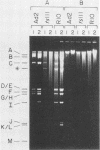
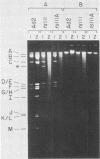
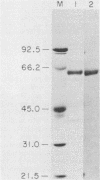
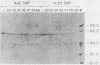
Abstract
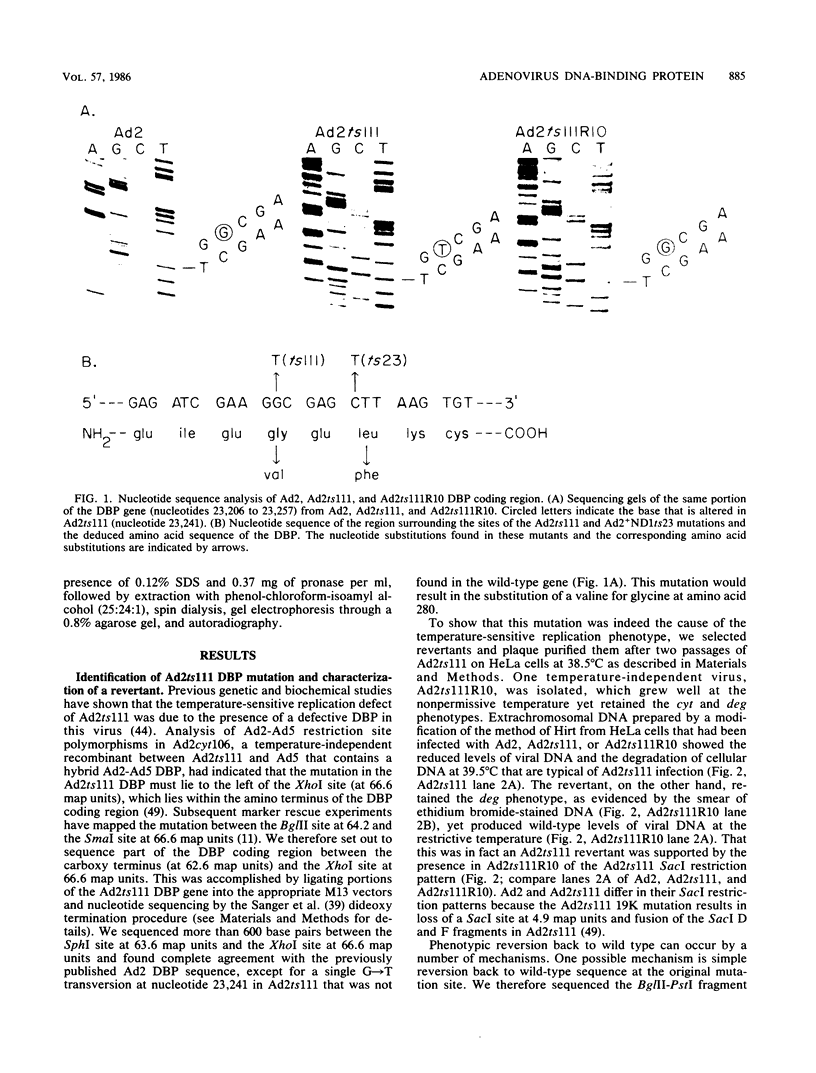
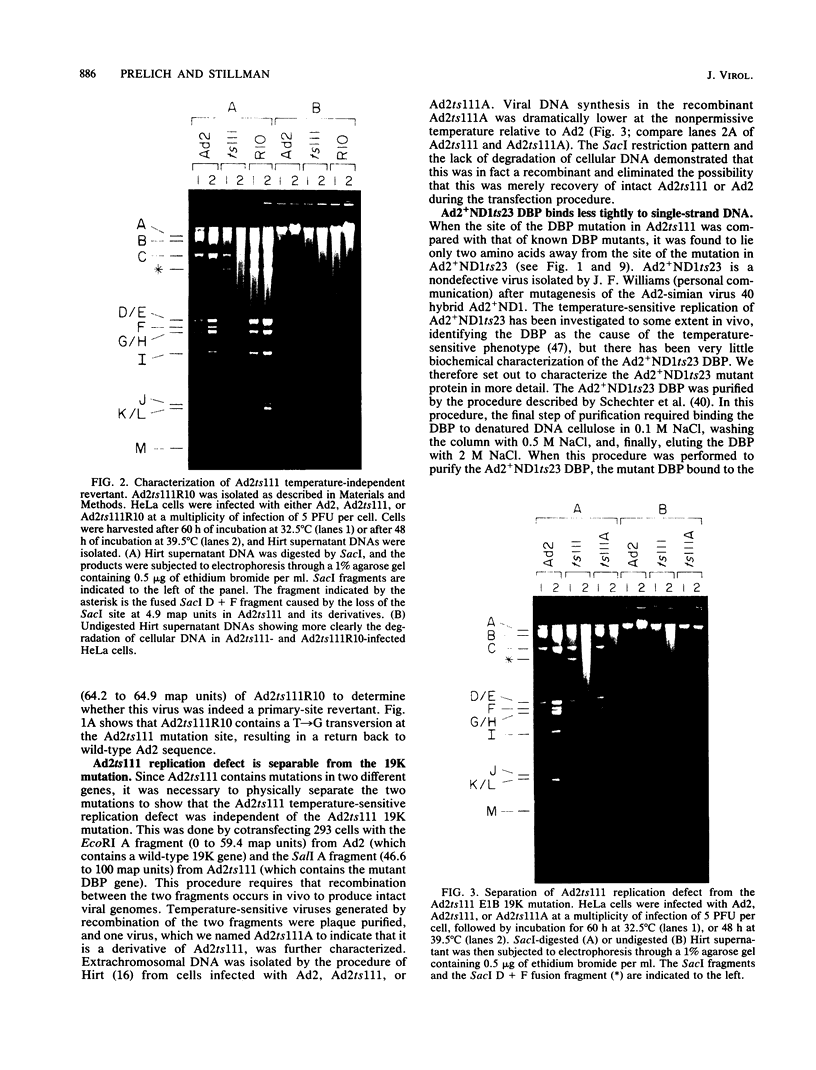
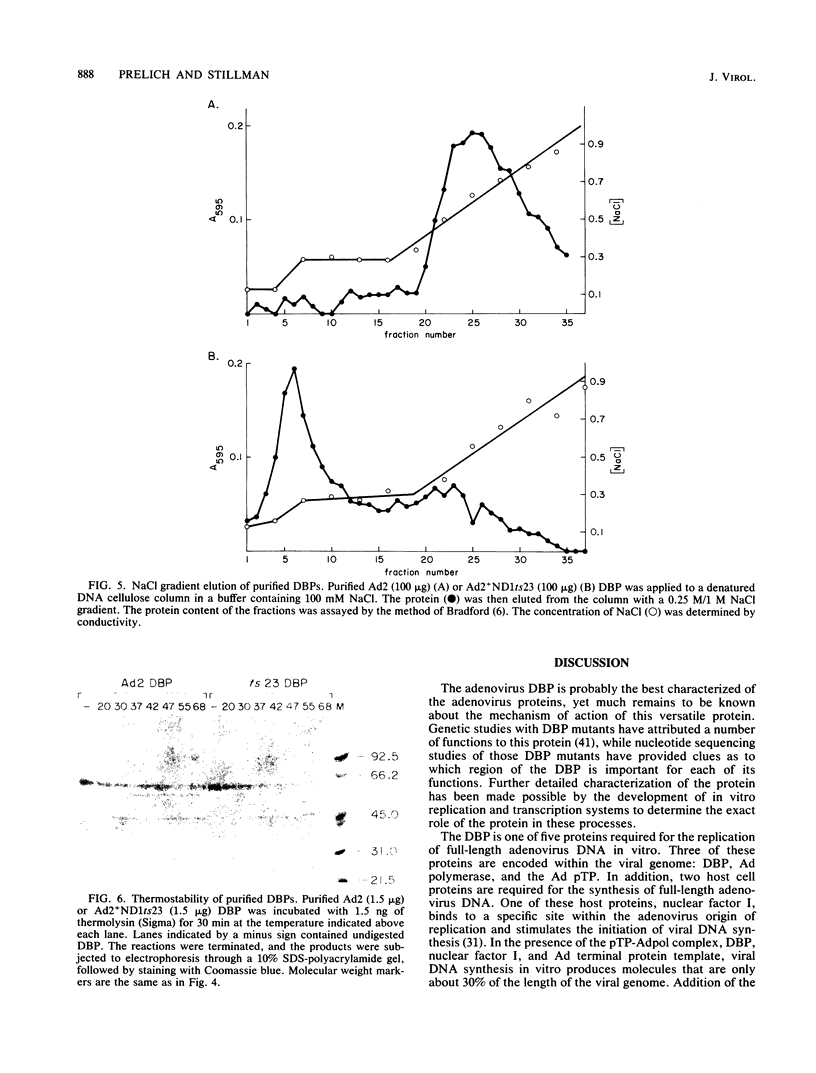
The human adenovirus type 2 (Ad2) mutant Ad2ts111 has previously been shown to contain two mutations which result in a complex phenotype. Ad2ts111 contains a single base change in the early region 1B (E1B) 19,000-molecular-weight (19K) coding region which yields a cyt deg phenotype and another defect which maps to the E2A 72K DNA-binding protein (DBP) coding region that causes a temperature-sensitive DNA replication phenotype. Here we report that the defect in the Ad2ts111 DBP is due to a single G----T transversion that results in a substitution of valine for glycine at amino acid 280. A temperature-independent revertant, Ad2ts111R10, was isolated, which reverts back to glycine at amino acid 280 yet retains the cyt and deg phenotypes caused by the 19K mutation. We physically separated the two mutations of Ad2ts111 by constructing a recombinant virus, Ad2ts111A, which contained a wild-type Ad2 E1B 19K gene and the gly----val mutation in the 72K gene. Ad2ts111A was cyt+ deg+, yet it was still defective for DNA replication at the nonpermissive temperature. The Ad2ts111 DBP mutation is located only two amino acids away from the site of the mutation in Ad2+ND1ts23, a previously sequenced DBP mutant. Biochemical studies of purified Ad2+ND1ts23 DBP showed that this protein was defective for elongation but not initiation of replication in a cell-free replication system consisting of purified Ad polymerase, terminal protein precursor, and nuclear factor I. Ad2+ND1ts23 DBP bound less tightly to single-strand DNA than did Ad2 DBP, as shown by salt gradient elution of purified DBPs from denatured DNA cellulose columns. This decreased binding to DNA was probably due to local conformational changes in the protein at a site that is critical for DNA binding rather than to global changes in protein structure, since both the Ad2+ND1ts23 and Ad2 DBPs showed identical cleavage patterns by the protease thermolysin at various temperatures.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson C. W., Hardy M. M., Dunn J. J., Klessig D. F. Independent, spontaneous mutants of adenovirus type 2-simian virus 40 hybrid Ad2+ND3 that grow efficiently in monkey cells possess indentical mutations in the adenovirus type 2 DNA-binding protein gene. J Virol. 1983 Oct;48(1):31–39. doi: 10.1128/jvi.48.1.31-39.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. W. Spontaneous mutants of the adenovirus-simian virus 40 hybrid, Ad2+ND3, that grow efficiently in monkey cells. Virology. 1981 May;111(1):263–269. doi: 10.1016/0042-6822(81)90670-x. [DOI] [PubMed] [Google Scholar]
