Abstract
Tetraalkylammonium (TAA) derivatives have been reported to serve as stabilizers of asymmetrical cyanine dyes in aqueous solutions and to increase the yield and efficiency of polymerase chain reaction (PCR) detected by end-point analysis. In this study, we compared the ability of various TAA derivatives (with alkyl chain ranging from 1 to 5 carbons) and some other compounds to serve as enhancers of real-time PCR based on fluorescence detection from intercalating dye SYBR Green I (SGI). Our data indicate that TAA chlorides and some other TAA derivatives serve as potent enhancers of SGI-monitored real-time PCR. Optimal results were obtained with 10–16 mM tetrapropylammonium chloride. The effect of TAA compounds was dependent on the nature of counter ions present and composition of the reaction mixtures used. Based on measurements of SGI-generated fluorescence signal in the presence of PCR-amplified DNA fragments, oligonucleotide primers and/or various additives, we propose that TAA-derivatives reduce the binding of SGI to oligonucleotide primers and thus enhance primer–template interactions during annealing phase. Furthermore, these compounds serve as stabilizers of SGI-containing PCR mixtures. The combined data indicate that TAA derivatives might be a new class of additives contributing to robustness of real-time PCR monitored by asymmetrical cyanine dye SGI.
INTRODUCTION
Quantitative real-time PCR is rapidly becoming an important research tool for a variety of analytical and diagnostic applications (1). Sequence-specific fluorescent probes and double-stranded (ds) DNA binding dyes have been used to continuously monitor PCR product formation. Sequence-specific probes allow highly sensitive detection of specific amplification products, but are relatively expensive. On the other hand, dsDNA-binding dyes, such as SYBR Green I (SGI), are inexpensive but also less specific, because they bind to all dsDNA present in PCR mixtures, including nonspecific products and primer-dimers. Although some nonspecific products and primer-dimers could be detected by analysis of melting curves, their presence reduces the sensitivity of real-time PCR monitored by intercalating dyes. To enhance the production of specific PCR products, optimization procedures are often employed (2). These include optimization of concentration of components in PCR mixtures, namely concentration of Mg2+, or DNA polymerase, and design of improved primer sets. However, in some cases nonspecific products are formed even when all PCR parameters are seemingly optimized. A variety of additives and enhancing agents have been tested to increase the specificity, yield and consistency of PCR amplification. These include dimethylsulfoxide (DMSO) (3–5), tetramethylene (TM) sulfone (6), TM sulfoxide (7), N,N,N-trimethylglycine monohydrate (betaine) (8,9), formamide (10–12), tetramethylammonium (TMA) chloride (13,14), TMA oxalate (15), ammonium sulfate (16), acetamide (12), nonionic detergents (17,18), glycerol (19) and trehalose (20,21). Most of these additives have been described to have beneficial effect on PCR amplifications as evaluated by end-point analysis. The efficiency of these agents in real-time PCR based on fluorescence detection from intercalating dyes is mostly unknown. It has been reported that inhibitory effect of SGI on PCR performance could be partially reversed by increased concentration of Mg2+ (22) or inclusion of DMSO (5). Other studies showed that quaternary compounds, such as tetrapentylammonium (TPA) hydroxide, TPA-bromide and tetrabutylammonium (TBA) hydroxide, stabilized fluorescence nucleic acid stains in aqueous solutions (23). Interestingly, tetraalkylammonium (TAA) derivatives also enhanced the stability of SGI in agarose gels used for electrophoresis (23,24). However, current knowledge does not make it possible to predict whether or not these agents would serve as enhancers and/or stabilizers of real-time PCR based on fluorescence detection from intercalating dye SGI.
In this study, we compared various TAA derivatives and some other compounds as to their ability to enhance the performance of SGI-monitored real-time PCR. Our data indicate that TAA derivatives could serve as a new class of additives contributing to robustness of real-time PCR monitored by asymmetrical cyanine dye SGI and serving as stabilizers of SGI-containing PCR reaction mixtures.
MATERIALS AND METHODS
Materials
TMA-Cl, TMA-OH, tetraethylammonium chloride (TEA-Cl), tetrapropylammonium chloride (TPrA-Cl), TPrA-OH, TBA-Cl, TBA-OH, TPA-Cl, TPA-OH, TM sulfoxide, DMSO, formamide, D-(+)-trehalose dihydrate, betaine monohydrate, and glycerol were obtained from Fluka Chemie GmbH (Buchs, Switzerland) or Sigma-Aldrich (Steinheim, Germany). TPA-Cl, TAA-oxalates or TAA-acetates were prepared by neutralizing the corresponding hydroxides with HCl, oxalic acid or acetic acid, respectively. The pH of all reagents used as additives was adjusted to 7.8–8.0. SGI was obtained from Invitrogen (Carlsbad, CA, USA). All other chemicals were from Sigma–Aldrich.
Real-time PCR conditions
Experiments were conducted in Mastercycler ep realplex (Eppendorf AG, Hamburg, Germany) according to the manufacturer's instructions. All reactions were performed in 10 μl reaction volumes in 96-well plates for PCR heat-sealed with heat sealing film (Eppendorf). A standard 1× real-time SGI-supplemented (SSG) PCR mixture contained 10 mM Tris–HCl, pH 8.0 (25°C), 50 mM KCl, 0.1% Triton X-100, 1.5 mM MgCl2, 200 μM each of dATP, dCTP, dGTP and dTTP (dNTPs), 5% DMSO, 25 U/ml of Taq DNA polymerase, protease inhibitors, 0.98 μM SGI (1:20 000 diluted stock of 19.6 mM; molar absorption coefficient of SGI at the absorption maximum (494 nm) is ∼73 000 M−1cm−1 (25)), 0.5 μM primers and cDNA or plasmid DNA. The alternative 1× real-time SGI-supplemented (ASG) PCR mixture contained 75 mM Tris–HCl, pH 8.0 (25°C), 20 mM (NH4)2SO4, 0.01% Tween 20, 2.5 mM MgCl2, 200 μM dNTPs, 6% DMSO, 25 U/ml Taq DNA polymerase, protease inhibitors, 0.98 μM SGI, 0.5 μM primers and cDNA or plasmid DNA. Also used was 1× iQ™ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA) containing 20 mM Tris–HCl, pH 8.4, 50 mM KCl, 3 mM MgCl2, 200 μM dNTPs, 25 U/ml iTaq DNA polymerase, unspecified amount of SGI, 20 nM fluorescein and unspecified stabilizers and 1× QuantiTect SYBR Green PCR kit (QIAGEN GmbH, Hilden, Germany) containing unspecified amount of HotStartTaq DNA polymerase, Tris–HCl, pH 8.7 (20°C), KCl, (NH4)2SO4, 2.5 mM MgCl2, dNTPs including dUTP, SYBR Green I and Rox passive reference dye.
