Abstract
In all organisms, translational initiation takes place on the small ribosomal subunit and two classes of methionine tRNA are present. The initiator is used exclusively for initiation of protein synthesis while the elongator is used for inserting methionine internally in the nascent polypeptide chain. The crystal structure of Escherichia coli initiator tRNAfMet has been solved at 3.1 Å resolution. The anticodon region is well-defined and reveals a unique structure, which has not been described in any other tRNA. It encompasses a Cm32•A38 base pair with a peculiar geometry extending the anticodon helix, a base triple between A37 and the G29-C41 pair in the major groove of the anticodon stem and a modified stacking organization of the anticodon loop. This conformation is associated with the three GC basepairs in the anticodon stem, characteristic of initiator tRNAs and suggests a mechanism by which the translation initiation machinery could discriminate the initiator tRNA from all other tRNAs.
INTRODUCTION
During translation initiation, the start codon of the message must be identified and aligned in the P-site of the 30S subunit, so that it basepairs with the anticodon of initiator tRNA. In Escherichia coli, this step involves initiation factor IF3 and requires the presence of three consecutive GC base pairs closing the anticodon loop. Interestingly, these GC base pairs are an exclusive hallmark of cytoplasmic initiator tRNAs from all living organisms (1). It has been shown for a long time that three consecutive GC pairs in the anticodon stem are a crucial feature that distinguishes initiator tRNA from elongator tRNAs during translational initiation (2–5). The presence of this specific sequence in the anticodon stem is associated with a modified reactivity towards nucleases and chemical reagents, suggesting that it induces structural differences related to the translation initiation process (2–4). However, currently available structures of initiator tRNA have so far provided little clues as to what this specific anticodon conformation might be, probably because this stem-loop is often less ordered in crystal structures (6,7). An NMR study comparing initiator and elongator stem-loop structures showed that they have closely similar solution structures (8). However, in the same study, NMR data collected at low temperature revealed a possible second conformation of the initiator stem-loop that could not be further analyzed for technical reasons. In addition, despite the large amount of structural data accumulated on the ribosome (9–11), it is not yet fully understood how the initiator tRNA is selected among all other tRNAs during initiation. Recent crystal structures of 70S ribosome functional complexes (9–11) provide a detailed description of how the ribosome interacts with its mRNA and tRNA partners [for a recent review, (12)]. Actually, the ribosome makes a number of contacts with the anticodon arm of tRNA in the P-site. Among these, the universally conserved nucleotides G1338 and A1339 which are located in the head of the small subunit form type II and type I A-minor interactions (13) with the GC base pairs 29:41 and 30:40 of initiator tRNA. By mutational studies (14,15), these interactions were demonstrated to help in differentiating between the initiator and the elongator tRNAs. However, displacement by IF3 of initiator-like tRNAs that contain these two GC pairs indicates that an additional feature allow to distinguish them from authentic initiator tRNAs (14,15). Additionally, although the three GC pairs are required for discrimination of initiation tRNA by IF3, they are not sufficient to specify the identity of initiator tRNA (4,16). These results are in favor with the formation of an additional interaction possibly specific to the anticodon region of the initiator tRNA that would favor its binding to the P-site in the presence of IF3.
We have determined the crystal structure of E. coli initiator tRNAfMet at 3.1 Å resolution. Overall, tRNAfMet has the classical L-shape geometry, with usual tertiary interactions. However, its structure reveals a unique conformation of the anticodon stem-loop, which explains previous biochemical data and provides information of its specific function in translation initiation.
MATERIALS AND METHODS
tRNA purification
The E. coli tRNAfMet was overexpressed from plasmid pBSTtRNAfMet in E. coli JM101TR using a protocol derived from that of Meinnel et al. (17). Briefly, after phenol extraction of RNAs from bacteria, total tRNA was separated by gel filtration on a HiLoad 26/60 Superdex 75 prepgrade chromatography column (Amersham Biosciences) equilibrated in 25 mM Tris–HCl pH 7.0. The overexpressed tRNAfMet was then separated from other tRNAs by an anion exchange step (Resource Q column) equilibrated in 25 mM Tris–HCl pH 7.0. tRNAs were eluted using a 350–550 mM NaCl gradient in the same buffer. The tRNAfMet was further purified by a hydrophobic interaction step (Phenyl Superose column) equilibrated in 10 mM ammonium acetate pH 6.5, 1.7 M ammonium sulfate. tRNAfMet was eluted using a 1.7–1.0 M ammonium sulfate reverse gradient. The fractions containing the purified tRNAfMet were pooled, dialyzed against 50 mM Tris–HCl pH 8.0, 100 mM KCl, concentrated using Amicon® Ultra (Millipore) and stored at –20°C.
