Abstract
During the highly conserved process of eukaryotic ribosome formation, RNA follows a maturation path with well-defined, successive intermediates that dynamically associate with many pre-ribosomal proteins. A comprehensive description of the assembly process is still lacking. To obtain data on the timing and order of association of the different pre-ribosomal factors, a strategy consists in the use of pre-ribsomal particles isolated from mutants that block ribosome formation at different steps. Immunoblots, inherently limited to only a few factors, have been applied to evaluate the accumulation or decrease of pre-ribosomal intermediates under mutant conditions. For a global protein-level description of different 60S ribosomal subunit maturation intermediates in yeast, we have adapted a method of in vivo isotopic labelling and mass spectrometry to study pre-60S complexes isolated from strains in which rRNA processing was affected by individual depletion of five factors: Ebp2, Nog1, Nsa2, Nog2 or Pop3. We obtained quantitative data for 45 distinct pre-60S proteins and detected coordinated changes for over 30 pre-60S factors in the analysed mutants. These results led to the characterisation of the composition of early, intermediate and late pre-ribosomal complexes, specific for crucial maturation steps during 60S assembly in eukaryotes.
INTRODUCTION
Eukaryotic ribosome biogenesis is a complex cellular pathway, which requires a large number of proteins, stably or transiently associated to many macromolecular complexes. This pathway results in the production of mature ribosomal subunits, the small one (40S), composed in yeast of 33 ribosomal proteins (r-proteins) and assembled with the 18S ribosomal RNA (rRNA) and the large one (60S) containing 48 r-proteins and the 25S, 5.8S and 5S rRNAs. The RNA component of the ribosomes is synthesized in the nucleolus as the 35S and 5S rRNA precursors. A large number of proteins and snoRNA associate co- or post-transcriptionally to the large 35S RNA and form, in association with U3 snoRNA, large particles known as 90S, or the SSU-processome (1,2). Pre-60S and pre-40S particles, formed from the initial large precursors, follow two distinct maturation pathways in the nucleoplasm and the cytoplasm. The pre-rRNAs are gradually modified and processed, and the subunits achieve their structure and translating capacity.
More than 300 proteins have been characterised or predicted to assist the pre-ribosomal particles during their maturation. Many of these factors are transiently associated with the 90S, pre-60S and pre-40S particles and are known as pre-ribosomal proteins. Functional studies on pre-ribosomal factors could be undertaken once the composition of pre-ribosomal complexes had been determined by affinity purification experiments (3–5). Such experiments gave limited information on the positioning of the identified proteins in the pathway. Affinity purifications of pre-ribosomal factors and their associated complexes result in the isolation of heterogeneous mixtures of intermediate particles because some factors associate very transiently with the pre-ribosomal particles, whereas others follow the process from the nucleolus to the cytoplasm. To isolate and characterise discrete intermediates in ribosome biogenesis, the purification of enriched complexes involved in a given step was performed in mutant strains (6–9). These intermediates were also analysed by testing the presence of known pre-rRNAs enriched in association with different pre-ribosomal or r-proteins (2). When antibodies are available, the presence of specific factors in the purified complexes under native and mutant conditions can be tested. The obtained results provide valuable, but limited information about the order of sequential association or dissociation of a number of factors during the assembly of 60S or 40S particles.
An alternative to the antibodies-based detection of proteins in the isolated complexes is quantitative mass spectrometry. One of its versions, using isotopically labelled peptides, the iTRAQ methodology (10), was used to compare the composition of complexes associated with Nop7-TAP under wild-type conditions and when the GTPase Nog1 was mutated (11). Only modest differences were observed, emphasizing the importance of the chosen mutant for this type of analysis. However, the results demonstrated that mass spectrometry is a suitable method for the quantitative analysis of the composition of pre-ribosomal complexes.
The present work aimed at establishing the assembly order of pre-60S factors during ribosome biogenesis, which would allow the composition of intermediate complexes at given steps to be defined. To this end, we analysed mutants that specifically accumulate different pre-rRNAs, and thus affect the pathway at early, intermediate or late steps, before the export of pre-60S particles to the cytoplasm. We compared the composition of the purified complexes under native or mutant conditions by semi-quantitative mass spectrometry, using a method derived from SILAC (Stable Isotope Labelling with Amino-acids in Cell culture) (12). The obtained results were in excellent correlation both with previously published data on specific factors and with the overall topology of the large-scale network of affinity-purification interactions, validating the approach. Moreover, we could define groups of early, intermediate and late associating proteins that accurately describe pre-60S assembly dynamics in yeast.
MATERIALS AND METHODS
Yeast strains
Yeast strains used in this work are listed in Supplementary Table S1. LMA335 (PGAL1-NOG1), LMA336 (PGAL1-NOG2), LMA337 (PGAL1-NSA2), LMA340 (PGAL1-EBP2) and LMA343 (PGAL1-POP3) conditional mutant strains were generated by homologous recombination using PCR products to transform BY4742 strain. PCR fragments containing the repressible GAL1 promoter flanked by sequence upstream and downstream the ATG of each gene respectively were synthesized from pFA6a-kanMX6-PGal1 plasmid (13). LMA368 was generated by mating LMA355 (MAK11-TAP) with LMA340. After sporulation and dissection, the spores were selected for histidine prototrophy and for the resistance to the Geneticin. LMA406 and LMA409 strains were obtained by transformation of LMA160 (RLP24-TAP) strain with a PCR product containing the repressible GAL1 promoter flanked by sequence upstream and downstream the ATG of either NOG2 or POP3 respectively.
Interaction data collection and network visualisation
A network of physical interactions involving known or putative pre-60S factors was built to assess visually the distribution of proteins that were showing significant changes in the purified complexes under mutant conditions. The factors that were either annotated as involved in ribosome biogenesis (SGD database) or were linked to pre-ribosomal factors by showing strong mRNA co-variation under various environmental conditions were selected. Co-purification data involving the selected 323 proteins were recovered from a total of 28 462 interactions available in the BioGrid database (14), corresponding to 32 individual publications, including large-scale TAP analyses (15–18). A network of interactions was obtained using factors showing at least 3 known interactions with other proteins involved in ribosome biogenesis and a sub-network, corresponding to pre-60S proteins was selected in Cytoscape (19). An automatic graph layout algorithm was used to distribute the proteins (nodes in the network) accordingly to their interactions (edges). Colours were assigned to nodes, depending on the observed variations in the SILAC experiments using a threshold of 2 to depict significant changes in protein levels under mutant conditions.
