Abstract
In the RecFOR pathway, the RecF and RecR proteins form a complex that binds to DNA and exerts multiple functions, including directing the loading of RecA onto single-stranded (ss) DNA regions near double-stranded (ds) DNA–ssDNA junctions and preventing it from forming a filament beyond the ssDNA region. However, neither the structure of the RecFR complex nor its DNA-binding mechanism was previously identified. Here, size-exclusion chromatography and small-angle X-ray scattering data indicate that Thermus thermophilus (tt) RecR binds to ttRecF to form a globular structure consisting of four ttRecR and two ttRecF monomers. In addition, a low resolution model shows a cavity in the central part of the complex, suggesting that ttRecR forms a ring-like tetramer inside the ttRecFR complex. Mutant ttRecR proteins lacking the N- or C-terminal interfaces that are required for tetramer formation are unable to form a complex with ttRecF. Furthermore, a ttRecFR complex containing the DNA-binding deficient ttRecR K23E/R27E double mutant, which contains mutations lying inside the ring, exhibits significantly reduced dsDNA binding. Thus, we propose that the ring-like ttRecR tetramer has a key role in tethering the ttRecFR complex onto dsDNA and that the ring structure may function as a clamp protein.
INTRODUCTION
Recombinational DNA repair is important for maintaining genomic integrity, since it exploits sequences homologous to the damaged DNA as a substrate for repair, thereby minimizing the loss of genetic information. The RecFOR and/or RecBCD pathways are required to initiate homologous DNA recombination in bacteria (1,2). The Escherichia coli RecFOR pathway is mainly used for single-stranded (ss) DNA gap repair, while the RecBCD pathway is responsible for double-stranded (ds) DNA break repair. However, the RecFOR pathway can also repair dsDNA breaks, as has been demonstrated in recBC sbcB mutants, which are deficient in the RecBCD pathway (3). The RecF, RecO and RecR proteins belong to a group of recombination mediator proteins, which mediate the loading of RecA protein onto ssDNA-binding protein (SSB)-coated ssDNA. RecA then forms a nucleoprotein filament on ssDNA that interacts with free homologous dsDNA to promote heteroduplex formation (4,5).
In contrast to the RecBCD pathway, a detailed mechanism has not been described for the RecFOR pathway. The RecR protein plays a critical role in recombinational DNA repair by forming complexes with RecO or RecF (6,7). The RecOR complex facilitates RecA filament formation on SSB-coated ssDNA and prevents the dissociation of RecA from ssDNA ends (8,9). The RecF protein alone binds dsDNA and has a weak dsDNA-dependent ATPase activity (10), and both of these activities are distinctively enhanced by RecR binding (7). The RecFR complex binds to dsDNA and attenuates the extension of RecA filaments beyond the region of ssDNA in gapped DNA (11). The specific loading of RecA onto dsDNA–ssDNA junctions is mediated by the RecF, RecO and RecR proteins in concert, and RecF, which may associate with RecR, is implicated in recognizing dsDNA–ssDNA junctions (12). Thus, the RecFR complex has multiple roles that regulate RecA filament distribution on ssDNA gap regions.
The crystal structure of RecR from Deinococcus radiodurans (drRecR) revealed it to be a ring-like tetramer that could encircle dsDNA (13), although T. thermophilus (tt) RecR and E. coli RecR are known to form dimers in solution (7,14). The crystal structure of drRecF was also determined recently (15). drRecF exhibits a high degree of structural similarity with the head domain of the recombination protein Rad50, implying that it may undergo ATP-dependent dimer formation coupled with dsDNA binding. However, the structure of the RecFR complex had not been determined, nor had the dsDNA-binding mechanism of the complex, which would be a critical aspect of the RecFR role in recombinational DNA repair.
Here, we analyze the solution structure of ttRecR, ttRecF and ttRecFR complex using the small-angle X-ray scattering (SAXS) technique. Based on SAXS data as well as biochemical evidence we present a ttRecFR model consisting of a ttRecR ring-like tetramer with two ttRecF monomers located near the center. We also found by mutagenesis and biochemical analysis that ttRecR tetramer formation is essential for the dsDNA-binding activity of ttRecFR complex, indicating that the ttRecR ring may function as a clamp on dsDNA.
EXPERIMENTAL PROCEDURES
Protein expression and purification
The ttRecR, ttRecR75–194, ttRecR1–173 and ttRecF proteins were prepared as previously described (16). To prepare the ttRecR K23E/R27E double mutants, point mutations were incorporated into an N-terminally His-tagged ttRecR expression plasmid using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) and confirmed by sequencing. The ttRecR K23E/R27E double mutant was overexpressed in E. coli BL21(DE3) and purified from cell extracts with the Magtration System 6GC (Precision System Science Co., LTD., Matsudo, Chiba, Japan) and Superdex 75 10/300 GL column chromatography (GE Healthcare, Amersham, Buckinghamshire, UK). The His tag was cleaved off by incubating the protein for 1 h at 37°C with thrombin protease (6 units/mg protein; GE Healthcare) before loading it onto the Superdex 75 column.