Crystallization and crystal characterization
tRNAfMet was prepared at a initial concentration of 3 mg/ml in 50 mM Tris–HCl pH 8.0, 100 mM KCl and 10 mM MgCl2 and crystallized by the vapor diffusion method at 19°C in several conditions. All conditions contained at least 2.0 M ammonium sulfate as a precipitating agent. Suitable crystals for diffraction data collection, were grown up at two different pH conditions: pH 4.6 (2.0 M ammonium sulfate, 100 mM AcOH/AcONa pH 4.6) and 8.0 (2.0 M ammonium sulfate, 50 mM Tris–HCl pH 8.0). Crystals grew up to 100 μm within 1 week. The hexagonal crystals belonged to space group P6422 with one tRNAfMet per asymmetric unit and with a particularly high solvent content of 79% (Table 1 for statistics).
Table 1.
Data collection and refinement statistics for the determination of tRNAfMet structure
| pH 4.6 (PDB ID 3CW5) | pH 8.0 (PDB ID 3CW6) | |
|---|---|---|
| Data collection statistics | ||
| Beamline (ESRF) | ID14–3 | ID29 |
| Wavelength (Å) | 0.931 | 0.918 |
| No. of frames (Δφ = 1°) | 90 | 90 |
| Space group | P6422 | P6422 |
| Unit-cell parameters (Å) | ||
| a | 93.79 | 93.80 |
| c | 219.36 | 218.59 |
| No. of measured reflections | 79 471 (10 063)a {96 111 (13 300)}b | 68 263 (4286) {82 534 (11 149)} |
| No. of unique reflections | 10 063 (899) {10 810 (1475)} | 8490 (875) {9240 (1318)} |
| Resolution limits (Å) | 40.00–3.10 (3.27–3.10) | 40.00–3.30 (3.48–3.30) |
| Rmergec (%) | 9.9 (47.7) {11.7 (81.0)} | 10.3 (67.4) {12.4 (137.2)} |
| Mean I/σ(I) | 15.7 (2.7) {15.3 (3.5)} | 13.9 (2.2) {13.5 (1.9)} |
| Rpimd (%) | 3.7 (27.4) {4.2 (27.7)} | 3.8 (32.8) {4.4 (49.2)} |
| Data completeness (%) | 91.4 (59.8) {97.9 (95.1)} | 91.4 (67.4) {99.5 (100.0)} |
| Multiplicity | 7.9 (3.2) {8.9 (9.0)} | 8.0 (4.9) {8.9 (8.5)} |
| Wilson B-value (Å2) | 31.3 {65.1} | 36.7 {56.7} |
| Refinement statistics | ||
| Resolution range | 40.00–3.10 (3.18–3.10) | 40.00–3.30 (3.39–3.30) |
| Number of reflexions | 9197 (468) | 7123 (268) |
| R-factor (%) | 24.1 (32.8) | 23.1 (28.1) |
| Rfree (%) | 27.0 (43.4) | 27.4 (39.5) |
| Number of tRNA atoms | 1645 | 1645 |
| RMSD (bonds) (Å) | 0.009 | 0.009 |
| RMSD (angles) (°) | 1.735 | 1.946 |
| Mean residual B-value (Å2)e | 19.5 | 82.3 |
| Solvent content (%) | 79 | 79 |
aValues for the outermost shell are given in parenthesis.
bValues before anisotropic treatment are given in brackets. Statistics for anisotropic data sets, used in structure refinement, were assessed on data sets including only reflections with I/σ(I) > 2.0. The lower completeness at the high-resolution limit reflects the diffraction anisotropy.
c , where the outer sum (h) is over the unique reflections, and the inner sum (i) is over the set of independent observations of each unique reflection.
, where the outer sum (h) is over the unique reflections, and the inner sum (i) is over the set of independent observations of each unique reflection.
d , where the outer sum (h) is over the unique reflections, the inner sum (i) is over the set of independent observations of each unique reflection, and N the redundancy of the measured reflections.
, where the outer sum (h) is over the unique reflections, the inner sum (i) is over the set of independent observations of each unique reflection, and N the redundancy of the measured reflections.
eAs TLS parameters were refined, B-value include only residual B-value.
