral mRNA consists of the M1 and NS1 mRNAs, which contain introns but do not undergo splicing. The M2 and NS2 mRNAs, which are produced by splicing, comprise the third type of viral mRNA. The mechanisms of the nuclear export of these three types of influenza A virus mRNA remain unknown.
Two pathways have been described that appear to be responsible for the export of viral mRNA (4). The first RNA export pathway was the CRM1 pathway, which is utilized by human immunodeficiency virus (HIV) through the mediation of the Rev protein (5). Herpes simplex virus (HSV) also utilizes CRM1 to export its mRNA (6). However, other studies showed that CRM1 may be not a major contributor to mRNA export in metazoans or yeast (7,8). The human protein TAP, and its yeast ortholog Mex67p, might be the best candidates for mRNA export receptors because they shuttle between the nucleus and cytoplasm, cross-link to poly(A)+ RNA, localize at the nuclear pores and interact directly with nucleoporins (9–12). The TAP pathway was reported to be used by HSV ICP27 to export its intronless mRNAs (4). Moreover, TAP protein could also promote the export of constitutive transport element (CTE) containing transcripts of some virus such as type D retrovirus (9,10,13). Influenza A virus mRNA may, therefore, be exported from the nucleus by the CRM1 dependent pathway or by the TAP/p15 pathway. Previous studies have shown that influenza virus NS1 protein could selectively inhibit cellular mRNA export by binding with CPSF and PABII (1), or by forming an inhibitory complex with cellular mRNA export factors TAP and p15 (14). Moreover, NS1 can also inhibit the splicing and export of its own mRNA, in an RNA binding-dependent manner (2). Nevertheless, the mechanisms by which influenza virus mRNAs are exported from the nucleus and the roles of viral NS1 protein in influenza A virus intronless mRNA export are still unclear.
The ability to accurately and repeatedly track mRNA in living mammalian cells would help us to fully understand the mRNA transport mechanism. There are a variety of tools currently used to visualize intracellular mRNAs, including molecular beacons (MBs) and fluorescently labeled oligonucleotide probes. MBs are a powerful and simple tool for cellular mRNA and viral RNA visualization in living cells (15–21). Live-cell imaging of mRNA could shed light on many fundamental processes, such as the kinetics of mRNA production, mRNA localization and transportation inside a cell and cellular responses to virus infection and to virus–host interaction. We, therefore, used MBs as a detection probe to track influenza A virus mRNA in living host cells, in order to explore the mechanisms of viral mRNA export.
In this study, we successfully visualized influenza A virus mRNA in living mammalian cells, and studied the dynamic behaviors of influenza virus mRNA by Confocal-FRAP experiments. By imaging experiments of living cells and protein immunofluorescence analysis in fixed cells, it was found that influenza A virus mRNAs could colocalize with viral NS1 and cellular TAP protein in cell nucleus. Moreover, coimmunoprecipitation experiments of influenza A virus mRNAs with NS1 and TAP protein revealed that NS1 and TAP protein could be physically associated with both intron-containing and intronless mRNAs of influenza A virus. By performing Actinomycin D (ActD) inhibition experiments in living cells we observed that RNA polymerase II (RNAP-II) and other factors might be involved in influenza virus mRNA export. Furthermore, Leptomycin B (LMB), a specific inhibitor of CRM1, could not inhibit influenza A virus mRNA export in living cells. Therefore, influenza A virus mRNA export may be independent of CRM1. Together, these results indicate that the cellular transport of influenza A virus mRNA may be energy dependent and utilize the cellular TAP/p15 transport pathway with the participation of viral NS1 protein and cellular RNAP-II.
MATERIALS AND METHODS
Cells, viruses, recombinant plasmids and other compounds
Influenza virus strains A/PR/8/34 (PR/8) was propagated in 10-day-old embryonated eggs for 3 days at 36.5°C. MDCK cells were grown in RPM1640 medium supplemented with 10% FBS, 100 U/ml of penicillin and 100 µg/ml of streptomycin. The 293T cells were grown in DMEM medium containing 10% FBS, 100 U/ml of penicillin and 100 µg/ml of streptomycin. Plasmid pEGFP-C1-NS1, which expresses the influenza virus strain PR/8 NS1 gene, was constructed by ligating NS1 cDNA in frame into vector pEGFP-C1. Plasmid pCDNA3.1-TAP-GFP was constructed by inserting the TAP gene, from PCS2-FLAG-TAP [kindly provided by Prof. R.M. Sandri-Goldin (University of California, Irvine, USA)] into vector pCDNA3.1-GFP. Full-length NS1 gene and TAP gene were also cloned into pCDNA3.1 vector. Rabbit anti-NS1 polyclonal antibody was generously provided by Prof. J. Ortín [Centro Nacional de Biotecnologý’a (CSIC), Madrid, Spain]. Mouse anti-TAP monoclonal antibody was purchased from Abcam (Jingmei, China). ActD (Sigma-Aldrich, St. Louis, MO, USA) was solubilized in DMSO to a stock of 1 mg/ml and used at 0.5 µg/ml. LMB (Sigma-Aldrich) was used at a concentration of 10 nM. Energy depletion experiments were performed by incubating cells in 20 mM 2-deoxy-d-glucose (Sigma-Aldrich) and 10 mM sodium azide for 15–30 min at 37°C.
Transfection and influenza A virus infection
For transfection and infection, cells were grown on coverslips placed on 35 mm dishes. Transfections were performed with Lipofectmine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. For virus infection, Influenza A virus [A/Puerto Rico/8/34(H1N1)] propagation solution was diluted in PBS (0.01 M, pH 7.4), containing 0.2% bovine serum albumin, and virus was added over cells at the indicated multiplicity of infection (MOI), then adsorbed for 60 min at 37°C. After removing the virus dilution, cells were maintained in infecting media (RPMI 1640, 4 ug/ml trypsin) at 37°C in 5% CO2.
Probe design, synthesis and delivery into living cells
Probes were designed based on genome sequence and structure of Influenza A virus [A/Puerto Rico/8/34(H1N1)] mRNAs using RNAdraw, Primer Premier 5.0 and NCBI BLAST. Probes specific to PR/8 mRNAs were synthesized and purified by Shanghai Sangon (Shanghai, China). The sequences of the MBs and control probes used in this study and the location of different labels are indicated in Table 1.
Table 1.
MBs and control probes used in this study
| MB |
| NA mRNA MB (nucleotides 594–618) |
| TAMRA-GCGACTTTCAGTTATTATGCCGTTGTATTTGTCGC-DABCYL |
| FAM-GCGACTTTCAGTTATTATGCCGTTGTATTTGTCGC-BHQ1 |
| M1 mRNA MB (nucleotides 526–550) |
| TAMRA-GCGACATGTCTGATTAGTGGGTTGGTTGTTGTCGC-DABCYL |
| M2 mRNA MB (nucleotides 912–936) |
| TAMRA-GCGACCTTCCCTCATAGACTTTGGCACTCCGTCGC-DABCYL |
| Random MB |
| TAMRA-CCAGCCTACAAACTAGCATTCTGGTGAGCTGG-DABCYL |
| Control probes (without 3′ quencher) |
| NA mRNA control probe (nucleotides 594–618) |
| TAMRA-GCGACTTTCAGTTATTATGCCGTTGTATTTGTCGC |
| M1 mRNA control probe (nucleotides 526–550; 203–227) |
| TAMRA-GCGACATGTCTGATTAGTGGGTTGGTTGTTGTCGC M1 (526–550) |
| FAM-GCGACCGGTGAGCGTGAACACAAATCCTAAGTCGC M1-2 (203–227) |
| M2 mRNA control probe (nucleotides 912–936) |
| TAMRA-GCGACCTTCCCTCATAGACTTTGGCACTCCGTCGC |
All sequences are listed from their 5′ end to their 3′ end. The underlined bases are bases added to create the stem domain of MBs. The nucleotides numbers of NA, M1 and M2 mRNA-specific MBs refer to positions in the NA mRNA sequence (accession number NC_002018) and M1 mRNA sequence (accession number NC_002016). The random MB that can not bind the mRNAs of influenza A virus [A/PR/8/34 (H1N1)] was used to verify the specificity of the MBs designed to target influenza A virus mRNAs in infected cells.
MBs targeting influenza A virus mRNAs, as well as control probes were delivered into living cells using a reversible permeabilization method with streptolysin O (SLO), as described previously (17,22,23). Specifically, SLO was activated first by adding 10 mM of dithiothreitol (DTT) to 1000 U/ml of SLO for 90 min at 37°C. Cells grown in 35 mm cell culture dishes were incubated for 30 min in 500 µl of Ca2+, Mg2+ free PBS containing 25 mM Hepes, 5 U/ml of activated SLO and 300 nM of each MB type. Cells were then resealed by adding 1.5 ml of the typical growth medium and incubated for 15 min at 37°C before performing fluorescence microscopy imaging.