Size-exclusion chromatography
ttRecF (30 μM), ttRecR (100 μM), ttRecF (30 μM) + ttRecR, ttRecR75–194 or ttRecR1–173 (60 μM) mixtures in buffer (25 mM HEPES-KOH pH 7.2, 150 mM NaCl) were applied to a Superdex-200 HR 10/300 column (GE Healthcare) and eluted at a flow rate of 0.5 ml/min in the same buffer.
Sample preparation for SAXS
Gel filtration chromatography was used to eliminate large aggregates in protein preparations and to ensure the highest quality SAXS data. Prior to SAXS experiments, proteins were purified by preparative gel filtration chromatography of Superdex 200 HR column chromatography resin (GE Healthcare) with a running buffer containing 25 mM HEPES (pH 7.3), 150 mM NaCl and 0.1 mM EDTA. After this treatment, the protein solution was concentrated, dialyzed and maintained on ice. The dialysis buffer was used for background measurements.
SAXS data collection and processing
All SAXS experiments were carried out at the RIKEN Structural Biology Beam line I (BL45XU) at SPring-8, Japan (17). The X-ray wavelength and the sample-to-detector distance were 0.9 Å and 2183 mm, respectively. The sample-to-detector length was calibrated by meridional reflections from collagen. The scattering profiles of protein samples and buffer were collected at 25°C using a cooled CCD equipped with an X-ray image intensifier (18). Buffer and sample were collected with 1 s of exposure time, which ensured that no radiation damage had occurred. For each sample, data were collected at several different protein concentrations, and the scattering data was processing as previously described (19). The radii of gyration, Rg, were determined by fitting the innermost portion of intensity profiles using the Guinier approximation: I(S) = I(0) exp(−4π2Rg2S2/3), where S is the reciprocal parameter that is equal to 2sinθ/λ (where 2θ is the scattering angle and λ is the X-ray wavelength), and I(0) is the forward scattering intensity at a zero angle. The fitting region was from 2 × 10−6 to 23.5 × 10−6 Å−2 of S2. Pair distribution functions, P(r), were calculated by indirect Fourier transformation using the GNOM package (20,21).
Model construction based on SAXS data
First, ab intio low resolution models were calculated using DAMMIN (22) as well as GASBOR (23). The averaged DAM models of the ttRecR dimer and ttRecFR were calculated under the constraints of P2 symmetry, as it was previously reported that ttRecR is a symmetrical homodimer and acts as a structural unit within the complex (14). The model of ttRecF was calculated with no symmetry restrictions. To obtain a representative model, 12 structures were superimposed and averaged using SUBCOMP and DAMAVER (24). Second, rigid body refinement of ttRecFR was done by using SASREF (ver 6.0) (25). The ring-like ttRecR tetramer was generated in reference to the drRecR tetramer as a template, since the ttRecR and drRecR amino acid sequences share 57% identity and 74% similarity. The ttRecR tetramer was fixed, and only the position and orientation of two drRecF molecules (2O5V.pdb) in place of ttRecF were optimized under the constraints of P2 symmetry to fit the data. The rigid body fittings were tried with the symmetry axis located in the ttRecR tetramer ring plane, or perpendicular to the ring. The former alignment resulted in the distribution of ttRecF monomers on each side of the ttRecR ring (trans model), while the other resulted in ttRecF monomers on one side of the ring (cis model). By introducing symmetry constraints, the differences between two models were more evident.
Agarose gel retardation assay
The dsDNA–ttRecFR complex formation was detected using an agarose gel assay according to a previously described protocol (14). Each substrate DNA was incubated with wild-type or mutant ttRecR (5, 10, 15, 20 μM) and ttRecF (2.5, 5, 7.5, 10 μM) in a 20 μl reaction mixture containing 20 mM Tris–HCl (pH 7.5), 150 mM NaCl and 1 mM EDTA at 37°C for 10 min. Samples were analyzed by electrophoresis in a 3% agarose gel with TAE buffer. DNA and DNA–protein complexes were visualized by Gel Star (Lonza Group Ltd., Basel, Switzerland). The nucleotide sequence of the dsDNA substrate is GCGATCCTTATTAAAACGGCAACTCCTCCTCCGGCGGAAAGTCTTCCAAGCCTTCGTCAATGTCCACCCC/GGGGTGGACATTGACGAAGGCTTGGAAGACTTTCCGCCGGAGGAGGAGTTGCCGTTTTAATAAGGATCGC.
