Abstract
Apicomplexans, including the pathogens Plasmodium and Toxoplasma, carry a nonphotosynthetic plastid of secondary endosymbiotic origin called the apicoplast. The P. falciparum apicoplast contains a 35 kb, circular DNA genome with limited coding capacity that lacks genes encoding proteins for DNA organization and replication. We report identification of a nuclear-encoded bacterial histone-like protein (PfHU) involved in DNA compaction in the apicoplast. PfHU is associated with apicoplast DNA and is expressed throughout the parasite's intra-erythocytic cycle. The protein binds DNA in a sequence nonspecific manner with a minimum binding site length of ∼27 bp and a Kd of ∼63 nM and displays a preference for supercoiled DNA. PfHU is capable of condensing Escherichia coli nucleoids in vivo indicating its role in DNA compaction. The unique 42 aa C-terminal extension of PfHU influences its DNA condensation properties. In contrast to bacterial HUs that bend DNA, PfHU promotes concatenation of linear DNA and inhibits DNA circularization. Atomic Force Microscopic study of PfHU–DNA complexes shows protein concentration-dependent DNA stiffening, intermolecular bundling and formation of DNA bridges followed by assembly of condensed DNA networks. Our results provide the first functional characterization of an apicomplexan HU protein and provide additional evidence for red algal ancestry of the apicoplast.
INTRODUCTION
The apicoplast is the nonphotosynthetic plastid of apicomplexan parasites that include the genera Plasmodium and Toxoplasma and is believed to have arisen from a secondary endosymbiotic event involving an ancestral protist and an alga (1). The apicoplast is essential for parasite survival; it is the site for Type II fatty acid biosynthesis, nonmevalonate pathway of isoprenoid biosynthesis, and synthesis of heme intermediates within the parasite (2,3). Biochemical pathways operative within this organelle provide novel sites for drug intervention against malaria. Due to its essentially prokaryotic nature the processes of DNA replication, transcription and translation within the apicoplast are also validated drug targets (4–6).
Each sporozoan cell of P. falciparum carries a single apicoplast with apicoplast DNA (plDNA) copy number estimates varying from 1 to 15 (7,8). The ∼35 kb, A+T-rich, circular double-stranded plDNA molecules of P. falciparum replicate via the D-loop/bi-directional ori mechanism at the late trophozoite-early schizont stage of the intraerythrocytic cycle. PlDNA replication origins (ori) localize within the inverted repeat (IR) region of the plDNA molecule (9–11). Although there is extensive sequence similarity between plDNA of P. falciparum and T. gondii they have distinct in vivo topology. The former is circular while the latter occurs as an oligomeric series of linear tandem arrays of the 35 kb genome. Light microscopy studies on T. gondii apicoplast genome have suggested that it is organized into a nucleoid that segregates into two equal portions during apicoplast division (8,12). The fact that a single plDNA circle is ∼12 µm in circumference and several molecules have to be packed into an organelle with a diameter of only ∼0.3 µm (8) as well as replicate and divide into daughter molecules without getting tangled is indicative of the involvement of a DNA-compacting protein in plDNA organization.
The apicoplast genome primarily encodes components of the transcription and translation machinery of the organelle (3,6) but lacks genes encoding any DNA organization or replication protein. Thus, all major protein components involved in plDNA organization/replication must be nuclear-encoded and imported into the organelle. There is accumulating evidence on the components of the plDNA replication machinery. A ∼220 kDa multi-domain polypeptide (PfPREX) that contains DNA polymerase as well as DNA primase, DNA helicase and 3′–5′ exonuclease activities (13) has been identified as a key enzyme for plDNA replication. Genes encoding putative apicoplast-targeted gyrase A and B have also been identified on Chr12 of P. falciparum and gyrase B has been functionally characterized (14,15). Ciprofloxacin and novobiocin, that target bacterial DNA gyrase, specifically inhibit replication of P. falciparum plDNA and also reduce parasite growth in culture (4,15) thus validating malarial apicoplast DNA replication as a drug target.
The dearth of information on proteins involved in organization of the P. falciparum apicoplast genome prompted us to investigate putative candidates from the parasite genome database. The prokaryotic nature and putative red algal origin of the apicoplast suggested the possible involvement of a histone-like protein (‘heat unstable’ or HU) (16) that is the primary organizational component of bacterial nucleoids, dinoflagellate chromosomes as well as red algal chloroplast genomes (17–19). A gene encoding a HU-ortholog that carries a conserved BHL domain (bacterial histone-like domain) together with a predicted N-terminal apicoplast targeting sequence was identified on Chr.9 of the P. falciparum nuclear genome. HU proteins are small basic proteins of prokaryotic origin that are structurally distinct from eukaryotic histones (20), belong to the DNABII family of DNA-binding proteins, and exhibit hetero- or homo-dimeric DNA binding. HU proteins also have regulatory effects on DNA replication, recombination and transcription (21–25). In bacteria, HU proteins together with the structurally related IHF (integration host factor), organize chromosomal DNA into periodic nucleosome-like structural units (26). Assuming an even distribution across the chromosome, in vivo concentrations of these proteins in Escherichia coli (27) provide for binding of one dimer only approximately every 125 bp, indicating that filament formation by HU could occur only locally (28). The presence of 10 other DNA-organization proteins such as histone-like nucleoid structuring protein (H-NS) and factor for inversion stimulation (Fis) that are also involved in DNA organization in E. coli (27) and their sequence preferences and co-operativity in binding DNA would further influence filament formation and condensation of E. coli DNA.
We report characterization of the P. falciparum nuclear-encoded bacterial histone-like protein (PfHU) and its interaction with apicoplast DNA. PfHU is expressed throughout the intra-erythrocytic phase of parasite growth, exhibits DNA binding and is capable of condensing DNA. The unique C-terminal domain of PfHU influences its interaction with DNA. Atomic Force Microscopic analysis shows that the protein is capable of forming both DNA bridges and bundles.
MATERIALS AND METHODS
Parasite culture
P. falciparum (strains 3D7 and NF54) was cultured in human RBCs maintained in RPMI-1640 supplemented with 0.5% AlbumaxII (Invitrogen). The parasites were synchronized with sorbitol (29).
Cloning, expression and purification of recombinant PfHU
The gene encoding the predicted 22.5 kDa P. falciparum bacterial histone-like protein (PlasmoDB ID PFI0230c) was amplified by PCR. DNA encoding unprocessed PfHUup (1–189 aa) was amplified using upstream (HU-f: 5′-CGCGGATCCTATGTAATATTATATTGTGTCCTT-3′) and downstream (HU-r: 5′-CGCGTCGACATAGATTAACTTTTCATTTACATTTTC-3′) primers carrying BamHI and SalI sites, respectively with P. falciparum cDNA as template. The sequence encoding predicted processed PfHU (PfHUp) (54–189 aa) was amplified using the forward primer HU-p (5′-CCGGGATCCATGTCACA GGCGATTACAAAG-3′) and HU-r as the reverse primer. Processed PfHU lacking 41 amino acids of the C-terminal end (PfHUΔC) was amplified using HU-p as the forward primer and ΔHU-r (5′-CGCGTCGACCTTTATTAACTCCTTGAAAACTTTTG-3′) as the reverse primer. The fragments were cloned into pQE30 (Qiagen, USA) and sequenced to confirm their identity. Soluble recombinant PfHUup, PfHUp and PfHUΔC proteins carrying the N-terminal RGS-6XHis tag were obtained after E. coli JM109 cells co-transformed with the RIG plasmid (gifted by Prof. W.G.J. Hol) were induced with 0.1 mM IPTG at 20°C for 18 h. The RIG plasmid carries tRNA genes whose transcripts recognize rare codons for the amino acids R, I and G in P. falciparum DNA expressed in E. coli (30). The recombinant proteins were purified on Ni-NTA Superflow (Qiagen, USA) and dialysed in a buffer containing 50 mM Tris–HCl (pH 7.6), 200 mM NaCl. PfHUp and PfHUΔC were further purified on a SP sepharose column. Final concentration of purified protein was determined by BCA assay. Far UV CD spectra analysis of purified PfHUp showed the presence of both α-helices and β-sheets and was comparable to other HU proteins thus indicating correct folding (data not shown). Chemical-crosslinking of PfHUp was carried out with 0.1% glutaraldehyde in 10 mM NaH2PO4 and 50 mM NaCl for 30 min at 37°C.
The gene encoding Bacillus subtilis HU (HBsu), was PCR-amplified from B. subtilis genomic DNA (upstream primer, 5′-CGGGGATCCATGAACAAAACAGAACTTATCAATG-3′ and downstream primer, 5′-CGTAAGCTTTTTTCCGGCAACTGCGTCTTTAAG-3′) and cloned in the pQE30 expression vector. The ∼12 kDa recombinant protein carrying a N-terminal His-tag was purified by Ni-NTA chromatography and used as positive control in DNA circularization assays.