RNA extraction, primer extension and northern blot
After a preculture in rich medium containing galactose, the cells were incubated in rich medium containing glucose for various times from 0 to 8 or 28 h. Total RNAs were extracted using standard glass beads and phenol procedure. Primer extension was done with 32P-labelled oligonucleotides CS10 (CGC CTA GAC GCT CTC TTC TTA) and MFR457 (GCT TAA AAA GTC TCT TCC CGT CC) specific of U2 snRNA used as an internal control; the products were separated on 5% polyacrylamide-urea gels.
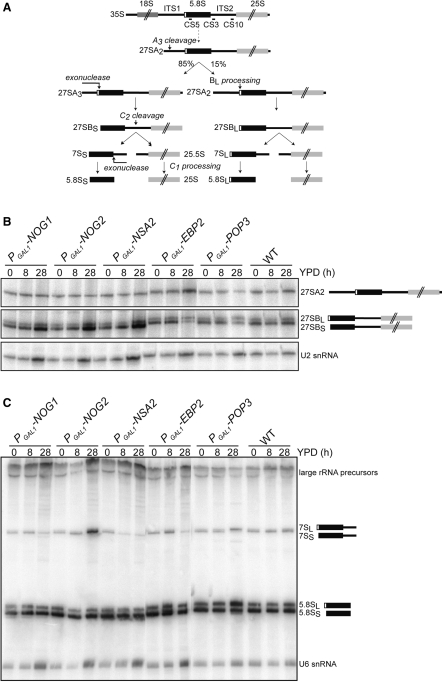
For northern blots, after separation of RNA on 5% polyacrylamide-urea gels and transfer on Hybond-N+ membranes, 7S, 5.8S and U6 snRNA were probed with 32P-labelled oligonucleotides CS3 (GGC CAG CAA TTT CAA GTT A), CS5 (CGG AAT TCT GCA ATT CAC ATT ACG) and MFR555 (AAC TGC TGA TCA TCT CTG) respectively. The position of pre-ribosomal probes is illustrated on Figure 1A.
Figure 1.
(A) Schematic representation of the rRNA maturation pathway. The oligonucleotides used in this study are indicated. (B) Total RNAs were extracted from the different strains after growth in galactose-containing, synthetic complete medium or after shift to glucose medium for either 8 or 28 h. 27SA2 and 27SB rRNA intermediates were detected by primer extension using oligonucleotide CS10, on a 5% polyacrylamide-urea gel. The U2 snRNA was used as a loading control. (C) 7S intermediate and 5.8S mature rRNA were separated on 5% polyacrylamide-urea gel and detected by northern blot using 32P-labelled oligonucleotides CS3 or CS5. The U6 snRNA was used as a loading control.
Purification of complexes for SILAC quantification
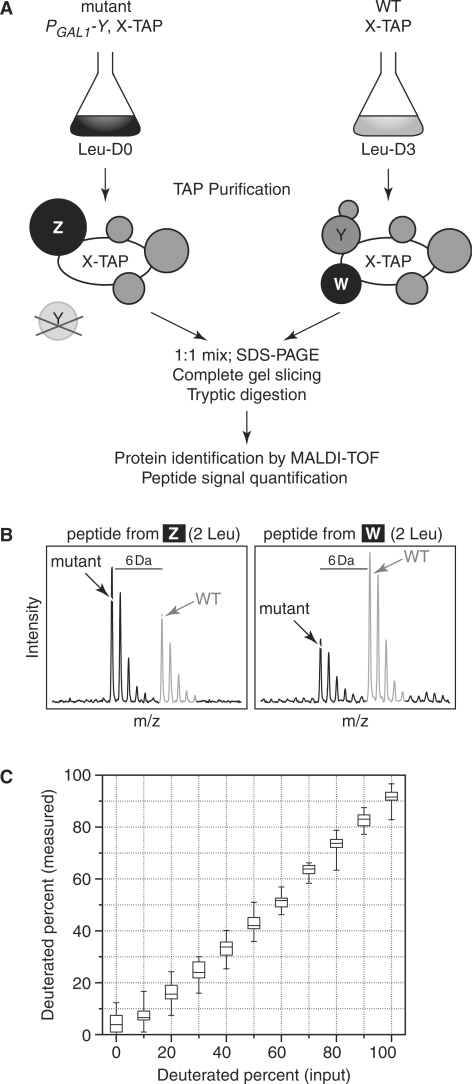
We adapted an isotopic labelling method (SILAC) based on mass spectrometry to distinguish quantitative changes in complexes composition under different conditions (12). For each experiment, we followed the protocol outlined in Figure 2A. We cultivated either the wild type or the mutant strains in deuterated leucine, glucose-containing minimal medium for at least 6 generations, so that all leucines were deuterated. The other strain was cultivated in normal leucine, glucose-containing minimal medium. The period of time for the shift to glucose was determined for each mutant according to its growth curve (Supplementary Figure S1). LMA182 (PGAL1-NOG1) was grown for 16 h in glucose-containing medium, LMA406 (PGAL1-NOG2) for 17 h, LMA272 (PGAL1-NSA2) for 16 h, LMA368 (PGAL1-EBP2) for 15 h, and finally LMA409 (PGAL1-POP3) for 15 h. The 600 nm optical densities of the recovered cultures were between 1 and 2.
Figure 2.