For real-time PCR amplification of genomic DNA fragments we used PCR mixture, denoted GSB, containing at a final 1× concentration 20 mM Tris–HCl, pH 8.8 (25°C), 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, 200 μM dNTPs, 25 U/ml Taq DNA polymerase, protease inhibitors, 0.65 μM SGI, 0.5 μM primers, genomic DNA and 22 nM anti-Taq DNA polymerase monoclonal antibody. Hybridoma cell line producing anti-Taq DNA polymerase antibody (clone 4/C7) was prepared after immunization of BALB/c mice with Thermus aquaticus DNA polymerase, fusion of spleen cells from immunized mice with SP2 myeloma cells as described (26), and selection and cloning of hybridoma cell line producing antibody specific for Taq DNA polymerase as determined by ELISA (27). The anti-Taq/4/C7 antibody, which is of the IgG1 subclass, inhibited the enzymatic activity of Taq DNA polymerase by >95%, as determined by DNA polymerase assay using activated salmon testes DNA as a substrate (27). A 200 bp fragment of human CD4 cloned in plasmid vector (Seegene, Seoul, Korea) was amplified with forward primer 5′-GTCTACCAGGCATTCGCTTCAT-3′ and reverse primer 5′-CTGTGAATGCTGCGACTACGAT-3′. In some experiments, a segment of 171 bps of rat actin cDNA was amplified using forward primer 5′-ACTCTTCCAGCCTTCCTTC-3' and reverse primer 5′-ATCTCCTTCTGCATCCTGTC-3'. For amplification of 864 bp genomic DNA fragment of mouse Thy-1 gene, a Thy-1 primer set was used, forward primer, 5′-ATGAACCCAGCCATCAGCG-3′ and reverse primer 5′-GGGTAAGGACCTTGATATAGG-3′. Thermal cycling consisted of an initial denaturation at 95°C for 2 min followed by 40 cycles of denaturation at 94°C for 15 s, annealing at various temperatures for 15 s, and extension at 72°C for 60 s (for genomic DNA) or 20 s for other amplifications. Commercial real-time PCR master mixes were used according to recommended conditions, including initial DNA polymerase activation step of 15 min at 95°C for QuantiTect SYBR Green PCR kit. Melting curve analysis was carried out from 70°C to 95°C with 0.2°C increments. In some experiments DNA amplicons were visualized on agarose gels stained with ethidium bromide (0.5 μg/ml). Threshold cycle (Ct) values were determined by automated threshold analysis. PCR efficiencies (E), were determined from dilutions of DNA and calculated from the slopes of the standard curves according to the equation, E = 10−1/a−1, where a is the slope of the corresponding standard curve. The specificity of PCR products was checked by agarose gel electrophoresis.
RNA extraction and cDNA synthesis
RNA was extracted from 2H3 clone of rat basophilic leukemia (RBL-2H3) cells cultured under standard conditions (28) using Tri reagent (Sigma–Aldrich). The amount of RNA was determined by spectrophotometer ND-1000 (NanoDrop Technologies, Inc., Wilmington, DE, USA). Single-stranded cDNA was synthesized by means of mouse moloney leukemia virus reverse transcriptase (Invitrogen) according to manufacturer's instructions using 10 µg of isolated RNA and 50 ng of random hexamers per reaction.
Isolation of PCR-generated DNA fragments
A 200 bp fragment of human CD4 was amplified by PCR as described above and isolated through its binding to diatomaceous earth particles in the presence of chaotropic agent guanidine thiocyanate (29). DNA was eluted from the particles with DNAse-free water. Concentration of PCR product was determined by spectrophotometer ND-100.
Genomic DNA
Genomic DNA was isolated from C57BL/6 mouse tails as described (30). The mass of the haploid mouse genome (C-value) is ∼3.3 pg (http://www.genomesize.com) and therefore 1 ng of mouse genomic DNA contains ∼330 copies of a single-copy gene. This number was used for generation of standard curve of Ct values from amplification plots versus log copy number (logQ).
SGI fluorescence measurements
Two-times concentrated SGI solution, containing 20 mM Tris–HCl, pH 8.0, 100 mM KCl, 0.2% Triton X-100 and 1.96 µM SGI, was supplemented with isolated PCR amplicons (final concentration 5 μg/ml), oligonucleotide primers (0.4 μM), DMSO (5%), H2O and/or TAA-derivatives (16 mM) to get 1× concentrated SGI solution. Ten microliter aliquots were transferred to 96-well PCR plates, heat-sealed, and fluorescence reading was carried out on Mastercycler ep realplex at 68°C or 55°C. Alternatively, fluorescence reading was carried out from 35°C to 80°C, recording fluorescence each 0.2°C increment. Fluorescence of SGI was tested with the primers described above, and several other primers including primer for mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH1), 5′-ATGACATCAAGAAGGTGGTG-3′, linker of activated T cells (LAT), 5′-CTGGGGAGCAGCCTTGAGTAG-3′, mouse/rat interleukin-6 (IL6), 5′-AAATAGTCCTTCCTACCCCAA-3′, mouse/human/rat actin 5′-ACTCTTCCAGCCTTCCTTC-3′, poly(A)20, poly(T)20. All primers were checked for hairpin and dimer formation by means of NetPrimer program (http://www.premierbiosoft.com). The primers were used at concentration 0.2 μM.
RESULTS
In initial experiments we used SGI-containing PCR mixture without DMSO (SSG-) and analyzed the effect of three previously described additives, DMSO (5), TM-sulfoxide (7) and glycerol (19), on real-time PCR amplification of CD4 DNA fragment. Ct values at various concentrations of the additives were determined and normalized to the lowest Ct value obtained in a given experiment. In the absence of any additive, PCR mixture based on SSG- failed to give any specific amplification signal indicating that 0.98 μM SGI inhibited the PCR. This conclusion was corroborated by agarose gel electrophoresis where the expected DNA amplicon was observed only in reactions without SGI (not shown). The observed inhibitory effect was not reversed by enhanced MgCl2 concentration up to 5 mM. As expected (5), addition of DMSO enhanced the PCR performance, as inferred from decreased Ct values in a broad range of concentrations (0.2–1.6 M) with the peak at ∼0.8 M (Figure 1). TM sulfoxide raised the PCR performance only slightly less than DMSO but within a much narrower range of concentrations (0.3–0.8 M) peaking at ∼0.4 M. Some improvement was also observed in PCR mixtures supplemented with a broad range of concentrations of glycerol (0.5–1.8 M) with maximum at ∼1 M.]