Antibody production, western blotting and immunoprecipitation
Antibodies against PfHUp were raised in rabbits and mice using purified PfHUp. The titer of the raised antiserum was determined by ELISA.
For preparation of total parasite lysate, parasites were released by 0.05% saponin lysis, washed with PBS, and suspended in 1× SDS loading buffer containing protease inhibitors (Protease Arrest, GBiosciences, USA). After brief sonication, the cell lysate was separated on a 15% SDS–polyacrylamide gel. Western blotting was carried out (6) and the blot was developed using a chemiluminescent system (Amersham Biosciences, UK).
For immunoprecipitation, parasite cultures at 6–8% parasitaemia were harvested when cells were predominantly at the late trophozoite stage. Cells were washed with PBS and parasites were released by 0.05% saponin lysis. The parasite pellet was washed with PBS and lysed in chilled immunoprecipitation (IP) buffer (30 mM Tris–HCl pH 8.0, 300 mM NaCl, 1 mM EDTA, 1% v/v Triton X-100, 1% v/v Igepal and protease inhibitor cocktail) on ice for 30 min. After brief sonication, the lysate was centrifuged at 12 500 r.p.m. for 10 min. The supernatant was precleared by addition of 3 mg Protein A sepharose CL-4B for 1 h. The cleared supernatant was incubated with primary antibody (rabbit anti-PfHUp serum) for 2 h on ice with concomitant mixing. After centrifugation at 10 000 r.p.m. for 2 min at 4°C, the supernatant was incubated overnight with 5 mg Protein A sepharose at 4°C with continuous mixing. Sepharose beads were pelleted at 12 000 r.p.m. for 2 min at 4°C and washed five times with chilled IP buffer followed by two PBS washes. Immunoprecipitated proteins were obtained by treating the beads with nonreducing SDS lysis buffer. The sample was electrophoresed on a 15% gel and transferred onto a nitrocellulose membrane. The membrane was probed with mouse anti-HUp serum as primary antibody and anti-mouse HRP conjugate as secondary antibody followed by development of the blot using a chemiluminescent detection system.
Electrophoretic mobility shift assay (EMSA)
Agarose gel electrophoresis was carried out with complexes of plasmid pBR322 and PfHUp or PfHUΔC as a function of protein concentration. Four hundred nanograms of supercoiled pBR322 (New England Biolabs, USA) or linear pBR322 DNA was used in the binding reaction with PfHUp or PfHUΔC in a reaction buffer containing 50 mM Tris–HCl pH 7.5, 0.1 mM EDTA. The reaction was incubated at 37°C for 40 min followed by electrophoresis on a 1% agarose gel at room temperature in 1× TAE buffer. The gel was stained with ethidium bromide (0.5 µg/ml) for 1 h, destained with 1× TAE and photographed.
For EMSAs on polyacrylamide gels, the binding reaction (20 µl) was carried out in 20 mM Tris–HCl, 0.1 mM EDTA and incubated for 30 min at room temperature. The gel was electrophoresed in 0.25× TBE. Kd determination was carried out by incubating 100 femtomoles of 5′-end-labeled 30 bp double-stranded oligo probe with increasing concentrations of protein as described in Ghosh and Grove (31). The region of a lane from the complex upto the free probe was taken as complex. Kd was calculated from the Hill equation, f = fmax × [PfHUp]n/(Kd + [PfHUp]n), where f is the fraction complex ([PfHUp]b/[DNA]t), fmax is fraction complex at maximal saturation and [PfHUp] is protein concentration. The Hill coefficient (n) was set to one for single-site binding. Signals for bound and free probe were quantitated using OptiQuant1 software in Cyclone phosphorimager (Packard). Kd was calculated by curve-fitting using nonlinear regression in GraphPad PRISM software.
Supercoiling assay
Negatively supercoiled pBR322 (200 ng/reaction) was relaxed with 2 units of E. coli topoisomerase I (New England Biolabs, USA) in 10 mM Tris–HCl, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA and 100 µg/ml BSA for 2 h at 37°C. The relaxed DNA was incubated with increasing concentrations of PfHUp or PfHUΔC and the volume was adjusted with 1× dilution buffer (20 mM Tris–HCl, pH 7.5, 0.1 mM EDTA, 100 µg/ml BSA) and incubated for 1 h at 37°C. Reactions were terminated with proteinase K (0.5 µg/µl) and 0.5% SDS and incubated at 37°C for 1 h. The DNA was electrophoresed on 0.8% agarose gel in 0.5 × TBE for 14 h.
In vivo condensation assay
Escherichia coli JM109 cells co-transformed with pQE30-HUp + RIG or the parent vector pQE30 + RIG as control were grown till the cultures reached an O.D. of 0.5. After induction with 0.5 mM IPTG for 3 h at 20°C, 1 ml culture was withdrawn and cells were washed twice with 1× TBS. Washed cells were suspended in 100 µl TBS, fixed with 0.5% glutaraldehyde and stained with 1.0 µg/ml DAPI for 30 min at 37°C. Cells were washed three times with 1× TBS and visualized in a fluorescence microscope (Leica DM5000B).
DNA-circularization assay
The ability of PfHUp to induce DNA circularization in the presence of T4 DNA ligase was assayed by incubating 2.5 ng of a 136 bp, 32P 5′-end-labeled DNA probe with or without PfHUp (or PfHUΔC) in a 20 µl reaction containing 1× ligase buffer at 25°C for 30 min. The 136 bp DNA was derived from a 369 bp PCR-amplified fragment from a Mycobacterium gene. The 369 bp fragment was digested with Taq1 to give the 136 bp probe containing 5′ overhangs at both ends. The incubation of the DNA probe with PfHU was followed by addition of 4.5 U of T4 DNA ligase (Promega, USA) and further incubation at 22°C for 2 h. If samples were to be treated with BAL31 exonuclease (Fermentas, USA) after ligation, the ligation reaction was added to an equal volume of 2× nuclease buffer containing 1U of BAL31. The digestion reaction was incubated at 30°C for 15 min. The DNA was extracted with phenol–chloroform, collected by ethanol precipitation and suspended in DNA loading dye. After electrophoresis on 7.5% 0.25× TBE-PA gel, the gel was dried and autoradiographed.
For the transformation assay, linear pBR322 (5 ng) was incubated with PfHUp and T4 DNA ligase as in the ligation assay above followed by protein removal by Proteinase K treatment (10 µg/reaction) and transformation of E. coli DH5α cells.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assay was performed as described by Gissot et al. (32) with some modifications. Thirty milliliters of trophozoite stage P. falciparum culture was subjected to protein–DNA crosslinking using 1% (v/v) formaldehyde at 37°C for 15 min. Parasites were released by 0.05% saponin lysis and the parasite pellet was suspended in 1 ml ChIP buffer [30 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.5% (v/v) Triton X-100 and 1% v/v Igepal]. Cells were incubated on ice for 20 min, sonicated nine times (Branson Digital Sonifier 450) at 30% amplitude for 10 s each with 1 min cooling between sonications. After centrifugation at 12 500 r.p.m. for 10 min at 4°C, the supernatant containing soluble chromatin was precleared with 50 µl of 50% Protein A sepharose CL-4B and 20 µg of sheared salmon sperm DNA for 2 h at 4°C. The beads were pelleted at 10 000 r.p.m. for 2 min. Preimmune serum or anti-PfHUp serum (1:75 dilution) was added to the supernatant and incubated at 4°C for 2 h. Forty microliters of 50% Protein A sepharose and 20 µg salmon sperm DNA was added to the mixture and incubated overnight at 4°C with continuous mixing. The sepharose beads were pelleted at 2000 r.p.m. at 4°C for 1 min and washed three times with ChIP buffer. This was followed by two washes with 1× TE and one wash with 1× TE and 0.01% SDS. Chromatin was eluted using 1× TE supplemented with 1% SDS. Chromatin was reverse cross-linked for 6 h at 65°C and treated with proteinase K (20 µg) for 2 h at 37°C. DNA was extracted with phenol–chloroform followed by ethanol precipitation. DNA from input and ChIP samples were resuspended in 100 and 20 µl TE, respectively. For PCR-amplification, 0.5 µl of input DNA and 2 µl of the ChIP sample were used as template. Primers for the nuclear gene PfHU and a plDNA sequence RIII (11) were used to amplify nuclear and apicoplast DNA, respectively. The PCR products were analysed on 1% agarose gel.