Data collection, structure solution and refinement
The crystals were harvested, soaked in a cryoprotectant solution [2.2–2.4 M ammonium sulfate, MgCl2 10 mM, glycerol 20% (v/v), and 50 mM AcOH/AcONa pH 4.6 or 50 mM Tris–HCl pH 8.0 depending the pH at which the crystals were grown up] for about 1–5 min and flash-frozen in liquid nitrogen before data collection. Diffraction data were collected at beam lines ID14–3 and ID29 of the European Synchrotron Radiation Facility (ESRF, Grenoble, France). All crystallographic calculations were performed using the CCP4 suite version 6 (18) as implemented in the graphical user interface (19). X-ray diffraction data were processed using MOSFLM (20) and scaled with SCALA (21). The structure of tRNAfMet was solved by molecular replacement using the program PHASER (22) and the coordinates of tRNAfMet from the E. coli tRNAfMet/formylase complex (PDB entry code 2FMT) as a model. Model and map visualizations for manual reconstruction were performed with the program COOT (23). In the last stages of refinement, TLS parameters (24) were refined using one group for the entire tRNA molecule. The acidic structure was refined as described in the following paragraph, and the model was then used to refined the basic structure.
Coordinates and structure factors of E. coli tRNAfMet have been deposited at the Protein Data Bank with the accession code 3CW5 and 3CW6.
RESULTS AND DISCUSSION
Structure determination
We have solved the crystal structure of the E. coli initiator tRNAfMet at two pH conditions (i.e. 4.6 and 8.0) using careful treatment of data anisotropy. Indeed, the diffraction patterns of the two crystal forms were severely anisotropic, with the acidic condition giving however higher resolution limits (Table 1). For instance, in the acidic condition, diffraction limits are 2.8 Å along the a* and b* directions, and 3.4 Å along the c* direction. We first attempted to refine the structure using the anisotropic scaling procedure applied by REFMAC (25). However, the obtained electron density maps were relatively featureless, especially around the anticodon stem-loop. To improve map quality, we then tried the procedure described by David Eisenberg's group (26) to deal with highly anisotropic data (http://www.doe-mbi.ucla.edu/~sawaya/anisoscale/). Briefly, data falling outside an ellipse centered at the reciprocal lattice origin and having vertices at 1/2.8, 1/2.8 and 1/3.4 Å−1 along a*, b* and c*, respectively, were removed. After this ellipsoidal truncation, anisotropic scale factors were applied. Lastly, a negative isotropic B factor was used to restore the magnitude of the high-resolution reflections diminished by anisotropic scaling. The statistics of the anisotropically scaled dataset are shown in Table 1. The structure was then refined to a resolution of 3.1 Å against the anisotropically scaled data using the program REFMAC (25). This yielded maps of very good quality including for the anticodon region (Figure 1 and Supplementary Figure 1). The final model shows a R-factor of 24.1% and a Rfree factor of 27.0% (Table 1). Data from crystals grown under basic conditions were processed similarly. The corresponding structure was refined to a resolution of 3.3 Å (R = 23.1%; Rfree = 27.4%). Otherwise stated, we will describe the tRNAfMet structure obtained in the acidic conditions that is solved at a higher resolution.
Figure 1.
Electron density maps around anticodon and TΨC arms of tRNAfMet structure in the acidic conditions. (a) The anticodon arm showing the A37 base triple interaction with G29-C41. (b) The TΨC arm showing well-defined base and phosphate position. The electron density map corresponds to a 2Fo–Fc contoured at 1σ.
Overall model and crystal packing
The overall tertiary structure of tRNAfMet matches the well-known L-shape structure first revealed in yeast tRNAPhe (27) with usual tertiary interactions (Figure 2): the reverse-Hoogsteen base pairs s4U8•A14 and T54•A58, the trans Watson–Crick G15•C48 (‘Levitt pair’), the one bond imino-4-carbonyl G18•Ψ55, the Watson–Crick base pair G19–C56 forming the corner of the L, the Watson–Crick like G26•A44, the ‘U-turn’ at U33 in the anticodon loop and at Ψ55 in the T loop, and finally the base triples A46•(G22–C13) and G45•(G10–C25). The base triple equivalent to the A9•(A23–U12) found in tRNAPhe, i.e. G9•(C23–G12) was not formed in tRNAfMet where N7-G9 is at 5.2 Å away from N4-C23.