RESULTS
Both N- and C-terminus regions of ttRecR are required to form the ttRecFR complex
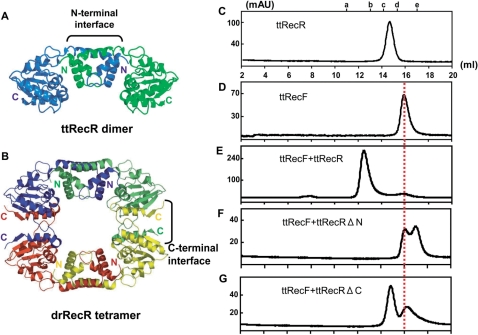
We previously reported that ttRecR forms a stable dimer and presented a ttRecR dimer structural model, which contains an N-terminal interface (16) as shown in Figure 1A. Meanwhile, the crystal structure of drRecR was revealed to be a characteristic ring-like tetramer, in which four subunits were interdigitated with each other in their N- and C-terminal interfaces (13) as described in Figure 1B. Since ttRecR and ttRecF form a 4 : 2 heterohexamer complex of 160 kDa (16), ttRecR may also be able to form a tetramer by forming a complex with ttRecF. To investigate this possibility, ttRecRΔN and ttRecRΔC mutants, which lack the N-terminal (1–74) or C-terminal (174–194) interface, respectively, were prepared. Size-exclusion chromatography was then performed to investigate whether these mutants can still form a ttRecFR complex. The ttRecR dimer, the ttRecF monomer and a ttRecFR complex eluted as single peaks, in general agreement with their calculated molecular masses of 42, 38 and 160 kDa, respectively (Figure 1C–E). When ttRecRΔN or ttRecRΔC was substituted for wild-type ttRecR and subjected to chromatography with ttRecF, 38 kDa and 19.2 kDa or 44.6 kDa peaks were observed, molecular masses that are identical to those of ttRecF and ttRecRΔN or ttRecRΔC, respectively (Figure 1F and G). The elution point of ttRecRΔN and of ttRecRΔC reflects monomer and dimer formation, respectively, since only the N-terminal interface is conserved in ttRecR dimer formation (16). These results indicate that both the N- and C-terminal interfaces corresponding to those of the drRecR tetramer are required to form a stable ttRecFR complex.
Figure 1.
Complex formation analysis of ttRecF, ttRecR and ttRecR deletion mutants by size-exclusion chromatography. (A) Ribbon model structure of the ttRecR dimer. The two subunits are colored in blue and green. (B) Crystal structure of the drRecR tetramer (Protein Data Bank, structure 1VDD). Individual subunits are colored in blue, green, red and yellow. (C–G) Elution profiles of (C) ttRecR, (D) ttRecF, (E) a ttRecF and ttRecR mixture, (F) a ttRecF and ttRecRΔN mutant mixture and (G) a ttRecF and ttRecRΔC mutant mixture determined by size-exclusion column chromatography. The elution volume is shown on the x-axis, and eluted proteins are detected by absorption at 280 nm. The elution points of molecular mass standards are labeled a–e. Molecular mass standards are (a) ferritin, 400 kDa; (b) catalase, 230 kDa; (c) aldolase, 130 kDa; (d) ovalbumin, 45 kDa and (e) chymotrypsin, 25 kDa. The elution point of ttRecF is indicated by a red line.
The association state and shape of ttRecR, ttRecF and ttRecFR complex
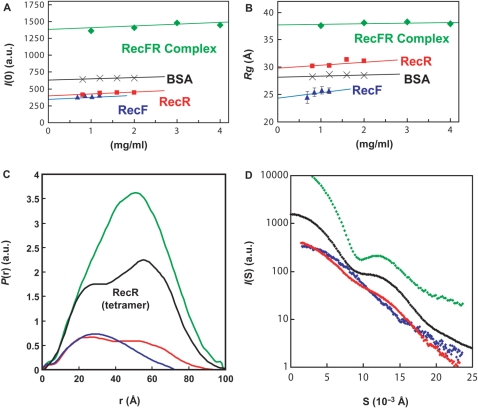
We examined the structural conformation of ttRecR, ttRecF and ttRecFR complex by using the SAXS method. Since SAXS yields information about molecular size and shape, it has been widely used to analyze the conformation of large protein complexes in solution (26,27). Table 1 shows structural parameters of ttRecR, ttRecF and ttRecFR complex obtained from SAXS data. The radius gyration, Rg, is defined as the second moment of the scattering particle, and the forward scattering parameter, I(0), is proportional to the molecular mass of the scattering particle and molecular weight was estimated relative to that of BSA (Supplementary Figure 1). The estimated molecular weights of ttRecF (37 kDa) and ttRecR (43 kDa) were in good agreement with theoretical values for a ttRecF monomer (37.8 kDa) and ttRecR dimer (42.4 kDa). The estimated molecular weight of the ttRecFR complex (142 kDa) was 89% of the sum of the values for four ttRecR and two ttRecF molecules (160 kDa) and is consistent with ttRecFR heterohexamer complex formation considering experimental error. These data are consistent with those of our previous report (16).