- Anderson K. P., Klessig D. F. Altered mRNA splicing in monkey cells abortively infected with human adenovirus may be responsible for inefficient synthesis of the virion fiber polypeptide. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4023–4027. doi: 10.1073/pnas.81.13.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariga H., Klein H., Levine A. J., Horwitz M. S. A cleavage product of the adenovirus DNA binding protein is active in DNA replication in vitro. Virology. 1980 Feb;101(1):307–310. doi: 10.1016/0042-6822(80)90510-3. [DOI] [PubMed] [Google Scholar]
- Babich A., Nevins J. R. The stability of early adenovirus mRNA is controlled by the viral 72 kd DNA-binding protein. Cell. 1981 Nov;26(3 Pt 1):371–379. doi: 10.1016/0092-8674(81)90206-3. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brough D. E., Rice S. A., Sell S., Klessig D. F. Restricted changes in the adenovirus DNA-binding protein that lead to extended host range or temperature-sensitive phenotypes. J Virol. 1985 Jul;55(1):206–212. doi: 10.1128/jvi.55.1.206-212.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter T. H., Blanton R. A. Autoregulation of adenovirus type 5 early gene expression II. Effect of temperature-sensitive early mutations on virus RNA accumulation. J Virol. 1978 Nov;28(2):450–456. doi: 10.1128/jvi.28.2.450-456.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg M. D., Kelly T. J., Jr Adenovirus DNA replication in vitro. Proc Natl Acad Sci U S A. 1979 Feb;76(2):655–659. doi: 10.1073/pnas.76.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C. N., Crawford L. V., Berg P. Simian virus 40 mutants with deletions at the 3' end of the early region are defective in adenovirus helper function. J Virol. 1979 Jun;30(3):683–691. doi: 10.1128/jvi.30.3.683-691.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Halluin J. C., Cousin C., Niel C., Boulanger P. Characterization of an early temperature-sensitive and cytocidal double mutant of adenovirus type 2. J Gen Virol. 1984 Aug;65(Pt 8):1305–1317. doi: 10.1099/0022-1317-65-8-1305. [DOI] [PubMed] [Google Scholar]
- Field J., Gronostajski R. M., Hurwitz J. Properties of the adenovirus DNA polymerase. J Biol Chem. 1984 Aug 10;259(15):9487–9495. [PubMed] [Google Scholar]