Figure 1.
The effect of additives on SGI-based qPCR performance. Standard SSG PCR mixtures, without DMSO, were supplemented with various concentrations of glycerol, TM-sulfoxide or DMSO and qPCRs were performed using cloned CD4 as a template. The lowest Ct value, obtained at a concentration of DMSO (0.8 M), was subtracted from Ct values of qPCR with additives at various concentrations (ΔCt). Means ± SD from three experiments are shown.
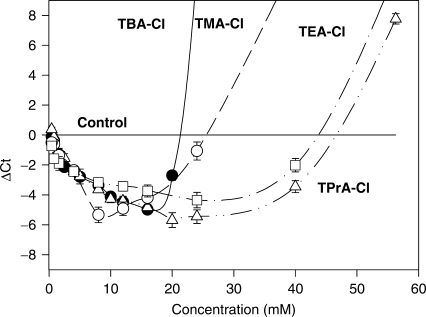
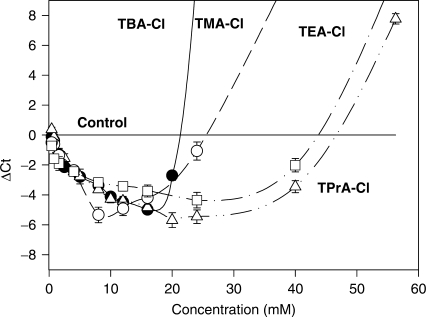
When the SSG- reaction mixtures were supplemented with various concentrations of TMA-Cl or TPrA-Cl, no amplification of DNA fragments was detected by enhanced fluorescence of SGI or agarose gel electrophoresis (not shown). However, if TMA-Cl was added to SSG mixture containing DMSO, a distinct decrease in Ct values was observed (Figure 2). With the size of alkyl chain increasing from 1 to 3 carbons, the range of concentrations of TAA-chlorides enhancing PCR performance rose. Best results were obtained with TPrA-Cl, which was found to enhance PCR performance within a large range of concentrations (0.5–40 mM) with a peak at ∼20 mM (Figure 2).
Figure 2.
Enhanced performance of SGI-based qPCR by TMA-Cl, TEA-Cl, TPrA-Cl or TBA-Cl. Five percent DMSO-containing SSG PCR mixtures were supplemented with various concentrations of the additives and qPCRs were performed using cloned CD4 as a template. The Ct values obtained in PCR mixtures without TAA-Cl additives were subtracted from the Ct values obtained in the presence of additives (ΔCt). Means ± SD from two experiments are shown.
Previously we have found that oxalate anion can enhance the specificity and efficiency of TMA enhancer as detected by PCR end-point analysis (15). In further work we therefore analyzed the real-time PCR performance of SSG mixtures supplemented with TAA derivatives with various anions and CD4 as a template. Data summarized in Table 1 indicate that all TAA-chlorides raised the real-time PCR efficiency. Comparable results were obtained with optimal concentrations of the TAA-Cls with 1–4 carbons in alkyl chain; however, with TPA-Cl, the performance decreased. In contrast, none TAA-oxalates were capable of decreasing Ct values. Acetate anions showed some improvement of PCR but only in TAA derivatives with 3 or 4 alkyl chains and were less potent than chloride anions. Inclusion of betaine also lowered the Ct values, but formamide and D-(+)-trehalose had no enhancing effect. Inclusion of additives at concentrations giving the lowest Ct values also reduced the melting temperature. The observed decrease was dependent on the number of alkyl chains; the lowest and highest decrease was seen with TMA-Cl and TPrA-acetate. However, there was no correlation between the drop in melting temperature and decline in Ct values. The reagents also differed in their ability to inhibit real-time PCR. As shown in Table 1, TPA-Cl inhibited PCR at relatively low concentrations (1.6 ± 0.6 mM), whereas TPrA-Cl was inhibitory at 50.4 ± 5.2 mM.
Table 1.
Effect of tested additives on efficiency and inhibition of real-time PCR
| Additive | ΔCta | ΔMT at maximum efficiencyb | Ct >40c |
|---|---|---|---|
| TMA-Cl | −4.1 ± 0.8 (8–16 mM) | −0.3 | 39.0 ± 7.3 mM |
| TMA-oxalate | N | ||
| TMA-acetate | N | ||
| TEA-Cl | −4.0 ± 0.5 (8–24 mM) | −1.0 | 43.9 ± 9.8 mM |
| TPrA-Cl | −4.6 ± 0.6 (10–40 mM) | −2.0 | 50.4 ± 5.2 mM |
| TPrA-oxalate | N | ||
| TPrA-acetate | −3.8 ± 0.1 (10–20 mM) | −2.4 | 40.5 ± 4.5 mM |
| TBA-Cl | −4.2 ± 0.5 (8–16 mM) | −2.2 | 45.0 ± 3.8 mM |
| TBA-oxalate | N | ||
| TBA-acetate | −2.8 ± 0.4 (2.5–8 mM) | −1.1 | 45.0 ± 5.0 mM |
| TPA-Cl | −1.6 ± 0.2 (0.3–1.25 mM) | −0.6 | 1.6 ± 0.6 mM |
| TPA-oxalate | N | ||
| TPA-acetate | N | ||
| Formamide | N | ||
| D-(+)-s dihydrate | N | ||
| Betaine monohydrate | −2.0 ± 0.4 (50–74 mM) | −0.8 | 84.0 ± 8.4 mM |
aΔCt represents difference between minimal Ct value obtained during amplification of CD4 DNA fragment in SSG PCR mixture supplemented with different concentrations of the tested additive and minimal Ct value in the absence of additives; an increase in negative ΔCt value is indicative of enhanced performance of PCR. Means ± SD were calculated from 4 to 11 experiments. N indicates no improvements of PCR performance at any concentration (range 0–100 mM) of the additive tested. Range of concentrations of the additive giving minimal Ct values in various experiments is shown in parenthesis.
bΔMT represents difference in melting temperature between sample with and without additive.
cFor PCR performance-enhancing additives, their concentrations which inhibit PCR performance reflected in an increase in Ct values exceeding 40 are shown; means ± SD were determined from at least three experiments.