Adaptation of SILAC to the quantitative analysis of pre-ribosomal complexes. (A) Schematics of the TAP-SILAC coupling for the quantitative analysis of protein composition changes. Two strains producing a TAP-tagged bait protein ‘X’, a wild type and a mutant expressing a glucose-repressible pre-ribosomal gene ‘Y’ are grown in glucose-containing minimal medium, either in presence of normal leucine, or in presence of deuterated leucine. X-TAP-associated complexes are purified by tandem affinity purification, then the wild type and mutant samples are mixed together and separated by SDS-PAGE. Specific protein bands are cut out from the gels and analysed by MALDI-TOF spectrometry. (B) The non-deuterated/deuterated ratio can be quantified for each identified peptide in a given protein, using the mass spectra. As an example, signals obtained with a peptide from protein ‘Z’, which accumulates in the mutant and a peptide from protein ‘W’, which is lost in complexes purified from the mutant strain were illustrated. (C) To test the performance of mass spectrometry quantification, total yeast protein extracts from cells grown in medium containing leucine or deuterated leucine were mixed in different ratios, with a percentage of deuterated proteins from 0% to 100%. After separation by SDS-PAGE, four bands at different molecular weights (range from 12–125 kDa) were recovered and proteins identified by MALDI-TOF mass spectrometry. Several peptide signals were quantified in the different samples (8–14 values, depending on the spectra quality) and a box and whiskers plot for the measured values were represented in correlation with the expected percentage of deuterated leucine-containing peptides. Boxes represent the upper and lower quartile, the horizontal line, the median and the bars represent the maximum and minimum measured values.
The complex purifications were performed as described (20), starting from 4 l of yeast cultures and using buffers containing 0.1 M NaCl. The same amount of purified complex, as estimated by the Bradford method or by Coomassie blue staining on analytical polyacrylamide gels, from either the wild type or the mutant strains were mixed and separated on a 5–20% polyacrylamide gradient-SDS gel. After electrophoresis, the proteins were stained with colloidal Coomassie blue, and then identified and quantified by mass spectrometry.
SILAC data analysis
All the results were obtained on a MALDI-TOF Voyager DE-STR mass spectrometer (Applied Biosystems). Protein identifications were obtained with MS-Fit (Baker, P.R. and Clauser, K.R. http://prospector.ucsf.edu, version 3.1.1) using a yeast proteome database to which common keratin contaminants and trypsin sequences had been added. All the calibrated spectra were exported to text files for quantitative analysis. For each spectrum, the peaks that corresponded to an identified protein were assigned to the corresponding peptide sequences. The peaks corresponding to leucine-containing peptides were manually analysed and the ratio between the deuterated and non-deuterated monoisotopic peak heights (with background subtracted) were further used in the analysis. Python scripts were used to extract relevant spectra region on the basis of MS-Fit proteins identifications, convert these spectra to a custom XML format and interactively validate the results. A summary of the quantification results is available in Supplementary Table S2, and more details about the number of quantified peptides in Supplementary Table S3.
Western blot analysis
Proteins were separated on a 10% polyacrylamide-SDS gel and transferred on a nitrocellulose membrane. Specific proteins were detected by indirect immunoblot, using specific polyclonal rabbit antibodies at a 1 : 5000 dilution. Secondary antibodies (Goat Anti-Rabbit-HRP Conjugate from Bio-Rad) were used at a 1 : 10 000 dilution.
RESULTS
Choosing mutants and tagged pre-60S factors to analyse different 60S biogenesis steps
To obtain an overview of the composition of the successive 60S precursor complexes, we defined a set of 83 proteins that are known or predicted to be involved in 60S formation and collected their physical interaction results (co-purification experiments) from the BioGrid database (14). Taken individually, each pre-60S factor that was used as a bait, co-purified with a number of other proteins, providing a profile of the factors that can be concomitantly found on pre-60S particles. To assign these factors to groups, we used the data to build a matrix of physical interactions, constituted of 49 tagged proteins, in rows, and 63 distinct associated proteins, in columns (Supplementary Figure S2). We expected proteins involved in the same pre-60S precursors to be grouped together and, as a result, to gain insights into the timing of association and dissociation of different pre-60S factors. Even though this kind of analysis proved previously to be efficient to distinguish 90S, pre-60S and pre-40S particles when using a large number of pre-ribosomal proteins and their associations (3), the obtained cluster of pre-60S factors was unable to further refine the involvement of many of the proteins at specific steps in the pathway.
To get a detailed description of the composition of pre-60S complexes along the pathway, other methods had to be set up. One such method uses quantitative mass-spectrometry to compare the composition of purified pre-ribosomal particles from mutant strains affected for 60S biogenesis. For this type of study, both the choice of mutants and tagged proteins used for the purification of the complexes were important. We chose 5 pre-60S factors (Ebp2, Nog1, Nsa2, Pop3 and Nog2) to block the biogenesis of the large ribosomal subunit at specific steps, defined by the accumulation of distinct pre-rRNA intermediates (6,21–24). As these factors are essential for cell viability, we generated strains where the genes encoding these factors were under the control of a glucose-repressed promoter (GAL1-10). All these strains showed a progressive growth defect when cultivated in glucose-containing medium (Supplementary Figure S1). The growth defect was associated with changes in the levels of different rRNA maturation intermediates (Figure 1A), tested by primer extensions (Figure 1B) and northern blotting (Figure 1C). As expected from previous reports on the role of these factors, we observed different effects of the proteins depletion on the steady-state levels of the pre-60S RNA components. Like the ebp2-1 mutant (21), the repression of EBP2 led to early defects in the 60S maturation pathway, with a 27SA2 increase and a decrease in 27SB and 7S pre-rRNA levels. Repression of POP3 expression induced changes in the 27SBL/27SBS, 7SL/7SS and 5.8SL/5.8SS ratios, which have been previously attributed to the role of the RNase MRP (Pop3 being one of its subunits) in the RNA cleavage at site A3 (22). As previously described (6,23), the repression of either NOG1 or NSA2 expression affected the cleavage of 27SB precursors at site C2, hence preventing the formation of 7S. Nog2 depletion had a different effect on 60S biogenesis at a late nuclear step with the accumulation of both 27SB and 7S species, as reported (24). Thus, using these five mutants, the 60S biogenesis could be blocked at early, intermediate or late nuclear steps.
We combined the glucose-repressible strains with genomic TAP-tagging of Rlp24, a factor that associates with pre-60S particles both in the nucleus and the cytoplasm (6), allowing the isolation of blocked complexes in the different mutants. We still needed a robust method for quantitative comparisons of the protein composition of these isolated complexes.
Adaptation of SILAC to the analysis of TAP complexes
Several methods of quantitative mass spectrometry, based on isotopic labelling of proteins or peptides have been described (25,26). To monitor the relative amounts of each purified protein in wild type and mutant samples, we chose SILAC, as a robust method of in vivo labelling that could be adapted to the quantification of simple MS spectra obtained with a MALDI-TOF mass spectrometer.