Fluorescent in situ hybridization and simultaneous labeling
The fluorescent in situ hybridization (FISH) of mRNA was carried out according to general procedure (24,25). Influenza A virus-infected MDCK cells in dishes were fixed with 4% paraformaldehyde in 1× PBS, pH 7.4 at room temperature for 20 min. After washing two times, cells were permeabilized using 0.5% (v/v) Triton-x-100 in PBS for 2 min followed by one PBS wash. Then cells were dehydrated in 75% ethanol overnight at 4°C. Next, the samples were rehydrated using 2× SSC, 50% formamide for 5 min and then incubated with the hybridization mixture containing 2× SSC, 1 mg/ml of tRNA, 10% dextran sulfate, 50% formamide, 0.01% BSA, 20 U/ml ribonuclease inhibitor and 500 nM of probes. After 4–16 h incubation at 37°C, cells were washed two times for 20 min with 50% formamide, 2× SSC and one time with 0.5× SSC for 10 min at room temperature. Simultaneous labeling of NS1, TAP protein and influenza A virus mRNAs were performed according to the protocol described previously (26). In brief, the infected MDCK cells were live-cell labeled with the influenza A virus mRNA specific MB. Thirty minutes after MB hybridization, cells were fixed with 4% formaldehyde in PBS for 15 min at room temperature, permeabilized in 0.2% Triton X-100 for 3 min at room temperature and preincubated with 2% BSA/PBS for 45 min at room temperature. They were then washed and incubated with anti-NXF1 antibody (Abcam, Cambridge, MA, USA) or anti-NS1 antibody for 60 min at 37°C. After washing, the cells were then incubated with FITC-labeled secondary antibody (Boster, Wuhan, China) for 1 h at 37°C. Finally, the cells were washed three times and stained with Hoechst for direct observation.
Image acquisition, data collection and analysis
All confocal-FRAP (fluorescence recovery after photobleaching) experiments were made using a TCS SP2 Leica laser scanning spectral confocal microscope equipped with a cooled CCD camera. For live-cell imaging, the prepared cell culture dishes were placed in a temperature-controlled incubator at 37 or 23°C and detection was made using a 100× oil objective (NA 1.32) with a 488 nm excitation beam. FRAP experiments were carried out with the standard Leica FRAP software (the advanced time-lapse software). The 488 nm lasers were set at 25% for imaging and 100% for bleaching. The nucleus was bleached for 4 s and the cytoplasm was bleached for 6 s. In addition, cells were scanned in 2D in time, with a pinhole setting of 2.4 airy. The diffusion coefficients (D-values) were calculated as described previously (27). Except for the FRAP experiments, all imaging experiments were carried out using an inverted wide-field fluorescent microscope (Axiovert 200, from Carl Zeiss, Germany) equipped with a cooled CCD camera (Model Cascade 512B, Photometrics, Tucson, AZ, USA). A filter set with 472/30 nm for excitation, 510 nm for dichroic beam splitter and 520/35 nm for emission viewed the green fluorescence from the EGFP channel. The red fluorescence from the TAMRA channel was viewed with an excitation filter of 531/40 nm, a dichroic beam splitter of 600 nm and an emission filter of 593/40 nm, while the blue fluorescence of Hoechst 33 258 was viewed with an excitation filter of 365 nm, a dichroic beam splitter of 395 nm and an emission filter of 420 nm (All filters are from Semrock, Rochester, NY, USA). In the double-labeling experiment, GFP and TAMRA were sequentially scanned to avoid cross talk. Image acquisition and analysis was performed using MetaMorph 6.0 software (Molecular Devices, Downingtown, PA, USA). And images were further analyzed using ImageJ (NIH) v.1.33u and Adobe Photoshop. For colocalization analysis, the merged images were used for measuring the colocalization coefficients, M1 and M2, the Pearson's correlation coefficients, Rp and the overlap coefficients, R by the ImageJ (NIH) v.1.33u software according to the manufacturer's instructions and the methods, as previously described (28).
Immunoprecipitation experiments and influenza A virus mRNA detection
The immunoprecipitation experiments were performed as described before (29) and the influenza virus-infected cell nuclear extracts were used for coimmunoprecipitation with NS1 or TAP protein by anti-NS1 or anti-TAP antibody. Monolayer cultures of MDCK cells or 293T cells grown in 100 mm dishes (4.8 × 106 cells) were infected with influenza A virus at a MOI of 10–100. Four to six hours after infection, the medium was removed and the monolayers were rinsed with cold PBS, overlaid with 1 ml per dish of cell lysis buffer [100 mM KCl, 5 mM MgCl2, 10 mM Hepes, pH 7.0, 0.5% NP-40, 1 mM TCEP, 100 U/ml ribonuclease inhibitor (Takara, Dalian, China), 2 mM vanadyl ribonucleoside complexes solution (Sigma), 25 μl/ml protease inhibitor mixture for mammalian tissues (Sigma)] and the cells were detached by scraping with a cell scraper and the cell lysate was transferred to a tube on ice. After incubation for 10 min at 4°C, the cultures were centrifuged at 5000 r.p.m. for 5 min and the supernatants were considered to be the cytoplasmic fraction while the pellet was the nuclear fraction. Then the pellet was resuspended in 500 µl of cold high salt lysis buffer [400 mM KCl, 5 mM MgCl2, 10 mM Hepes, pH 7.0, 0.5% NP-40, 1 mM DTT, 100 U/ml ribonuclease inhibitor, 2 mM vanadyl ribonucleoside complexes solution, 25 μl/ml protease inhibitor cocktail (Sigma)] and the tubes were vigorously rocked at 4°C for 45 min on a shaking platform. The supernatant was saved as nuclear extract after centrifugation at 10 000 × g for 30 min and used for the immunoprecipitation experiments.
For immunoprecipitation analysis, the nuclear extracts were precleared by incubation with 35 µl of protein A agarose beads at 4°C for 1 h with gently rotation. After centrifugation at 2500 r.p.m. for 5 min, the supernatant was collected and incubated with 2 µl of the anti-NS1 or anti-TAP antibody at 4°C overnight. Then 60 μl of protein A agarose beads was added to each tube and the tubes were rotated at 4°C for 3 h. Then the beads were washed four times with cell lysis buffer and were separated into two aliquots. One of them was used for detection of influenza A virus mRNAs and the other one was used for protein analysis. The RNA present in the immunoprecipitates was extracted as described before (29), and the isolated RNA pellet was resuspended in 45 μl of nuclease-free water and analyzed with the One Step RT–PCR kit (Takara) using primers specific for either influenza A virus NA, M1 and PB1 mRNA or cellular β-actin and H2a mRNA. The following primers were used: NA-Forward, TACAGCAAAGACAATAGCATA and NA-Reverse, CCAAACACCATTACCATACCT; corresponding to NA mRNA (NC_002018) sequences 273–293 and 1029–1049, respectively; M1-Forward, ACCGTGCCCAGTGAGCGAGGA and M1-Reverse, ATGTCTGATTAGTGGGTTGGT; corresponding to M1 mRNA (NC_002016) sequences 224–244 and 530–550, respectively; PB1-Forward, ACCGAAACTGGAGCACCGCA and PB1-Reverse, TGTTCAGGGTCAATGCTCTA; corresponding to PB1 mRNA (EF467819) sequences 199–218 and 681–700, respectively; Actin-Forward, AGCGGGAAATCGTGCGTGAC and Actin-Reverse, CGTCATACTCCTGCTTGCTG; corresponding to β-actin mRNA (NM_001101) sequences 698–717 and 1155–1174, respectively. H2a-Forward, ATGTCTGGACGAGGGAAG and H2a-Reverse, ACTTGCTTTGGGCTTTATGG; corresponding to H2a mRNA (NM_170745) sequences 36–53 and 410–429, respectively. A portion of the immunoprecipitated sample was also subjected to SDS- polyacrylamide gel electrophoresis for analysis of bound proteins. The proteins were detected by western blotting with anti-NS1 or anti-TAP antibody.
RESULTS
Visualizing influenza A virus mRNA in living MDCK cells
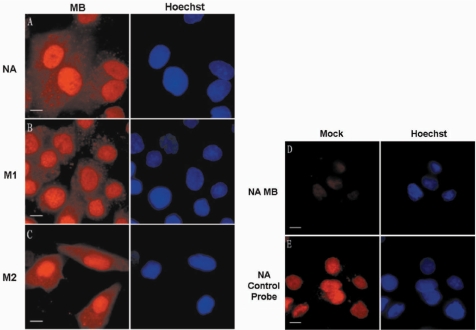
To visualize influenza A virus mRNA in living cells, MBs were delivered into influenza virus-infected MDCK cells by SLO at various time points postinfection (p.i.). NA mRNA (intronless), M1 mRNA (intron-containing but unspliced) and M2 mRNA (produced by splicing) were chosen as models for the three different types of influenza A virus mRNAs and visualized in living host cells using three specific MBs (NA, M1 and M2) (Table 1). First, NA, M1 and M2 MBs were delivered into influenza A virus-infected MDCK cells at 3.5 h p.i., and the resulting fluorescence signals were observed under TAMRA excitation (543 nm) about 1 h after beacon delivery. As shown in Figure 1A–C, the MB fluorescence signals that represented influenza A virus NA, M1 and M2 mRNAs at 4.5 h p.i. appeared in both the nucleus and the cytoplasm, and the fluorescence signals in the cytoplasm mainly appeared with a granular morphology. Noninfluenza virus-infected MDCK cells showed little fluorescence 60 min after the addition of SLO and MB mixture (Figure 1D), and still had very low fluorescence 1.5 h after MB delivery. Moreover, when control probes, without the 3′ quencher, were added into noninfluenza virus-infected MDCK cells, the fluorescence signals were found to be distributed mainly in the nuclei of living MDCK cells without obvious cytoplasmic distribution (Figure 1E), indicating that MB molecules in noninfected cells and the excessive MBs in influenza virus-infected cells could both distribute in cell nuclei. Many studies have reported that both oligonucleotides and MBs can be rapidly taken up by the nucleus, although the mechanism is not well understood (16,20,30). These results ensured that the fluorescent signals in the nucleus and cytoplasm of influenza virus-infected cells really came from the hybridization of mRNA with MBs, and were not caused by the degradation of MBs.