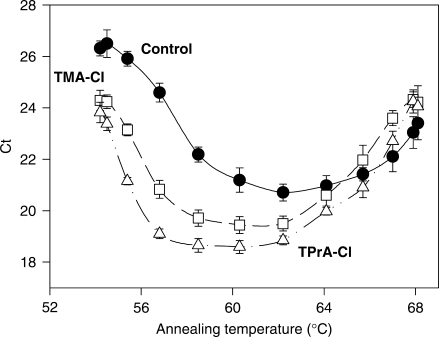
Using temperature gradient function of Mastercycler ep realplex, we also studied the changes in optimal annealing temperatures in SSG PCR mixtures with various additives (Figure 3). When DNA fragment of CD4 was amplified, optimal annealing temperature was found to be 62°C. Inclusion of 16 mM TMA-Cl or TPrA-Cl caused not only a decrease in Ct values but also a broader range of optimal annealing temperatures.
Figure 3.
The effect of additives on optimal annealing temperature. Standard SSG PCR mixtures without TAA-additives (filled circle; Control) or supplemented with 16 mM TMA-Cl (open square) or 16 mM TPrA-Cl (open triangle) were run using cloned CD4 as a template and qPCR temperature gradient function from 54°C to 68°C. Means ± SD from three experiments are shown.
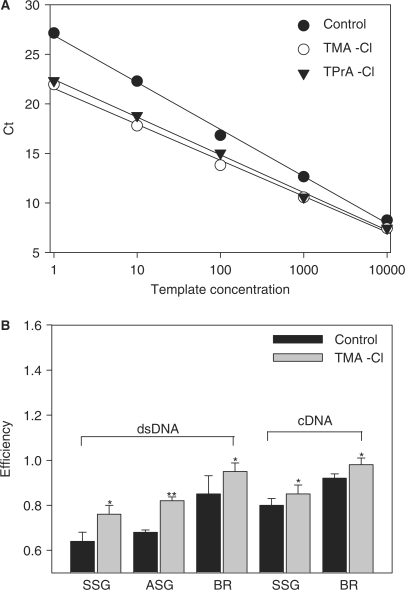
Next we analyzed the changes in PCR efficiencies under different conditions. For these experiments the CD4 template was serially diluted and added to master mixes differing in composition. As shown in Figure 4A, SSG PCR mixtures supplemented with 16 mM TMA-Cl or TPrA-Cl exhibited enhanced efficiencies, respectively, E = 0.85 and E = 0.90, compared to controls supplemented with vehicle (water) alone (E = 0.65). Data from four such experiments showed that TMA-Cl-mediated increase in PCR efficiency was significant (Figure 4B, SSG, dsDNA). Similar increase was also observed with ASG master mixes or with iQ SYBR Green Supermixes from Bio-Rad Laboratories. When actin cDNA was amplified using SSG PCR mixture or iQ™ SYBR Green Supermix, both supplemented with 16 mM TMA-Cl, a significant increase in PCR efficiency was also found (Figure 4B, cDNA). These data indicate that addition of cDNA mixture in reverse transcriptase reaction buffer does not interfere with TMA-Cl performance; in fact, actin cDNA was amplified in general with higher efficiency, indicating better optimized conditions for amplification of this template.
Figure 4.
Enhanced efficiencies of SSG PCR mixtures supplemented with TMA-Cl or TPrA-Cl. (A) Tenfold serial dilutions of the CD4 DNA template were amplified with the corresponding primers in triplicates in standard SSG mixtures without additives (filled circle; Control) or supplemented with 16 mM TMA-Cl (open circle) or 16 mM TPrA-Cl (inverted filled triangle). Standard curves were obtained for control (E = 0.65; R2 = 0.997), TMA-Cl-supplemented (E = 0.85; R2 = 0.995) and TPrA-Cl-supplemented (E = 0.90; R2 = 0.997) PCR mixtures. Typical experiments from 4 (Control and TMA-Cl) or 2 (TPrA-Cl) performed are shown. (B) Tenfold serial dilutions of the CD4 plasmid template (dsDNA) or cDNA from RBL-2H3 cells (cDNA) were amplified with primers for CD4 or actin, respectively, in triplicates in SSG, ASG or iQ SYBR Green Supermix from Bio-Rad (BR) Laboratories PCR mixtures, supplemented (grey column) or not (black column) with 16 mM TMA-Cl. Efficiencies were calculated from standard curves. Means ± SD were calculated from at least three independent experiments. Statistical significance of the differences between controls and TMA-Cl-supplemented samples is also shown; *P ≤ 0.05; **P ≤ 0.01.
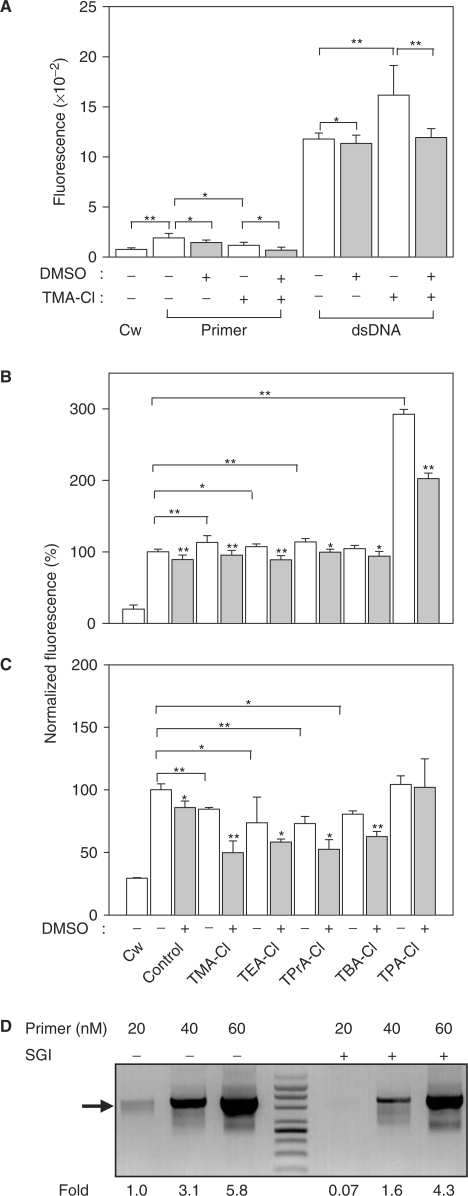
In an attempt to explain the enhancing effect of TAA-derivatives on SGI-monitored real-time PCR, we evaluated SGI fluorescence under various conditions. In initial experiments we measured fluorescence at annealing temperature (55°C) in SGI solutions supplemented with forward CD4 primer or isolated 200 bp CD4 dsDNA amplicon. Data presented in Figure 5A show that addition of oligonucleotide primer significantly increased SGI fluorescence. When DMSO (5% final concentration) or TMA-Cl (16 mM final concentration) were added, fluorescence was significantly inhibited. Oligonucleotide primer-induced SGI fluorescence was completely inhibited when both TMA-Cl and DMSO were added together. Addition of CD4 dsDNA amplicons to SGI solution resulted in a dramatically enhanced fluorescence, which could be weakly but reproducibly inhibited by DMSO. In contrast to oligonucleotide primers, however, SGI fluorescence was enhanced by TMA-Cl, and DMSO reduced this enhanced fluorescence to levels observed in samples without additives. The effect of TAA-Cls and DMSO on fluorescence of SGI-dsDNA and SGI-oligonucleotide primer complexes was further analyzed in experiments in which all TAA-Cls employed in this study were successively used. Data presented in Figure 5B show that TAA-Cls with 1–3 carbons enhanced fluorescence of SGI-dsDNA, whereas TBA-Cl was without effect. Interestingly, TPA-Cl enhanced fluorescence of SGI-dsDNA substantially more than did other TAA-Cls. In all cases, DMSO partially inhibited the fluorescence. In contrast, TAA-Cls had different effect on fluorescence of SGI-oligonucleotide primers; fluorescence was inhibited by TAA-Cl with 1–4 carbons and this inhibition was potentiated by DMSO, whereas TPA-Cl alone or in combination with DMSO had no inhibitory effect (Figure 5C). Inhibitory effect of TMA-Cl and DMSO was observed with several other primers tested, including a primer for GAPDH and reverse primer for CD4 (not shown).
