Abstract
XRCC1 is a critical scaffold protein that orchestrates efficient single-strand break repair (SSBR). Recent data has found an association of XRCC1 with proteins causally linked to human spinocerebellar ataxias—aprataxin and tyrosyl-DNA phosphodiesterase 1—implicating SSBR in protection against neuronal cell loss and neurodegenerative disease. We demonstrate herein that shRNA lentiviral-mediated XRCC1 knockdown in human SH-SY5Y neuroblastoma cells results in a largely selective increase in sensitivity of the nondividing (i.e. terminally differentiated) cell population to the redox-cycling agents, menadione and paraquat; this reduced survival was accompanied by an accumulation of DNA strand breaks. Using hypoxanthine–xanthine oxidase as the oxidizing method, XRCC1 deficiency affected both dividing and nondividing SH-SY5Y cells, with a greater effect on survival seen in the former case, suggesting that the spectrum of oxidative DNA damage created dictates the specific contribution of XRCC1 to cellular resistance. Primary XRCC1 heterozygous mouse cerebellar granule cells exhibit increased strand break accumulation and reduced survival due to increased apoptosis following menadione treatment. Moreover, knockdown of XRCC1 in primary human fetal brain neurons leads to enhanced sensitivity to menadione, as indicated by increased levels of DNA strand breaks relative to control cells. The cumulative results implicate XRCC1, and more broadly SSBR, in the protection of nondividing neuronal cells from the genotoxic consequences of oxidative stress.
INTRODUCTION
Base excision repair (BER) operates as the primary pathway for resolving small base modifications, abasic sites and several forms of DNA single-strand breaks (SSBs) (1). Such damage arises upon oxidation or alkylation of DNA, or by spontaneous hydrolytic decay (e.g. deamination of cytosine to uracil). BER is typically initiated by removal of a target base by a specific DNA glycosylase, which generates an abasic site product (AP site). The AP site is then processed either by an AP endonuclease, which incises the phosphodiester bond 5′ to the lesion to create a SSB with an abnormal 5′-deoxyribose phosphate (dRP) residue and a conventional 3′ hydroxy (OH) terminus, or by a bifunctional DNA glycosylase, which cleaves 3′ to the abasic site to produce a SSB with a normal 5′ phosphate (P) group and an atypical 3′ α,β unsaturated aldehyde or 3′ P fragment. The resulting termini are processed accordingly to generate 3′ OH and 5′ P ends to allow gap-filling and nick ligation (2,3).
The polymerization step of BER involves either a short- or long-patch synthetic mechanism (4). In short-patch BER, DNA polymerase β (Pol β) adds a single nucleotide to fill the gap and removes the 5′-dRP group at the SSB site. The nick is then sealed by DNA ligase IIIα (LIG3) in complex with the scaffold protein XRCC1 (X-ray cross-complementing 1) to re-generate an intact strand. Long-patch BER involves the synthesis of a patch of 2–10 nt and the formation of a displaced 5′-flap structure. This pathway engages the proliferating cell nuclear antigen (PCNA)-dependent polymerases Pol δ/ɛ, or Pol β, in conjunction with the 5′-flap endonuclease (Fen1) and DNA ligase I.
An important aspect of BER is the processing of abnormal DNA 3′ or 5′ terminal blocking groups. Such lesions can arise by free radical attack of DNA, or as products or intermediates of specific enzymatic reactions (see for instance above) (3). In order for repair to be successfully completed, abnormal SSB ends, which prevent polymerization and/or ligation, must be converted to 5′ P and 3′ OH groups. Often considered a subpathway of BER, SSBR and its complement of proteins, has evolved to remove abnormal DNA terminal ends. XRCC1 is a key component of SSBR, as cells defective in this nonenzymatic scaffold protein show reduced SSBR, enhanced sensitivity to DNA-damaging agents that produce SSBs, and increased sister chromatid exchange (5). Notably, XRCC1 has been shown to possess important and distinct roles in SSBR in both the G1 and S phase of the cell cycle, presumably reflecting its contribution to global and replication-coupled repair, respectively (6,7). Mice devoid of XRCC1 die early in embryogenesis, indicating an essential role for this protein in animal development (8).
XRCC1 coordinates several key enzymatic factors at sites of SSBs, including the DNA polynucleotide kinase–phosphatase (PNKP), Polβ and LIG3α, via direct physical interactions (9). Recent evidence also indicates critical associations of XRCC1 with two proteins, aprataxin and tyrosyl-DNA phosphodiesterase 1 (TDP1), which are defective in the recessive hereditary spinocerebellar ataxias, ataxia with oculomotor apraxia (AOA1) and spinocerebellar ataxia with axonal neuropathy (SCAN1), respectively (10–14). Aprataxin is responsible for removing obstructive 5′ adenylate groups that arise as abortive DNA ligation intermediates (10), as well as 3′ phosphate and 3′ phosphoglycolate groups that are products of free radical attack of DNA (15). TDP1 functions to excise abortive 3′ topoisomerase 1 (Top1)-DNA intermediates, which occur when Top1 becomes irreversibly trapped on DNA during its strand cleavage reaction, most frequently at sites of nearby DNA damage (16,17).
As the above neurodegenerative disorders are characterized by cerebellar ataxia and peripheral axonal neuropathy without nonneurological symptoms, such as cancer predisposition (18,19), it has been proposed that SSBR defects give rise to neurological dysfunction selectively because nondividing cells, particularly neuronal cells which are highly metabolically active, accumulate a large number of SSBs that have the potential to block transcription and induce apoptosis (20). Conversely, SSBR-deficient dividing cells could potentially resolve SSBs efficiently and accurately using alternative mechanisms, such as replication-dependent homologous recombination (HR). Synergism between HR and SSBR has recently been demonstrated, as HR-deficient BRCA mutant cell lines are inviable upon inhibition of the strand break response protein, poly (ADP) ribose polymerase (PARP-1) (21–23). These observations have prompted the hypothesis that SSBR may have a more prominent role in nondividing tissue. Using XRCC1 as a representative of the global SSBR response, we investigated herein the contribution of this process to oxidative stress resistance in both dividing and nondividing neural cell populations.
MATERIALS AND METHODS
Differentiation of SH-SY5Y cells
Twenty to thirty percent confluent SH-SY5Y cells in a T-75 flask were treated with 10 μM retinoic acid (RA) in high glucose DMEM media (Gibco/Invitrogen, Carlsbad, California, USA) with 1% penicillin–streptomycin and 10% fetal bovine serum (FBS) for 5 days. The RA media was replaced with serum free DMEM with 10 ng/ml brain derived neurotrophic factor (BDNF, Alamone Labs, Jerusalem, Israel), and differentiation was allowed to continue for another 5 days.