SILAC is based on the differential labelling of two cultures before the analysis, for example, a wild type and a mutant strain. Proteins from one of the samples incorporate a deuterated amino acid, leucine with 3 deuterium atoms in our case, which leads to a shift in the m/z for all the peptides coming from that sample, when compared with a sample coming from a culture grown with non-deuterated leucine (12). For each single-charged peptide containing a leucine, this results in a shift of 3 Da in the mass spectrum. Assignment of the peptide peaks to identified proteins and measurement of the pairs (deuterated/non-deuterated) peak heights in the MALDI-TOF spectra provide a relative quantitative measure for the initial content of the protein in the two samples. To limit the analysis to the changes in composition of specific complexes, an additional TAP purification step was required; the whole procedure is illustrated in Figure 2A. The results obtained for different proteins can be explained by following the example shown in Figure 2B. Proteins that are no longer co-purified with the tagged protein in the mutant strain generate spectra profiles resembling that shown for the protein marked ‘W’. In contrast, proteins that are enriched in the mutant complexes result in peptide profiles like the one shown as example ‘Z’. Both shown peptides have a single charge and contain 2 leucines, resulting in a 6 Da shift in the detected m/z.
Since our method only relies on the identification of proteins by their peptide mass fingerprint, we tested the reproducibility and precision of quantitative measures that are based on MALDI-TOF MS spectra. We mixed in different proportions total protein extracts from strains grown with deuterated or non-deuterated leucine and obtained an excellent correlation between our measures and the expected fraction of deuterated proteins (Figure 2C).
Defining the association of proteins to early or late 60S precursors
The depletion of many pre-60S factors results in a relative 27SB pre-rRNA accumulation (3,5). To assign these different proteins to groups that would predict their early or late association to the ribosome precursors, we first tested the effects of Nog1 depletion on the composition of Rlp24-TAP associated complexes. Our previous data, based on immunoblots, indicated that under these conditions several important changes in the composition of the purified complexes occur, like the accumulation of Mak11 or the depletion of Nog2 (6). Characterisation of Mak11 and Nog2 confirmed their involvement in early or late steps of nuclear 60S maturation, respectively (24,27). We thus began our investigation of the order of assembly of 60S ribosomal subunits with an analysis of the changes in the composition of Rlp24-TAP-associated complexes when Nog1 was depleted.
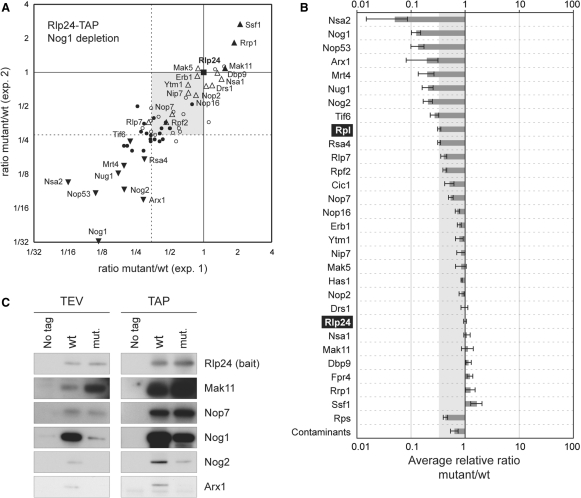
In addition to large subunit r-proteins and usual contaminants (Ssb, translation elongation factors or small subunit r-proteins), we identified and quantified the relative abundance of 28 pre-60S factors in the purified complexes (Figure 3A and B). As expected, one of the proteins showing the largest decrease in the particles isolated from the mutant strain, was the depleted protein Nog1. Other factors, like Nog2 or Nug1 showed a similar behaviour, in good correlation with previous descriptions of the late association of these factors to pre-60S complexes (24,28). Conversely, proteins like Ssf1, described as acting early in the 60S biogenesis (29), were enriched in the complexes isolated under mutant conditions.
Figure 3.
Rlp24-TAP combined with Nog1 depletion defines classes of pre-60S factors. (A) Scatter plot of the mutant/wild-type ratio obtained for each protein in two independent experiments where, in turn, the wild type or the mutant strain were cultivated in deuterated leucine-containing medium. Bait (Rlp24) is indicated by a filled square; Proteins enriched in the complexes purified from the mutant strain when using the bait protein as reference are indicated as upward pointing filled triangles; Proteins enriched or in similar amounts in the wild type and mutant complexes when using ribosomal proteins (Rpl) as reference are indicated as empty triangles; Proteins showing decreased levels in the complexes purified from the mutant strain (downward pointing filled triangles); Large ribosomal subunit proteins (filled circles); Contaminants (empty circles). Dashed lines indicate the average ratios obtained for large subunit r-proteins. (B) Quantifications of the mutant/wild-type ratios for proteins identified in Rlp24-TAP-associated complexes purified from strains expressing NOG1 or not. Boxes represent the average value of the ratio for all quantified peptides in a given protein in two distinct experiments. Error bars indicate 95% confidence intervals. (C) Immunoblot validation of the SILAC quantifications. Aliquots from Rlp24-TAP-associated complexes recovered from the wild type or the PGAL1-NOG1 strain were separated by SDS-PAGE and the amounts of Rlp24, Mak11, Nop7, Nog1, Nog2 or Arx1 were assessed by western blot using specific antibodies. Both the eluate from the first purification step on IgG-sepharose (TEV) and the final eluate (TAP) were analysed (left and right panels, respectively).
Many quantified pre-60S factors (located in the grey area on Figure 3A and B) were decreased when compared to the levels of Rlp24-TAP, the protein used for the isolation of the particles, but were enriched in the complexes, when compared to the average level of the quantified ribosomal proteins (indicated by dashed lines in Figure 3A). In the absence of an absolute standard, we arbitrarily fixed the measured ratio for Rlp24-TAP to 1 and reported the results for the other proteins accordingly. If we assume that the ribosomal proteins decreased under mutant conditions in the complexes isolated in association with Rlp24-TAP, a straightforward explanation would be that a fraction of the Rlp24-TAP bait was no longer associated with large complexes when ribosome biogenesis was blocked. While changing the reference has an impact on what can be considered as an increase or a decrease in protein amounts, it does not modify the classification of the ratios for the different pre-60S factors.