Figure 5.
Interaction of SYBR with single-stranded oligonucleotide primers or dsDNA fragments. (A) SYBR-containing solutions were supplemented with water alone (Cw), CD4 forward primer (Primer) or PCR amplified CD4 fragment (dsDNA), DMSO (+, filled columns, final concentration 5%) and/or TMA-Cl (+, final concentration 16 mM). Fluorescence of the samples was determined in Mastercycler ep realplex at 55°C. (B) SYBR-containing solutions were supplemented with water (Cw), or dsDNA-amplified CD4 fragment, DMSO (+, filled columns, final concentration 5%) and the indicated TAA-Cls at final concentration 16 mM. Fluorescence was determined at 68°C and normalized to samples without additives (Control). (C) SYBR-containing solutions were supplemented and analyzed as in (B) except that Thy-1.2 primer was used and fluorescence was determined at 55°C. Means ± SD were calculated from at least three independent experiments performed in triplicates or quadruplicates. Statistical significance of the differences between samples is also shown; *P ≤ 0.05, **P ≤ 0.01. Asterisks over columns indicate statistical significance of differences between samples with and without DMSO. (D) Fragment of Thy-1 genomic DNA was amplified in standard real-time PCR mixture with different concentrations of Thy-1 primers (20, 40 or 60 nM) in the absence (−) or presence (+) of 0.98 μM SYBR. After 30 cycles, PCR amplicons were analyzed by agarose gel electrophoresis. Position of DNA markers in the middle of the gel is also indicated. Arrows indicate position of 864 bps Thy-1 amplicon. Relative amounts of Thy-1 fragments generated during PCR were determined by densitometry and normalized to the amount of Thy-1 amplicons produced in PCR with 20 nM primers but without SYBR (Fold). A typical experiment of two performed is shown.
To decide whether SGI-mediated inhibition of PCR is dependent on primers concentration, we analyzed by agarose gel electrophoresis production of PCR amplicons in reaction mixtures with or without SGI and various concentrations of primers. Data shown in Figure 5D indicate that, at low concentration of primers (20 nM), specific PCR amplicons were formed in the absence of SGI, but were completely inhibited in its presence. Increasing concentrations of primers to 40 and 60 nM resulted in enhanced production of PCR amplicons, which remained lower in SGI-supplemented reaction mixtures; thus SGI could interfere with annealing of primers and initiation of PCR.
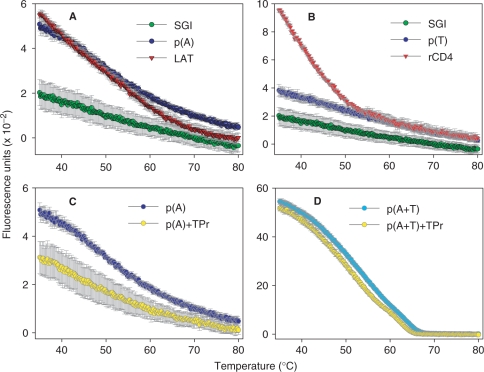
The observed enhanced fluorescence of SGI in the presence of oligonucleotide primers was surprising and could be explained by suboptimal design of primers forming hairpins and/or dimers. In further experiments we therefore evaluated that SGI fluorescence in the presence of such individual primers exhibit no secondary structures and/or dimers as determined by NetPrimer analysis. Data presented in Figure 6A show that rising temperature caused a slight decrease of SGI fluorescence. If SGI-containing solutions were supplemented with LAT primer (Figure 6A), reverse CD4 primer (Figure 6B) or GAPDH1, IL-6 or actin primer (not shown), enhanced fluorescence of SGI was observed at 35°C. Rising temperature caused a gradual decrease in SGI fluorescence; at 65°C and above the primers contributed only very little to enhanced SGI fluorescence. Surprisingly, poly(A)20 oligonucleotide (Figure 6A) and poly(T)20 oligonucleotide (Figure 6B) also enhanced SGI fluorescence, indicating that SGI could directly interact with ssDNA. When poly(A)20 oligonucleotide was mixed with 16 mM TPrA-Cl, a clear decrease in SGI fluorescence was observed (Figure 6C). As expected, mixing poly(A)20 with poly(T)20 induced a dramatic increase in SGI fluorescence, and TPrA-Cl at temperature range 35–65°C decreased it (Figure 6D). The combined data indicate that even homopolymeric oligonucleotides could interact with SGI, and thus strengthen the concept that SGI could interact with ssDNA oligonucleotide primers.
Figure 6.
The effect of temperature on SGI fluorescence in the presence of single- or double-stranded oligonucleotides and additives. SGI-containing solutions were supplemented or not with various oligonucleotides and additives, and temperature-dependent changes in SGI fluorescence were determined. (A) poly(A)20 [p(A)] or oligonucleotide primer LAT was used. (B) poly(T)20 [p(T)] or reverse oligonucleotide primer (rCD4) was used. (C) Poly(A)20 was used alone or mixed with TPrA-Cl (TPr; 16 mM final concentration]. (D) Poly(A)20 and Poly(T)20 [p(A+T)] were mixed and analyzed alone or after addition of TPrA-Cl. In A and B temperature-dependent changes in fluorescence of SGI alone are also shown. Means ± SD from 2 to 4 experiments performed in triplicates or quadruplicates are shown.