Preparation of lentivirus stock and infection of SH-SY5Y cells
An XRCC1-specific TRC shRNA-pLKO.1 plasmid construct (clone ID TRCN0000007913; Open Biosystems, Huntsville, Alabama, USA) or a negative control scramble shRNA-pLKO.1 construct (plasmid #1864; Addgene, Cambridge, Massachusetts, USA) was co-transfected with the packaging plasmid pCMV-dr8.2 dvpr (plasmid #8455; Addgene) and the envelop plasmid pCMV-VSV-G (plasmid #8454; Addgene) into human embryonic kidney 293T cells maintained in the media above using lipofectamine (Invitrogen) and Optimem (Gibco). Media was replaced after 24 h and virus-containing media was collected at 48 and 72 h posttransfection, and filtered using a 0.45-micron PVDF membrane filter (Millipore, Billerica, Massachusetts, USA).
A total of 1 ml or 0.5 ml of the above filtered viral stock was added per well of a 6-well plate to 70% confluent SH-SY5Y cells with 4 μg/ml hexadimethrine bromide (a.k.a. polybrene, Sigma, St. Louis, Missouri, USA). After 24 h, viral media was removed and fresh high glucose DMEM media with 1% penicillin–streptomycin and 10% FBS was added. SH-SY5Y cells were selected after another 24 h in 2.1 μg/ml puromycin (Sigma). Resistant colonies were periodically checked for XRCC1 expression using standard western blotting procedures. Antibodies used were XRCC1 ab-1 or ab-3 (NeoMarkers-Thermo Fisher Scientific Inc., Fremont, California, USA), and β-actin or Lamin AC (Santa Cruz Biotechnology, Santa Cruz, California, USA).
Survival assay for SH-SY5Y cells
Fifty thousand cells/well in a 96-well plate were treated with menadione (1 h), MMS (1 h), arsenic trioxide (24 h), methyl viologen dichloride or paraquat (24 h), or hypoxanthine (at the indicated concentration, see Figures) plus 0.1 U/ml xanthine oxidase (15 min) in standard DMEM media for replicating SH-SY5Y cells or in serum free-BDNF media for the differentiated cells. Following treatment, cells were incubated with 1:10 concentration of WST-1 cell proliferation reagent (Roche, Pleasanton, California, USA) in appropriate media for 4 h at 37°C. Viability was scored using a BioRad microplate spectrophotometer, with the number of viable cells proportional to the OD 450 nm reading. Percent survival for each dose was calculated relative to the OD 450 nm for the untreated samples, which was assumed to represent 100% viability.
SSB measurement
The TUNEL assay, which employs terminal transferase to detect all strand breaks, was modified to use DNA Polymerase I (Pol I) to detect mainly SSBs (24). Pol I-mediated biotin-dUTP nick-end labeling (PUNEL) of SSBs was carried out as outlined in the NeuroTACS™ II in situ apoptosis detection kit (Trevigen Inc., Gaithersburg, Maryland, USA). Briefly, differentiated SH-SY5Y cells, mouse cerebellar granule cells or human fetal brain neuronal–glial cells were fixed in 3.7% formaldehyde or 4% paraformaldehyde, and permeablized for 10 min at room temperature with neuropore solution (Trevigen). Samples were incubated with Pol I (New England Biolabs, Ipswich, Massachusetts, USA) and dNTPs, including biotin-dUTP (Trevigen), for 1 h at 37°C, followed by streptavidin–HRP solution and diaminobenzidine (DAB) or streptavidin–fluor for detection of DNA breaks. Slides were counterstained, dehydrated, mounted in Permount and visualized by light microscopy. Damage was estimated based on the intensity of DAB staining, with increased intensity correlating with greater damage. At least 100 cells were counted per treatment.
Extraction of primary cerebellar granule neurons from mice
Six- to eight-day-old pups from a timed mating between an XRCC1 heterozygous male and a C57BL/6 wild-type female mouse were sacrificed and the cerebellum isolated. The cerebellum was then placed into Hibernate A medium (Brain Bits LLC, Springfield, Illinois, USA) and minced into fine pieces. The minced tissue was digested in Earle's balanced salt solution (EBBS) with 0.1% trypsin (Gibco), 4 mg DNase (Sigma) and 3 mM magnesium sulphate (Sigma) for 15 min at 37°C. Following digestion, the tissue was triturated by passing multiple times through a 22.5G sterile needle until a fine suspension was obtained, which was left at room temperature for 30 min to sediment. The supernatant, containing the single cell solution, was spun at 1000 r.p.m. for 5 min. The pellet was resuspended in neurobasal medium (Gibco) with 10% FBS, 10% horse serum, 0.5 mM glutamine, 25 mM potassium chloride and 2% B-27 supplement. Cells were counted using a Beckman Coulter Z1 Particle counter and plated at 65 000 cells per chamber of an 8-well poly-D-lysine coated chamber slide (Becton Dickinson, Franklin Lakes, New Jersey, USA), and incubated for 2 h at 37°C to allow cells to attach. The media was then changed to serum-free neurobasal medium containing 25 mM KCl, 0.5 mM l-glutamine and 2% B27 supplement and cells were maintained in culture as specified.
Neuronal cell viability
Differentiated mouse cerebellar granule cells in neurobasal maintenance medium (see above) for 7 days were treated with varying concentrations of menadione for 1 h in EBBS, and differentially stained with 15 μg/ml fluorescein diacetate, 5 μg/ml propidium iodide (PI) and 2 μg/ml Hoechst in maintenance media for 5 min at 37°C. The staining solution was washed off with PBS, and cells were mounted in prolong gold antifade reagent (Invitrogen), cured for 24 h in the dark at room temperature, and visualized using a Zeiss Axiovert microscope at 20–40× magnification. Blue Hoechst was used to detect all cells in the frame. Cells with a green fluorescein staining and without PI staining were counted as viable. Cells with red PI staining were counted as dead. Approximately 50 cells were analyzed per group.