Confocal microscopy
P. falciparum cultures were processed for immunofluorescence labeling and confocal microscopy according to the method of Tonkin et al. (33). For mitochondrial labeling, live cells were incubated in 25 ng/ml MitoTracker Deep Red 633 (Molecular Probes) in PBS for 20 min at 4°C prior to fixation. Cells were washed with PBS and fixed in solution using 4% paraformaldehyde and 0.0075% glutaraldehyde in PBS for 30 min. After one wash with PBS, fixed cells were permeabilized with 0.1% Triton X-100 in PBS for 10 min. After another PBS wash, cells were treated with ∼0.1 mg/ml sodium borohydride in PBS for 10 min. Cells were washed once with PBS, blocked in 3% BSA/PBS for 1 h and incubated overnight with rabbit anti-PfHUp serum (1:50 dilution in PBS containing 3% BSA) at 4°C. After three washes (10 min for each wash) with PBS, the cells were incubated with AlexaFluor514-tagged anti-rabbit secondary antibody for 2 h at room temperature and allowed to settle onto coverslips coated with poly-l-lysine (100 μg/ml). The coverslips were then washed three times in PBS and mounted in anti-fade mounting media (Oncogene, USA). For apicoplast co-localization studies, the D10-ACPleader-GFP line in which GFP is an apicoplast marker was used. Mouse anti-PfHUp Ab (1:25) or rabbit anti-GFP Ab (1 : 1000) (Molecular Probes) was used as primary antibody with Texas Red tagged anti-mouse Ab (Molecular Probes) or Alexa Fluor488 tagged anti-rabbit Ab (Sigma) as secondary antibodies. The slides were viewed in a confocal laser-scanning microscope (Zeiss LSM 510) under a 63× oil immersion lens.
Atomic Force Microscopy (AFM)
Atomic force microscopy of DNA–PfHU complexes was carried out by using freshly cleaved mica treated with vapors of 3-aminopropyl triethoxysilane (APTES) at room temperature for 2 h for immobilization of samples (18). For sample preparation, PfHUp and linear pBR322 DNA were mixed at different dimer/bp ratios in 20 µl of AFM buffer (20 mM Tris–HCl, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 1 mM EDTA). The final concentration of DNA in the mix was ∼1 ng/µl. After incubation for 10 min at room temperature, 10 µl of sample was dropped on APTES-coated mica and allowed to bind for 2 min at room temperature. The mica was washed extensively with deionized water, blotted at the edge and air-dried.
AFM imaging was carried out using the PicoSPM equipment (Molecular Imaging, AZ, USA). Images were obtained in the AAC mode with 225 μm long cantilevers that had resonance frequency of 65 kHz and force constant of 2.8 N/m. DNA contour length (in absence of protein) was calculated to be 1.403 µm (SD = 0.156 µm, n = 95), which is close to the expected contour length (1.439 µm) of the 4361 bp pBR322.
RESULTS
Structure modeling of PfHU reveals extensive fold conservation
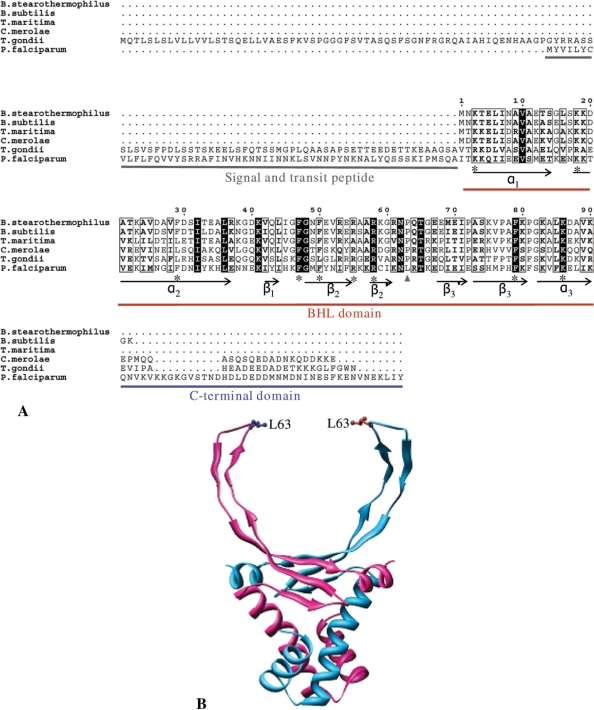
The nuclear DNA-encoded 22.5 kDa P. falciparum protein (PlasmoDB ID PFI0230c) was identified as a candidate protein for DNA organization in the apicoplast due to the presence of a predicted bipartite apicoplast-targeting sequence that is the characteristic of nuclear-encoded proteins targeted to the organelle (34). The sequence has also been recently annotated as ‘bacterial histone-like protein, putative’ in the Plasmodium database (www.plasmodb.org). PfHU has a predicted 53 aa long signal and transit peptide with a high score in the apicoplast-targeting sequence prediction software Plasmo AP. This is followed by a conserved bacterial histone-like (BHL) domain found in histone-like proteins in bacteria, dinoflagellates, red algal chloroplasts as well as in the related apicomplexan T. gondii (35) (Figure 1). PfHU also carries a 42 aa unconserved C-terminal extension beyond the BHL domain.
Figure 1.
Sequence alignment and structure model of PfHU. (A) ClustalW alignment of PfHU with bacterial (Bacillus stearothermophilus, B. subtilis, Thermotoga maritima), red algal chloroplast (Cyanidioschyzon merolae) and apicomplexan (Toxoplasma gondii) HU proteins. Conserved residues described in the text are marked with asterisk. (B) Structure of the PfHU dimer modeled on the crystal structure of B. stearothermophilus HU. The position of the leucine residue (L63) that replaces the conserved proline of other HU proteins is indicated. The 42 aa C-terminal domain could not be modeled on any known protein structure and is not depicted in the figure.
Alignment of the PfHU sequence with selected HU homologs from other organisms demonstrates significant sequence conservation within the BHL domain (Figure 1A) suggesting overall similarity of structure and function with other HU proteins. PfHU exhibits maximum homology with the T. gondii HU (30% identity) followed by the chloroplast HU of the red alga Cyanidioschyzon merolae (28% identity). Hydrophobic residues in the BHL core, notably phenylalanine residues at positions 29, 47, 50 and 79, are conserved in PfHU. These residues are part of the dimerization signal and are involved in the formation of an aromatic hydrophobic core involving inter-subunit stacking (36,37). PfHU has an additional F54 residue that may also contribute to formation of the hydrophobic core. Structural analysis has shown that a number of arginine residues (R55, R58, R61) within the DNA-binding arms participate in hydrogen bonding or electrostatic contacts to the DNA (38). Of these, R55 and R58 are conserved in PfHU while R61 is replaced by a lysine. Surface-exposed lysine residues (K3, K18, K86) that line the body of the protein and have been shown to contribute to DNA binding (39) are also conserved in PfHU suggesting their contribution to wrapping of DNA around the protein.
Structure modeling of PfHU on the Bacillus Stearothermophilus structure (PDB ID: 1huu) (40) was carried out by SWISS-MODEL using the alignment mode. Extensive fold conservation in the PfHU BHL domain was revealed. The 42 aa C-terminal extension could not be modeled as it lacked significant homology with any known protein and contained regions of high disorder predicted using DisEMBL (41). The structure model of the PfHU homodimer is shown in Figure 1B. The predicted homodimer structure is comprised of a largely α-helical body with the α1- and α2-helices being formed by residue K3–T14 and K18–E37 of the predicted processed protein, respectively. β1 (residue I42–I44), β2 (residue G48–R55) and β3 (residue S73–F81) strands form the saddle-like β sheet structure. 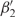 and
and 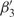 (R58–K61 and D68–E72, respectively) are part of the DNA-binding arm of PfHU. The DNA-binding arm of the PfHU monomer (residues 53–75) contains eight positively charged residues but lacks the highly conserved proline (P63) that is implicated in induction and/or stabilization of DNA bending by intercalating between base pairs (42). The DNA-binding arms of HU proteins have been described as disordered in crystal structures (36,43) and NMR studies show that the homodimer DNA-binding arms are folded at the tips and are flexible in solution (44).
(R58–K61 and D68–E72, respectively) are part of the DNA-binding arm of PfHU. The DNA-binding arm of the PfHU monomer (residues 53–75) contains eight positively charged residues but lacks the highly conserved proline (P63) that is implicated in induction and/or stabilization of DNA bending by intercalating between base pairs (42). The DNA-binding arms of HU proteins have been described as disordered in crystal structures (36,43) and NMR studies show that the homodimer DNA-binding arms are folded at the tips and are flexible in solution (44).