Figure 2.
Tertiary interactions on the secondary structure of E. coli tRNAfMet. Solid lines represent base–base tertiary interaction through hydrogen bonds, and dashed lines represent base stacking interactions.
The crystal packing is quite remarkable. First, the acceptor arm of the tRNA molecule interacts with the major groove of the acceptor stem of a symmetry-related tRNA molecule, thereby forming a triple helix (Figure 3a and b). Namely, nt 72–76 of one tRNA molecule are basepaired in an antiparallel manner to G2-G6 of the acceptor stem of another tRNA molecule. The C1 nucleotide is very mobile and is not visible in electron density maps. However the triple helix formation necessarily rejects C1 in the solvent with G2 rather stacks on A72. Therefore, the always mismatched 1 and 72 bases, characteristic of bacterial initiator tRNAs, are not face-to-face. Another packing region involves anticodon–anticodon base pairing (Figure 3c and d). Anticodon loops interact through pairing between C34A35U36A38 and the four same bases in a symmetry-related tRNA molecule, thereby forming two Watson–Crick AU pairs and two non-canonical CA pairs. The same packing interaction at the level of the anticodon was previously observed in the E. coli tRNAfMet/formylase complex (PDB entry code 2FMT) and in the yeast initiator tRNA crystal structure (PDB entry code 1YFG). In tRNAAsp, the crystal packing also involves anticodon–anticodon interactions (28). We cannot exclude that the crystal packing at the level of the anticodon loop, in particular contacts between C34 and A38, could contribute to the stabilization of the anticodon loop conformation, in particular stabilisation of the non-canonical Cm32•A38 base pair described in the following paragraph.
Figure 3.
Packing interaction in the crystal structure of tRNAfMet. (a) and (b) in the acceptor stem, (c) and (d) in the anticodon loop.
Although the overall structure globally resembles that of elongator tRNAs, there are however striking features at the level of the anticodon arm that deserve discussion.
The anticodon stem-loop of initiator tRNA adopts a non-canonical conformation
The structure of the anticodon stem-loop region can be described by three specific features (Figure 4): (i) an additional Cm32•A38 unusual base pair extending the anticodon stem (Figure 4c). (ii) Formation of a base triple between A37 in the anticodon loop and G29-C41 in the stem (Figure 4d). This induces a large turn in the phosphate backbone just after the anticodon triplet. (iii) An unusual stacking pattern for the anticodon loop: A38 is directly stacked onto the last base of the anticodon (U36), whereas it is stacked on nt 37 in all other tRNA structures (Figure 4a and b). Overall, this induces a more compact and constrained geometry, stabilized by additional specific interactions involving the first and the last initiator-characteristic GC pairs. The Cm32•A38 wobble-like base pair, not encountered in other tRNA structures (29), is stabilized by stacking onto G31-C39 and by a crystal contact between A38 and C34. It involves a polar contact between N1(A38) and O2(Cm32) in addition to the N3(Cm32)–N6(A38) hydrogen bond and suggests that the N1 group of A38 is protonated (Figure 4c). The particular Cm32•A38 wobble-like base pair is probably formed because of the acidic pH of the crystallization conditions. Unfortunately, the lower resolution of the model refined from crystals grown at pH 8.0 did not allow us to confirm this proposal, since the prediction on the protonation state from the structure depends on small variations in base pair orientation that could not be strongly reliable at this resolution. In the structure of the E. coli tRNAfMet/formylase complex (30), which was determined using crystals grown at pH 6.6, the Cm32•A38 base pair is less tight than in the present structure. However, the two bases are also facing each other. Such an (A+)•C base pair was already shown to occur, with a pKa of about 6.0, in the NMR structures of hypomodified tRNA3Lys anticodon stem-loop (31). In tRNAfMet, as a consequence of the (A+)•C pairing, the anticodon stem extends to Cm32•A38.
Figure 4.
Unique conformation of the anticodon arm of the E. coli initiator tRNAfMet: (a) the anticodon arm of tRNAfMet, (b) for comparison tRNAPhe anticodon arm (PDB ID 1EHZ), (c) the Cm32•A38 wobble-like base pair observed in crystal structure of tRNAfMet and (d) the A37• (G29-C41) base triple observed in crystal structure of tRNAfMet.