To validate the results of mass spectrometry quantifications we performed immunoblots on TEV and TAP purifications, using antibodies that recognise specifically Rlp24, Mak11, Nop7, Nog1, Nog2 or Arx1 (Figure 3C). The results were in good agreement with the mass-spectrometry-based measurements, hence validating the SILAC analysis.
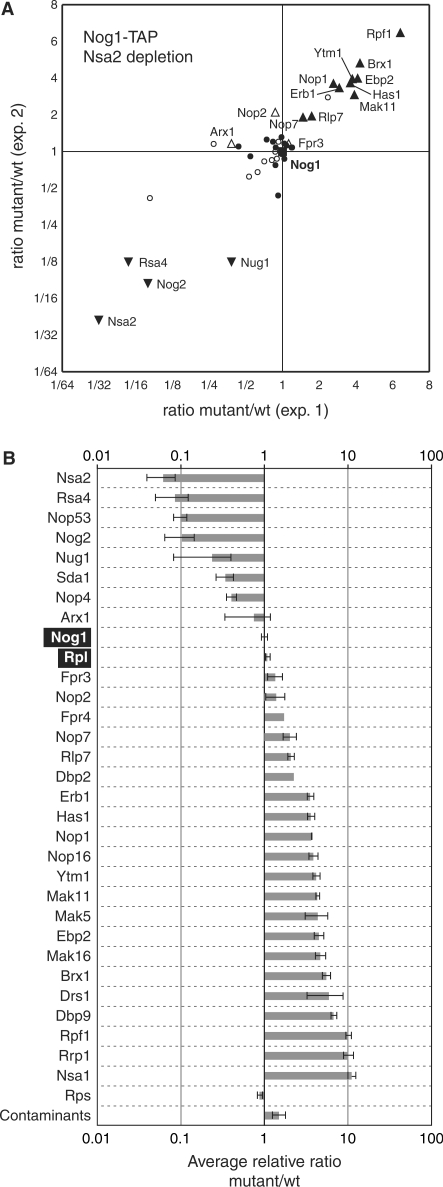
To confirm and reinforce the data from our first experiments, we used another mutant having a similar pre-rRNA phenotype and another protein as bait. We chose Nsa2 depletion as mutant condition since it led to the same pre-rRNA species accumulation as the depletion of Nog1 (Figure 1B). In addition, Nsa2 was the protein showing the strongest decrease in the Rlp24-TAP complexes when Nog1 was depleted (Figure 3A and B), an effect explained by the observation of Nsa2 destabilisation under these conditions (23). Nog1-associated proteins are very similar to those found in association with Rlp24 (6). We obtained quantitative data, by TAP-SILAC for Nog1-TAP under wild type or Nsa2 depletion conditions for 30 pre-60S factors (Figure 4A and B). Twenty-three proteins were quantified in both Rlp24-TAP, Nog1 depletion and Nog1-TAP, Nsa2 depletion experiments (Supplementary Figure S3), with a very good Kendall tau rank correlation coefficient (t = 0.727) of the observed changes (p = 1.5 × 10−6 that the values are not correlated). The results obtained in two independent experiments with different mutants and different TAP-tagged proteins allowed the definition of two groups of pre-60S factors. An early one is composed by proteins that are likely to be present on pre-60S particles before the step blocked in the absence of either Nog1 or Nsa2, and is composed of at least 20 different proteins, including factors like Ssf1 or Mak11. Proteins that showed reproducibly diminished levels in the complexes under mutant conditions (Nop53, Nog2, Nug1 or Rsa4) are likely to associate to the 60S precursors at a later stage. The different abundance of proteins in the early and late group of pre-60S factors is correlated with the progressive simplification of pre-60S particles during their maturation (30).
Figure 4.
Nog1-TAP combined with the depletion of Nsa2 defines C2-specific 60S precursors. (A, B) Legends are as for Figure 3, except that the bait was Nog1, and the depleted protein, Nsa2.
The early pre-60S factors can be separated in subgroups
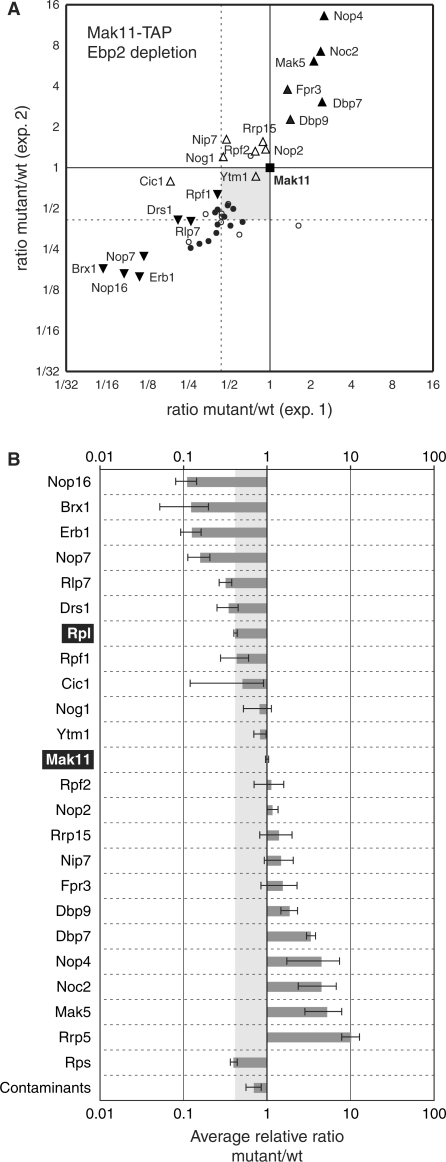
In view of the successful SILAC-based measurements of pre-60S factors redistribution under Nog1 and Nsa2 depletion, we sought to refine the results by testing the effects of depleting an earlier acting factor (as defined by pre-rRNA accumulation studies), Ebp2. No protein could be observed accumulating in association with Rlp24 under Ebp2 mutant conditions, when compared with the wild type (Supplementary Figure S4A and B). These results suggest that Ebp2 action precedes the association of Rlp24 to the pre-60S particles and that an earlier protein should be used to isolate the complexes that accumulate in the absence of Ebp2. As we have previously shown that Mak11 might already be present on the complexes at a moment preceding Rlp24 association to pre-60S particles (27), we tested the influence of Ebp2 depletion on Mak11-TAP-associated complexes composition. A number of proteins, and especially Noc2, Nop4 and Rrp5, where present in higher amounts in the Mak11-associated complexes in the mutant (Figure 5A and B), in good agreement with the function of these proteins early in the pathway (1,31,32). On the contrary, some of the proteins that were classified as ‘early’ in respect with Nog1 or Nsa2 function were lost when Ebp2 was depleted, for example Nop16, Erb1 or Nop7.