PfHU specifically binds apicoplast DNA
The processed form of PfHU (PfHUp) was expressed as a soluble protein in E. coli. Although the expected size of the 6XHis-tagged processed protein is ∼18 kDa, purified PfHUp ran at ∼23 kDa on SDS–PA gels (Figure 2A) with the intact protein comprising ∼93% of the purified fraction. The observed difference in size may be attributed to the presence of nonglobular domains in the protein or an excess of positively charged residues; such differences between expected and observed sizes on SDS–PA gels have also been observed for other P. falciparum proteins (15). The unprocessed form of PfHU (PfHUup) carrying the apicoplast-targeting sequence was also expressed in E. coli and resolved at ∼25 kDa on SDS–PA gels (Figure 2A). Purified recombinant processed PfHU lacking the 42 aa C-terminal domain (PfHUΔC) ran at ∼15 kDa, above its expected size of ∼13 kDa, together with some truncated products. The truncated products were recognized by anti-His Ab in western blots (data not shown) indicating that they contain deletions from the C-terminal end. The major 13.5 kDa band would lack the α3-helix in addition to the C-terminal domain but is likely to retain DNA binding. The two minor truncation products would additionally lack both the β3 and β3 strands and are thus unlikely to bind DNA. The 15 kDa band and its major truncated product of ∼13.5 kDa that together comprised ∼85% of the total purified protein are referred to as the ‘C-terminal deletion’ (PfHUΔC) and were used for all calculations for quantitation of PfHUΔC for DNA-binding experiments.
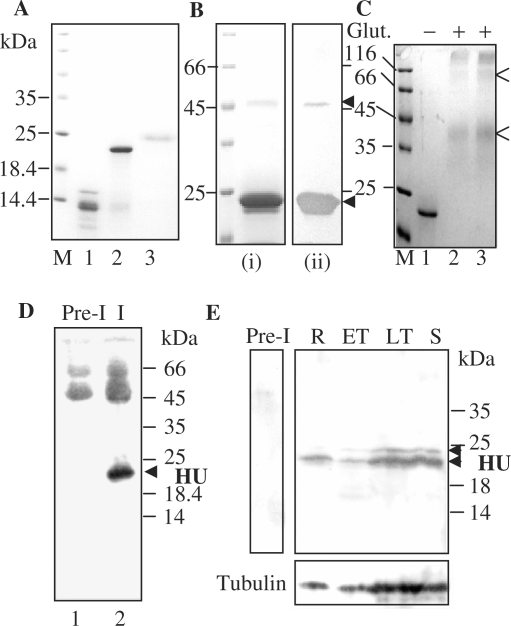
Figure 2.
Expression of recombinant PfHU and its detection in P. falciparum. (A) Purified recombinant proteins PfHUΔC (lane 1), PfHUp (lane 2) and PfHUup (lane 3). M denotes marker lane. (B) Multimeric forms of purified PfHUp seen in Coomassie-stained SDS–PA gels (i) and detected by anti-6XHis antibody in Western blots (ii). (C) Chemical-crosslinking of 5 µg (lane 2) and 7 µg (lane 3) PfHUp indicates dimerization of the protein in solution. Glut., glutaraldehyde. Dimer and tetramer forms are indicated by arrows. (D) Detection of processed HU protein in P. falciparum lysates immunoprecipitated with rabbit anti-PfHUp antibody followed by detection using mouse anti-PfHUp antibody in a Western blot. Lane 1 represents immunoprecipitation with rabbit preimmune serum. (E) Expression of PfHU in P. falciparum intra-erythrocytic stages. The upper and lower panels are Western blots using anti-PfHUp antibody and α, β-tubulin antibodies (Sigma), respectively. R, rings; ET, early trophozoites; LT, late trophozoites; S, schizonts. Pre-I, lysate probed with preimmune serum. Arrows indicate unprocessed and processed forms of PfHU.
When purified recombinant PfHUp was subjected to SDS–PAGE analysis, an additional band of ∼44 kDa was observed together with the ∼23 kDa monomeric form at high protein concentration (Figure 2B, i). The 44 kDa band corresponds to the size expected for the dimeric form of PfHUp. Anti-His-antibody recognized the band in western blot confirming that it was the PfHUp dimer that either remained associated even after boiling in the presence of 0.1% SDS (Figure 2B, ii) or the PfHUp monomers reassociated in 0.1% SDS after removal from heat. Treatment of purified PfHUp with 8 M urea caused dissociation of the dimer (data not shown) suggesting that hydrophobic interactions play a major role in dimerization of the protein in vitro. Glutaraldehyde-mediated crosslinking of PfHUp demonstrated that the protein existed predominantly as a dimer in solution (Figure 2C). A minor tetrameric form was also seen upon crosslinking.
Antibodies raised against recombinant PfHUp specifically recognized a ∼22 kDa protein in western blots together with a fainter band at ∼24 kDa (Figure 2E). This size difference corresponds to the difference observed for recombinant PfHUp and PfHUup indicating that the lower and upper bands represent processed and unprocessed forms of PfHU, respectively. Rabbit anti-PfHUp serum immunoprecipitated the processed form of PfHU that was specifically recognized by mouse anti-PfHUp serum (Figure 2D). A specific RT-PCR product of the length expected after cleavage of the single intron in the PfHU gene was also amplified from total parasite RNA (data not shown). These results confirm that PfHU is translated in the parasite. PfHU was also detected in parasite lysates from synchronized cells at different intra-erythrocytic stages (Figure 2E). Expression of the protein was observed at all stages and comparison with parasite tubulin levels indicated constitutive PfHU expression during the P. falciparum erythrocytic cycle.
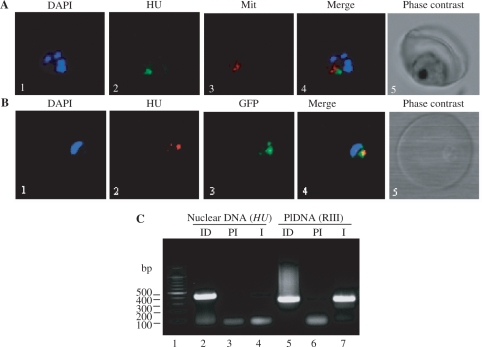
P. falciparum carries three DNA genomes- nuclear, apicoplast and mitochondrial (45). To address the possibility of PfHU also serving as a mitochondrial/nuclear DNA organization protein, we carried out immunofluorescence detection of PfHU using anti-PfHUp antibody in confocal microscopy. PfHU localized to an organellar structure close to, but distinct from, the mitochondria stained with Mitotracker Red (Figure 3A). PfHU signal was also not observed in nuclei that were stained with DAPI. Additionally, PfHU co-localized with apicoplast-targeted GFP in the D10 ACPleader-GFP line with GFP as an apicoplast marker (Figure 3B). These data confirm apicoplast-specific localization of PfHU in P. falciparum as also reported for the HU protein of T. gondii (46). In order to investigate whether PfHU interacts with P. falciparum apicoplast DNA in vivo, we carried out chromatin immunoprecipitation (ChIP) of total P. falciparum DNA using anti-PfHUp antibody. This was followed by PCR-amplification of immunoprecipitated DNA using primer pairs for amplification of a 411 bp internal region of the PfHU gene representing nuclear DNA and a 332 bp fragment (RIII) (11) representing the apicoplast genome. While both primer pairs amplified the corresponding fragments from input DNA (Figure 3C, lanes 2 and 5), amplification of only the apicoplast-specific fragment was observed when DNA precipitated with the anti-PfHUp immune serum was used as template (lane 7). Neither fragment was amplified from DNA immunoprecipitated using preimmune serum (lanes 3 and 6). The clear recovery of apicoplast DNA in the ChIP assay indicated that PfHU binds specifically to the apicoplast genome thus strengthening its candidature as a key protein in plDNA organization.
Figure 3.
Localization of PfHU. (A) Immunolocalization of PfHU using confocal microscopy. Panels show nuclear DNA staining with DAPI (1), PfHU fluorescence (2), MitoTracker Red signal (3) and their overlap (4) in a late trophozoite. The corresponding phase–contrast scan is shown in (5). (B) Co-localization of PfHU and apicoplast-targeted GFP. Nuclear DNA stained with DAPI (1), PfHU signal (2), GFP signal (3) and their overlay (4) are shown. (C) Anti-PfHUp antibody specifically precipitates apicoplast DNA (plDNA) in a ChIP assay. Lanes 2–4 show PCR products obtained using primers for nuclear DNA (HU sequence) while lanes 5–7 show PCR products obtained using plDNA-specific primers for RIII. ID, input DNA; PI, preimmune serum; I, immune serum.