Additionally, A37 participates in a base triple within the major groove of the anticodon stem. The Watson–Crick edge of A37 interacts with the Hoogsteen edge of G29 and is stabilized by an hydrogen bond between A37 O4′ and C39 N4 (Figure 2d). This triple interaction is probably strengthened by protonation of A37 N1. The A37•(G29-C41) interaction has never been described and is clearly observed in the present structures obtained from crystals at two pH conditions (see Supplementary Figure 2 for the basic structure). As a consequence, the major groove of the anticodon stem is severely obstructed at the level of the three GC pairs. This could explain the previously reported unreactivity of the N7 groups of G29, G30, G31 in the major groove of E. coli initiator tRNA anticodon stem (32). In particular, the N7 group of G30 which is reactive in all elongator tRNAs does not react with dimethyl-sulfate in tRNAfMet. In conclusion, two features are undoubtedly conserved in the tRNAfMet structures solved at two pH conditions: the peculiar stacking of bases in the anticodon loop and the base triple made by A37.
E. coli initiator tRNA was previously crystallized in complex with methionyl–tRNAfMet transformylase (30). The anticodon stem-loop region does not interact with the protein. In this structure, A37, although not involved in a base-triple with G29-C41, also fails to stack between U36 and A38 (Figure 3a and b), in contrast to what is observed in all available elongator tRNA structures (27,33–38). Differences observed for the position of A37 in the present structure and in the tRNAfMet/formylase complex structure are in favor of its mobility. In this context, it should be noted that in the structure of tRNAfMet/formylase complex the position of the adenine ring of base 37 was only tentatively modelled (30). Increased mobility of A37 in the tRNAfMet/formylase complex is also supported by an increased RNase sensitivity at phosphate G29, G30 upon binding of tRNAfMet to the formylase (39). Finally, although in yeast initiator tRNA structure the anticodon region is poorly defined (6), a similar organization of the anticodon bases was observed, with U36 stacked on A38 and A37 rejected in the solvent. Therefore, in available X-ray structures, initiator tRNAs appear consistently different from elongator tRNAs at the level of the base stacking of the anticodon loop. This corroborates their unique S1 nuclease cleavage pattern (2,3,40). Indeed, all initiator tRNAs (E. coli, yeast and mammalian) are cleaved after C34 and A35 (2), while the anticodon loops of elongator tRNAs, including elongator tRNAMet, are much more sensitive, being cleaved after U33, C34, A35 and U36 (2,40). Interestingly, in yeast, the anticodon loop sequences of initiator and elongator tRNAMet are identical, including for the post-transcriptional base modification. These observations led to the proposal that the three conserved GC base pairs of the initiator anticodon stem were the key determinants for an unusual conformation of the anticodon loop (2–4). This hypothesis is in full agreement with the structure reported here.
Relevance for initiator tRNA discrimination
Mutagenesis of the anticodon or stem-loop showed that the S1 cleavage pattern correlates with the initiator function and selection by IF3 (3,4). This strongly suggests that the peculiar conformation of the anticodon loop of initiator tRNAs is required for their initiator function. This raises the question of the detailed underlying mechanism. In the structure of the Thermus thermophilus 70S ribosome, tRNAfMet is paired with the initiation codon in the P-site (9), and the canonical conformation of the anticodon loop is restored, with A37 stacked between U36 and A38 (Figure 3c). Such a conformation was also seen in the structure of E. coli ribosome with a tRNAfMet anticodon stem-loop paired to mRNA in the P-site (41). This conformation is stabilized by a number of contacts within the P-site. Among these, universally conserved bases G1338 and A1339 make types I and II A-minor interactions with G29-C41 and G30-C40, respectively (9–11). Although the GC pairs are crucial for IF3-dependent tRNAfMet discrimination, they are too far from its 30S subunit binding site to be directly inspected by IF3, according to the current understanding of the position of IF3 (42). This led these authors to propose that under the influence of IF3 binding, G1338 and A1339 might check the identity of tRNAfMet through their minor groove interactions with these conserved GC. By mutational studies, these interactions were actually shown to play a role in the IF3-dependent tRNAfMet discrimination (14,15). However, tRNAs that contain two consecutive GC pairs at positions 29–41 and 30–40 are not resistant to rejection by IF3, indicating that discrimination must involve recognition of at least one additional feature of the tRNAfMet anticodon stem-loop (15). The singular structure of the anticodon stem-loop that we report here strongly supports this idea.