Figure 5.
Early 60S precursors, isolated in association with Mak11-TAP are enriched after Ebp2 depletion. (A, B) Legends are as for Figure 3, except that the bait was Mak11, and the depleted protein, Ebp2.
We conclude that Ebp2 is required for the progression of the 60S assembly pathway at an early stage and affects the specific association of a large number of pre-60S factors to the nascent particles.
Nog2 is required for the association of late pre-60S factors
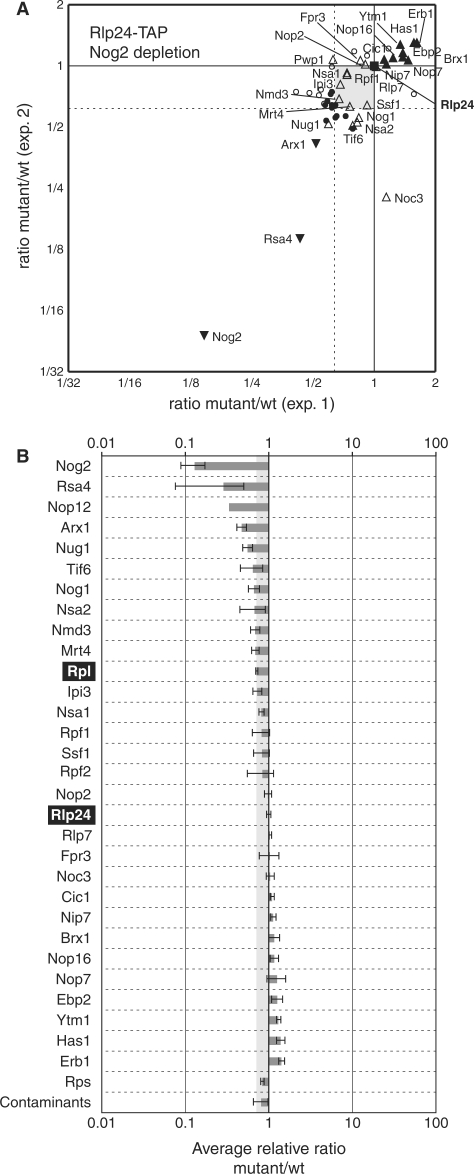
Since we were able to define subgroups of pre-60S factors that associate early in the pathway, we wondered if we could distinguish different classes of late, nuclear-associating pre-60S proteins. Among the different proteins that we defined as late-associating, we chose Nog2 depletion as the mutant condition coupled with the isolation of pre-60S complexes in association with Rlp24-TAP for further experiments. Unlike Arx1 or Tif6, Nog2 is strictly nuclear and its depletion leads to the accumulation of the last nuclear pre-rRNA intermediate, 7S (Figure 1C) and also blocks the export of the 60S precursors to the cytoplasm.
Little change was observed in Rlp24-TAP-associated complexes between the wild-type conditions and the depletion of Nog2 (Figure 6A and B). The fact that both 7S and 27S species are accumulated in Nog2-depleted cells might provide an explanation for the lack of amplitude of the observed effects. Still, Rsa4 and Arx1 levels decreased in these complexes, suggesting that their association to late pre-60S depends on Nog2 presence. Rsa4 was previously described as a factor required for late pre-60S biogenesis (33), but its precise binding step had not been specifically addressed. Recently, Arx1 was described as a nuclear export receptor for the 60S precursors (34,35); its decrease in the Nog2 mutant is thus in good agreement with a late role of Arx1 in 60S biogenesis. Changes in the composition of 60S particles, detected by quantitative mass spectrometry, can be thus useful to orient further functional characterisation of specific pre-60S factors.
Figure 6.
Modest changes in the composition of late nuclear pre-60S complexes are induced by Nog2 depletion. (A, B) Legends are as for Figure 3, with Rlp24 as the tagged protein and Nog2 as the depleted factor.
The two distinct routes of 27SB formation involve a similar set of pre-60S factors
Our experiments were based on the hypothesis that proteins association to the different intermediates follows, as the RNA maturation, a linear order, with every pre-60S particle deriving from a preceding one. We wondered whether protein composition may have an influence on the pre-rRNA processing and if particles of similar pre-rRNA components may contain different protein sets. To this end, we explored the effects of an RNase MRP mutant on Rlp24-TAP-associated complexes composition. After A2 cleavage that leads to the 27SA2 precursor, processing of 27S species can follow two distinct routes (Figure 1A): the major one involves a cleavage at site A3 by RNase MRP; the minor one consists of a 5′-3′ exonucleolytic trimming by a yet unidentified ribonuclease, and leads to the production of 5′-extended species (27SBL, 7SL precursors and 5.8SL rRNA). The repression of POP3 impairs RNase MRP function, leading to a complete redistribution of the processing towards the minor pathway. The analysis of Rlp24-TAP complexes in the presence or absence of Pop3 revealed hardly any change in protein composition (Supplementary Figure S4C and D), despite a clear change in the ratio of the large and short form of the 5.8S rRNA levels (Supplementary Figure S5), suggesting that except for 27SA3 processing, both the major and the minor processing pathways involve the same set of pre-60S factors.
DISCUSSION
Ribosome biogenesis in eukaryotes involves a large number of factors, many of them identified during recent large-scale co-purification studies (16–18). While these results were crucial to define the number of different proteins involved in this conserved pathway, they are not suitable for a detailed characterisation of the different intermediate ribosome precursors. Novel approaches are needed to address this question and one of them is the study of changes in the protein composition of the intermediate particles under mutant conditions (2,6,8,9,23,36,37). We show here that coupling TAP purifications with carefully chosen mutants and novel methods of quantitative mass-spectrometry like SILAC (12) allows the description of groups of pre-ribosomal factors that are likely to be functionally related. The definition of such groups may lead to oriented experiments that would help the comprehension of the involved molecular mechanisms.