Neuronal cell apoptosis
Differentiated mouse cerebellar granule cells were treated with menadione as above and then incubated with a carboxyfluorescein labeled fluormethyl ketone peptide inhibitor of caspase (carboxyfluorescein FLICA apoptosis detection kit, Immunochemistry Technologies LLC, Bloomington, Minneapolis, USA) for 90 min at 37°C. Cells were washed for 30 min at 37°C and stained with 0.5% V/V Hoechst for 5 min at 37°C. Slides were fixed in 3.7% buffered formaldehyde, mounted in Permount (Fisher Scientific, Pittsburgh, Pennsylvania, USA) and visualized using fluorescence and light microscopy. Activation of caspases in apoptotic cells results in the binding of the inhibitor peptide and emission of green fluorescence. Cells stained with both carboxyfluorescein and Hoechst were scored as apoptotic, and cells stained with Hoechst only were scored as nonapoptotic. At least 100 cells per pup, per dose were counted.
Culture of mixed human fetal brain neuronal–glial cultures
Human fetal neuronal–glial cultures were prepared as described previously (25) from human fetal brain specimens of 12–15 weeks gestational age, with consent from women undergoing elective termination of pregnancy, as approved by the Johns Hopkins University Institutional Review Board. Isolated fetal neurons were resuspended in Optimem with 5% heat inactivated FBS, 0.2× N-2 supplement, penicillin G (100 units/ml), streptomycin (100 μg/ml) and amphotericin B (0.25 μg/ml) added. Cells were maintained in tissue culture flasks for at least 4 weeks, then plated into 6-well plates and 35 mm glass bottom dishes (MatTek Corp., Ashland, Massachusetts, USA). Lentiviral infection was performed for 24 h in a 6-well plate essentially as above, except that polybrene was omitted. Human fetal brain cells were maintained for three additional days in regular maintenance medium (Optimem plus supplements) and then fixed with 4% paraformaldehdye in PBS for 15 min at room temperature for immunofluorescence detection of XRCC1. One batch of cells was treated with varying doses of menadione for 1 h, before fixation and used in a PUNEL assay for detection of DNA damage (see above).
Immunocytochemistry of human fetal brain neuronal-glial cultures
Paraformaldehyde fixed neurons were blocked in 5% goat serum at room temperature for 30 min and separately incubated with either a primary XRCC1 antibody (clone 33-2-5; NeoMarkers-Thermo Fisher Scientific Inc.) or a Tuj-1 (neuronal class III β-tubulin) antibody (Sigma) at room temperature for 45 min. Cells were washed with PBS and incubated at room temperature for an additional 45 min with either an Alexa fluor 488 goat-anti mouse secondary antibody (Invitrogen) for detection of XRCC1 or an Alexa fluor goat anti-mouse 546 secondary antibody (Invitrogen) for identification of neurons. Excess secondary antibody was washed off with PBS, samples were mounted in prolong gold antifade reagent with DAPI (Invitrogen) and visualized at 20–40× magnification using a Zeiss Axiovert microscope with detection filters for DAPI, FITC and Cy3 fluorescence. Samples stained with Tuj-1 were further examined for strand breaks and apoptosis using the TUNEL assay (Trevigen) as described above, except that a streptavidin-fluor secondary antibody was used for detection of damage and apoptosis.
RESULTS
The SH-SY5Y cell line is a third generation subclone of the SK-N-SH line established from a bone marrow biopsy of a neuroblastoma patient. The cells are neuronal in origin, arising from the neural crest region of the sympathetic nervous system with both dividing and nondividing sub-populations. The dividing cells can be morphologically and functionally differentiated into a nondividing neuron-like state upon treatment with various reagents such as 12-O-tetradecanoyl-phorbol-13-acetate, RA or staurosporine (26,27). This feature makes this cell line especially suitable for studies on the differential role of specific pathways in dividing versus nondividing (a.k.a. terminally differentiated) neural cell populations.
Morphologically, differentiation is characterized by the extension of neurites with motile terminal growth cones, which on contact with neighboring cells form an intricate network or grid of connections. We initially chose RA to induce differentiation (28), but were unable to achieve complete differentiation, as indicated by a sub-population of cells that continued to divide following several days of RA treatment (data not shown). A subsequent treatment after RA with BDNF in serum free media [as reported in (29)] was therefore adapted and yielded a population of almost exclusively nondividing cells used for subsequent studies (Figure 1A).
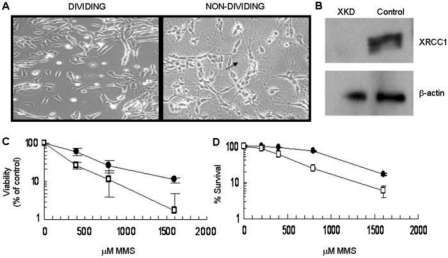
Figure 1.
Differentiation of SH-SY5Y cells and generation of XKD cells. (A) Cellular differentiation of the SH-SY5Y cells was established using a combination of RA and BDNF (see Materials and methods section). Differentiated cells (right) stop dividing, are morphologically distinct in appearance from undifferentiated dividing cells (left), and form distinct neurite processes (arrow) that create an intricate network of neuronal-type connections. (B) Dividing SH-SY5Y cells were infected with a lentivirus bearing an XRCC1-targeting shRNA sequence or a control scramble sequence. Infected cells were selected on the basis of puromycin resistance and expression of XRCC1 was analyzed regularly. Shown is a representative western blot of XRCC1 protein expression in dividing XKD and control cells. β-actin was used as a loading control. (C) Functional down-regulation of SSBR was verified via a WST-1 survival assay following treatment of dividing SH-SY5Y cells with MMS at the indicated doses for 1 h (see Materials and methods section). Percentage survival for XKD (open square) and control scramble (filled circle) cells was calculated as a percentage of survival of untreated cells. Averages and SDs plotted are from two separate runs of three repeats each. (D) Down-regulation of SSBR was independently verified using a colony formation assay. XKD (open square) or control (filled circle) SH-SY5Y dividing cells, which were plated at 500 cells per well, were treated with the indicated doses of MMS for 1 h. Following treatment, cells were washed with 1X PBS and allowed to form colonies under standard growth conditions for about 15 days. Percentage survival was calculated as a percentage of colonies formed relative to the untreated sample. Averages and SDs plotted are from three repeats.
Methods for down-regulation of XRCC1 were established using an XRCC1 specific TRC shRNA-pLKO.1 plasmid construct and a lentiviral package and delivery system as described in Materials and Methods section. A scramble-shRNA (30) was used in the preparation of a control virus for the simultaneous infection of a separate batch of SH-SY5Y cells. Figure 1B shows the expression of XRCC1 protein in the control (scramble) and XRCC1-knockdown (XKD) shRNA replicating cells. Stable suppression of the XRCC1 protein was achieved using the targeting shRNA system, while the scramble-shRNA infected cells retained XRCC1 expression.