- Friefeld B. R., Krevolin M. D., Horwitz M. S. Effects of the adenovirus H5ts125 and H5ts107 DNA binding proteins on DNA replication in vitro. Virology. 1983 Jan 30;124(2):380–389. doi: 10.1016/0042-6822(83)90354-9. [DOI] [PubMed] [Google Scholar]
- Handa H., Kingston R. E., Sharp P. A. Inhibition of adenovirus early region IV transcription in vitro by a purified viral DNA binding protein. Nature. 1983 Apr 7;302(5908):545–547. doi: 10.1038/302545a0. [DOI] [PubMed] [Google Scholar]
- Hecht M. H., Nelson H. C., Sauer R. T. Mutations in lambda repressor's amino-terminal domain: implications for protein stability and DNA binding. Proc Natl Acad Sci U S A. 1983 May;80(9):2676–2680. doi: 10.1073/pnas.80.9.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Horwitz M. S., Ariga H. Multiple rounds of adenovirus DNA synthesis in vitro. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1476–1480. doi: 10.1073/pnas.78.3.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman J. A., Kornberg A., Alberts B. M. Stimulation of T4 bacteriophage DNA polymerase by the protein product of T4 gene 32. J Mol Biol. 1971 Nov 28;62(1):39–52. doi: 10.1016/0022-2836(71)90129-x. [DOI] [PubMed] [Google Scholar]
- Kaplan L. M., Ariga H., Hurwitz J., Horwitz M. S. Complementation of the temperature-sensitive defect in H5ts125 adenovirus DNA replication in vitro. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5534–5538. doi: 10.1073/pnas.76.11.5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein H., Maltzman W., Levine A. J. Structure-function relationships of the adenovirus DNA-binding protein. J Biol Chem. 1979 Nov 10;254(21):11051–11060. [PubMed] [Google Scholar]
- Klessig D. F., Grodzicker T. Mutations that allow human Ad2 and Ad5 to express late genes in monkey cells map in the viral gene encoding the 72K DNA binding protein. Cell. 1979 Aug;17(4):957–966. doi: 10.1016/0092-8674(79)90335-0. [DOI] [PubMed] [Google Scholar]
- Kruijer W., Nicolas J. C., van Schaik F. M., Sussenbach J. S. Structure and function of DNA binding proteins from revertants of adenovirus type 5 mutants with a temperature-sensitive DNA replication. Virology. 1983 Jan 30;124(2):425–433. doi: 10.1016/0042-6822(83)90358-6. [DOI] [PubMed] [Google Scholar]
- Kruijer W., Van Schaik F. M., Sussenbach J. S. Nucleotide sequence of the gene encoding adenovirus type 2 DNA binding protein. Nucleic Acids Res. 1982 Aug 11;10(15):4493–4500. doi: 10.1093/nar/10.15.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijer W., van Schaik F. M., Speijer J. G., Sussenbach J. S. Structure and function of adenovirus DNA binding protein: comparison of the amino acid sequences of the Ad5 and Ad12 proteins derived from the nucleotide sequence of the corresponding genes. Virology. 1983 Jul 15;128(1):140–153. doi: 10.1016/0042-6822(83)90325-2. [DOI] [PubMed] [Google Scholar]