PfHUp binds and condenses DNA
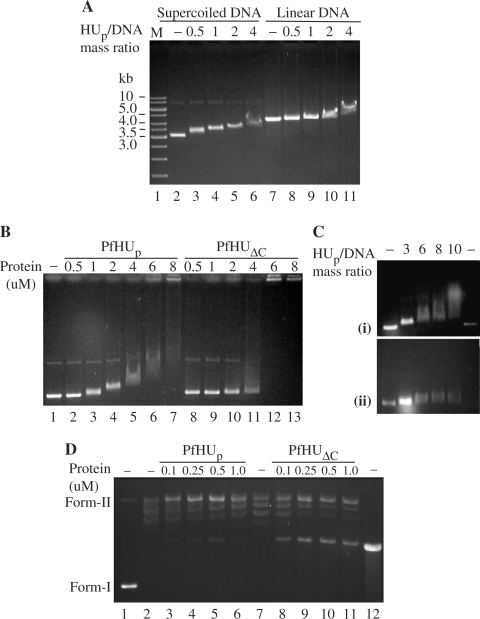
The DNA-binding activity of recombinant PfHUp was investigated by electrophoretic mobility shift assays (EMSAs) in agarose gels using supercoiled and linear pBR322. The mobility of the supercoiled form was retarded sharply by PfHUp starting at the protein/DNA mass ratio of 0.5 while retardation of linear DNA was clearly visible only beyond the protein/DNA mass ratio of 2 (Figure 4A). This indicated that PfHUp exhibits preferential condensation of supercoiled DNA compared to linear DNA. EMSA with increasing concentrations of PfHUp and PfHUΔC incubated with supercoiled pBR322 (Figure 4B) revealed a difference in their DNA condensing properties. While PfHUp condensed DNA in vitro at a concentration of 8 µM (protein/DNA mass ratio ≥10) when nearly all the DNA was retained in the well, complete condensation of DNA by PfHUΔC was observed at 6 µM (protein/DNA mass ratio = 6.2). Additionally, in contrast to PfHUp, no intermediate retardation forms were observed with PfHUΔC. A minor retardation with 2 µM PfHUΔC was followed by complete condensation of DNA as the protein was increased from 4 to 6 µM. These results indicate a role of the 42 aa C-terminal domain in influencing the DNA-condensation properties of PfHUp.
Figure 4.
DNA-binding properties of PfHUp. (A) EMSA depicting binding of PfHUp to supercoiled and linear pBR322. Four hundred nanograms of supercoiled (lanes 2–6) or linear (lanes 7–11) plasmid was incubated with PfHUp at different protein/DNA mass ratios. M, DNA marker. (B) EMSA with increasing concentrations of PfHUp (lanes 2–7) and PfHUΔC (lanes 8–13) using supercoiled pBR322 DNA. Lane 1 is free DNA. (C) Binding of PfHUp to supercoiled DNA in the presence of 50 mM (i) and 100 mM (ii) NaCl. (D) DNA supercoiling assay with increasing concentrations of PfHUp (lanes 3–7) and PfHUΔC (lanes 8–12). Lane 1 is naked DNA (negatively supercoiled pBR322), lane 2 is pBR322 partially relaxed with topoisomerase I, lane 12 is linearized pBR322. Form-I, supercoiled DNA; form-II, relaxed DNA.
The effect of salt on DNA–protein interaction was assayed by addition of 50 or 100 mM NaCl to the binding reaction at different PfHU/DNA mass ratios. Partial inhibition of binding was observed with 50 mM NaCl as a much greater amount of PfHUp was required to cause the same level of retardation (Figure 4C). Binding was completely inhibited by 100 mM NaCl and no retardation was observed even at the protein/DNA mass ratio of 10 indicating that interaction between PfHUp and DNA was primarily electrostatic.
The effect of increasing concentrations of PfHUp and PfHUΔC on DNA supercoiling (Figure 4D) showed that the supercoiling ability of both the products was negligible. This contrasts with reports for other HU proteins (47) that hinder relaxation by topoisomerase I and constrain negative supercoils in a concentration-dependent manner.
The effect of over-expression of PfHUp on E. coli DNA was examined by fluorescence microscopy of DAPI-stained E. coli cells that were transformed with the expression vector pQE30 or the PfHUp expression vector pQE30-HUp + RIG plasmid and induced with IPTG (RIG overcomes codon bias for expression of P. falciparum genes in E. coli). Compared with control cells, cells expressing PfHUp exhibited very slow growth upto 3 h after induction. Additionally, control cells lacking PfHUp exhibited uniform DNA distribution while extensive condensation of the E. coli nucleoid was visible in cells expressing PfHUp (Figure 5). This result indicates that an excess of PfHUp causes greater compaction of E. coli nucleoids. Similar compaction of E. coli nucleoids was also observed with overexpression of PfHUΔC (data not shown).
Figure 5.
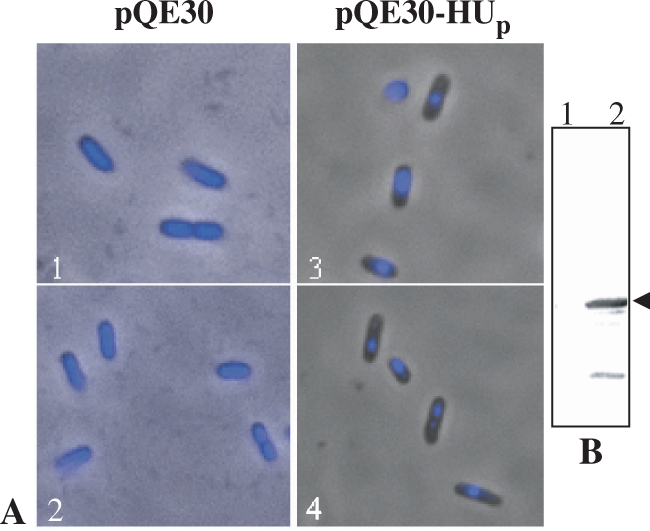
PfHUp-mediated condensation of E. coli nucleoids. (A) Fluorescence images of DAPI-stained, IPTG-induced E. coli cells that had been co-transformed with pQE30 + RIG plasmid (1 and 2) or pQE30-HUp + RIG plasmid (3 and 4). (B) Western blot using anti-PfHUp antibody to detect expression of PfHUp in E. coli cells co-transformed with RIG and pQE30 (lane 1) or pQE30-HUp (lane 2) followed by induction with IPTG. Arrow indicates the PfHUp band.
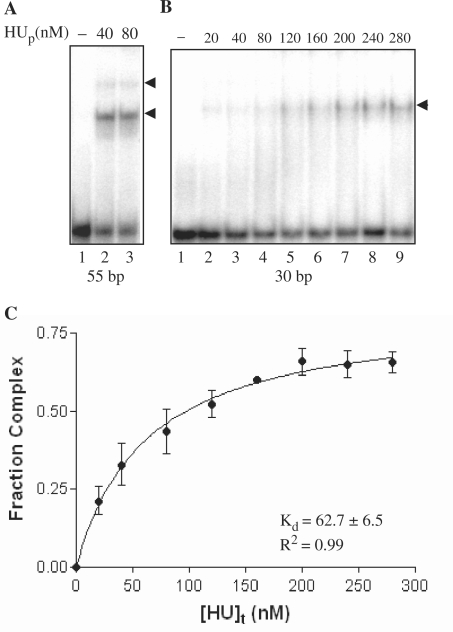
The minimal binding-site length of PfHU was determined by using double-stranded oligonucleotide probes of 23, 30 and 55 bp in binding reactions resolved in EMSAs on PA gels. A very faint smeared complex was observed with the 23 bp probe (data not shown) while a single clear complex was obtained with the 30 bp probe (Figure 6B). Two complexes were obtained when the 55 bp probe was used (Figure 6A) indicating a binding site of between 24 and 27 bp. The 55 bp probe containing two PfHUp dimer binding sites was also used to determine the active fraction of PfHUp by titrating 125 nM of PfHUp dimer with increasing concentrations (2.5–160 nM) of the 55 bp DNA probe in EMSA (data not shown). The protein saturated at ∼60 nM DNA indicating that >95% of the protein was active. The affinity of the PfHUp dimer for DNA was determined by calculation of Kd of the single complex obtained with the 30 bp probe (Figure 6C). The Kd value obtained for PfHUp was 62.7 ± 6.5 nM.
Figure 6.
Binding site length and affinity of PfHUp for DNA. (A) EMSA showing binding of PfHUp with end-labeled 55 bp probe. (B) EMSA of PfHUp with a 30 bp probe. (C) The Kd of the single complex obtained in (B) was determined from its binding isotherm by curve-fitting using nonlinear regression. The mean Kd value from three repeat experiments was estimated as 62.7 ± 6.5 nM.