Figure 1.
Imaging influenza A virus mRNA in living influenza virus-infected MDCK cells. (A–C) Influenza A virus NA, M1, M2 mRNAs, detected with TAMRA-labeled MBs in influenza A virus-infected MDCK cells at 4.5 h p.i., were found in both the nucleus and cytoplasm. (D) Control noninfected MDCK cells show no obvious fluorescence after being delivered with NA MBs. (E) Control probe of NA mRNA shows concentrated fluorescence signals in the nucleus after being delivered into noninfected MDCK cells. Bars, 10 µm.
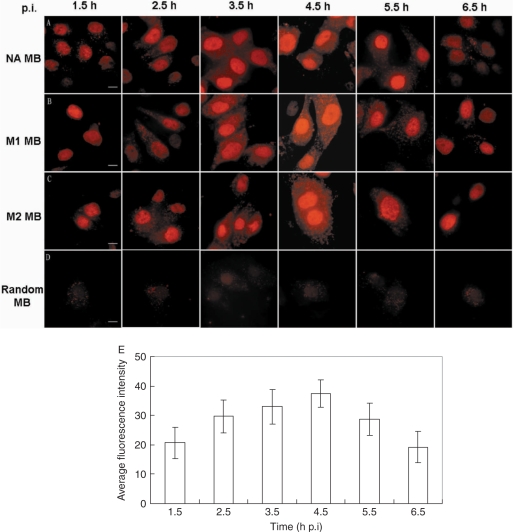
To explore the mechanisms of influenza virus mRNA export, MBs were delivered into influenza A virus-infected MDCK cells at different p.i. time points and the fluorescence signals were observed at 1.5, 2.5, 3.5, 4.5, 5.5, 6.5 h p.i. (Figure 2). At 1.5 h p.i., the fluorescence signals were very weak and only found in the nucleus (Figure 2A–C). The fluorescence signals in the cytoplasm and in the nucleus became stronger from 2.5 h p.i. to 4.5 h p.i. After that, from 5.5 h p.i., the fluorescence signals became weaker in both cytoplasm and nucleus. At 6.5 h p.i., there were virtually no fluorescence signals in the cytoplasm and signals in the nucleus also became very weak. These different distribution patterns may be associated with the kinetics of mRNA synthesis and mRNA export. To further confirm the specificity of MBs designed to target influenza A virus mRNAs, a random MB that can not bind the mRNAs of influenza A virus [A/PR/8/34 (H1N1)] (Table 1) was delivered into influenza A virus-infected MDCK cells at different p.i. time points (Figure 2D). The cells delivered with this random MB at indicated time points all showed very low fluorescence compared with the cells delivered with NA, M1 or M2 MB in Figure 2A–C, thereby demonstrating the high specificity of the influenza A virus mRNA-targeting MBs. As shown in Figure 2E, the average fluorescence intensity of M1 MB per unit area of infected cells at different p.i. time points was calculated by MetaMorph 6.0 software (Molecular Devices, USA) and the bar chart of average fluorescence intensity, averaged from 20 repeats, showed the fluorescence intensity peak of M1 mRNA was between 3.5 h and 4.5 h p.i. It has been reported that the synthesis of segment 7-specific mRNA showed a peak at 4 h p.i. and continued later at a slower rate (31), which may explain why the fluorescence signals appeared strongest at about 4 h p.i. (between 3.5 and 4.5 h p.i.). There was little difference among the fluorescence distribution patterns of NA, M1 and M2 mRNAs at indicated p.i. time points.
Figure 2.
Visualizing influenza A virus mRNAs in living influenza virus-infected MDCK cells at different p.i. time points. (A) NA mRNA was detected with TAMRA-labeled MBs delivered into influenza A virus-infected MDCK cells at different time p.i. (B) M1 mRNA was detected with TAMRA-labeled MBs delivered into influenza A virus-infected MDCK cells at different time p.i. (C) M2 mRNA was detected with TAMRA-labeled MBs delivered into influenza A virus-infected MDCK cells at different time p.i. (D) The random control MB was delivered into influenza A virus-infected MDCK cells at different time p.i. and showed very low fluorescence at indicated time points. (E) The average fluorescence intensity of M1 MBs at different time points, p.i., is indicated by the bar chart. The average fluorescence intensity was measured by MetaMorph 6.0 software (Molecular Devices, USA) to calculate average intensity per unit area of cells in integrated cell regions of different images (n > 20). The integrated cell regions including cell nucleus and cytoplasm were selected and manually traced by MetaMorph 6.0. All the images were acquired with an exposure time of 5000 ms and a γ-value of 1.5. Bars, 10 µm.
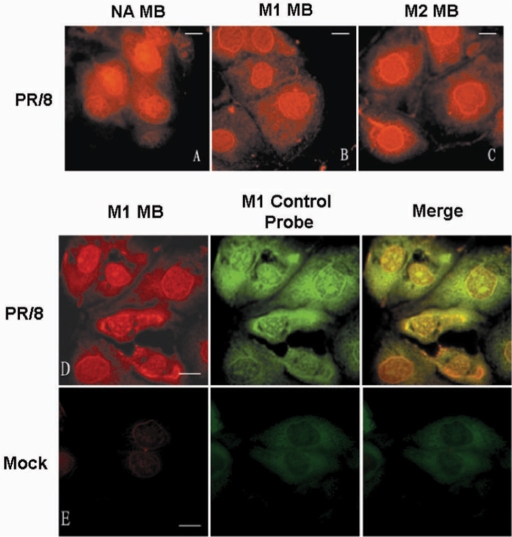
To corroborate the live-cell imaging results, FISH assays were performed targeting influenza A virus NA, M1 and M2 mRNAs in fixed influenza virus-infected MDCK cells. As shown in Figure 3A–C, the fluorescence images obtained from the FISH assay of NA, M1 and M2 mRNAs in influenza virus-infected MDCK cells at 4.5 h p.i. showed a localization pattern similar to the live-cell imaging results, shown in Figure 1A–C. To further test whether the MBs can be specifically hybridized to target viral mRNAs in influenza A virus-infected cells, FISH experiments were performed using MBs and FAM-labeled probes targeting different regions of influenza A virus mRNAs (Table 1). Figure 3D shows that the M1 MBs and M1 control probe, which target different regions of M1 mRNA, were colocalized in fixed influenza A virus-infected MDCK cells at 4.5 h p.i. Although some noise–signal from the M1 control probe was occasionally found in the nucleus of noninfected cells, there was very low signal from M1 MBs in noninfected cells (Figure 3E). Together, these results show that MBs could be specifically hybridized to influenza A virus mRNAs and used for viral mRNA imaging in influenza A virus-infected cells.
Figure 3.
Localization of NA, M1 and M2 MBs in influenza A virus-infected MDCK cells by FISH. (A–C) Localization of NA, M1 and M2 MBs in influenza A virus-infected MDCK cells at 4 h p.i. by FISH. (D) Colocalization of M1 MBs and M1 control probes in influenza virus-infected MDCK cells at 4 h p.i. by FISH. (E) Low fluorescence background of M1 MBs in nonvirus-infected MDCK cells at 4 h p.i. by FISH. Bars, 10 µm.