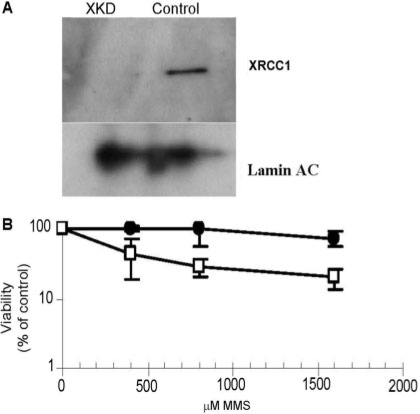
To determine the functional consequence of XRCC1 down-regulation, we evaluated methyl methanesulfonate (MMS) sensitivity. Figure 1C shows the results from a WST-1 viability assay, in which XKD dividing cells were hypersensitive to MMS, a phenotype that is characteristic of XRCC1-deficient mammalian cells (5). A 3- to 6-fold hypersensitivity of the XKD cells to MMS when comparing LD50 values (i.e. the concentration of genotoxin to induce 50% cell killing) was confirmed independently by a colony survival assay (Figure 1D), and is consistent with previous siRNA targeting studies in human cells (13,31,32). Growth, morphology and proliferation rates were also monitored for control and XKD cells and found to be similar (Kulkarni and Wilson, unpublished observation). We point out that the nondividing XKD cells were down-regulated significantly for XRCC1 protein expression and exhibited ∼4-fold increase in sensitivity to MMS as well (Figure 2). Finally, we note that Narciso et al. (33) recently reported down-regulation of XRCC1 in differentiated human skeletal muscle cells; however, analysis of expressional changes of several DNA damage response genes following RA-BDNF differentiation of SH-SY5Y neuroblastoma cells did not reveal down-regulation of XRCC1 protein expression (Kulkarni and Wilson, unpublished observations), suggesting a cell type-specific regulation of this gene product.
Figure 2.
MMS sensitivity of nondividing XKD SH-SY5Y cells. (A) Following establishment of undifferentiated dividing XKD SH-SY5Y cells, a population of these cells was differentiated with RA and BDNF as described in Materials and methods section. XRCC1 knock-down was maintained in these nondividing cells as shown by western blotting (XKD). The control represents the scramble shRNA–lentivirus infected SH-SY5Y cells. Denoted is the position of XRCC1 protein and the Lamin AC loading control. (B) Differentiated, nondividing XKD (open square) or control (filled circle) cells plated at 50 000 cells/well in a 96-well plate were treated with the indicated doses of MMS for 1 h. Following treatment, cells were incubated with the WST-1 reagent for 4 h as described in Materials and methods section. Viability at each dose was determined and calculated as a percent of the reading obtained for untreated cells. Averages and SDs represent three repeats of a representative experiment.
Role of XRCC1 in oxidative stress resistance in dividing and nondividing cells
Following establishment of the above methods, we examined the contribution of XRCC1 to oxidative stress resistance in dividing and nondividing SH-SY5Y cells. We chose oxidative stress as the damaging agent, since neuronal cells, on account of their higher energy needs, are likely to be subject to elevated oxidative tension in vivo and hence increased oxidative DNA damage. Both dividing and nondividing SH-SY5Y cell populations were initially tested with the oxidizing agent menadione, a redox-cycling quinone that undergoes a two step reduction that leads to the production of superoxide radicals (34). As seen from Figure 3A (WST-1 assay), the dividing population of XKD or control cells showed a similar survival response following menadione challenge, suggesting that XRCC1 plays no observable role in oxidative stress resistance in replicating SH-SY5Y cells. On the other hand, the nondividing XKD cells (LD50 of ∼16 μM) showed about 6-fold greater sensitivity to menadione as compared to the control cells with an LD50 value of over 100 μM (Figure 3A). This finding suggests that the lack of XRCC1 and functional SSBR is selectively detrimental to the nondividing cell population, at least in the case of menadione. We also note that, we observed a ≥2-fold increase in sensitivity of nondividing, differentiated XKD cells following menadione exposure when a 24 -h recovery period was incorporated as opposed to the immediate response measured in the assays above (Supplementary Figure 1).
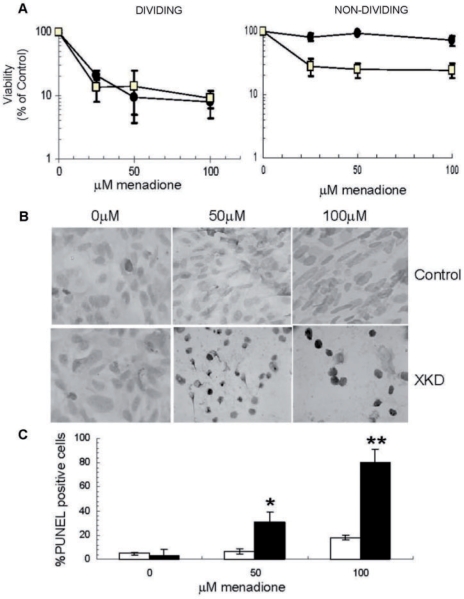
Figure 3.
Role of XRCC1 in stress resistance in dividing and nondividing cells. (A) Survival of XKD (open square) and control cells (filled circle) in dividing (left) and nondividing (right) states, as estimated by a WST-1 survival assay following a 1 h exposure to menadione as described in Materials and methods section. Averages and SDs plotted represent a composite of 6–12 repeats. (B) PUNEL assay carried out on nondividing cells after treatment for 1 h with indicated doses of menadione and recovery in drug-free media for 75 min. XKD and control cells were stained with DAB, which produces a dark brown signal that is indicative of an accumulation of SSBs. (C) Percentage PUNEL positive XKD (black bar) or scramble control (open bar) cells were estimated as a percent of total nondividing cells counted in each frame (B). Percentage PUNEL positive cells represent the number of PUNEL positive cells relative to total cells counted. Averages and SDs plotted represent at least 50 cells scored per dose over three repeats. *P = 0.007 and **P = 0.0004.
To confirm that the above difference in sensitivity is related to a SSBR defect, we examined the recovery of XKD and control nondividing cell populations to DNA damage introduced by menadione. In particular, cells were allowed to recover following menadione treatment in drug-free media for ∼75 min, at which time they were stained for SSB accumulation using the adapted PUNEL assay (see Materials and Methods section). These studies showed little or no DAB staining (dark brown cells) in the RA-BDNF differentiated control cells (Figure 3B), suggesting efficient repair and nearly complete recovery from any DNA damage incurred; this observation is consistent with the near 100% viability observed for these cells after menadione treatment (Figure 3A). The nondividing XKD cells, on the other hand, stained intensely for labeled SSBs, indicating persistent DNA damage and nonfunctional repair (Figure 3B). Additionally, these cells appeared rounded up, and in some cases shrunken or shriveled, suggesting that they might be apoptotic or necrotic. Quantification of the DAB-stained cells as a percent of the total cells showed about a 4-fold increase in the frequency of PUNEL positive cells (and by extension SSBs) in the XKD nondividing cells relative to the control cells at both menadione concentrations (Figure 3C).