Table 1.
Structural parameters for the ttRecR, ttRecF and ttRecFR complex, determined by SAXS experiments
| I(0)a (a.u.) | Rg (Å)b | Dmax (Å)c | Theoretical Mw (kDa)d | Estimated Mw (kDa)e | Association state | |
|---|---|---|---|---|---|---|
| ttRecR | 420 ± 10 | 31.1 ± 0.7 | 85 ± 5 | 42.4 | 43 | Dimer |
| ttRecF | 360 ± 10 | 23.5 ± 2.1 | 65 ± 5 | 37.8 | 37 | Monomer |
| ttRecFR complex | 1390 ± 30 | 38.0 ± 1.5 | 105 ± 5 | 160.4 | 142 | Four ttRecR and two ttRecF |
aForward scattering intensities divided by weight concentration at infinite dilution (Figure 2A).
bRadii of gyration at infinite dilution (Figure 2B).
cLongest linear distances estimated using GNOM (Figure 2D).
dMolecular mass calculated from the amino acid sequence.
eMolecular mass determined with the SAXS experiments using I(0) = 650 obtained for BSA.
Figure 2A and B illustrates the concentration dependence of the scattering intensity of I(0) and Rg. It shows that the oligomerization state of each component was stable within this protein concentration range. The P(r) function, the distribution of interatomic distances within the scattering particle, contains information relevant to the shape of molecule (Figure 2C). The ttRecF P(r) function showed that ttRecF has an elongated shape (blue line in Figure 2C). The P(r) function of the ttRecR dimer is bimodal, with a maximum peak at 25 Å, which points to a dumbbell-like structure (red line in Figure 2C). The bell shape of the P(r) function of the ttRecFR complex indicates that the ttRecFR complex forms a globular shape with a maximum dimension of about 100 Å (green line in Figure 2C).
Figure 2.
SAXS profiles of ttRecR, ttRecF and ttRecFR complexes. (A) Concentration dependence of normalized forward intensity, I(0), determined by Guinier analysis. Green diamonds, ttRecFR complex; red squares, ttRecR; blue triangles, ttRecF and black crosses, BSA. The continuous lines show a linear extrapolation to an infinite dilute solution. (B) Concentration dependence for the apparent radius of gyration, Rg. The zero-extrapolated values of I(0) and Rg are listed in Table 1. (C) Pair distribution functions, P(r), of the ttRecR dimer (Red), ttRecF (blue) and the ttRecFR complex (green). A black line indicates the theoretical curve of the ttRecR tetramer. (D) SAXS profiles. Red, blue and green diamonds represent scattering curves for the ttRecR dimer, ttRecF and the ttRecFR complex, respectively. Black diamonds indicate the theoretical SAXS curve calculated from the crystal structure of drRecR using the program CRYSOL (32).