Dynamic character of influenza A virus mRNA inside the nucleus and the cytoplasm
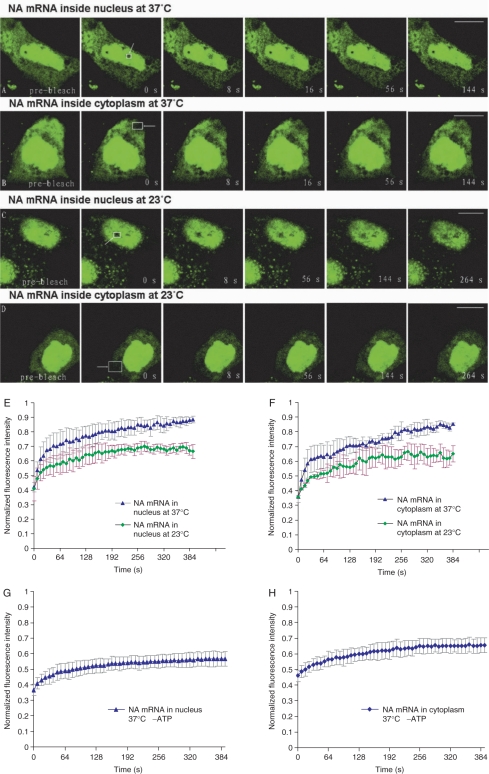
To study the dynamic character of influenza A virus mRNA inside virus-infected MDCK cells, confocal-FRAP (Fluorescence recovery after photobleaching) experiments were performed. Most influenza A virus mRNAs are intronless, so we studied the mobility characteristics of intronless NA mRNA to explore if there were differences between influenza A virus mRNAs and cellular intron-containing mRNAs. First, the kinetic behaviors of NA mRNA in both nucleoplasm and cytoplasm were studied by FRAP at 37°C. FRAP images of typical acquisitions in nucleoplasmic and cytoplasmic areas of virus-infected MDCK cells, delivered with FAM-labeled NA MBs 2 h p.i., are shown in Figure 4A and B. After the mean intensity of the regions of interest were recorded and quantified for a series of FRAP images, recovery curves averaged from six repeats were drifted and the mobile fractions and diffusion coefficients were calculated (Figure 4E and F). The results of NA mRNA in the nucleus at 37°C showed that the t1/2 of recovery was about 22 s, and the diffusion coefficient (D) was 0.027 µm2/s. However, the recovery was slightly slower in the cytoplasm: t1/2 was about 30 s, D-value was 0.018 µm2/s (Table 2). The lower diffusion rate of NA mRNA in the cytoplasm compared with that in the nucleus may due to the association of NA mRNA with cytoplasmic microtubules and the anchoring of NA mRNA on the ER membrane for translation. Furthermore, it was found that the fluorescence in both nucleus and cytoplasm during FRAP experiments could only recover about 50% of the initial fluorescence intensity. These results suggested that about 50% of NA mRNA could diffuse freely while the remaining 50% appeared relatively immobile or diffused very slowly.
Figure 4.
Diffusion dynamics of influenza A virus NA mRNA as revealed by FRAP in influenza virus-infected MDCK cells. Confocal-FRAP measurements were carried out in influenza virus-infected MDCK cells using a 100·objective at 3 h p.i. (A and B) Serial FRAP images of NA mRNA at 37°C in influenza virus-infected MDCK cells. (C and D) Serial FRAP images of NA mRNA at 23°C in influenza virus-infected MDCK cells. (E) Typical recovery curves (n = 6) of the FRAP measurements of NA mRNA in influenza virus-infected MDCK cell nuclei at 23 and 37°C, respectively. (F) Typical recovery curves (n = 6) of the FRAP measurements of NA mRNA in influenza virus-infected MDCK cell cytoplasm at 23 and 37°C, respectively. (G) Typical recovery curves (n = 6) of the FRAP measurements of NA mRNA in influenza virus-infected MDCK cell nuclei at 37°C after ATP depletion. (H) Typical recovery curves (n = 6) of the FRAP measurements of NA mRNA in influenza virus-infected MDCK cell cytoplasm at 37°C after ATP depletion. The bleached areas are indicated by arrowheads and rectangles. Bars, 10 µm.
Table 2.
Diffusion times and coefficients measured for influenza A virus NA mRNA
| Samples | t1/2 recovery (s) | D (µm2/s) |
|---|---|---|
| NA mRNA (37°C) in nucleus | 22 | 0.027 ± 0.016 |
| NA mRNA (37°C) in cytoplasm | 30 | 0.018 ± 0.003 |
| NA mRNA (23°C) in nucleus | 30 | 0.013 ± 0.007 |
| NA mRNA (23°C) in cytoplasm | 35 | 0.009 ± 0.003 |
| NA mRNA (37°C) in nucleus—ATP depleted | 30 | 0.014 ± 0.005 |
| NA mRNA (37°C) in cytoplasm—ATP depleted | 38 | 0.008 ± 0.003 |
Values for t1/2 and D were determined as described in Materials and methods section.
D-values represent mean ± SEM.
To investigate whether the transport of influenza A virus mRNA was energy dependent or not, FRAP experiments were also performed for influenza A virus NA mRNA in influenza A virus-infected MDCK cells maintained at 23°C, according to methods previously described (27,32). Images of typical acquisitions are shown in Figure 4C and D. The recovery curves, each averaged from six repeats, showed a slower recovery of fluorescence in both nucleus and cytoplasm at 23°C compared with 37°C (Figure 4E and F). Significantly, not >30% fluorescence recovery was measured in both nucleus and cytoplasm at 23°C. Furthermore, the recovery rate appeared a little slower: t1/2 ≈ 30 s, D ≈ 0.013 µm2/s in the nucleus; t1/2 ≈ 35 s, D ≈ 0.009 µm2/s in the cytoplasm (Table 2). This result clearly indicates that the mobility of influenza A virus intronless NA mRNA is sensitive to temperature conditions and may suggest that the intracellular transport of influenza A virus mRNA also requires energy. To further establish that the mobility of influenza A virus mRNA is mediated by active process, we reduced ATP levels by treating cells with 10 mM sodium azide plus 20 mM 2-deoxyglucose in the absence of glucose for 15–30 min. Then FRAP experiments were performed at 37°C and the recovery curves, each averaged from six repeats, showed only about 20% fluorescence recovery in both nucleus and cytoplasm (Figure 4G and H). Furthermore, the recovery rate appeared significantly slower: t1/2 ≈ 30 s, D ≈ 0.014 µm2/s in the nucleus; t1/2 ≈ 38 s, D ≈ 0.008 µm2/s in the cytoplasm (Table 2). Thus, the results in Figure 4E–H together clearly indicate that active transport processes may be responsible for the transport of influenza A virus mRNAs in living cells.
ActD treatment could inhibit influenza A virus mRNA export in living cells
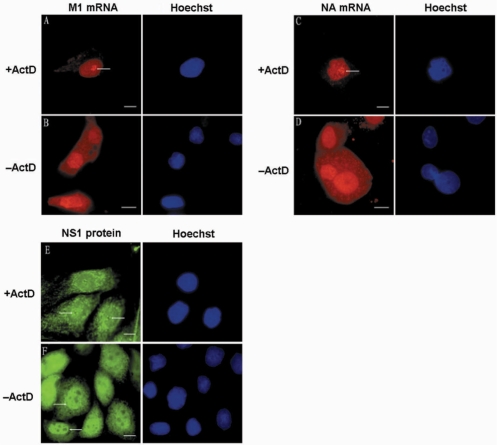
It is well established that influenza A virus replication depends on the function of host cell RNAP-II. Some drugs that can inhibit the functions of RNAP-II, such as DRB and ActD, have been reported to inhibit influenza A virus segment 7 mRNA export from cell nucleus (25). Here, we investigated the effect of ActD treatment on influenza A virus mRNA export in living cells. First, MDCK cells were infected by PR/8 virus with a MOI of 1.0–10.0, and ActD was added at 1.5 h p.i. Cells were then delivered with M1 MB by SLO at 3.5 h p.i., and imaged at 4.5 h p.i. As shown in Figure 5A and B, M1 mRNA distributed in both the cytoplasm and the nucleus of the untreated cells, while in cells treated with ActD, M1 mRNA was only located in the nucleus, particularly in the perinucleoli area. This result showed that ActD largely blocked the nuclear export of M1 mRNA. Furthermore, after 2 h of treatment with ActD, the nuclear export of influenza A virus intronless NA mRNA was also inhibited and NA mRNA also concentrated in the nucleoli cap (Figure 5C and D). This suggests that the nucleocytoplasmic transport of both influenza A virus intron-containing and intronless mRNAs should be related to the function of RNAP-II. As the viral NS1 protein was reported involved in cellular intron-containing mRNA splicing and nuclear export, we also examined its cellular distribution in ActD-treated MDCK cells to investigate if it was also associated with influenza A virus mRNAs. Influenza A virus-infected MDCK cells were treated with ActD from 1.5 to 3.5 h p.i., and the cells observed following immunofluorescence analysis of viral NS1 protein. As shown in Figure 5E and F, after 2 h treatment of ActD, NS1 was concentrated in the peri-nucleoli area, which was very similar to the localization of influenza A virus NA and M1 mRNAs after ActD treatment. Besides that, the levels of NS1 in the nucleoplasm were largely decreased compared with that in nondrug-treated cells, which indicates that the import of NS1 was also inhibited by ActD treatment. The relocalization of NS1 protein after ActD treatment might also relate to the inhibition effect of ActD on influenza A virus mRNA export.
Figure 5.
ActD treatment could inhibit influenza A virus mRNA export and change virus mRNA localization. (A and B) MDCK cells were infected with PR/8 and incubated without drug or with ActD from 90 min p.i., until 3.5 h p.i. when they were delivered with M1 MBs for influenza virus M1 mRNA detection. (C and D) MDCK cells were infected with PR/8 and incubated without drug or with ActD from 90 min p.i., until 3.5 h p.i. when they were delivered with NA MBs for influenza virus NA mRNA detection. (E and F) Distribution of viral NS1 protein in fixed influenza virus-infected MDCK cells without drug or with 2 h ActD treatment. The nucleoli cap positions are indicated by arrowheads. Bars, 10 µm.