PfHUp promotes DNA concatenation
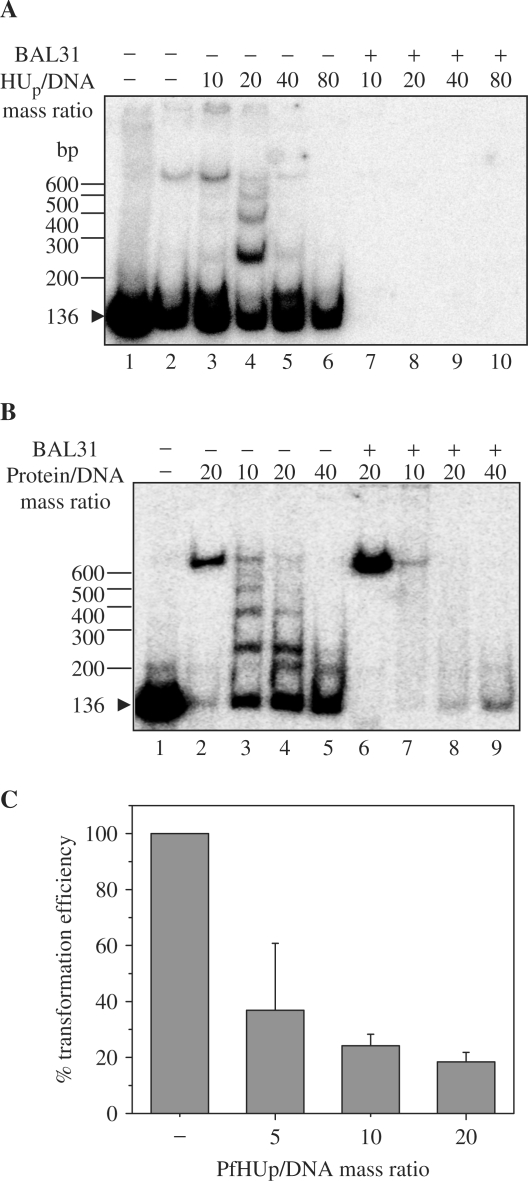
Bacterial HU is capable of mediating DNA ring closure (48) while its dinoflagellate counterpart promotes concatenation of DNA fragments and inhibits DNA circularization (18). The ability of PfHUp to promote circularization of short-length DNA fragments as a result of protein-mediated DNA bending was tested in a T4 ligase-mediated DNA ligation assay. Ligation of a 136 bp DNA fragment was carried out in the presence of increasing concentrations of PfHUp and PfHUΔC (Figure 7A and B). HBsu (B. subtilis encoded HU homolog) was included as positive control for cyclization (49). The ligation products were also treated with exonuclease to identify exonuclease-resistant circularized DNA. Unlike bacterial HU and similar to the dinoflagellate HCc3, PfHUp promoted concatenation of DNA as evident from the increase in exonuclease-sensitive linear multimers of the 136 bp DNA fragment upto PfHUp/DNA mass ratio of 20 (Figure 7A). Higher concentrations of protein (PfHUp/DNA mass ratio of 40 and 80) inhibited concatenation. Similar results were obtained with PfHUΔC indicating that the C-terminal extension was not responsible for mediating DNA concatenation (Figure 7B). PfHUp and PfHUΔC also promoted concatenation and inhibited circularization of a longer 207 bp DNA fragment in a concentration-dependent manner (data not shown).
Figure 7.
DNA concatenation by PfHUp. (A) DNA ligation of a 136 bp labeled DNA fragment carried out in the presence of increasing concentrations of PfHUp (lanes 3–6). Lane 1 is free DNA while lane 2 is DNA probe ligated in the absence of PfHUp. Ligation reactions were treated with BAL31 nuclease to detect circularized DNA products (lanes 7–10). (B) Ligation of the 136 bp fragment in the presence of increasing concentrations of PfHUΔC (lanes 3–5 and 7–9). Lane 1 is free DNA while lanes 2 and 6 are DNA probe ligated in the presence of HBsu as positive control for DNA circularization. (C) Percentage transformation efficiency of ligation reactions of linear pBR322 carried out in the presence of PfHUp. Transformation efficiency was calculated as percentage of that obtained with pBR322 ligated in the absence of PfHUp. Mean and SE of repeat determinations is plotted.
PfHUp also inhibited DNA circularization as evident from results of the transformation assay (Figure 7C) where linear pBR322 (5 ng) was incubated with PfHUp and T4 DNA ligase as in the ligation assay above followed by transformation of E. coli DH5α cells. PfHUp exhibited dose-dependent inhibition of DNA circularization. Upto 80% inhibition was observed at PfHUp/DNA mass ratio of 20, which corresponds to the protein concentration at which maximum concatenation is seen in Figure 7A.
AFM studies
Atomic Force Microscopy of PfHUp–DNA complexes at different PfHUp-dimer/bp ratios provided further evidence for DNA condensation mediated by PfHUp (Figure 8). At lower concentration of protein (dimer/bp ratio 1 : 750) stiffening of DNA strands was evident (Figure 8B, i). Intermolecular DNA bundling with two DNA strands forming a tight bundle was also observed at this concentration (Figure 8B, ii). PfHUp-mediated formation of DNA loops (Figure 8C, ii) was seen at the dimer/bp ratio of 1 : 500 and assembly of condensed DNA complexes with a small number of foci and extruding DNA loops was initiated (Figure 8C, i and iii). This data indicated that PfHUp also forms DNA bridges. Formation of larger PfHUp–DNA complexes was seen at higher protein concentration (dimer/bp ratio 1 : 250) (Figure 8D). These complexes appeared to be formed by the assembly of large DNA bundles/bridges brought together by intermolecular interactions between PfHUp subunits (Figure 8D, ii and iii).
Figure 8.
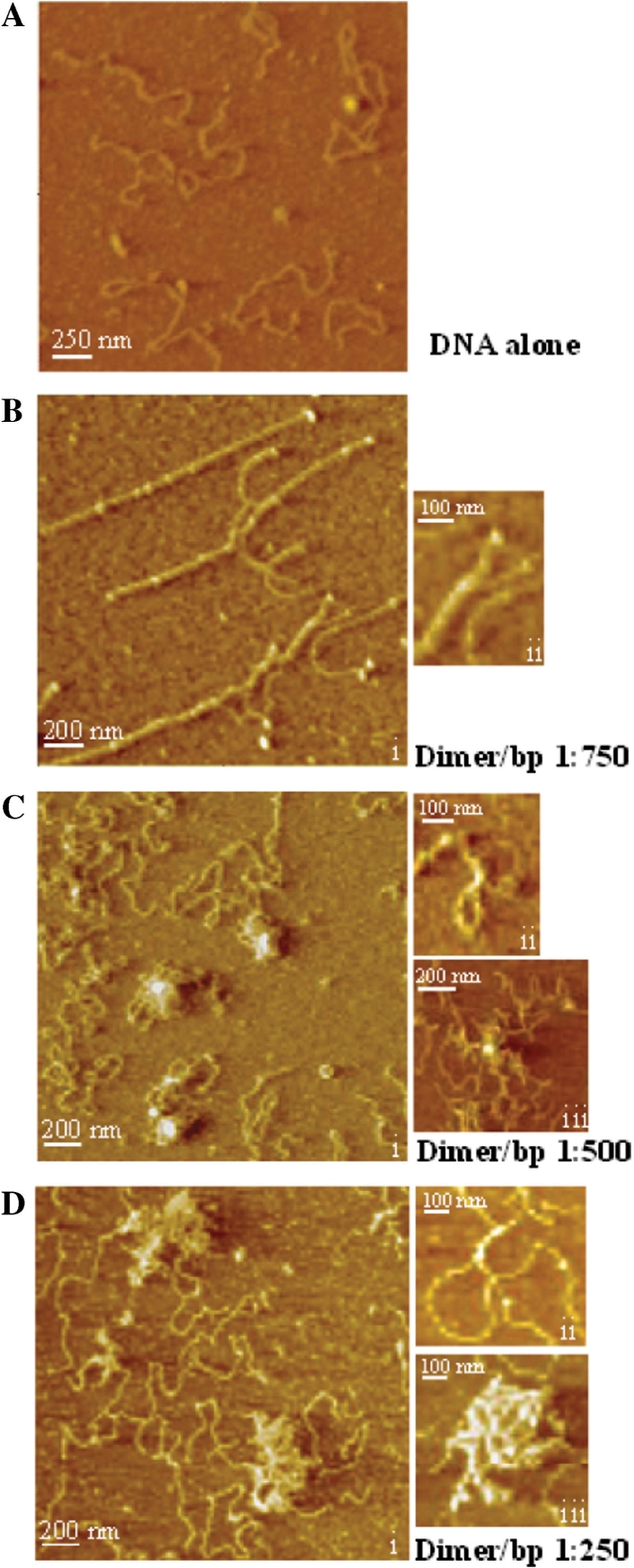
AFM images of PfHUp-linear pBR322 DNA complexes with increasing dimer/bp ratio. (A) DNA in the absence of protein. (B) Dimer/bp ratio of 1 : 750. Stiffened DNA strands (i) as well as DNA bundles (ii) are shown. (C) Dimer/bp ratio of 1 : 500. Formation of complexes with a small number of foci and extruding DNA loops (i), a DNA bridge resulting in DNA looping (ii) and a nucleoprotein complex with a single focus (iii) are shown. (D) Dimer/bp ratio of 1 : 250. Large complexes (i and iii) are formed by the assembly of DNA bundles (ii) and bridges.