Solution structure model of ttRecR, ttRecF and ttRecFR complex
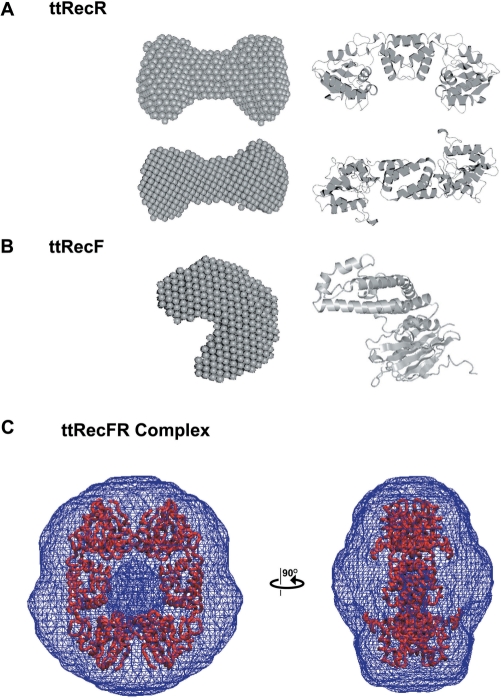
These SAXS data were further analyzed and ab intio low resolution models were created using the DAMMIN and GASBOR programs. The averaged model of ttRecR in solution was well fitted to the ttRecR dimer model, in which the N-terminal region forms the dimer interface (Figure 3A). The ttRecF monomer model was a peanut shape, which is also similar to the structure reported for the drRecF monomer (Figure 3B). The orientation angle of the two domains was slightly different, in that the ttRecF model is more bent compared with the drRecF structure. All ttRecFR models had a large cavity in the center as shown in Figure 3C. To determine whether this feature is a computational artifact, the P(r) function and scattering profile of the ttRecR ring structure were constructed based on the drRecR ring structure (black line in Figure 2C and D). The scattering profiles of both the ttRecFR complex and the ttRecR ring structure (shown as green and black dots in Figure 2D, respectively) showed a common broad peak (S = 0.013 Å−1), which is characteristic for a hollow shape. In addition, the simulated maximum dimension of the ttRecR ring structure was identical to that of the ttRecFR complex (Figure 2C), in which the ttRecR tetramer appears to have a drRecR-like ring structure (see also Figure 3C). Considering that the N- and C-terminal interfaces required for tetramer formation in the case of drRecR are also highly conserved in ttRecR, this large cavity is probably the bent hole of the ttRecR tetramer in the ttRecFR complex. The pore size of ttRecFR looks narrower compared with that of the drRecR tetramer, which may be overlapped by ttRecF. Further structural interpretations of the ttRecFR complex are addressed in the Discussion section.
Figure 3.
Dummy atom models (DAM) of ttRecR, ttRecF and ttRecFR complex. (A) Two orthogonal views of a ttRecR averaged DAM model (left) and the ttRecR dimer structure (right). (B) Averaged DAM model of ttRecF. (C) Averaged GASBOR model of the ttRecFR complex. The averaged GASBOR model was constructed under P2 symmetry constraints with the symmetry axis perpendicular to the long axis. The model is represented in wireframe style, converted by the situs software package, pdb2vol (33). For comparison, the ttRecR tetramer model was superimposed on the GASBOR averaged structure. The dense wire frame in the center corresponds to the cavity.
The ttRecR K23E/R27E double mutation abrogates the interaction of ttRecFR with dsDNA
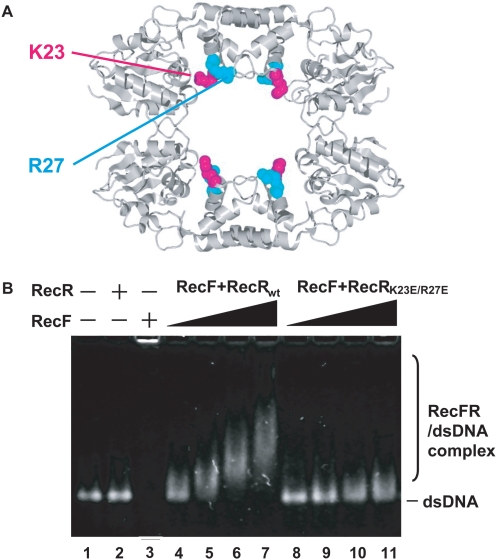
The drRecR tetramer containing the drRecR K23E/R27E double mutant was previously reported to lack dsDNA-binding ability (13). To investigate the role of ttRecR, which may form a ring-like tetramer inside the ttRecFR complex, we generated the ttRecR K23E/R27E double mutant and analyzed the effect of these mutations on dsDNA binding using an agarose gel retardation assay. The conserved K23 and R27 residues lie inside the cavity of the ttRecR tetramer as depicted in Figure 4A. In the assay, the dsDNA-binding activity of the ttRecR dimer was not observed because of its weak affinity (Figure 4B, lane 2). ttRecF was capable of binding dsDNA, but most ttRecF formed aggregates that were trapped in the well of the agarose gel (Figure 4B, lane 3). When ttRecF, ttRecR and dsDNA were all present, ttRecR formed a complex with ttRecF and bound dsDNA in a concentration-dependent manner (Figure 4B, lanes 4–7). In contrast, when the K23E/R27E double mutant was used instead of wild-type ttRecR, the band corresponding to the ttRecFR–dsDNA complex was significantly attenuated, if not abolished (Figure 4B, lanes 8–11). This subtle change may be due to the dsDNA-binding ability of ttRecF. These results support the idea that the K23 and R27 residues face the cavity of the ttRecR tetramer where dsDNA penetrates and binds the ttRecFR complex. This interpretation was confirmed by size-exclusion chromatography, which showed that the ttRecR mutant could form a ttRecFR complex as well as wild-type ttRecR (data not shown). There were no ttRecF–dsDNA aggregates in lanes 8–11, which also supports the conclusion that the mutant retains the ability to bind to ttRecF.
Figure 4.