Influenza A virus mRNA export might not depend on the function of CRM1
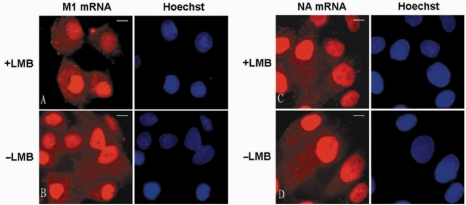
CRM1, an evolutionarily conserved protein, is a receptor for leucine rich nuclear export signal-dependent protein transport and is also involved in mRNA export in eukaryotic cells (33). In addition, CRM1 is also involved in some viral mRNA export, such as HIV mRNA nucleocytoplasmic transport mediated by REV protein (34,35). In this study, using LMB to inhibit the function of CRM1 in living cells, we investigated the role of CRM1 in influenza A virus mRNA export. First, MDCK cells were infected by PR/8 virus with a MOI of 1.0–10.0, and LMB then added at 0.5 h p.i. At 3.5 h p.i., cells were delivered with M1 MBs by SLO, and were imaged at about 4.5 h p.i. As shown in Figure 6A and B, M1 mRNA distributed in both nucleus and cytoplasm after LMB treatment, in the same way as in nondrugs-treated cells. This indicates that the nucleocytoplasmic transport of influenza A virus M1 mRNA could not be inhibited by LMB treatment. In addition, LMB inhibition of influenza A virus intronless NA mRNA also showed that the NA mRNA distribution in LMB-treated MDCK cells was not significantly different compared with that in untreated cells (Figure 6C and D). Hence, the nucleocytoplasmic transport of influenza A virus NA mRNA was not affected by the inhibition of CRM1. Moreover, immunoprecipitation experiments of CRM1 protein were performed in vitro, and showed that influenza A virus NA and M1 mRNA could not be detected in the coimmunoprecipitation samples (data not shown). The results indicate that there was no obvious physical association between CRM1 and influenza A virus NA and M1 mRNAs. Together, these results suggest that the nuclear export of intron-containing and intronless influenza A virus mRNAs may not depend on the CRM1 pathway of the host cell.
Figure 6.
Influenza virus mRNA export was not dependent on CRM1. (A and B) MDCK cells were infected with PR/8 and incubated without drug or with LMB from 30 min p.i., until 3.5 h p.i. when they were delivered with M1 MBs for influenza virus M1 mRNA detection. (C and D) MDCK cells were infected with PR/8 and incubated without drug or with LMB from 30 min p.i., until 3.5 h p.i. when they were delivered with NA MBs for influenza virus NA mRNA detection. Bars, 10 µm.
All three types of influenza A virus mRNA could colocalize with NS1 protein in the nucleus
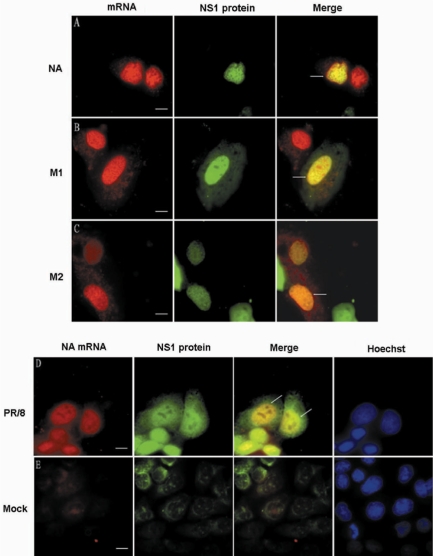
Influenza A virus NS1 protein can inhibit the splicing and nucleocytoplasmic transport of cellular mRNA and might also play roles in the splicing and transport of viral intron-containing NS1 mRNA (2,14,36–38). Our experiments above showed that NS1 protein and influenza A virus mRNA concentrated in the nucleolar cap in ActD-treated MDCK cells. Here, combining the live-cell imaging of both viral mRNAs and NS1 protein in influenza A virus-infected MDCK cells, we investigated the association between NS1 protein and influenza A virus mRNAs. First, the MDCK cells were transiently transfected with plasmid pEGFP-C1-NS1, which contains the coding sequence for NS1 protein. Sixteen hours after transfection, the MDCK cells were infected with PR/8 virus at a MOI of 1.0–10.0. MBs were then delivered into the influenza virus-infected cells by SLO at 3.5 h p.i. About 1 h after MB transfection, the fluorescence of NS1 protein and MBs hybridized with influenza A virus mRNAs were imaged in the same cells. As shown in Figure 7A–C, influenza A virus NA, M1 and M2 mRNAs were all colocalized with NS1 protein in the nucleus. The colocalization coefficients were measured by software ImageJ (NIH) v.1.33u according to methods previously described (28), and the high Pearson's correlation coefficients indicated good colocalization (Table 3). The colocalization of NS1 protein and M2 mRNA might be correlated with NS1 inhibition of the splicing and export of intron-containing mRNAs, but the colocalization of intronless NA mRNA with NS1 protein was very surprising. Simultaneous labeling of both NA mRNA and NS1 protein in fixed influenza A virus-infected MDCK cells was also carried out to confirm the results of the live-cell imaging. Thirty minutes after live-cell MB hybridization with NA mRNA, cells were fixed and NS1 protein was labeled by immunofluorescence. As shown in Figure 7D, the NA mRNA could also be found colocalized with NS1 in the cell nucleus of fixed influenza A virus-infected MDCK cells. Furthermore, as expected, the M1 and M2 mRNAs could also be colocalized with NS1 protein in fixed MDCK nuclei (data not shown). As a control, there was very little signal of both influenza A virus NA mRNA and NS1 protein in fixed nonvirus-infected MDCK cells (Figure 7E). Together, these results show that not only intron-containing but also intronless mRNA of influenza A virus may interact with NS1 protein in cell nucleus.
Figure 7.
Colocalization of influenza A virus mRNA with NS1 protein in influenza virus-infected MDCK nuclei. (A–C) NA, M1 and M2 mRNAs could colocalize with NS1 protein in living influenza A virus-infected MDCK cells. (D) NA mRNA could colocalize with NS1 protein in fixed influenza A virus-infected MDCK cells. (E) Low fluorescence background of NA mRNA and NS1 protein in fixed nonvirus-infected MDCK cells. Bars, 10 µm.
Table 3.
Colocalization and correlation coefficients for influenza A virus mRNAs with viral NS1 protein and cellular TAP proteina
| Objects | M1b | M2c | Rpd | Re |
|---|---|---|---|---|
| NA mRNA and NS1 protein | 0.448 | 0.923 | 0.696 | 0.708 |
| M1 mRNA and NS1 protein | 0.622 | 0.966 | 0.718 | 0.767 |
| M2 mRNA and NS1 protein | 0.440 | 1.000 | 0.440 | 0.542 |
| NA mRNA and TAP protein | 0.851 | 0.927 | 0.936 | 0.944 |
| M1 mRNA and TAP protein | 0.352 | 0.998 | 0.740 | 0.732 |
| M2 mRNA and TAP protein | 1.000 | 0.655 | 0.953 | 0.945 |
aThe colocalization of influenza A virus mRNAs with NS1 protein or TAP protein was analyzed using ImageJ (NIH) v.1.33u software as described in Materials and methods section.
bThe colocalization coefficients for red channel (NA, M1 or M2 mRNA).
cThe colocalization coefficients for green channel (NS1 protein or TAP protein).
dThe Pearson's correlation coefficients for each object colocalization.
eThe overlap coefficients for each object colocalization.
All three types of influenza A virus mRNA could colocalize with cellular TAP protein in the nucleus
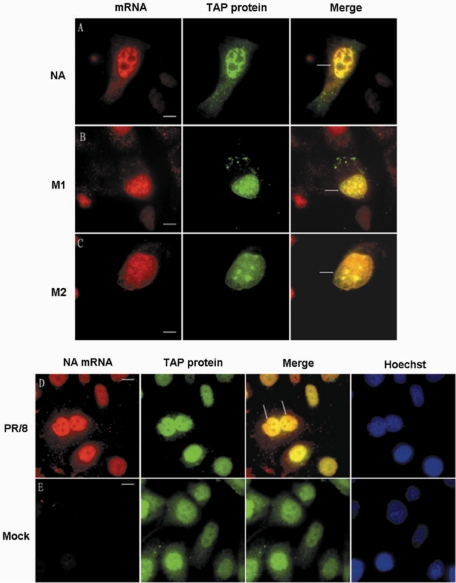
We discussed above that CRM1 may not be involved in influenza A virus mRNA export, so influenza A virus mRNAs may utilize another cellular mRNA export pathway, the TAP/p15 pathway. TAP protein is a very important factor involved in cellular mRNA export and some virus mRNA export (3–5,39). Influenza A virus spliced NS2 and M2 mRNA may be able to use the standard EJC/TAP-p15-dependent mRNA export pathway, just as cellular intron-containing mRNAs do (2). To further uncover the roles of TAP protein in influenza A virus mRNA export, especially intronless mRNA export, we visualized the cellular localization of influenza A virus mRNA and TAP protein in same living influenza A virus-infected MDCK cells. First, the MDCK cells were transiently transfected with plasmid pCDNA3.1-TAP-GFP, which contains the TAP coding sequence. Sixteen hours after transfection, the MDCK cells were infected with PR/8 virus and delivered with NA, M1 or M2 MBs by SLO at 3.5 h p.i. As shown in Figure 8A–C, the three kinds of influenza A virus mRNA, including the intronless NA mRNA, could all colocalize with TAP protein in the nucleus. The colocalization was also analyzed quantitatively using software ImageJ (NIH) v.1.33u (Table 3). As in living cells, the influenza virus mRNA could be found colocalized with TAP protein in fixed influenza A virus-infected MDCK cells (Figure 8D). In noninfected MDCK cells, there was obvious signal of TAP protein and only very little background signal from NA mRNA in fixed cell nuclei (Figure 8E). These results indicate that TAP protein may interact with both intron-containing and intronless mRNAs of influenza A virus in cell nucleus.