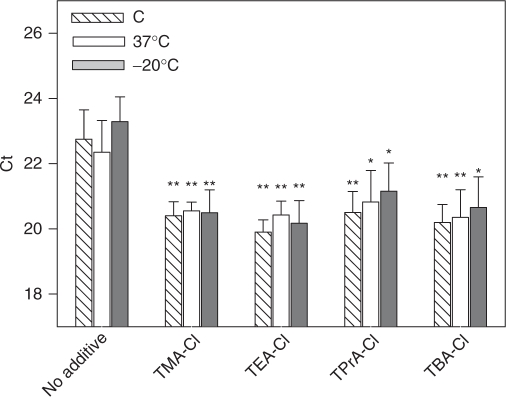
TAA-Cls at concentrations enhancing RT–PCR performance were also tested for their contribution to stability of SSG PCR master mixes. Two-times concentrated master mixes were supplemented or not with 32 mM TMA-Cl, TEA-Cl, TPrA-Cl or TBA-Cl and stored at −20 or 37°C in the dark. After 2 weeks they were supplemented with CD4 template DNA, primers and water and used for real-time PCR. Freshly prepared SSG PCR mixes with or without additives were prepared and also used for PCR as controls. Data presented in Figure 7 indicate that all master mixes supplemented with TAA-Cls showed significantly lower Ct values than those without additives. Interestingly, storage of SSG master mixes at −20°C or even at 37°C for two weeks had no negative effect on their PCR performance.
Figure 7.
The effect of additives on performance of PCR mixtures stored under different conditions. Two-times concentrated SSG PCR mixes were supplemented or not with various TAA-Cls at 32 mM concentration and stored for two weeks at 37 or −20°C. As controls (C) freshly prepared SSG PCR mixes with or without additives were also analyzed. Real-time PCRs were performed using cloned CD4 DNA as a template and corresponding primers, and Ct values were determined. Means ± SD were calculated from three experiments performed in triplicates. Statistical significance of the differences between samples with no additives and samples supplemented with additives is also shown; *P ≤ 0.05, **P ≤ 0.01.
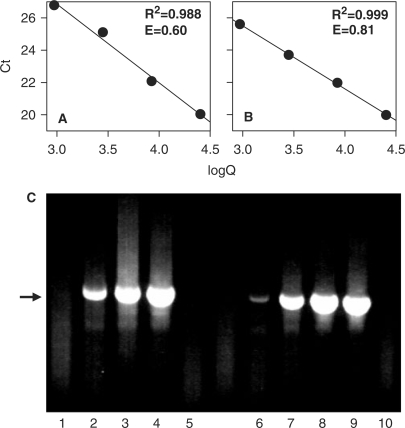
Finally, we checked whether TAA derivatives could improve SGI-monitored amplification of difficult templates. For these experiments we used mouse genomic DNA and amplified 864 bps fragment of mouse Thy-1 gene. In pilot experiments we found that Thy-1 amplicons were not detectable by SGI fluorescence measurement or agarose gel electrophoresis when SSG, ASG, iQ™ SYBR Green Supermix or QuantiTect SYBR Green PCR kit under optimized conditions were used. Removal of SGI from SSG or ASG master mixes resulted in production of the expected 864 bps amplicon, confirming the inhibitory effect of SGI in this system. Addition of TMA-Cl or TPrA-Cl at concentrations 5–30 mM to various SGI-based PCR mixtures did not enhance production of the 864 bp Thy-1 amplicons as detected by agarose gel electrophoresis. To solve this problem we performed a series of optimization experiments and prepared a new DMSO-free PCR mixture, denoted GSB, the composition of which is described in ‘Materials and Methods’ section. When Thy-1 fragment was amplified in GSB-based mixture, PCR efficiency was 0.60 ± 0.12 (mean ± SD, n = 3, Figure 8A). In the presence of TPrA-Cl at optimal concentration 10 mM, efficiency was increased to 0.81 ± 0.08 (n = 4; Figure 8B). Agarose gel electrophoresis after real-time PCR confirmed generation of the corresponding band and enhanced production of the Thy-1 amplicons in TPrA-Cl-supplemented PCR mixtures (Figure 8C). It should be noted that addition of TPrA-Cl also decreased nonspecific amplification as reflected by lower ‘background’. Similar results were observed when genomic fragments of Thy-1 and three other genes were amplified in the presence of 10 mM TPrA-Cl, TMA-Cl or TEA-Cl (not shown). The combined data indicate that addition of TAA-Cls improves SGI-monitored real-time PCR performance even when difficult genomic fragments are amplified. Key factor for amplification of large genomic fragments in the presence of SGI is to use GSB PCR mixture.
Figure 8.
Effect of TPrA-Cl on PCR performance using genomic DNA as a template. (A and B) Genomic DNA was 3-fold diluted and used as a template for real-time PCR using GSB reaction mixes without (A) or with (B) 10 mM TPrA-Cl. Thy-1 fragment of 864 bps was amplified using the corresponding primer set. Ct values were obtained from amplification curves and plotted against calculated copy numbers (logQ). Typical experiments from 3 (A) or 4 (B) performed. (C) After 40 cycles of PCR amplification the samples were analyzed by agarose gel electrophoresis. Lines 1–5 and 6–10 correspond to samples run, respectively, in the absence or presence of TPrA-Cl. Lines 1 + 6, 2 + 7, 3 + 8 and 4 + 9 correspond to samples containing template DNA at concentrations, respectively, 0.3, 0.9, 2.7 and 8.1 ng/μl. Lines 5 + 10 did not contain template DNA.
DISCUSSION
Data presented in this study indicate that TAA derivatives could serve as potent enhancers of SGI-monitored real-time PCR. The effect of TAA derivatives depended on the nature of ions present. The strongest effect was observed with TAA-cations with 1–4 carbons in alkyl chain combined with chloride as anion. TPA-Cl with 5 carbons in alkyl chain was less potent. As concerns TAA derivatives with acetate as anion, the enhancing effect was confined only to TPrA-acetate and TBA-acetate (Table 1). In contrast to previous results showing that TMA-oxalate enhances the specificity and yield of PCRs evaluated by end-point analysis (15), all TAA-oxalates failed to act as SGI-monitored real-time PCR enhancers. Although melting temperature slightly decreased with increasing size of alkyl chain (up to 3 carbons), optimal annealing temperature was not dramatically changed, thus corroborating previous findings (13).