Effects of the ttRecR K23E/R27E mutation on dsDNA binding by the ttRecFR complex. (A) The two conserved basic residues K23 and R27 of ttRecR are labeled and depicted by a space-filling model in pink and cyan, respectively, on the drRecR tetramer. (B) The agarose gel retardation assay used to detect the binding of dsDNA by ttRecFR complexes. ttRecF (5 μM), ttRecR (10 μM) or a mixture of ttRecF (5 μM) and ttRecR (10 μM) were incubated for 10 min in the presence of a 70 bp dsDNA (20 μM) and ATP (1 mM), and each mixture was then subjected to electrophoresis as described in Experimental procedures section.
DISCUSSION
Here both biochemical and structural results suggested that the ttRecR dimer forms a ring-like tetramer inside the ttRecFR complex and that the ttRecR tetramer has a predominant role in the dsDNA-binding activity of the complex. RecR proteins can form a dimer or tetramer, and their oligomeric state is known to differ from species to species (7,13,14). Our finding of a dimer-to-tetramer transition of ttRecR indicates that RecR tetramer formation might be globally conserved. Moreover, the DNA-binding deficient ttRecR K23E/R27E double mutant significantly reduced the dsDNA-binding activity of ttRecFR complex, suggesting a role for the ttRecR tetramer ring in tethering the ttRecFR complex to DNA through its central pore. This clamp-like role of the ttRecR tetramer readily explains why it can enhance dsDNA binding by ttRecF, whereas ttRecR alone has only weak DNA-binding activity (Supplementary Figure 2). In E. coli, a similar RecR-mediating enhancement of RecF–dsDNA binding is also observed, and it has also been shown that RecR prevents ATP hydrolysis-dependent RecF dissociation from dsDNA (7,28).
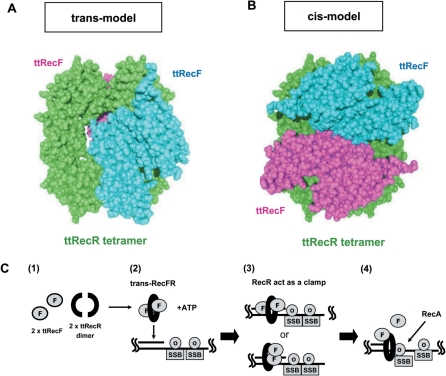
A SAXS model of the ttRecFR complex revealed a globular particle containing a central pore. However, these ab initio models could not clearly show the configuration of ttRecF and the ttRecR tetramer in the complex. Since the ttRecR and ttRecF structures were similar to those of drRecR and drRecF, rigid body refinement, i.e. optimizing the positions/orientations of those components to fit the scattering profile, enabled us to incorporate crystal structural data into the ttRecFR complex model. This refinement procedure assumes that the structure of the ttRecR tetramer in the complex is same as that of the drRecR tetramer and that differences in shape between drRecF and ttRecF can be ignored. The refined ttRecFR complex models fell into two classes. In one model, two ttRecF monomers are distributed on each side of the ttRecR ring (trans model) (χ2 = 21) (Figure 5A). In the other, both ttRecF monomers are on one side of the ring (cis model) (χ2 = 23) (Figure 5B). Although the χ-values cannot completely rule out the possibility of a cis model, trans models always gave a better fitting to the experimental data. These results strongly suggest that the ttRecFR complex exists as a trans complex. As expected from P(r) and ab initio models, none of the models specifies a broad cavity that allows DNA to penetrate. Both ttRecF monomers are located near the center of the ring, which causes the RecFR complex to have a globular shape.
Figure 5.
A rigid body model structure of the ttRecFR complex and proposed scheme of its function. (A) A trans model of the ttRecFR complex constructed by the rigid body model method. Two ttRecF monomers (cyan and pink) bind to the opposite side of the ttRecR tetramer (green). (B) A cis model of the ttRecFR complex constructed by the rigid body model method. Two ttRecF monomers bind to the same side of the ttRecR tetramer. (C) Schematic diagram of the dsDNA clamp model of the ttRecR tetramer in ttRecFOR assembly at dsDNA–ssDNA junctions. dsDNA–ssDNA junctions are generated as a consequence of DNA damage. (1) Two ttRecR dimers form a ring-like tetramer with two ttRecF monomers, resulting in a trans-RecFR complex. (2) ATP causes large conformational changes of the ttRecFR complex to tether the complex to dsDNA. (3) A trans (above) or cis (below) ttRecFR complex recognizes or binds to the junction site and then interacts with ttRecO localized on ssDNA regions containing the SSB protein. (4) The ttRecOR complex, which may associate with ttRecF, facilitates the loading of RecA onto SSB-coated ssDNA. DNA damage is subsequently repaired by homologous recombination mediated by RecA.