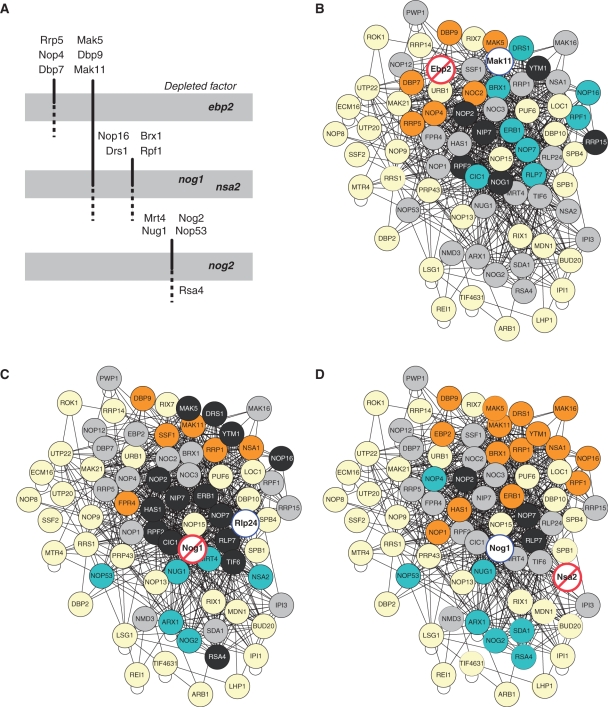
We integrated our results about the assembly of pre-ribosomal factors on pre-60S particles in a simple schematic (Figure 7A). Several groups of pre-60S factors could be defined, on the basis of the three analysed mutant effects. The earliest group of pre-60S factors is defined by the proteins that accumulate during Ebp2 depletion. Since this mutant accumulates 27SA2 pre-rRNA, the first form of RNA that is specific to the 60S assembly pathway and signals the separation from pre-40S particles, proteins of this group are likely to be already present on the earliest nascent 60S precursors.
Figure 7.
SILAC-based definition of pre-60S sub-complexes. (A) Based on the results of our analysis we deduced possible association and dissociation steps that define several groups of pre-60S factors, relative to the involvement of Ebp2, Nog1/Nsa2 and Nog2 in the rRNA maturation pathway. (B, C, D) The results of the SILAC quantifications were colour-coded and superposed on a Cytoscape representation of pre-60S proteins, showing identified bait–prey interactions from published affinity purification experiments. We first extracted a 60S-specific sub-network from the larger pre-ribosomal interactions network. The network layout was obtained by applying the yFiles organic algorithm in Cytoscape. The represented experiments are (B) Mak11-TAP, depletion of Ebp2, (C) Rlp24-TAP, depletion of Nog1 and (D) Nog1-TAP, depletion of Nsa2. The bait is framed by a blue line, and the depleted factor in a white disk dashed in red. Factors which were enriched in complexes isolated from the mutant strain at least by a factor of 2 are shown in orange, factors which were decreased by >50% are in cyan, factors of unchanged levels are in dark grey disks. Light grey disks indicate factors that we identified in at least one of our experiments. Light yellow disks correspond to known pre-60S factors which were not identified in the SILAC experiments.
One of the most interesting findings of this study was that some pre-60S factors, like Nop16, decreased in the purified complexes when Ebp2 was depleted while they accumulated under Nog1 or Nsa2 depletion, whatever the protein used for the precursors purification. The simplest explanation for this behaviour is that proteins from this intermediate group associate to the 60S precursors at a step that precedes the one blocked by the absence of Nog1 or Nsa2, and follows the one blocked by Ebp2 absence (Figure 7A). The proteins that, like Mrt4, associate to the nuclear pre-60S particles at late steps depending on the action of Nog1 and Nsa2 can be further classified in two sub-groups, including in the latest one the factors that also depend on Nog2 for association to the pre-60S particles (for instance, Rsa4).
Our data cannot indicate with precision the moment of dissociation for most of the identified proteins because the lack of identification of a protein in a given complex is not an absolute proof for its absence from that complex. However, when a protein could be identified under some conditions in a complex but was absent from similar complexes purified with another bait protein, such information was pragmatically used to differentiate subgroups of pre-60S factors. With the above limitations in mind, we distinguished the proteins that looked specific to the early Mak11-associated complexes (for instance Dbp7) from those that were also present on later Rlp24 or Nog1-associated pre-60S particles (for instance Mak5). Except for known shuttling factors that are exported to the cytoplasm, the estimation of the point where other factors leave the particles would need further validation by specific experiments.
Our analysis provides new insights into the assembly scenario for the eukaryotic large ribosomal subunit in yeast, by describing the protein composition of distinct assembly intermediates along the pathway. The results were in excellent correlation with the topology of the network of physical association of pre-60S factors. Proteins that were enriched or decreased in precursor particles under mutant conditions were clustered in specific regions of the graph, depending on the analysed protein depletion (Figure 7B, C and D). Our results are also in agreement with previously published data about the pre-60S factors function or association to the pre-ribosomal complexes. For instance, Dbp7, Dbp9, Nop4, Noc2, Rpf2, Ebp2, Rrp1 and Ssf1 were known for their participation in early steps of the large ribosomal subunit biogenesis (8,21,29,31,32,38–42), and Rrp5 is a component of the SSU-processome (1). Nug1 was characterised for its functions in the late steps of biogenesis, at the same time as Nog2 (28) and Arx1 are both involved in the export of pre-60S particles from the nucleus (34,35).