Molecular mechanism of the enhancing effect of TAA-derivatives on real-time PCR is unknown. Several possibilities may be considered. First, TMA-Cl is known to increase thermal stability of AT base pair (31,32). The enhanced AT base-pair stability could thus contribute to a more efficient binding of primers to the template and to a more efficient amplification in the presence of TAA-Cls. Our finding that optimal annealing temperature was similar in control and TAA-Cls-supplemented PCR mixtures, however, suggests that contribution of the additives to the base-pairs stability is small. It should also be noted that the effect of TMA-Cl on base-pairs stability was in previous studies observed at relatively high concentrations of TMA-Cl (3 M), whereas the enhancing effect on real-time PCR was effected at much lower concentrations (∼16 mM); higher concentrations (>32 mM) were inhibitory. Second, additives could interfere with the inhibitory effect of SGI on PCR. This possibility is supported by our finding that in the absence of fluorescence dye the template amplification, evaluated by end-point analysis, was of comparable extent in both control and TAA-Cl-supplemented samples. The structure of SGI and its positive charges (25) should allow its interaction with negative electrostatic potential in the minor groove. Furthermore, van der Waals interactions within the boundaries of the minor groove are likely to contribute to its high affinity for dsDNA. These interactions could be affected by TAA-containing solutions. The highest SGI-derived fluorescence signal was observed in solution supplemented with TPA-Cl (Figure 5B). However, TPA-Cl had only small effect on RT–PCR performance, a finding suggesting that enhanced SGI binding rather inhibits than promotes PCR amplification. Interestingly, the enhanced fluorescence observed in samples with TAA derivatives was decreased in samples supplemented with DMSO. Thus, DMSO could contribute to enhanced PCR performance by partially reducing the binding of SGI to DNA. Yet these effects were relatively small and do not explain those dramatic changes induced by DMSO and TAA derivatives. Third, it is known that SGI binds with low efficiency to single-stranded DNA (25). In fact, addition of primers to SGI solution enhanced significantly fluorescence signal measured at annealing temperature. Interestingly, the signal was inhibited by TAA-derivatives, and DMSO further enhanced the inhibitory effect. Disparate effects of TAA-derivatives and DMSO on SGI binding to primers and dsDNA suggest different underlying binding mechanisms. When samples containing SGI and oligonucleotide primers were supplemented with TPA-Cl and DMSO or vehicle (water), comparable fluorescence was observed (Figure 5C); these data suggest that DMSO and TAA-derivatives do not interfere with fluorescence signal detection, but rather with binding of SGI to primers and generation of fluorescence signal. Unexpectedly, enhanced SGI fluorescence was also observed in samples with poly(A)20 and poly(T)20, which have no possibility to form complementary secondary structures and/or duplexes, and again TPrA-Cl inhibited this fluorescence. In view of these findings we propose that SGI could inhibit amplification through its binding to primers by a distinct mechanism, one which interferes with their annealing and/or initiation of polymerase reaction. This conclusion was corroborated by other experimental data indicating that production of electrophoretically detectable amplicons at limiting concentrations of primers was inhibited in the presence of SGI. Obviously, it is still possible that SGI could also affect PCR efficiency by some other mechanism(s).
In initial experiments we used SGI-supplemented home-made or commercial real-time PCR master mixes, which performed well when relatively short (100–400 bp) DNA or cDNA fragments were amplified. When TAA-Cls (up to four carbons) were added, PCR efficiency during amplification of such fragments was enhanced. However, with larger genomic fragments (>800 bps) in the presence of SGI, amplification was inhibited in all tested SGI-supplemented master mixes regardless of the presence or absence of various concentrations of TAA-Cls. Evidently, SGI was the inhibitor because its withdrawal led to easily detectable products end-point agarose gel electrophoresis. To solve this problem we formulated new SGI-based PCR mixture, GSB, which allowed amplification of longer genomic fragments in the presence of SGI. PCR performance in GSB-based mixtures was also enhanced by TAA-Cls, confirming general suitability of these additives for SGI-based real-time PCR analysis. At the same time, addition of TAA-Cls decreased nonspecific amplicons often observed when genomic fragments are amplified even under seemingly optimal conditions. It has been reported previously that SGI is not chemically stable for >3 weeks (33). Furthermore, Karsai and co-workers found that SGI degradation products are potent inhibitors of PCR (34). Our data that TAA derivatives protect SGI-containing PCR mixes from aging, comply with and somewhat extend the previous findings that TAA derivatives stabilize SGI in agarose gels (23). Inclusion of TAA derivatives not only enhances the efficiency of SGI-containing PPP master mixes but also contributes to their increased stability even at 37°C. This should facilitate their handling.
ACKNOWLEDGEMENTS
We thank Lukáš Kocanda and Hanka Mrázová for skilled technical assistance. Funding was provided by the Academy of Sciences of the Czech Republic (1QS500520551, KAN200520701, Institutional project AVOZ50520514). Funding to pay the Open Access publication charges for this article was provided by the Academy of Sciences of the Czech Republic, project KAN200520701.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bustin SA. A-Z of Quantitative PCR. La Jolla: International University Line; 2004. [Google Scholar]
- 2.Dieffenbach CW, Dveksler GS. PCR primer. A laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- 3.Baskaran N, Kandpal RP, Bhargava AK, Glynn MW, Bale A, Weissman SM. Uniform amplification of a mixture of deoxyribonucleic acids with varying GC content. Genome Res. 1996;6:633–638. doi: 10.1101/gr.6.7.633. [DOI] [PubMed] [Google Scholar]
- 4.Sidhu MK, Liao MJ, Rashidbaigi A. Dimethyl sulfoxide improves RNA amplification. BioTechniques. 1996;21:44–47. doi: 10.2144/96211bm08. [DOI] [PubMed] [Google Scholar]
- 5.Jung M, Muche JM, Lukowsky A, Jung K, Loening SA. Dimethyl sulfoxide as additive in ready-to-use reaction mixtures for real-time polymerase chain reaction analysis with SYBR Green I dye. Anal. Biochem. 2001;289:292–295. doi: 10.1006/abio.2000.4931. [DOI] [PubMed] [Google Scholar]