Figure 8.
Colocalization of influenza A virus mRNA with TAP protein in influenza virus-infected MDCK cell nuclei. (A–C) NA, M1 and M2 mRNAs could colocalize with TAP protein in living influenza A virus-infected MDCK cells. (D) NA mRNA could colocalize with TAP protein in fixed influenza A virus-infected MDCK cells. (E) Obvious fluorescence distribution of cellular TAP protein and very low fluorescence background of virus NA mRNA in fixed nonvirus-infected MDCK cells. Bars, 10 µm.
Coimmunoprecipitation of influenza A virus mRNAs with NS1 or TAP protein
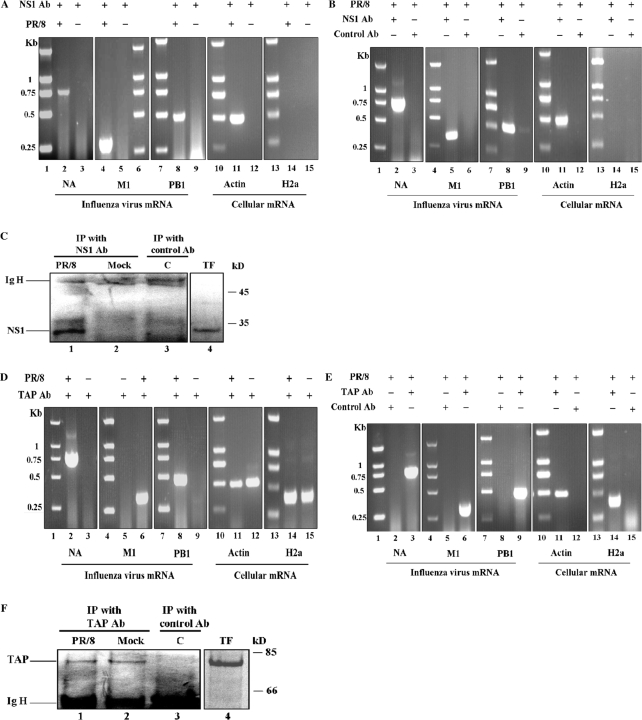
To find more direct evidence that NS1 or TAP protein were involved in the transport of influenza A virus mRNAs, immunoprecipitation experiments of mRNA–protein complexes were performed using anti-NS1 and anti-TAP antibodies. For this analysis, nuclear extracts were prepared from influenza A virus-infected MDCK cells at 5 h p.i. and immunoprecipitated with anti-NS1 antibody. The RNA extracted from the immunoprecipitation samples was used for RT–PCR analysis of the presence of influenza A virus NA, M1 and PB1 mRNAs. The results in Figure 9A and B indicated that influenza A virus NA, M1 and PB1 mRNA could be detected from the infected coimmunoprecipitation sample pulled down by anti-NS1 antibody but not from the sample pulled down by control antibody. There was also no influenza A virus mRNA present in the noninfection coimmunoprecipitation samples pulled down by anti-NS1 antibody. In contrast, not cellular intronless H2a mRNA but spliced β-actin mRNA was detected from the infected coimmunoprecipitation sample pulled down by anti-NS1 antibody (Figure 9A and B). The association of NS1 protein with spliced β-actin mRNA may be due to the interaction of NS1 protein with splicing complexes in cell nucleus. Nuclear extracts were also prepared from influenza A virus-infected 293T cells at 5 h p.i. and immunoprecipitated with anti-TAP antibody to investigate the association of influenza A virus mRNA with TAP. And as shown in Figure 9D and E, the results were very similar to that of coimmunoprecipitation by anti-NS1 antibody. Influenza A virus NA, M1 and PB1 mRNA could only be detected from the infected coimmunoprecipitation sample pulled down by anti-TAP antibody. There were no influenza A virus NA, M1 and PB1 mRNAs in noninfection or negative control coimmunoprecipitation samples. As control, cellular spliced β-actin mRNA and intronless H2a mRNA which were reported to be exported by TAP/p15 pathway (40), could all be detected from the infected or noninfected coimmunoprecipitation samples pulled down by anti-TAP antibody. Moreover, the presence of NS1 or TAP protein pulled down by anti-NS1 antibody or by anti-TAP antibody in infected coimmunoprecipitation samples was confirmed by western blot analysis, as shown in Figure 9C and F. These results further confirmed that influenza A virus NA, M1 and PB1 mRNAs could all be associated with influenza A virus NS1 protein and cellular TAP protein in cell nucleus.
Figure 9.
Coimmunoprecipitation of influenza A virus mRNAs with viral NS1 protein or cellular TAP protein. (A and B) Influenza A virus NS1 protein was pulled down with anti-NS1 antibody from nuclear extract of influenza A virus-infected MDCK cells. Then RNA was purified from the immunoprecipitated complexes and used for RT–PCR analysis of the presence of NA, M1 and PB1 mRNAs. Molecular size standards in kilobase are indicated to the left. (C) Western blot analysis to show specificity of anti-NS1 antibody used in this study. All protein samples were separated by SDS–15% PAGE and analyzed by western blotting. C: IP with control antibody in PR/8 virus-infected cell nuclear extract; PR/8: IP with anti-NS1 antibody in PR/8 virus-infected cell nuclear extract; Mock: IP with anti-NS1 antibody in nonvirus-infected cell nuclear extract. TF: Total extract of MDCK cells transfected with pCDNA3.1-NS1. (D–E) TAP protein was pulled down with anti-TAP antibody from nuclear extract of influenza A virus-infected 293T cells. Then RNA was purified from the immunoprecipitated complexes and used for RT–PCR analysis of the presence of NA, M1 and PB1 mRNAs. Molecular size standards in kilobase are indicated at the left. (F) Western blot analysis to show specificity of anti-TAP antibody used in this study. All protein samples were separated by SDS–10% PAGE and analyzed by western blotting. C: IP with control antibody in PR/8 virus-infected cell nuclear extract; PR/8: IP with anti-TAP antibody in PR/8 virus-infected cell nuclear extract; Mock: IP with anti-TAP antibody in nonvirus-infected cell nuclear extract. TF: Total extract of 293T cells transfected with pCDNA3.1-TAP. The position of heavy chain of the IgG used for immunoprecipitation is indicated to the left and the position of molecular weight marker (in kilodalton) is indicated to the right. All DNA markers were DL2000 (Takara).
DISCUSSION
Elucidating the dynamic behaviors of influenza A virus mRNAs in living cells is crucial to better understanding the mechanisms of its transport. In this report, by delivering specific MBs into living influenza A virus-infected MDCK cells, influenza A virus NA, M1 and M2 mRNAs were successfully visualized in living host cells. Consistent with previous studies, MBs were testified to be a powerful tool for imaging viral RNA in living cells. The reversible permeabilization of cells by SLO was also confirmed to be a good method to deliver MBs into living cells, as previously described (23,26). The mRNA imaging was mainly focused on the initial phase of probe uptake by cells, within 1 h, to ensure the reliability of the results, and we also optimized the dose of MBs and the virus MOI to improve the signal-to-background ratio. Moreover, the MBs targeted for influenza A virus mRNA might also detect the positive sense virus cRNA in nucleus, but the cRNA only represents a minor proportion (5–10%) of the total positive sense RNA in infected cells (25) and cRNA might be encapsidated with viral NP protein (41), so the virus cRNA is unlikely to significantly interfere with viral mRNA detection in living cells. The distribution changes of influenza A virus mRNA was imaged at different p.i. time points and the results indicated that influenza A virus NA, M1 and M2 mRNA have a peak of production at about 4.5 h p.i. (Figure 2A–C and E), which was in concern with the results described earlier (31).