DISCUSSION
Proteins that mediate organization of the Plasmodium organellar genomes are yet to be identified and functionally characterized. While core histones (H2A, H2B, H3 and H4) for nuclear DNA assembly are encoded in the genome together with a few histone variants (50), two nucleosome assembly proteins, PfNAPS and PfNAPL, which preferentially interact with H3–H4 tetramer histones have also been characterized (51). Although the high mobility group (HMG) protein Abf2 is an abundant basic protein that supercoils mitochondrial DNA in yeast (52), the HMG homologs on the P. falciparum genome lack predicted mitochondrial-targeting signals. On the other hand, a mitochondrial-targeting sequence is predicted for a putative histone 2B (PF11_0062) encoded by the nuclear genome. Of the 517 nuclear-encoded proteins predicted to target to the apicoplast, PfHU (PFI0230c) is the only one with a dsDNA-binding BHL domain similar to HU-like DNA organization proteins of plastids. Our results identify PfHU as a component of the apicoplast and confirm its interaction with apicoplast DNA. The DNA-binding characteristics of PfHU and its effect on bacterial nucleoid condensation are also indicative of its role as a plDNA architecture protein in P. falciparum.
Sequence-function analyses of PfHU reveal some interesting features. While the DNA-intercalating proline (P63) and surrounding residues are conserved in most HU proteins, a few exceptions such as bacteriophage SPO1–encoded TF1 and HU from Thermotoga, Thermus, Aquifex, Deinococcus and Mycoplasma have R61 replaced by V or M residues (53). R61 is replaced by a K residue in P. falciparum while the P63 residue is replaced by L, thus converting the conserved HU RNP motif into a KNL motif. These observed substitutions may be explained by the low G+C content (∼75% A+T) of the P. falciparum genome which results in a bias against residues G, P, A and R. Significantly, the T. gondii HU sequence (T. gondii genome has 47% A+T) retains conserved R61 and P63 residues. P63 is implicated in induction and/or stabilization of DNA bending and the absence of this residue in PfHU may explain the inability of the protein to induce DNA circularization. An alternative possibility is that PfHU may bend DNA, but association of multiple PfHU molecules results in out-of-phase bending. HCc3, a likely constituent of permanently condensed crystalline chromosomes of the dinoflagellate Crypthecodinium cohnii, also lacks P63 and fails to induce DNA circularization in vitro; similar to PfHU, HCc3 promotes DNA concatenation (18). Assaying DNA circularization by PfHU after mutagenesis to restore P63 would be required to confirm the role of L63 in preventing DNA bending by the protein. Analysis of the αP64L mutant of IHF (position corresponding to P63 in HU) that interacts with DNA in a site-specific manner has demonstrated that the substitution affects binding specificity of the protein (54,55).
As opposed to other HU proteins, PfHUp is unable to constrain negative supercoils. The P63 residues in the E. coli HU dimer intercalate into the minor groove of DNA 9 bp apart and induce two DNA kinks. These kinks are not co-planar and result in negative supercoiling (underwinding) of ∼31° per bp (56). The observed absence of DNA-bending activity of PfHU offers an explanation for its inability to constrain negative supercoils in the DNA double-helix.
HU proteins bind to random DNA sequences with Kd values ranging from 5 to 2500 nM with E. coli HU exhibiting a Kd value of ∼200–300 nM (53,57). The Kd value of PfHU–DNA interaction is ∼63 nM indicating slightly higher affinity of the protein for DNA compared to E. coli HU. HU proteins have been reported to interact with DNA with binding sites of between 9 and 42 bp (57). It has been suggested that variations in HU-binding site lengths are determined by the presence or absence of amino acids capable of forming salt bridges distal to sites of kinking (58). In HU homologs with shorter binding sites, K3 is proposed to form a salt bridge with D26. In TF1, which lacks D26, K3 contacts DNA 8–9 bp away from the DNA kink leading to a longer binding site of 37 bp. Similarly, the absence of D26 in PfHU may explain its binding site length of 24–27 bp.
There is a difference in patterns of DNA shifts obtained with increasing concentrations of PfHUp and PfHUΔC, with sudden transition to highly condensed DNA observed with the latter. The 42 aa C-terminal domain present in PfHUp has a predominance of acidic residues (net PI of PfHUp is 8.81). PfHUΔC, which lacks the domain, has a PI of ∼9.5. The DNA shifts observed in EMSAs with the two proteins indicate that rapid condensation of DNA follows an initial ‘nucleation’ step with increasing PfHUΔC/DNA ratio, while stepwise assembly of intermediate forms is observed with the full-length PfHUp. Although the precise mode for this is unclear, the negatively charged C-terminal domain unique to PfHUp may directly influence interaction of the protein with DNA. Alternatively, rapid DNA condensation by PfHUΔC may be caused by enhanced cooperativity of binding of PfHUΔC dimers to DNA and/or increased intramolecular attraction between PfHUΔC dimers bound at different sites on DNA. The C-terminal domain also seems to influence stability of PfHUp as removal of the domain results in greater protease-sensitivity. In vivo, the intrinsically unstructured C-terminal domain may interact with other parasite proteins (59) and influence PfHU activity.
AFM analysis of DNA–protein complexes formed with PfHUp showed formation of DNA bundles and bridges. DNA bundling is a property exhibited by the RecA protein (60) and also reported for the dinoflagellate HCc3 (18). DNA bundling by PfHUp may bring the ends of two or more DNA strands closer thus promoting ligase-mediated concatenation observed with PfHUp. The bundling of two strands would be mediated by the interaction between two PfHUp dimers, each of which binds to a single DNA strand. Indeed, formation of such tetrameric PfHUp forms is indicated by chemical-crosslinking of the protein in solution. The clustering of long DNA bundles and bridges to form large complexes observed at high PfHUp concentration could be mediated by intermolecular interactions between PfHUp tetramers. Thick rigid DNA–protein filaments as observed for high concentrations of E. coli HU (1 dimer per 1.8 bp) (28) are seen at much lower concentrations of PfHUp (1 dimer per 750 bp). Unlike E. coli HU, whose AFM analysis shows DNA bending mediated by single dimers at one dimer per 92 bp (28), DNA bending was not observed with PfHUp consistent with our observation of inhibition of DNA circularization with the protein. DNA stiffening and bundling by PfHUp may explain the inhibition of circularization observed with the protein. DNA loop formation and bridging by PfHUp is also clearly seen in AFM images. Formation of DNA bridges by the bacterial nucleoid structuring protein, H-NS, which is structurally distinct from HU, has been reported (61). DNA bridging resulting in formation of loops may have implications not only in DNA compaction mediated by PfHUp but also suggests a possible mechanism by which it may influence transcription processes in the apicoplast.
The origin of apicomplexan plastids has been of recent interest and their rhodophyte versus chlorophyte ancestry has been debated (62,63). The presence of a functional HU-like protein in apicomplexan plastids provides further support for red algal ancestry of the apicoplast; HU-like proteins are found in plastid genomes of red algal lineage but not in those of green alga (64). Additionally, the sequence of PfHU is closest to red algal plastid HUs of C. merolae and Guillardia theta and the apicomplexan and red algal HUs cluster with cyanobacteria HUs (35,65). The presence of a nuclear gene encoding apicoplast-targeted HU in P. falciparum indicates that the gene was acquired by a secondary endosymbiotic event from a red alga with subsequent transfer from the red algal plastid to the host nuclear genome.
Our results provide evidence for the involvement of a HU-like protein in DNA organization and compaction in the plastid of an apicomplexan. Apart from being major components of nucleoids, HU proteins in bacteria also play important roles in initiation of DNA replication and regulation of transcription (21,24,66). Although the specific roles that HU may play in apicoplast DNA replication and/or regulation of transcription remain to be elucidated, the reported missegregation of the T. gondii apicoplast genome upon over-expression of TgHU (67) together with the plDNA-specific interaction and DNA condensation properties of PfHU described here are indicative of its significance in the process of apicoplast DNA replication and organellar division.