On the basis of the analysis of three distinct 60S biogenesis steps (depending on Ebp2, Nog1/Nsa2 and Nog2) we were thus able to obtain an unbiased panorama of the association timing for 30 pre-60S factors during ribosome biogenesis. These results, integrated into the physical association map for all the previously identified pre-60S proteins, and combined with data about the molecular function of each pre-60S factor in the biogenesis, are useful to build a view of protein dynamics in the ribosome biogenesis pathway.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to Françoise Stutz (Centre Médical Universitaire, Genève, Switzerland) for the pFA6a-TAP-Tag-His3MX6 vector. We thank Yanhua Yao and Tommaso Villa for critical reading of the manuscript. This work was supported by the Ministère délégué à l’Enseignement Supérieur et à la Recherche (ACI-BCM0089-2003). Funding to pay the Open Access publication charges for this article was provided by the Ministere delegue a l'enseignement superieur et a la recherche, France in the form of the ACI-BCM0089-2003 grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferreira-Cerca S, Poll G, Kuhn H, Neueder A, Jakob S, Tschochner H, Milkereit P. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol. Cell. 2007;28:446–457. doi: 10.1016/j.molcel.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 3.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 4.Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 5.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol. Life Sci. 2008 doi: 10.1007/s00018-008-8027-0. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saveanu C, Namane A, Gleizes PE, Lebreton A, Rousselle JC, Noaillac-Depeyre J, Gas N, Jacquier A, Fromont-Racine M. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell Biol. 2003;23:4449–4460. doi: 10.1128/MCB.23.13.4449-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miles TD, Jakovljevic J, Horsey EW, Harnpicharnchai P, Tang L, Woolford J.L., Jr Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol. Cell. Biol. 2005;25:10419–10432. doi: 10.1128/MCB.25.23.10419-10432.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, Rout MP, Hiley SL, Hughes T, Woolford J.L., Jr Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007;21:2580–2592. doi: 10.1101/gad.1569307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Fernandez J, Roman A, De Las Rivas J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell Biol. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Fuentes JL, Datta K, Sullivan SM, Walker A, Maddock JR. In vivo functional characterization of the Saccharomyces cerevisiae 60S biogenesis GTPase Nog1. Mol. Genet. Genomics. 2007;278:105–123. doi: 10.1007/s00438-007-0233-1. [DOI] [PubMed] [Google Scholar]
- 12.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 13.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 14.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 16.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 17.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 18.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 19.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 21.Huber MD, Dworet JH, Shire K, Frappier L, McAlear MA. The budding yeast homolog of the human EBNA1-binding protein 2 (Ebp2p) is an essential nucleolar protein required for pre-rRNA processing. J. Biol. Chem. 2000;275:28764–28773. doi: 10.1074/jbc.M000594200. [DOI] [PubMed] [Google Scholar]
- 22.Dichtl B, Tollervey D. Pop3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. EMBO J. 1997;16:417–429. doi: 10.1093/emboj/16.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lebreton A, Saveanu C, Jacquier A, Fromont-Racine M. Nsa2 is an unstable, conserved factor required for the maturation of 27 SB pre-rRNAs. J. Biol. Chem. 2006;281:27109–27116. doi: 10.1074/jbc.M602199200. [DOI] [PubMed] [Google Scholar]
- 24.Saveanu C, Bienvenu D, Namane A, Gleizes PE, Gas N, Jacquier A, Fromont-Racine M. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 2001;20:6475–6484. doi: 10.1093/emboj/20.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 26.Mueller LN, Brusniak MY, Mani DR, Aebersold R. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J. Proteome Res. 2008;7:51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- 27.Saveanu C, Rousselle JC, Lenormand P, Namane A, Jacquier A, Fromont-Racine M. The p21-activated protein kinase inhibitor Skb15 and its budding yeast homologue are 60S ribosome assembly factors. Mol. Cell Biol. 2007;27:2897–2909. doi: 10.1128/MCB.00064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 29.Fatica A, Cronshaw AD, Dlakic M, Tollervey D. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell. 2002;9:341–351. doi: 10.1016/s1097-2765(02)00458-6. [DOI] [PubMed] [Google Scholar]
- 30.Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun C, Woolford J.L., Jr The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 1994;13:3127–3135. doi: 10.1002/j.1460-2075.1994.tb06611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milkereit P, Gadal O, Podtelejnikov A, Trumtel S, Gas N, Petfalski E, Tollervey D, Mann M, Hurt E, Tschochner H, et al. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell. 2001;105:499–509. doi: 10.1016/s0092-8674(01)00358-0. [DOI] [PubMed] [Google Scholar]
- 33.de la Cruz J, Sanz-Martinez E, Remacha M. The essential WD-repeat protein Rsa4p is required for rRNA processing and intra-nuclear transport of 60S ribosomal subunits. Nucleic Acids Res. 2005;33:5728–5739. doi: 10.1093/nar/gki887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hung NJ, Lo KY, Patel SS, Helmke K, Johnson AW. Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast. Mol. Biol. Cell. 2008;19:735–744. doi: 10.1091/mbc.E07-09-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradatsch B, Katahira J, Kowalinski E, Bange G, Yao W, Sekimoto T, Baumgartel V, Boese G, Bassler J, Wild K, et al. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol. Cell. 2007;27:767–779. doi: 10.1016/j.molcel.2007.06.034. [DOI] [PubMed] [Google Scholar]
- 36.Lebreton A, Saveanu C, Decourty L, Rain JC, Jacquier A, Fromont-Racine M. A functional network involved in the recycling of nucleo-cytoplasmic pre-60S factors. J. Cell Biol. 2006;173:349–360. doi: 10.1083/jcb.200510080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pertschy B, Saveanu C, Zisser G, Lebreton A, Tengg M, Jacquier A, Liebminger E, Nobis B, Kappel L, van der Klei I, et al. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol. Cell Biol. 2007;27:6581–6592. doi: 10.1128/MCB.00668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Marchis ML, Giorgi A, Schinina ME, Bozzoni I, Fatica A. Rrp15p, a novel component of pre-ribosomal particles required for 60S ribosome subunit maturation. RNA. 2005;11:495–502. doi: 10.1261/rna.7200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berges T, Petfalski E, Tollervey D, Hurt EC. Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J. 1994;13:3136–3148. doi: 10.1002/j.1460-2075.1994.tb06612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daugeron MC, Kressler D, Linder P. Dbp9p, a putative ATP-dependent RNA helicase involved in 60S-ribosomal-subunit biogenesis, functionally interacts with Dbp6p. RNA. 2001;7:1317–1334. doi: 10.1017/s1355838201010640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daugeron MC, Linder P. Dbp7p, a putative ATP-dependent RNA helicase from Saccharomyces cerevisiae, is required for 60S ribosomal subunit assembly. RNA. 1998;4:566–581. doi: 10.1017/s1355838298980190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horsey EW, Jakovljevic J, Miles TD, Harnpicharnchai P, Woolford J.L., Jr Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA. 2004;10:813–827. doi: 10.1261/rna.5255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.