The confocal-FRAP measurements of influenza A virus intronless NA mRNA revealed that about 50% of NA mRNA diffused freely within the nucleus and cytoplasm, with a diffusion coefficient (∼0.027 μm2/s) in the nucleoplasm similar to that of poly(A)+ RNA (∼0.04 μm2/s) (27). The fluorescence recovery rates in the cytoplasm were a little slower than those in nucleus, with only a diffusion coefficient of about 0.018 µm2/s (Figure 4A, B, E and F). These differences of recovery rates in cytoplasm and nucleus are very similar to those of cellular endogenous RNAs (42). These differences may be due to NA mRNA anchoring to microtubules or to the ER membrane for translation in the cytoplasm, just as cellular mRNAs with signal peptides do. FRAP experiments with NA control probe in noninfected MDCK cells were also performed and the results revealed the free NA control probe moved much faster through the nucleus (D ≈ 0.22 ± 0.06 µm2/s) compared with the NA MB in influenza A virus-infected cells. These results strongly indicated that the mobility characters measured by FRAP analysis truly reflect the dynamic behaviors of influenza A virus NA mRNA. In addition, we also found that active transport may be required for intracellular transport of influenza A virus intronless NA mRNA by investigating effect of low temperature and ATP depletion on the mobility of NA mRNA. The very low diffusion coefficient (0.013 µm2/s in nucleoplasm; 0.009 µm2/s in cytoplasm) and not >30% fluorescence recovery at 23°C (see Figure 4C–F) together suggested that influenza A virus NA mRNA transport in both nucleus and cytoplasm is sensitive to temperature conditions. ATP depletion also significantly decreased the portion of mobile fraction (only 20%) and reduced the mobility of NA mRNA (0.014 µm2/s in nucleoplasm; 0.008 µm2/s in cytoplasm) in both nucleus and cytoplasm. Some previous researches reported that the mobility of mRNP particles in nucleus was not directed but was governed by simple diffusion within interchromatin spaces and ATP is only required for the complexes to resume their motion after they become stalled at dense chromatin (42–44). The decreased mobility of influenza A virus NA mRNA in nucleus at low temperature might be related to the increased viscosity of nuclei to some extent. But the ATP depletion could decrease both mobile fraction and diffusion coefficients of influenza virus NA mRNA in both nucleus and cytoplasm was different from that of cellular mRNP particles described previously (42). So the transport mechanism of influenza A virus mRNA in nucleus may differ from that of some cellular mRNP particles. In another words, the transport of influenza A virus mRNA in both nucleus and cytoplasm is an energy-dependent process as is that of some poly(A)+ RNAs reported before (27,45).
It has been reported that influenza A virus segment 7 mRNA nuclear export requires host cell RNAP-II function (25). In this study, we also investigated the function of RNAP-II in influenza A virus mRNA export by using ActD inhibition of RNAP-II, combined with live-cell imaging of influenza A virus mRNAs. The nuclear export of both intron-containing M1 mRNA and intronless NA mRNA were all largely blocked in living virus-infected MDCK cells treated with ActD. Moreover, PB1 mRNA was also inhibited to some extent by ActD treatment (data not shown). This suggests that the export of both influenza A virus intron-containing mRNA and intronless mRNA is related to the function of RNAP-II. There may be the possibility that some cellular or viral proteins needed for efficient transport of viral mRNAs are recruited to the mRNAs in the presence of functional RNAPII but not in its absence. It has been reported that a group of nucleoplasmic proteins, mostly RNA-binding proteins, relocalize from the nucleoplasm to a specific nucleolar cap during transcriptional inhibition (46). Viral NS1 protein was found concentrated to the nucleoli cap after ActD treatment in influenza virus-infected MDCK cells and the import of NS1 was also inhibited by ActD treatment (Figure 5E and F). The relocalization of NS1 protein after ActD treatment might relate to the inhibition of ActD on influenza A virus mRNAs export.
The CRM1 pathway was the first RNA export pathway to be elucidated, and is involved in mRNA export in eukaryotic cells (5,33). In this study, we investigated if the CRM1 pathway was involved in influenza A virus mRNA export by using LMB inhibition in living cells. The nuclear export of both influenza A virus intron-containing M1 mRNA and intronless NA mRNA could not be inhibited by LMB treatment in living cells (Figure 5A–D). The export of intronless PB1 mRNA also could not be blocked by LMB treatment (data not shown). Moreover, the results of in vitro CRM1 immunoprecipitaion indicated that CRM1 could not associate with influenza A virus NA and M1 mRNAs (data not shown). All these results suggest that both intron-containing and intronless influenza A virus mRNAs are exported from MDCK cell nuclei by a CRM1 independent pathway.
NS1 is a very important multifunctional nonstructural protein, which can inhibit cellular intron-containing mRNA splicing and export by binding with CPSF and PABPII, or by forming an inhibitory complex with TAP/p15 (1,14). Moreover, NS1 protein can also inhibit the splicing and export of its own mRNA in an RNA binding-dependent manner (2). In this report, we combined the live-cell imaging of both influenza A virus mRNAs and NS1 protein to explore the roles of NS1 in the transport of influenza A virus mRNA, especially in the export of intronless mRNAs. The results showed that NS1 could colocalize with all three types of influenza A virus mRNA including the intronless NA mRNA (Figure 7A–C). Simultaneous labeling of virus mRNAs and NS1 protein in fixed cells further confirmed colocalization. Furthermore, influenza A virus NA, M1 and PB1 mRNAs could be coimmunoprecipitated with NS1 protein by pull-down with anti-NS1 antibody (Figure 9A and B). The coimmunoprecipitation experiments further testified the association of NS1 with influenza A virus mRNA in the nucleus. NS1 protein was reported to interact with influenza A viral transcription–replication complexes in infected cells (47) and be cross-linked to influenza A mRNAs in cytoplasm for enhancement of viral protein translation (48,49). Recently, NS1 protein was also found to be able to selectively bind to influenza A virus NS1 mRNA (2). Together with our results, these might suggest that NS1 protein could also associate with all three kinds of influenza A virus mRNAs in the nucleus and this association may start from the initiation of virus transcription by first interacting with viral transcription–replication complexes. Moreover, the association of NS1 with influenza A virus intronless mRNAs in the nucleus could not be simply interpreted by the inhibition of NS1 on virus mRNA, like the roles of NS1 protein in NS1 and NS2 mRNAs export. NS1 protein might also play some active roles in the export of virus intronless mRNAs by promoting the recruitment of cellular mRNA transport factors to influenza A virus intronless mRNAs. Lastly, the roles of NS1 protein in influenza A virus mRNA transport may be different at different infection times.
TAP/NXF1 is, to date, the most important receptor involved in the export of mRNA from the nucleus to the cytoplasm (12,39,50). Some DNA and RNA viruses also use the TAP/p15 pathway to transport their own mRNA from the nucleus to the cytoplasm, such as HSV (5). We also investigated whether TAP protein is involved in the nuclear export of influenza A virus mRNA. Live-cell imaging of both influenza A virus mRNA and TAP protein revealed that TAP could colocalize not only with intron-containing mRNAs but also with intronless NA mRNA (Figure 8A–C). Simultaneous labeling of virus mRNA and TAP protein in fixed cells also confirmed that TAP could colocalize with influenza A virus mRNA. Moreover, coimmunoprecipitation of TAP protein and influenza virus mRNPs with anti-TAP antibody indicated physical association of TAP and influenza virus mRNA (Figure 9D and E). Together, these data suggest the association of TAP protein with the three kinds of influenza A virus mRNAs. However, considering that influenza A virus intronless mRNAs cannot recruit TAP protein by splicing complexes like intron-containing cellular mRNAs do, there may be some viral or cellular factors bridging TAP to the influenza virus intronless mRNAs, in a similar manner to the role of ICP27 in HSV intronless mRNA export (4). In conclusion, it was revealed that TAP protein might be involved in influenza A virus mRNA export by live-cell imaging and by in vitro analysis, but how influenza A virus intronless mRNAs recruit TAP protein and the accurate mechanisms of transport need to be further elucidated.
In conclusion, the distribution and dynamic behaviors of influenza A virus mRNAs in living virus-infected cells were successfully visualized. The export of influenza A virus mRNA was found to be independent of the CRM1 pathway, while RNAP-II is involved. Imaging analysis and coimmunoprecipitation experiments uncovered that influenza A virus mRNAs associate with viral NS1 and cellular TAP protein in the nucleus. These findings suggest that the influenza A virus mRNA may be exported from the nucleus by the cellular TAP/p15 pathway with NS1 protein and RNAP-II participation.