It has been reported that the RecFR complex is also required for reassembly of the replication holoenzyme after recombinational DNA repair at replication forks (29). By analogy of DNA replicating clamp proteins such as PCNA (27), the RecR tetramer ring might function as a docking station on DNA for other DNA-modulating enzymes. A recent structural study shows that drRecR and drRecO form a drRecOR heterohexamer complex and that two drRecO monomers are positioned on either side of the drRecR tetramer ring. The positioning of two drRecO monomers next to the drRecR ring resembles our trans model structure of the ttRecFR complex, except that drRecO almost obscures the central cavity of the drRecR ring and inhibits drRecR binding to dsDNA (30). We previously reported that ttRecR preferentially binds ttRecF rather than ttRecO and that the ttRecF-binding site on ttRecR overlaps with that of ttRecO (16). Nevertheless, formation of the RecOR complex is indispensable for mediating RecA filament nucleation on SSB-coated ssDNA, as reported for E. coli (8,31) and T. thermophilus (9). Our previous results also indicate that ttRecO displaces SSB from ssDNA that it forms the ssDNA–RecO–SSB complex and that upon interacting with ttRecR to form the ttRecOR complex it stimulates RecA filament formation (9). Therefore, the ttRecFR complex would deliver the ttRecR ring to ttRecO associated with ssDNA regions near dsDNA–ssDNA junctions, an important target in the RecFOR pathway, as depicted in Figure 5C.
The ttRecR tetramer in the trans model possesses a symmetrical interface for ttRecF on both sides, which provides a mechanism for the stoichiometry of the ttRecR4/ttRecF2 complex. Thus, the trans model appears to be more biochemically appropriate, which is consistent with the rigid body refinement. On the other hand, the cis model leaves open space for new proteins on the opposite side, which may be used for binding of ttRecF, ttRecO or other proteins to exert additional functions. In either case the effects of ATP on ttRecF conformation are expected, since ttRecF contains the conserved Walker-A/Walker-B motifs. Indeed, ATP-dependent dimerization has been reported for drRecF, and it has been proposed that the drRecF dimer forms a symmetrical crab-claw that cradles dsDNA within its cleft (15). We also confirmed the ATP-dependent dimerization of ttRecF by using the SAXS method (Supplementary Figure 3). Thus, ATP-dependent ttRecF dimer formation probably causes large conformational rearrangements such as the conversion of a trans to a cis configuration of the ttRecFR complex, which would be important for dsDNA binding. Further studies on structural changes of the ttRecFR complex coupled with ATP binding and/or hydrolysis analyses are necessary to clarify how it binds to and translocates on dsDNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Prof. S.Kuramitsu for providing expression plasmids for the T. thermophilus RecF pathway proteins used in this study. This work was supported in part by Grants-in-Aid from the Japan Society for the Promotion of Science (JSPS) to T.S. and T.M. Funding to pay the Open Access publication charges for this article was provided by the JSPS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 2.Cox MM. Regulation of Bacterial RecA Protein Function. Crit. Rev. Biochem. Mol. Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- 3.Lloyd RG, Buckman C. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J. Bacteriol. 1985;164:836–844. doi: 10.1128/jb.164.2.836-844.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shibata T, Cunningham RP, DasGupta C, Radding CM. Homologous pairing in genetic recombination: complexes of recA protein and DNA. Proc. Natl Acad. Sci. USA. 1979;76:5100–5104. doi: 10.1073/pnas.76.10.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinstock GM, McEntee K, Lehman IR. ATP-dependent renaturation of DNA catalyzed by the recA protein of Escherichia coli. Proc. Natl Acad. Sci. USA. 1979;76:126–130. doi: 10.1073/pnas.76.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Umezu K, Kolodner RD. Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J. Biol. Chem. 1994;269:30005–30013. [PubMed] [Google Scholar]
- 7.Webb BL, Cox MM, Inman RB. An interaction between the Escherichia coli RecF and RecR proteins dependent on ATP and double-stranded DNA. J. Biol. Chem. 1995;270:31397–31404. doi: 10.1074/jbc.270.52.31397. [DOI] [PubMed] [Google Scholar]