We next tested the response of dividing and nondividing control and XKD SH-SY5Y cells to other generators of ROS, namely paraquat, arsenic and a xanthine oxidase-hypoxanthine (XOH) system. As in the case of menadione, treatment with paraquat, another redox cycling agent that generates primarily superoxide (35), led to a more pronounced increase in sensitivity of the XKD nondividing cells (∼6-fold based on LD50), although elevated sensitivity of the dividing population was seen at the highest paraquat concentration (Figure 4A). Arsenic treatment yielded a similar response, whereby the nondividing XKD cells were ∼4-fold more sensitive than the control cells, with no hypersensitivity, and perhaps a slight increased resistance, for the dividing cells (Figure 4B). Interestingly, XOH treatment showed a disparate pattern of response in that both dividing and nondividing XKD cells were more sensitive to oxidative stress, with the greatest sensitivity seen in the former situation (Figure 4C). As gleaned from the survival data, it is noteworthy that terminal differentiation (and perhaps the nondividing status) led to a general increase in stress resistance (to menadione, arsenic and XOH treatments), an observation that is consistent with the reported improvement in resistance of RA treated SH-SY5Y cells to certain drugs and DNA damaging agents (36,37).
Figure 4.
Response of XKD SH-SY5Y cells to other forms of oxidative stress. XKD (open square) and control (filled circle) cells in dividing (left column) and nondividing (right column) states were treated with the indicated concentrations of paraquat-24 h (B), arsenic-24 h (B) and hypoxanthine (for XOH system)-15 min (C) and then examined for viability using the WST1 assay as described in Materials and methods section. Viability for each dose was calculated as a percentage of the OD 450 nm reading relative to the untreated cells. Averages and SDs plotted represent a composite of 4–6 repeats.
Effect of XRCC1 deficiency on primary mouse and human neuronal cell survival
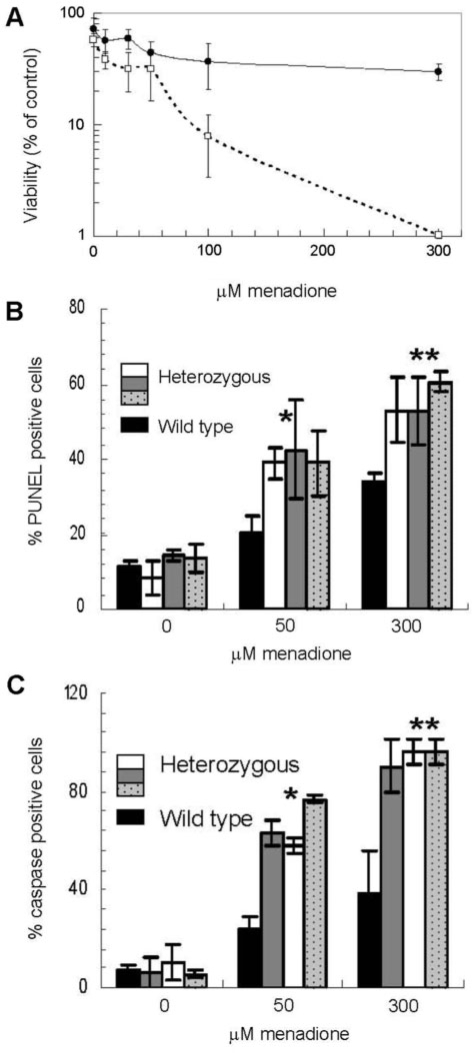
To further investigate the role of XRCC1 in oxidative stress resistance in nondividing neuronal cells, we examined the response of XRCC1-deficient primary cells from both mouse and human in culture. First, we determined the effect of menadione exposure on relative survival, DNA damage accumulation and apoptosis of primary cerebellar granule neurons from wild type or XRCC1 heterozygous 6- to 8-day-old mice. Mouse cerebellar granule neurons are primary cells that are nondividing by nature, and after 7 days in culture, have a phenotype of fully differentiated neurons with an intricate neurite network and synaptic contacts. We examined independent cell populations isolated from four wild-type and three haploinsufficient littermates in the initial experiment. The combined results indicate that XRCC1 heterozygous neurons are hypersensitive to menadione relative to their wild-type counterparts (Figure 5A). At higher doses, the difference in sensitivity of the heterozygous cells is most pronounced, with ∼5-fold hypersensitivity at 100 μM and a 30-fold increase in PI staining frequency at 300 μM menadione. Using a separate set of experimental animals (one wild-type and three haploinsufficient pups in this litter), we also measured apoptosis and the accumulation of DNA damage in isolated primary mouse granule neurons. Consistent with the survival data, these studies revealed an increase in both SSB accumulation (∼1.6- to 1.9-fold, comparing the average percentage of PUNEL positive cells in the heterozygous animals to the wild-type counterpart at 50 or 300 μM menadione; Figure 5B) and apoptosis (∼2.2- to 2.8-fold, derived as above using percentage of caspase positive cells; Figure 5C) in the XRCC1 heterozygous cells challenged with menadione. We note that untreated wild-type and heterozygous cells exhibited similar growth and cell viability for up to 4 weeks in culture at 20% oxygen (Kulkarni, McNeill and Wilson, unpublished observation).
Figure 5.
Role of XRCC1 in stress resistance in wild type and heterozygous mouse neuronal cells. (A) Primary mouse cerebellar granule cells were treated with the oxidizing agent menadione for 1 h at 37°C at the indicated concentrations, and then differentially stained for living and dead cells. XRCC1 heterozygous cells (open square) showed reduced viability as compared to wild type (filled circle). Viability for each treatment was calculated as the percentage of live cells scored per total cells scored. Averages and SDs plotted represent data from three heterozygous and four wild-type pups with at least 50 cells scored per dose, per pup. (B) Mouse cerebellar granule cells were tested for accumulation of SSBs using the PUNEL assay after exposure to menadione. ‘% PUNEL positive cells’ was scored as a percentage of the total cells visualized in each frame. Values plotted represent averages and SDs from at least 100 cells counted over three repeats. P-values were determined using the t-test function in Excel, comparing the percentage PUNEL positive cells for the single wild-type animal to the combined measurements of the three heterozygous mice. *P = 0.002 and **P = 2.8E−05 at the indicated menadione concentration. (C) Mouse cerebellar granule cells were tested for apoptosis following treatment with menadione. Activated caspases were visualized using fluorescence microscopy and ‘% caspase positive cells’ was calculated as percentage of total cells scored. At least 100 cells were counted per treatment for the same pups in panel B to arrive at the plotted averages and SDs. P-values were determined as above, except comparing percentage caspase positive cells of the single wild-type animal to the combined measurements of the three heterozygous mice. *P = 7.2E−05 and **P = 0.0018 (B).