ACKNOWLEDGEMENTS
The authors would like to thank Prof. J. Ortín for the kind gift of anti-NS1 polyclonal antibody. We thank Prof. R.M. Sandri-Goldin., Prof. B.R. Cullen. and Prof. M.L. Hammarskjöld for generously providing plasmids encoding human TAP. Z.-Q.C. is supported by National Basic Research Program of China (2006CB933102) and the National Natural Science Foundation of China (30700169). Z.C. is supported by National Basic Research Program of China (2005CB523007 and 2006CB933102). The others are supported by the Nano Projects of the Chinese Academy of Sciences (kjcx2-sw-h12). Funding to pay the Open Access publication charges for this article was provided by Chinese Academy of Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chen Z, Krug RM. Selective nuclear export of viral mRNAs in influenza-virus-infected cells. Trends Microbiol. 2000;8:376–383. doi: 10.1016/s0966-842x(00)01794-7. [DOI] [PubMed] [Google Scholar]
- 2.Garaigorta U, Ortín J. Mutation analysis of a recombinant NS replicon shows that influenza virus NS1 protein blocks the splicing and nucleo-cytoplasmic transport of its own viral mRNA. Nucleic Acids Res. 2007;35:4573–4582. doi: 10.1093/nar/gkm230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 4.Chen I-HB, Sciabica KS, Sandri-Goldin RM. ICP27 Interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 2002;76:12877–12889. doi: 10.1128/JVI.76.24.12877-12889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandri-Goldin RM. Viral regulation of mRNA Export. J. Virol. 2004;78:4389–4396. doi: 10.1128/JVI.78.9.4389-4396.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soliman TM, Silverstein SJ. Herpesvirus mRNAs are sorted for export via CRM1-dependent and -independent pathways. J. Virol. 2000;74:2814–2825. doi: 10.1128/jvi.74.6.2814-2825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullen BR. Nuclear RNA export pathways. Mol. Cell. Biol. 2000;20:4181–4187. doi: 10.1128/mcb.20.12.4181-4187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neville M, Rosbash M. The NES-Crm1 export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 1999;18:3746–3756. doi: 10.1093/emboj/18.13.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bear J, Tan W, Zolotukhin AS, Tabernero C, Hudson EA, Felber BK. Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol. Cell. Biol. 1999;19:6306–6317. doi: 10.1128/mcb.19.9.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang Y, Cullen BR. The human tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katahira J, Straber K, Podtelejnikov A, Mann M, Jung JU, Hurt E. The Mex-67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachi A, Braun IC, Rodrigues JP, Pante N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Gorlich D, Carmo-Fonseca M, et al. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bacchi A, Wilm M, Felber BK, Izaurralde E. TAP, the human homologue of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 14.Satterly N, Tsai PL, Deursen JV, Nussenzveig DR, Wang YM, Faria PA, Levay A, Levy DE, Fontoura BMA. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl Acad. Sci. USA. 2007;104:1853–1858. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bratu DP, Cha B, Mhlanga MM, Kramer FR, Tyagi S. Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl Acad. Sci. USA. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyagi S, Alsmadi O. Imaging Native b-Actin mRNA in Motile Fibroblasts. Biophys. J. 2004;87:4153–4162. doi: 10.1529/biophysj.104.045153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santangelo PJ, Nix B, Tsourkas A, Bao G. Dual FRET molecular beacons for mRNA detection in living cells. Nucleic Acids Res. 2004;32:e57. doi: 10.1093/nar/gnh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nitin N, Santangelo PJ, Kim G, Nie SM, Bao G. Peptide-linked molecular beacons for efficient delivery and rapid mRNA detection in living cells. Nucleic Acids Res. 2004;32:e58. doi: 10.1093/nar/gnh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui ZQ, Zhang ZP, Zhang XE, Wen JK, Zhou YF, Xie WH. Visualizing the dynamic behavior of poliovirus plus-strand RNA in living host cells. Nucleic Acids Res. 2005;33:3245–3252. doi: 10.1093/nar/gki629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mhlanga MM, Vargas DY, Fung CW, Kramer FR, Tyagi S. tRNA-linked molecular beacons for imaging mRNAs in the cytoplasm of living cells. Nucleic Acids Res. 2005;33:1902–1912. doi: 10.1093/nar/gki302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santangelo P, Nitin N, LaConte L, Woolums A, Bao G. Live-cell characterization and analysis of a clinical isolate of bovine respiratory syncytial virus, using molecular beacons. J. Virol. 2006;80:682–688. doi: 10.1128/JVI.80.2.682-688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walev I, Bhakdi SC, Hofmann F, Djonder N, Valeva A, Aktories K, Bhakdi S. Delivery of proteins into living cells by reversible membrane permeabilization with streptolysin-O. Proc. Natl Acad. Sci. USA. 2001;98:3185–3190. doi: 10.1073/pnas.051429498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faria M, Spiller DG, Dubertret C, Nelson JS, White MRH, Scherman D, Hélène C, Giovannangeli C. Phosphoramidate oligonucleotides as potent antisense molecules in cells and in vivo. Nat. Biotechnol. 2001;19:40–44. doi: 10.1038/83489. [DOI] [PubMed] [Google Scholar]
- 24.Singer RH. In situ hybridization of mammalian cells (RNA and Oligonucleotide probes) 1998. http://www.singerlab.org/protocols.
- 25.Amorim MJ, Reed EK, Dalton RM, Medcalf L, Digard P. Nuclear Export of Influenza A virus mRNAs Requires Ongoing RNA Polymerase II Activity. Traffic. 2007;8:1–11. doi: 10.1111/j.1600-0854.2006.00507.x. [DOI] [PubMed] [Google Scholar]
- 26.Santangelo PJ, Bao G. Dynamics of filamentous viral RNPs prior to egress. Nucleic Acids Res. 2007;35:3602–3611. doi: 10.1093/nar/gkm246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molenaar C, Abdulle A, Gena A, Tanke HJ, Dirks RW. Poly(A)+ RNAs roam the cell nucleus and pass through speckle domains in transcriptionally active and inactive cells. J. cell Biol. 2004;165:191–202. doi: 10.1083/jcb.200310139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura T, Hashimoto I, Nagase T, Fujisawa J-I. CRM1-dependent, but not ARE-mediated, nuclear export of IFN-a1mRNA. J. Cell Sci. 2004;117:2259–2270. doi: 10.1242/jcs.01076. [DOI] [PubMed] [Google Scholar]
- 29.Peritz T, Zeng FY, Kannanayaka TJ, Kilk K, Eiríksdóttir E, Langel U, Eberwine J. Immunoprecipitation of mRNA-protein complexes. Nat. protocols. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- 30.Leonetti JP, Mechti N, Degols G, Gagnor C, Lebleu B. Intracellular distribution of microinjected antisense oligonucleotide. Proc. Natl Acad. Sci. USA. 1991;88:2702–2706. doi: 10.1073/pnas.88.7.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valcárcel J, Portela A, Ortín J. Regulated M1 mRNA splicing in influenza virus-infected cells. J. Gen. Virol. 1991;72:1301–1308. doi: 10.1099/0022-1317-72-6-1301. [DOI] [PubMed] [Google Scholar]
- 32.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe M, Fukuda M, Yoshida M, Yanagida M, Nishida E. Involvement of CRM1, a nuclear export receptor, in mRNA export in mammalian cells and fission yeast. Genes Cells. 1999;4:291–297. doi: 10.1046/j.1365-2443.1999.00259.x. [DOI] [PubMed] [Google Scholar]
- 34.Malim MH, Cullen BR. HIV-1 structural gene expression requires the binding of multiple rev monomers to the viral RRE: implications for latency. Cell. 1991;65:241–248. doi: 10.1016/0092-8674(91)90158-u. [DOI] [PubMed] [Google Scholar]
- 35.Fischer U, Meyer S, Teufel M, Heckel C, Luhrmann R, Rautmann G. Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J. 1994;13:4105–4112. doi: 10.1002/j.1460-2075.1994.tb06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fortes P, Beloso A, Ortín J. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 1994;13:704–712. doi: 10.1002/j.1460-2075.1994.tb06310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu Y, Krug RM. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A) J. Virol. 1994;68:2425–2432. doi: 10.1128/jvi.68.4.2425-2432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen ZY, Li YZ, Krug RM. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 1999;18:2273–2283. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cullen BR. Nuclear RNA export. J. Cell Sci. 2003;116:587–597. doi: 10.1242/jcs.00268. [DOI] [PubMed] [Google Scholar]
- 40.Lai MC, Tarn WY. Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J. Biol. Chem. 2004;279:31745–31749. doi: 10.1074/jbc.C400173200. [DOI] [PubMed] [Google Scholar]
- 41.Portela A, Digard P. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 2002;83:723–734. doi: 10.1099/0022-1317-83-4-723. [DOI] [PubMed] [Google Scholar]
- 42.Vargas DY, Raj A, Marras SAE, Kramer FR, Tyagi S. Mechanism of mRNA transport in the nucleus. Proc. Natl Acad. Sci. USA. 2005;102:17008–17013. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Politz JC, Tuft RA, Pederson T, Singer RH. Movement of nuclear poly(A) RNA throughout the interchromatin space in living cells. Curr. Biol. 1999;9:285–291. doi: 10.1016/s0960-9822(99)80136-5. [DOI] [PubMed] [Google Scholar]
- 44.Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Dynamics of Single mRNPs in Nuclei of Living Cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dargemont C, Kühn LC. Export of mRNA from microinjected nuclei of Xenopus laevis oocytes. J. Cell Biol. 1992;118:1–9. doi: 10.1083/jcb.118.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marión RM, Zürcher T, de la Luna S, Ortín J. Influenza virus NS1 protein interacts with viral transcription–replication complexes in vivo. J. Gen. Virol. 1997;78:2447–2451. doi: 10.1099/0022-1317-78-10-2447. [DOI] [PubMed] [Google Scholar]
- 48.Park YW, Katze MG. Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J. Biol. Chem. 1995;270:28433–28439. doi: 10.1074/jbc.270.47.28433. [DOI] [PubMed] [Google Scholar]
- 49.Burgui I, Aragón T, Ortín J, Nieto A. PABP1 and eIF4GI associate with influenza virus NS1 protein in viral mRNA translation initiation complexes. J. Gen. Virol. 2003;84:3263–3274. doi: 10.1099/vir.0.19487-0. [DOI] [PubMed] [Google Scholar]
- 50.Reed R, Hurt E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell. 2002;108:523–531. doi: 10.1016/s0092-8674(02)00627-x. [DOI] [PubMed] [Google Scholar]