- Kruijer W., van Schaik F. M., Sussenbach J. S. Structure and organization of the gene coding for the DNA binding protein of adenovirus type 5. Nucleic Acids Res. 1981 Sep 25;9(18):4439–4457. doi: 10.1093/nar/9.18.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levinson A. D., Postel E. H., Levine A. J. In vivo and in vitro phosphorylation of the adenovirus type 5 single strand-specific DNA-binding protein. Virology. 1977 Jun 1;79(1):144–159. doi: 10.1016/0042-6822(77)90341-5. [DOI] [PubMed] [Google Scholar]
- Lewis J. B., Atkins J. F., Baum P. R., Solem R., Gesteland R. F., Anderson C. W. Location and identification of the genes for adenovirus type 2 early polypeptides. Cell. 1976 Jan;7(1):141–151. doi: 10.1016/0092-8674(76)90264-6. [DOI] [PubMed] [Google Scholar]
- Lichy J. H., Horwitz M. S., Hurwitz J. Formation of a covalent complex between the 80,000-dalton adenovirus terminal protein and 5'-dCMP in vitro. Proc Natl Acad Sci U S A. 1981 May;78(5):2678–2682. doi: 10.1073/pnas.78.5.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. R., Warocquier R., Cousin C., D'Halluin J. C., Boulanger P. A. Isolation and phenotypic characterization of human adenovirus type 2 temperature-sensitive mutants. J Gen Virol. 1978 Nov;41(2):303–314. doi: 10.1099/0022-1317-41-2-303. [DOI] [PubMed] [Google Scholar]
- Nagata K., Guggenheimer R. A., Enomoto T., Lichy J. H., Hurwitz J. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6438–6442. doi: 10.1073/pnas.79.21.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K., Guggenheimer R. A., Hurwitz J. Adenovirus DNA replication in vitro: synthesis of full-length DNA with purified proteins. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4266–4270. doi: 10.1073/pnas.80.14.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R., Winkler J. J. Regulation of early adenovirus transcription: a protein product of early region 2 specifically represses region 4 transcription. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1893–1897. doi: 10.1073/pnas.77.4.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas J. C., Ingrand D., Sarnow P., Levine A. J. A mutation in the adenovirus type 5 DNA binding protein that fails to autoregulate the production of the DNA binding protein. Virology. 1982 Oct 30;122(2):481–485. doi: 10.1016/0042-6822(82)90249-5. [DOI] [PubMed] [Google Scholar]