Second, we determined the effect of XRCC1-deficiency on the DNA damage response of primary human fetal neuronal cells using the PUNEL assay after exposure to menadione. Immunocytochemical analysis of a mixed glial–neuronal cell population infected with the XRCC1-targeting shRNA lentivirus system (see above) showed a significant knockdown of the XRCC1 protein relative to the control virus (scramble-shRNA) (Figure 6A). Neuronal cells were distinguished from both glial cells and feeder layer astrocytes using a neuron specific antibody probe (Figure 6B, ‘Tuj-1’ column). As seen in Figure 6B, at the lowest dose of menadione tested (25 μM), XKD human neuronal cells, showed ∼4-fold greater frequency of DNA damage staining relative to the control cells following a 75 min recovery period in drug-free media (100 versus 27%, see Figure 6B). At higher doses of 100 μM (data not shown) and 400 μM (Figure 6B), the damage was found to be about equal for the control and XKD cells, apparently reflecting a near maximal level of DNA damage for both cell populations.
Figure 6.
Oxidative stress response in XKD human fetal brain neurons. (A) Human fetal brain neuronal-glial cell populations were treated with shRNA targeting XRCC1 or a scramble control shRNA via a lentivirus delivery system (see Materials and methods section). Three days post-infection, cells were fixed and stained with DAPI (left column, ‘total cells’) and for XRCC1 protein expression using an XRCC1-specific antibody and a FITC-labeled secondary antibody (center column, ‘XRCC1’). Image shows decreased intensity of FITC staining in XRCC1 shRNA targeted cells (lower panel), indicating significant knockdown of XRCC1. The merge of the DAPI and FITC signals is shown to the right. (B) XKD or control human fetal brain neuronal–glial cell populations were treated with the indicated concentrations of menadione for 1 h (left), allowed to recover in drug free media for 75 min, fixed and then examined for DNA damage and apoptosis using the PUNEL assay (see Materials and methods section). The Tuj-1 neuronal marker, coupled with a cy3 detection system, was used to identify neuronal cells (left column, ‘Tuj-1’). FITC fluorescence indicates DNA damage accumulation and apoptotic cells (center column). For untreated cells, out of 25 XKD cells counted nine were PUNEL positive (36%) as compared to four of 27 (15%) scored for the control cells (P = 0.172). At 25 μM menadione, out of 25 XKD cells counted, all were PUNEL positive (100%), as compared to seven of 26 (27%) scored for the control cells (P = 0.008). P-values were determined via chi-square contingency table analysis at http://www.physics.csbsju.edu/stats/contingency_NROW_NCOLUMN_form.html.
DISCUSSION
XRCC1 is a critical scaffold protein that promotes efficient SSBR by coordinating the enzymatic processing of nonconventional 3′ or 5′ DNA termini and SSB gaps (5,9). Recent evidence has indicated that a defect in SSBR gives rise to the neurodegeneration associated with the hereditary disorders AOA1 and SCAN1 (9,12,18,38–42). Using XRCC1 as a representative of global SSBR, we assessed the role of this response in cellular protection against the deleterious consequences of oxidative stress in both dividing (undifferentiated) and nondividing (differentiated) neural cell populations. Prior work had demonstrated that XRCC1 deficient CHO cells are hypersensitive to hydrogen peroxide (5), an observation that is consistent with the sensitivity data herein using the XOH system (Figure 4C, left), which creates a high level of extracellular hydrogen peroxide as the toxic agent (35,43,44). The studies within are the first to determine the consequences of an XRCC1 defect on cellular sensitivity to the oxidizing agents menadione, paraquat and arsenic.
Using the SH-SY5Y neuroblastoma cell line, which can exist in both a dividing and nondividing state, we examined the effects of lentiviral-mediated shRNA knockdown of XRCC1 on cellular sensitivity to oxidative stress induced by menadione, paraquat, arsenic and XOH. In the case of the first three agents, enhanced sensitivity was seen primarily in the differentiated, nondividing cell population, and was apparently related to a defect in SSBR as evident from the inability of the menadione-treated XKD cells to recover from induced DNA damage (Figure 3B). The only exception to this ‘sensitivity profile’ was the XOH treatment, which caused reduced viability of both dividing and nondividing XKD cells relative to the comparable scramble shRNA controls (Figure 4C). Nonetheless, the typically higher sensitivity of nondividing XRCC1 deficient cells to oxidative stress is consistent with such cells lacking the alternative mechanisms present in dividing cells to resolve SSBs (20).
The disparate profile seen with the different agents above could stem from a difference in the types of ROS formed and thus the types of DNA lesions introduced. Both menadione and paraquat are redox-cycling agents that generate significant levels of intracellular superoxide anions, as well as hydrogen peroxide and other radical species, during their metabolism (34,35,44–46). Arsenic induces the formation of primarily superoxide, singlet oxygen, the peroxyl radical, nitric oxide, hydrogen peroxide, dimethylarsinic peroxyl radicals and the dimethylarsinic radical (47–49). For the XOH system, which produces an extracellular flux of superoxide anion radical and hydrogen peroxide, the molecule most likely responsible for the induction of oxidative DNA damage is the latter diffusible species and its intracellular decomposition products, mainly the hydroxyl radical (35,50,51). Significantly, it has been found that catalase, but not superoxide dismutase, functions to suppress the chromosomal instability associated with XOH, whereas catalase has no effect on, and superoxide dismutase increases, the instability seen with paraquat (35,43,44–51). These findings provide evidence that the differing array of ROS (possibly superoxide versus hydrogen peroxide) can give rise to a disparate spectrum of genotoxic damage, and that the constellation of DNA lesions (base modifications, sugar damage and SSBs), in combination with the platform of active repair responses, will dictate the overall response of XRCC1 deficient cells.