- 8.Bork JM, Cox MM, Inman RB. The RecOR proteins modulate RecA protein function at 5′ ends of single-stranded DNA. EMBO J. 2001;20:7313–7322. doi: 10.1093/emboj/20.24.7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue J, Honda M, Ikawa S, Shibata T, Mikawa T. The process of displacing the single-stranded DNA-binding protein from single-stranded DNA by RecO and RecR proteins. Nucleic Acids Res. 2008;36:94–109. doi: 10.1093/nar/gkm1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madiraju MV, Clark AJ. Evidence for ATP binding and double-stranded DNA binding by Escherichia coli RecF protein. J. Bacteriol. 1992;174:7705–7710. doi: 10.1128/jb.174.23.7705-7710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Webb BL, Cox MM, Inman RB. Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell. 1997;91:347–356. doi: 10.1016/s0092-8674(00)80418-3. [DOI] [PubMed] [Google Scholar]
- 12.Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell. 2003;11:1337–1347. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 13.Lee BI, Kim KH, Park SJ, Eom SH, Song HK, Suh SW. Ring-shaped architecture of RecR: implications for its role in homologous recombinational DNA repair. EMBO J. 2004;23:2029–2038. doi: 10.1038/sj.emboj.7600222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honda M, Rajesh S, Nietlispach D, Mikawa T, Shibata T, Ito Y. Backbone 1H, 13C, and 15N assignments of a 42 kDa RecR homodimer. J. Biomol. NMR. 2004;28:199–200. doi: 10.1023/B:JNMR.0000013821.02982.01. [DOI] [PubMed] [Google Scholar]
- 15.Koroleva O, Makharashvili N, Courcelle CT, Courcelle J, Korolev S. Structural conservation of RecF and Rad50: implications for DNA recognition and RecF function. EMBO J. 2007;26:867–877. doi: 10.1038/sj.emboj.7601537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honda M, Inoue J, Yoshimasu M, Ito Y, Shibata T, Mikawa T. Identification of the RecR Toprim domain as the binding site for both RecF and RecO. A role of RecR in RecFOR assembly at double-stranded DNA-single-stranded DNA junctions. J. Biol. Chem. 2006;281:18549–18559. doi: 10.1074/jbc.M512658200. [DOI] [PubMed] [Google Scholar]
- 17.Fujisawa T, Inoue K, Oka T, Iwamoto H, Uruga T, Kumasaka T, Inoko Y, Yagi N, Yamamoto M, Ueki T. Small-angle X-ray scattering station at the SPring-8 RIKEN beamline. J. Appl. Crystallog. 2000;33:797–800. [Google Scholar]
- 18.Fujisawa T, Inoko Y, Yagi N. The use of a Hamamatsu X-ray image intensifier with a cooled CCD as a solution X-ray scattering detector. J. Synchrotron Rad. 1999;6:1106–1114. [Google Scholar]
- 19.Yamada S, Akiyama S, Sugimoto H, Kumita H, Ito K, Fujisawa T, Nakamura H, Shiro Y. The signaling pathway in histidine kinase and the response regulator complex revealed by X-ray crystallography and solution scattering. J. Mol. Biol. 2006;362:123–139. doi: 10.1016/j.jmb.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Svergun DI. Mathematical methods in small-angle scattering data analysis. J. Appl. Crystallog. 1991;24:485–492. [Google Scholar]
- 21.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallog. 1992;25:495–503. [Google Scholar]
- 22.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dmitri I, Svergun DI, Petoukhov MV, Koch MHJ. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl.Crystallog. 2003;36:860–864. [Google Scholar]
- 25.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pascal JM, Tsodikov OV, Hura GL, Song W, Cotner EA, Classen S, Tomkinson AE, Tainer JA, Ellenberger T. A flexible interface between DNA ligase and PCNA supports conformational switching and efficient ligation of DNA. Mol. Cell. 2006;24:279–291. doi: 10.1016/j.molcel.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 27.Shell SS, Putnam CD, Kolodner RD. The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. Mol. Cell. 2007;26:565–578. doi: 10.1016/j.molcel.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Webb BL, Cox MM, Inman RB. ATP hydrolysis and DNA binding by the Escherichia coli RecF protein. J. Biol. Chem. 1999;274:15367–15374. doi: 10.1074/jbc.274.22.15367. [DOI] [PubMed] [Google Scholar]
- 29.Courcelle J, Carswell-Crumpton C, Hanawalt PC. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc. Natl Acad. Sci. USA. 1997;94:3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timmins J, Leiros I, McSweeney S. Crystal structure and mutational study of RecOR provide insight into its mode of DNA binding. EMBO J. 2007;26:3260–3271. doi: 10.1038/sj.emboj.7601760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Umezu K, Chi NW, Kolodner RD. Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc. Natl Acad. Sci. USA. 1993;90:3875–3879. doi: 10.1073/pnas.90.9.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svergun DI, Barberato C, Koch MHJ. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallog. 1995;28:768–773. [Google Scholar]
- 33.Wriggers W, Chacon P. Using Situs for the registration of protein structures with low-resolution bead models from X-ray solution scattering. J. Appl. Crystallog. 2001;34:773–776. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.