- Nicolas J. C., Sarnow P., Girard M., Levine A. J. Host range temperature-conditional mutants in the adenovirus DNA binding protein are defective in the assembly of infectious virus. Virology. 1983 Apr 15;126(1):228–239. doi: 10.1016/0042-6822(83)90474-9. [DOI] [PubMed] [Google Scholar]
- Quinn C. O., Kitchingman G. R. Sequence of the DNA-binding protein gene of a human subgroup B adenovirus (type 7). Comparisons with subgroup C (type 5) and subgroup A (type 12). J Biol Chem. 1984 Apr 25;259(8):5003–5009. [PubMed] [Google Scholar]
- RABSON A. S., O'CONOR G. T., BEREZESKY I. K., PAUL F. J. ENHANCEMENT OF ADENOVIRUS GROWTH IN AFRICAN GREEN MONKEY KIDNEY CELL CULTURES BY SV40. Proc Soc Exp Biol Med. 1964 May;116:187–190. doi: 10.3181/00379727-116-29197. [DOI] [PubMed] [Google Scholar]
- Reuben R. C., Gefter M. L. A DNA-binding protein induced by bacteriophage T7. Proc Natl Acad Sci U S A. 1973 Jun;70(6):1846–1850. doi: 10.1073/pnas.70.6.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T., Williams J. Genetic analysis of adenoviruses. Curr Top Microbiol Immunol. 1984;111:1–39. doi: 10.1007/978-3-642-69549-0_1. [DOI] [PubMed] [Google Scholar]
- Stillman B. W., Tamanoi F., Mathews M. B. Purification of an adenovirus-coded DNA polymerase that is required for initiation of DNA replication. Cell. 1982 Dec;31(3 Pt 2):613–623. doi: 10.1016/0092-8674(82)90317-8. [DOI] [PubMed] [Google Scholar]
- Stillman B. W., White E., Grodzicker T. Independent mutations in Ad2ts111 cause degradation of cellular DNA and defective viral DNA replication. J Virol. 1984 May;50(2):598–605. doi: 10.1128/jvi.50.2.598-605.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamanoi F., Stillman B. W. Function of adenovirus terminal protein in the initiation of DNA replication. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2221–2225. doi: 10.1073/pnas.79.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsernoglou D., Tucker A. D., Van der Vliet P. C. Crystallization of a fragment of the adenovirus DNA binding protein. J Mol Biol. 1984 Jan 15;172(2):237–239. doi: 10.1016/s0022-2836(84)80042-x. [DOI] [PubMed] [Google Scholar]
- Van Der Vliet P. C., Levine A. J., Ensinger M. J., Ginsberg H. S. Thermolabile DNA binding proteins from cells infected with a temperature-sensitive mutant of adenovrius defective in viral DNA synthesis. J Virol. 1975 Feb;15(2):348–354. doi: 10.1128/jvi.15.2.348-354.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E., Grodzicker T., Stillman B. W. Mutations in the gene encoding the adenovirus early region 1B 19,000-molecular-weight tumor antigen cause the degradation of chromosomal DNA. J Virol. 1984 Nov;52(2):410–419. doi: 10.1128/jvi.52.2.410-419.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]