XRCC1 deficiency in primary, nondividing mouse cerebellar granule and fetal human neuronal cells resulted in impaired SSBR and reduced viability under conditions of oxidative stress. Behavioral studies (i.e. rotorod tests) carried out with 19- to 20-month-old XRCC1 heterozygous mice have not, however, revealed spontaneous (i.e. noninduced) motor incoordination relative to age-matched wild-type animals (Paul Pistell, McNeill and Wilson, unpublished data), consistent with the fact that untreated mouse xrcc1+/− neurons display a normal basal level of DNA damage (Figure 5B). Katyal et al. recently reported that mice deleted for tdp1, which results in defective SSBR in neurons, exhibit an age-dependent progressive cerebelar atrophy, but lack the severe motor incoordination seen with SCAN1 patients, suggesting dissimilarity in the mouse and human responses. Additional behavioral and histopathological studies using both XRCC1-deficient and control mice following an external insult are necessary to investigate further the contribution of XRCC1 and functional SSBR in the presentation or acceleration of mammalian neurodegenerative disease. Notably, human XKD fetal brain neurons showed a trend, though not statistically significant, towards increased DNA damage in the untreated samples relative to the controls (see Figure 6B), suggesting that there could exist a set of ataxia patients that arise from genetic defects in XRCC1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank P. Pistell (Pennington Research Institute) for supervising the animal behavioral studies, A. Nath (Johns Hopkins) for providing the human mixed glial-neuronal cell populations, H. Nguyen (Stanford University) for assisting in some of the early mouse brain cell studies, and Z. Guo (NIA) for helpful discussion. We also thank Drs L.Weissman and J-L.Yang (NIA) for critical reading of the manuscript. Funding to pay Open Access publication charges for this article was provided by the Intramural Research Program of the NIH, National Institute on Aging.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wilson DM, III, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst) 2007;6:544–559. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 2.Dianov GL, Parsons JL. Co-ordination of DNA single strand break repair. DNA Repair (Amst) 2007;6:454–460. doi: 10.1016/j.dnarep.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Wilson DM., III Processing of nonconventional DNA strand break ends. Environ. Mol. Mutagen. 2007;48:772–782. doi: 10.1002/em.20346. [DOI] [PubMed] [Google Scholar]
- 4.Fortini P, Dogliotti E. Base damage and single-strand break repair: mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair (Amst) 2007;6:398–409. doi: 10.1016/j.dnarep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Thompson LH, West MG. XRCC1 keeps DNA from getting stranded. Mutat. Res. 2000;459:1–18. doi: 10.1016/s0921-8777(99)00058-0. [DOI] [PubMed] [Google Scholar]
- 6.Fan J, Otterlei M, Wong HK, Tomkinson AE, Wilson DM., III XRCC1 co-localizes and physically interacts with PCNA. Nucleic Acids Res. 2004;32:2193–2201. doi: 10.1093/nar/gkh556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor RM, Thistlethwaite A, Caldecott KW. Central role for the XRCC1 BRCT I domain in mammalian DNA single-strand break repair. Mol. Cell. Biol. 2002;22:2556–2563. doi: 10.1128/MCB.22.8.2556-2563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tebbs RS, Flannery ML, Meneses JJ, Hartmann A, Tucker JD, Thompson LH, Cleaver JE, Pedersen RA. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev. Biol. 1999;208:513–529. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- 9.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 10.Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 11.Clements PM, Breslin C, Deeks ED, Byrd PJ, Ju L, Bieganowski P, Brenner C, Moreira MC, Taylor AM, Caldecott KW. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair (Amst) 2004;3:1493–1502. doi: 10.1016/j.dnarep.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 12.El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 13.Luo H, Chan DW, Yang T, Rodriguez M, Chen BP, Leng M, Mu JJ, Chen D, Songyang Z, Wang Y, et al. A new XRCC1-containing complex and its role in cellular survival of methyl methanesulfonate treatment. Mol. Cell. Biol. 2004;24:8356–8365. doi: 10.1128/MCB.24.19.8356-8365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plo I, Liao ZY, Barcelo JM, Kohlhagen G, Caldecott KW, Weinfeld M, Pommier Y. Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the repair of topoisomerase I-mediated DNA lesions. DNA Repair (Amst) 2003;2:1087–1100. doi: 10.1016/s1568-7864(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi T, Tada M, Igarashi S, Koyama A, Date H, Yokoseki A, Shiga A, Yoshida Y, Tsuji S, Nishizawa M, et al. Aprataxin, causative gene product for EAOH/AOA1, repairs DNA single-strand breaks with damaged 3′-phosphate and 3′-phosphoglycolate ends. Nucleic Acids Res. 2007;35:3797–3809. doi: 10.1093/nar/gkm158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McClendon AK, Osheroff N. DNA topoisomerase II, genotoxicity, and cancer. Mutat Res. 2007;623:83–97. doi: 10.1016/j.mrfmmm.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 18.Moreira MC, Barbot C, Tachi N, Kozuka N, Mendonca P, Barros J, Coutinho P, Sequeiros J, Koenig M. Homozygosity mapping of Portuguese and Japanese forms of ataxia-oculomotor apraxia to 9p13, and evidence for genetic heterogeneity. Am. J. Hum. Genet. 2001;68:501–508. doi: 10.1086/318191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 20.Wilson DM, III, Mattson MP. Neurodegeneration: nicked to death. Curr. Biol. 2007;17:R55–R58. doi: 10.1016/j.cub.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 22.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 23.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Jin K, Chen M, Pei W, Kawaguchi K, Greenberg DA, Simon RP. Early detection of DNA strand breaks in the brain after transient focal ischemia: implications for the role of DNA damage in apoptosis and neuronal cell death. J. Neurochem. 1997;69:232–245. doi: 10.1046/j.1471-4159.1997.69010232.x. [DOI] [PubMed] [Google Scholar]