Relevance for translation initiation
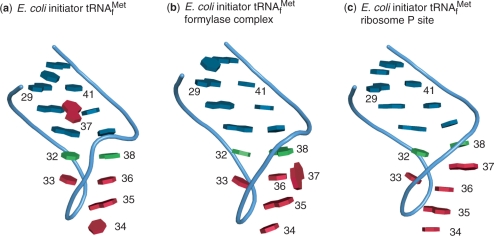
Clearly, the ‘37-unstacked’ conformation observed in the anticodon loop of free initiator tRNA is markedly different from the ‘37-stacked’ observed in P-site-bound tRNAfMet. Therefore, the available data suggests that an initiator tRNA must switch between the two conformations during the initiation process. An attractive hypothesis would be that a ‘37-unstacked’ conformation is required at the first stages of initiator tRNA accommodation by the ribosomal P-site and IF3. Recognizing a ‘37-unstacked’ conformation would help to reject elongator tRNAs (Figure 5). In a second step, stabilization of the ‘37-stacked’ conformation would require correct pairing with the AUG initiation codon. Such a conformational transition could be used as a ‘sensor’ that a correct initiation codon has been found. Interestingly, in the present structure as well as in that of the formylase/tRNAfMet complex and in that of yeast initiator tRNA, crystal packing involves only two Watson–Crick base pairs of the anticodon (A35 and U36; see Supplementary Figure 2). In the context of the above hypothesis, such a structure may mimic pairing with an incorrect start codon, unable to trigger a ‘37-stacked’ conformation of the anticodon loop.
Figure 5.
Comparison of the anticodon loop base stacking. (a) E. coli tRNAfMet, (b) E. coli tRNAfMet in the complex with formylase (PDB ID 2FMT), (c) in the P-site (PDB ID 2J00). The ‘37-unstacked’ conformation refers to (a) and (b) structures and the ‘37-stacked’ to the (c) one.
SUPPLEMENTARY DATA
Supplementary Data are available on NAR Online.
ACKNOWLEDGEMENTS
The authors acknowledge the European Synchrotron Radiation Facility for providing the synchrotron radiation facilities on beamlines ID14-3 and ID29. P.B. was supported by a studentship from the ‘Ministère de la recherche’. We thank the CNRS for financial support. Funding to pay the Open Access publication charges for this article was provided by CNRS, French National Agency of Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wrede P, Woo NH, Rich A. Initiator tRNAs have a unique anticodon loop conformation. Proc. Natl Acad. Sci. USA. 1979;76:3289–3293. doi: 10.1073/pnas.76.7.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seong BL, RajBhandary UL. Escherichia coli formylmethionine tRNA: mutations in GGGCCC sequence conserved in anticodon stem of initiator tRNAs affect initiation of protein synthesis and conformation of anticodon loop. Proc. Natl Acad. Sci. USA. 1987;84:334–338. doi: 10.1073/pnas.84.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartz D, Binkley J, Hollingsworth T, Gold L. Domains of initiator tRNA and initiation codon crucial for initiator tRNA selection by Escherichia coli IF3. Genes Dev. 1990;4:1790–1800. doi: 10.1101/gad.4.10.1790. [DOI] [PubMed] [Google Scholar]
- 5.Varshney U, Lee CP, RajBhandary UL. From elongator tRNA to initiator tRNA. Proc. Natl Acad. Sci. USA. 1993;90:2305–2309. doi: 10.1073/pnas.90.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basavappa R, Sigler PB. The 3 A crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination. EMBO J. 1991;10:3105–3111. doi: 10.1002/j.1460-2075.1991.tb07864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo NH, Roe BA, Rich A. Three-dimensional structure of Escherichia coli initiator tRNAfMet. Nature. 1980;286:346–351. doi: 10.1038/286346a0. [DOI] [PubMed] [Google Scholar]
- 8.Schweisguth DC, Moore PB. On the conformation of the anticodon loops of initiator and elongator methionine tRNAs. J. Mol. Biol. 1997;267:505–519. doi: 10.1006/jmbi.1996.0903. [DOI] [PubMed] [Google Scholar]
- 9.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 10.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 11.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 12.Korostelev A, Noller HF. The ribosome in focus: new structures bring new insights. Trends Biochem. Sci. 2007;32:434–441. doi: 10.1016/j.tibs.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdi NM, Fredrick K. Contribution of 16S rRNA nucleotides forming the 30S subunit A and P sites to translation in Escherichia coli. RNA. 2005;11:1624–1632. doi: 10.1261/rna.2118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lancaster L, Noller HF. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol. Cell. 2005;20:623–632. doi: 10.1016/j.molcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Mangroo D, Limbach PA, McCloskey JA, RajBhandary UL. An anticodon sequence mutant of Escherichia coli initiator tRNA: possible importance of a newly acquired base modification next to the anticodon on its activity in initiation. J. Bacteriol. 1995;177:2858–2862. doi: 10.1128/jb.177.10.2858-2862.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meinnel T, Mechulam Y, Fayat G. Fast purification of a functional elongator tRNAMet expressed from a synthetic gene in vivo. Nucleic Acids Res. 1988;16:8095–8096. doi: 10.1093/nar/16.16.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 19.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 20.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr. D. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 21.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 22.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim SH, Quigley GJ, Suddath FL, McPherson A, Sneden D, Kim JJ, Weinzierl J, Rich A. Three-dimensional structure of yeast phenylalanine transfer RNA: folding of the polynucleotide chain. Science. 1973;179:285–288. doi: 10.1126/science.179.4070.285. [DOI] [PubMed] [Google Scholar]
- 28.Moras D, Dock AC, Dumas P, Westhof E, Romby P, Ebel JP, Giege R. Anticodon-anticodon interaction induces conformational changes in tRNA: yeast tRNAAsp, a model for tRNA-mRNA recognition. Proc. Natl Acad. Sci. USA. 1986;83:932–936. doi: 10.1073/pnas.83.4.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auffinger P, Westhof E. Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J. Mol. Biol. 1999;292:467–483. doi: 10.1006/jmbi.1999.3080. [DOI] [PubMed] [Google Scholar]
- 30.Schmitt E, Panvert M, Blanquet S, Mechulam Y. Crystal structure of methionyl-tRNAfMet transformylase complexed with the initiator formyl-methionyl-tRNAfMet. EMBO J. 1998;17:6819–6826. doi: 10.1093/emboj/17.23.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durant PC, Davis DR. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J. Mol. Biol. 1999;285:115–131. doi: 10.1006/jmbi.1998.2297. [DOI] [PubMed] [Google Scholar]
- 32.Wakao H, Romby P, Westhof E, Laalami S, Grunberg-Manago M, Ebel JP, Ehresmann C, Ehresmann B. The solution structure of the Escherichia coli initiator tRNA and its interactions with initiation factor 2 and the ribosomal 30 S subunit. J. Biol. Chem. 1989;264:20363–20371. [PubMed] [Google Scholar]
- 33.Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A. Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature. 1974;250:546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- 34.Suddath FL, Quigley GJ, McPherson A, Sneden D, Kim JJ, Kim SH, Rich A. Three-dimensional structure of yeast phenylalanine transfer RNA at 3.0angstroms resolution. Nature. 1974;248:20–24. doi: 10.1038/248020a0. [DOI] [PubMed] [Google Scholar]
- 35.Jovine L, Djordjevic S, Rhodes D. The crystal structure of yeast phenylalanine tRNA at 2.0 A resolution: cleavage by Mg(2+) in 15-year old crystals. J. Mol. Biol. 2000;301:401–414. doi: 10.1006/jmbi.2000.3950. [DOI] [PubMed] [Google Scholar]
- 36.Shi H, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: a classic structure revisited. RNA. 2000;6:1091–1105. doi: 10.1017/s1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Westhof E, Dumas P, Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J. Mol. Biol. 1985;184:119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- 38.Benas P, Bec G, Keith G, Marquet R, Ehresmann C, Ehresmann B, Dumas P. The crystal structure of HIV reverse-transcription primer tRNA(Lys,3) shows a canonical anticodon loop. RNA. 2000;6:1347–1355. doi: 10.1017/s1355838200000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramesh V, Mayer C, Dyson MR, Gite S, RajBhandary UL. Induced fit of a peptide loop of methionyl-tRNA formyltransferase triggered by the initiator tRNA substrate. Proc. Natl Acad. Sci. USA. 1999;96:875–880. doi: 10.1073/pnas.96.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wrede P, Rich A. Stability of the unique anticodon loop conformation of E.coli tRNAfMet. Nucleic Acids Res. 1979;7:1457–1467. doi: 10.1093/nar/7.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berk V, Zhang W, Pai RD, Cate JH. Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl Acad. Sci. USA. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dallas A, Noller HF. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell. 2001;8:855–864. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.