- 6.Chakrabarti R, Schutt CE. The enhancement of PCR amplification by low molecular-weight sulfones. Gene. 2001;274:293–298. doi: 10.1016/s0378-1119(01)00621-7. [DOI] [PubMed] [Google Scholar]
- 7.Chakrabarti R, Schutt CE. Novel sulfoxides facilitate GC-rich template amplification. BioTechniques. 2002;32:866–873. doi: 10.2144/02324rr04. [DOI] [PubMed] [Google Scholar]
- 8.Weissensteiner T, Lanchbury JS. Strategy for controlling preferential amplification and avoiding false negatives in PCR typing. BioTechniques. 1996;21:1102–1108. doi: 10.2144/96216rr03. [DOI] [PubMed] [Google Scholar]
- 9.Hengen PN. Optimizing multiplex and LA-PCR with betaine. Trends Biochem. Sci. 1997;22:225–226. doi: 10.1016/s0968-0004(97)01069-4. [DOI] [PubMed] [Google Scholar]
- 10.Sarkar G, Kapelner S, Sommer SS. Formamide can dramatically improve the specificity of PCR. Nucleic Acids Res. 1990;18:7465. doi: 10.1093/nar/18.24.7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comey CT, Jung JM, Budowle B. Use of formamide to improve amplification of HLA DQ alpha sequences. BioTechniques. 1991;10:60–61. [PubMed] [Google Scholar]
- 12.Chakrabarti R, Schutt CE. The enhancement of PCR amplification by low molecular weight amides. Nucleic Acids Res. 2001;29:2377–2381. doi: 10.1093/nar/29.11.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chevet E, Lemaitre G, Katinka MD. Low concentrations of tetramethylammonium chloride increase yield and specificity of PCR. Nucleic Acids Res. 1995;23:3343–3344. doi: 10.1093/nar/23.16.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hung T, Mak K, Fong K. A specificity enhancer for polymerase chain reaction. Nucleic Acids Res. 1990;18:4953. doi: 10.1093/nar/18.16.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovárová M, Dráber P. New specificity and yield enhancer of polymerase chain reactions. Nucleic Acids Res. 2000;28:e70. doi: 10.1093/nar/28.13.e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe M, Abe K, Aoki M, Kameya T, Itoyama Y, Shoji M, Ikeda M, Iizuka T, Hirai S. A reproducible assay of polymerase chain reaction to detect trinucleotide repeat expansion of Huntington's disease and senile chorea. Neurol. Res. 1996;18:16–18. doi: 10.1080/01616412.1996.11740370. [DOI] [PubMed] [Google Scholar]
- 17.Pomp D, Medrano JF. Organic solvents as facilitators of polymerase chain reaction. BioTechniques. 1991;10:58–59. [PubMed] [Google Scholar]
- 18.Bachmann B, Luke W, Hunsmann G. Improvement of PCR amplified DNA sequencing with the aid of detergents. Nucleic Acids Res. 1990;18:1309. doi: 10.1093/nar/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varadaraj K, Skinner DM. Denaturants or cosolvents improve the specificity of PCR amplification of a G + C-rich DNA using genetically engineered DNA polymerases. Gene. 1994;140:1–5. doi: 10.1016/0378-1119(94)90723-4. [DOI] [PubMed] [Google Scholar]
- 20.Carninci P, Nishiyama Y, Westover A, Itoh M, Nagaoka S, Sasaki N, Okazaki Y, Muramatsu M, Hayashizaki Y. Thermostabilization and thermoactivation of thermolabile enzymes by trehalose and its application for the synthesis of full length cDNA. Proc. Natl Acad. Sci USA. 1998;95:520–524. doi: 10.1073/pnas.95.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spiess A-N, Muellr N, Ivell R. Trehalose is a potent PCR enhancer: lowering of DNA melting temperature and thermal stabilization of Taq polymerase by the disaccharide trehalose. Clin. Chem. 2004;50:1256–1259. doi: 10.1373/clinchem.2004.031336. [DOI] [PubMed] [Google Scholar]
- 22.Nath K, Sarosy JW, Hahn J, Di Como CJ. Effects of ethidium bromide and SYBR Green I on different polymerase chain reaction systems. J. Biochem. Biophys. Methods. 2000;42:15–29. doi: 10.1016/s0165-022x(99)00033-0. [DOI] [PubMed] [Google Scholar]
- 23.Wu M, White HW, Kusukawa N, Stein TM. Stabilization of highly sensitive nucleic acid stains in aqueous solutions, US6365341. 2002 [Google Scholar]
- 24.Zeng Z, Clark SM, Mathies RA, Glazer AN. Improved stability and electrophoretic properties of preformed fluorescent cationic dye-DNA complexes in a taps-tetrapentylammonium buffer in agarose slab gels. Anal. Biochem. 1997;252:110–114. doi: 10.1006/abio.1997.2303. [DOI] [PubMed] [Google Scholar]
- 25.Zipper H, Brunner H, Bernhagen J, Vitzthum F. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. Nucleic Acids Res. 2004;32:e103. doi: 10.1093/nar/gnh101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dráber P, Zikán J, Vojtíšková M. Establishment and characterization of permanent murine hybridomas secreting monoclonal anti-Thy-1 antibodies. J. Immunogenet. 1980;7:455–474. doi: 10.1111/j.1744-313x.1980.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 27.Scalice ER, Sharkey DJ, Daiss JL. Monoclonal antibodies prepared against the DNA polymerase from Thermus aquaticus are potent inhibitors of enzyme activity. J. Immunol. Methods. 1994;172:147–163. doi: 10.1016/0022-1759(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 28.Dráberová L, Dráber P. Functional expression of the endogenous Thy-1 gene and the transfected murine Thy-1.2 gene in rat basophilic leukemia cells. Eur. J. Immunol. 1991;21:1583–1590. doi: 10.1002/eji.1830210703. [DOI] [PubMed] [Google Scholar]
- 29.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der NJ. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolff R, Gemmill R. Purifying and Analyzing Genomic DNA. In: Birren B, Green ED, Klapholz S, Myers RM, Roskams J, editors. Genome Analysis: A Laboratory Manual. Cold Spring Harbor, New York, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 1–81. [Google Scholar]
- 31.Melchior WB, Jr, Von Hippel PH. Alteration of the relative stability of dA-dT and dG-dC base pairs in DNA. Proc. Natl Acad. Sci USA. 1973;70:298–302. doi: 10.1073/pnas.70.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobs KA, Rudersdorf R, Neill SD, Dougherty JP, Brown EL, Fritsch EF. The thermal stability of oligonucleotide duplexes is sequence independent in tetraalkylammonium salt solutions: application to identifying recombinant DNA clones. Nucleic Acids Res. 1988;16:4637–4650. doi: 10.1093/nar/16.10.4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monis PT, Giglio S, Saint CP. Comparison of SYTO9 and SYBR Green I for real-time polymerase chain reaction and investigation of the effect of dye concentration on amplification and DNA melting curve analysis. Anal. Biochem. 2005;340:24–34. doi: 10.1016/j.ab.2005.01.046. [DOI] [PubMed] [Google Scholar]
- 34.Karsai A, Muller S, Platz S, Hauser MT. Evaluation of a homemade SYBR green I reaction mixture for real-time PCR quantification of gene expression. BioTechniques. 2002;32:790–796. doi: 10.2144/02324st05. [DOI] [PMC free article] [PubMed] [Google Scholar]