- 25.Magnuson DS, Knudsen BE, Geiger JD, Brownstone RM, Nath A. Human immunodeficiency virus type 1 tat activates non-N-methyl-D-aspartate excitatory amino acid receptors and causes neurotoxicity. Ann. Neurol. 1995;37:373–380. doi: 10.1002/ana.410370314. [DOI] [PubMed] [Google Scholar]
- 26.Vaughan PF, Peers C, Walker JH. The use of the human neuroblastoma SH-SY5Y to study the effect of second messengers on noradrenaline release. Gen. Pharmacol. 1995;26:1191–1201. doi: 10.1016/0306-3623(94)00312-b. [DOI] [PubMed] [Google Scholar]
- 27.Jalava A, Heikkila J, Lintunen M, Akerman K, Pahlman S. Staurosporine induces a neuronal phenotype in SH-SY5Y human neuroblastoma cells that resembles that induced by the phorbol ester 12-O-tetradecanoyl phorbol-13 acetate (TPA) FEBS Lett. 1992;300:114–118. doi: 10.1016/0014-5793(92)80176-h. [DOI] [PubMed] [Google Scholar]
- 28.Prasad KN. Differentiation of neuroblastoma cells: a useful model for neurobiology and cancer. Biol. Rev. Camb. Philos. Soc. 1991;66:431–451. doi: 10.1111/j.1469-185x.1991.tb01148.x. [DOI] [PubMed] [Google Scholar]
- 29.Encinas M, Iglesias M, Liu Y, Wang H, Muhaisen A, Cena V, Gallego C, Comella JX. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J. Neurochem. 2000;75:991–1003. doi: 10.1046/j.1471-4159.2000.0750991.x. [DOI] [PubMed] [Google Scholar]
- 30.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 31.Fan J, Wilson PF, Wong HK, Urbin SS, Thompson LH, Wilson DM., III XRCC1 down-regulation in human cells leads to DNA-damaging agent hypersensitivity, elevated sister chromatid exchange, and reduced survival of BRCA2 mutant cells. Environ. Mol. Mutagen. 2007;48:491–500. doi: 10.1002/em.20312. [DOI] [PubMed] [Google Scholar]
- 32.Brem R, Hall J. XRCC1 is required for DNA single-strand break repair in human cells. Nucleic Acids Res. 2005;33:2512–2520. doi: 10.1093/nar/gki543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narciso L, Fortini P, Pajalunga D, Franchitto A, Liu P, Degan P, Frechet M, Demple B, Crescenzi M, Dogliotti E. Terminally differentiated muscle cells are defective in base excision DNA repair and hypersensitive to oxygen injury. Proc. Natl Acad. Sci. USA. 2007;104:17010–17015. doi: 10.1073/pnas.0701743104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe N, Dickinson DA, Liu RM, Forman HJ. Quinones and glutathione metabolism. Methods Enzymol. 2004;378:319–340. doi: 10.1016/S0076-6879(04)78024-6. [DOI] [PubMed] [Google Scholar]
- 35.Afanas’ev IB. Superoxide Ion: Chemistry and Biological Implications. CRC Press; 1991. [Google Scholar]
- 36.Lasorella A, Iavarone A, Israel MA. Differentiation of neuroblastoma enhances Bcl-2 expression and induces alterations of apoptosis and drug resistance. Cancer Res. 1995;55:4711–4716. [PubMed] [Google Scholar]
- 37.Shen JH, Zhang Y, Wu NH, Shen YF. Resistance to geldanamycin-induced apoptosis in differentiated neuroblastoma SH-SY5Y cells. Neurosci. Lett. 2007;414:110–114. doi: 10.1016/j.neulet.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 38.El-Khamisy SF, Caldecott KW. TDP1-dependent DNA single-strand break repair and neurodegeneration. Mutagenesis. 2006;21:219–224. doi: 10.1093/mutage/gel024. [DOI] [PubMed] [Google Scholar]
- 39.El-Khamisy SF, Hartsuiker E, Caldecott KW. TDP1 facilitates repair of ionizing radiation-induced DNA single-strand breaks. DNA Repair (Amst) 2007;6:1485–1495. doi: 10.1016/j.dnarep.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 40.Gueven N, Becherel OJ, Kijas AW, Chen P, Howe O, Rudolph JH, Gatti R, Date H, Onodera O, Taucher-Scholz G, et al. Aprataxin, a novel protein that protects against genotoxic stress. Hum. Mol. Genet. 2004;13:1081–1093. doi: 10.1093/hmg/ddh122. [DOI] [PubMed] [Google Scholar]
- 41.Gueven N, Chen P, Nakamura J, Becherel OJ, Kijas AW, Grattan-Smith P, Lavin MF. A subgroup of spinocerebellar ataxias defective in DNA damage responses. Neuroscience. 2007;145:1418–1425. doi: 10.1016/j.neuroscience.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 42.Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, Mendonca P, Costa M, Barros J, Yanagisawa T, et al. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat. Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 43.Phillips BJ, James TE, Anderson D. Genetic damage in CHO cells exposed to enzymically generated active oxygen species. Mutat. Res. 1984;126:265–271. doi: 10.1016/0027-5107(84)90006-x. [DOI] [PubMed] [Google Scholar]
- 44.Sofuni T, Ishidate M., Jr Induction of chromosomal aberrations in cultured Chinese hamster cells in a superoxide-generating system. Mutat. Res. 1984;140:27–31. doi: 10.1016/0165-7992(84)90067-8. [DOI] [PubMed] [Google Scholar]
- 45.Moody CS, Hassan HM. Mutagenicity of oxygen free radicals. Proc. Natl Acad. Sci. USA. 1982;79:2855–2859. doi: 10.1073/pnas.79.9.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yonei S, Noda A, Tachibana A, Akasaka S. Mutagenic and cytotoxic effects of oxygen free radicals generated by methylviologen (paraquat) on Escherichia coli with different DNA-repair capacities. Mutat. Res. 1986;163:15–22. doi: 10.1016/0027-5107(86)90053-9. [DOI] [PubMed] [Google Scholar]
- 47.Shi H, Hudson LG, Ding W, Wang S, Cooper KL, Liu S, Chen Y, Shi X, Liu KJ. Arsenite causes DNA damage in keratinocytes via generation of hydroxyl radicals. Chem. Res. Toxicol. 2004;17:871–878. doi: 10.1021/tx049939e. [DOI] [PubMed] [Google Scholar]
- 48.Shi H, Shi X, Liu KJ. Oxidative mechanism of arsenic toxicity and carcinogenesis. Mol. Cell. Biochem. 2004;255:67–78. doi: 10.1023/b:mcbi.0000007262.26044.e8. [DOI] [PubMed] [Google Scholar]
- 49.Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr. Med. Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- 50.Emerit I, Keck M, Levy A, Feingold J, Michelson AM. Activated oxygen species at the origin of chromosome breakage and sister-chromatid exchanges. Mutat. Res. 1982;103:165–172. doi: 10.1016/0165-7992(82)90024-0. [DOI] [PubMed] [Google Scholar]
- 51.Iwata K, Shibuya H, Ohkawa Y, Inui N. Chromosomal aberrations in V79 cells induced by superoxide radical generated by the hypoxanthine-xanthine oxidase system. Toxicol. Lett. 1984;22:75–81. doi: 10.1016/0378-4274(84)90048-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.