ACKNOWLEDGEMENTS
We thank Dr Samir Sawant and Amol Ranjan for help with confocal microscopy, Anita Mann for AFM analysis, Ashutosh and Dr Amogh Sahasrabuddhe for helpful discussion and J.P. Srivastava for technical assistance. E.V.S.R.R. and R.N. are recipients of research fellowships from the Council of Scientific and Industrial Research and Department of Biotechnology, respectively. This is CDRI communication no. 7456. Funding by the Council of Scientific and Industrial research (NWP-0038 and NMITLI grant TLP-0010 to SH) is acknowledged. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wilson RJM. Progress with parasite plastids. J. Mol. Biol. 2002;319:257–274. doi: 10.1016/S0022-2836(02)00303-0. (Iain) [DOI] [PubMed] [Google Scholar]
- 2.Foth BJ, McFadden GI. The apicoplast: a plastid in Plasmodium falciparum and other apicomplexan parasites. Int. Rev. Cytol. 2003;224:57–110. doi: 10.1016/s0074-7696(05)24003-2. [DOI] [PubMed] [Google Scholar]
- 3.Wilson RJM. Parasite plastids: approaching the endgame. Biol. Rev. 2005;80:129–153. doi: 10.1017/s1464793104006591. (Iain) [DOI] [PubMed] [Google Scholar]
- 4.Fichera ME, Roos DS. A plastid organelle as a drug target in apicomplexan parasites. Nature. 1997;390:407–409. doi: 10.1038/37132. [DOI] [PubMed] [Google Scholar]
- 5.Gardner MJ, Williamson DH, Wilson RJM. A circular DNA molecule in malaria parasites encodes an RNA polymerase like that of prokaryotes and chloroplasts. Mol. Biochem. Parasitol. 1991;44:115–124. doi: 10.1016/0166-6851(91)90227-w. [DOI] [PubMed] [Google Scholar]
- 6.Chaubey S, Kumar A, Singh D, Habib S. The apicoplast of Plasmodium falciparum is translationally active. Mol. Microbiol. 2005;56:81–89. doi: 10.1111/j.1365-2958.2005.04538.x. [DOI] [PubMed] [Google Scholar]
- 7.Kohler S, Delwiche CF, Denny PW, Tilney LG, Webster P, Wilson RJM, Palmer JD, Roos DS. A plastid of probable green algal origin in apicomplexan parasites. Science. 1997;275:1485–1489. doi: 10.1126/science.275.5305.1485. [DOI] [PubMed] [Google Scholar]
- 8.Matsuzaki M, Kikuchi T, Kojima S, Koroiwa T. Large amounts of apicoplast nucleoid DNA and its segregation in Toxoplasma gondii. Protoplasma. 2001;218:180–191. doi: 10.1007/BF01306607. [DOI] [PubMed] [Google Scholar]
- 9.Williamson DH, Preiser PR, Moore PW, McCready S, Strath M, Wilson, Wilson R.J.M. The plastid DNA of the malaria parasite Plasmodium falciparum is replicated by two mechanisms. Mol. Microbiol. 2002;45:533–542. doi: 10.1046/j.1365-2958.2002.03033.x. (Iain) [DOI] [PubMed] [Google Scholar]
- 10.Singh D, Chaubey S, Habib S. Replication of the Plasmodium falciparum apicoplast DNA initiates within the inverted repeat region. Mol. Biochem. Parasitol. 2003;126:9–14. doi: 10.1016/s0166-6851(02)00251-7. [DOI] [PubMed] [Google Scholar]
- 11.Singh D, Kumar A, Raghu Ram EVS, Habib S. Multiple replication origins within the inverted repeat region of the Plasmodium falciparum apicoplast genome are differentially activated. Mol. Biochem. Parasitol. 2005;139:99–106. doi: 10.1016/j.molbiopara.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 12.Striepen B, Crawford M, Shaw M, Tilney LD, Seeber F, Roos DS. The plastid of Toxoplasma gondii is divided by association with the centrosomes. J. Cell. Biol. 2000;151:1423–1434. doi: 10.1083/jcb.151.7.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seow F, Sato S, Janssen CS, Riehle MO, Mukhopadhyay A, Phillips RS, Wilson RJM, Barrett MP. The plastidic DNA replication enzyme complex of Plasmodium falciparum. Mol. Biochem. Parasitol. 2005;141:145–153. doi: 10.1016/j.molbiopara.2005.02.002. (Iain) [DOI] [PubMed] [Google Scholar]
- 14.Dar MA, Sharma A, Mondal N, Dhar SK. Molecular cloning of apicoplast-targeted Plasmodium falciparum DNA gyrase genes: unique intrinsic ATPase activity and ATP-independent dimerization of PfGyrB subunit. Euk. Cell. 2007;6:398–412. doi: 10.1128/EC.00357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ram EVSR, Kumar A, Biswas S, Kumar A, Chaubey S, Siddiqi MI, Habib S. Nuclear gyrB encodes a functional subunit of the Plasmodium falciparum gyrase that is involved in apicoplast DNA replication. Mol. Biochem. Parasitol. 2007;154:30–39. doi: 10.1016/j.molbiopara.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Varshavsky AJ, Nedospasov SA, Bakayev VV, Bakayeva TG, Georgiev GP. Histone-like proteins in the purified Escherichia coli deoxyribonucleoprotein. Nucleic Acids Res. 1977;4:2725–2745. doi: 10.1093/nar/4.8.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drlica K, Rouviere-Yaniv J. Histone-like proteins of bacteria. Microbiol. Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan Y-H, Wong ATY. Concentration-dependent organization of DNA by the dinoflagellate histone-like protein HCc3. Nucleic Acids Res. 2007;35:2573–2583. doi: 10.1093/nar/gkm165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi T, Takahara M, Miyagishima S, Kuroiwa H, Sasaki N, Ohta N, Matsuzaki M, Kuroiwa T. Detection and localization of a chloroplast-encoded HU-like protein that organizes chloroplast nucleoids. Plant Cell. 2002;14:1579–1589. doi: 10.1105/tpc.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christodoulou E, Rypniewski WP, Vorgias CE. High-resolution X-ray structure of the DNA-binding protein HU from the hyper-thermophilic Thermotoga maritima and the determinants of its thermostability. Extremophiles. 2003;7:111–122. doi: 10.1007/s00792-002-0302-7. [DOI] [PubMed] [Google Scholar]
- 21.Skarstad K, Baker TA, Kornberg A. Strand separation required for initiation of replication at the chromosome origin of E. coli is facilitated by a distant RNA-DNA hybrid. EMBO J. 1990;9:2341–2348. doi: 10.1002/j.1460-2075.1990.tb07406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kano Y, Ogawa T, Ogura S, Hiraga T, Okazaki T, Imamoto F. Participation of histone-like protein HU and of IHF in minichromosomal maintenance in Escherichia coli. Gene. 1991;103:25–30. doi: 10.1016/0378-1119(91)90386-p. [DOI] [PubMed] [Google Scholar]
- 23.Dri AM, Moreau PL, Rouviere-Yaniv J. Role of the histone-like proteins OsmZ and HU in homologous recombination. Gene. 1992;120:11–16. doi: 10.1016/0378-1119(92)90003-8. [DOI] [PubMed] [Google Scholar]
- 24.Manna D, Gowrishankar J. Evidence for involvement of protein HU and RpoS in transcription of the osmoresponsive proU operon in Escherichia coli. J. Bacteriol. 1994;176:5378–5384. doi: 10.1128/jb.176.17.5378-5384.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kar S, Choi EJ, Guo F, Dimitriadis EK, Kotova SL, Adhya S. Right-handed DNA supercoiling by an octameric form of histone-like protein HU: modulation of cellular transcription. J. Biol. Chem. 2006;281:40144–40153. doi: 10.1074/jbc.M605576200. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Yoshimura SH, Hizume K, Ohniwa RL, Ishihama A, Takeyasu K. Fundamental structural units of the Escherichia coli nucleoid revealed by atomic force microscopy. Nucleic Acids Res. 2004;32:1982–1992. doi: 10.1093/nar/gkh512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talukder AA, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;18:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: Formation of flexible hinges and rigid filaments. Proc. Natl Acad. Sci. USA. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambros C, Vandenberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- 30.Baca AM, Hol WG. Overcoming codon bias: a method for high-level over expression of Plasmodium and other AT-rich parasite genes in Escherichia coli. Int. J. Parasitol. 2000;30:113–118. doi: 10.1016/s0020-7519(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 31.Ghosh S, Grove A. The Deinococcus radiodurans encoded HU protein has two DNA binding domains. Biochemistry. 2006;45:1723–1733. doi: 10.1021/bi0514010. [DOI] [PubMed] [Google Scholar]
- 32.Gissot M, Briquet S, Refour P, Boschet C, Vaquero C. PfMyb1, a Plasmodium falciparum transcription factor, is required for intraerythrocytic growth and controls key genes for cell cycle regulation. J. Mol. Biol. 2005;346:29–42. doi: 10.1016/j.jmb.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 33.Tonkin CJ, van Dooren GG, Spurck TP, Struck NS, Good RT, Handman E, Cowman AF, McFadden GI. Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol. Biochem. Parasitol. 2004;137:13–21. doi: 10.1016/j.molbiopara.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 34.Foth BJ, Ralph SA, Tonkin CJ, Struck NS, Fraunholz M, Roos DS, Cowman AF, McFadden GI. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science. 2003;299:705–708. doi: 10.1126/science.1078599. [DOI] [PubMed] [Google Scholar]
- 35.Chan YN, Kwok ACM, Tsang JSH, Wong JTY. Alveolata histone-like proteins have different evolutionary origins. J. Evol. Biol. 2006;19:1717–1721. doi: 10.1111/j.1420-9101.2006.01089.x. [DOI] [PubMed] [Google Scholar]
- 36.Christodoulou E, Rypniewski WR, Vorgias CE. High-resolution X-ray structure of the DNA-binding protein HU from the hyperthermophilic Thermotoga maritima and the determinants of its thermostability. Extremophiles. 2003;7:111–122. doi: 10.1007/s00792-002-0302-7. [DOI] [PubMed] [Google Scholar]
- 37.Burley SK, Petsko GA. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- 38.Rice PA, Yang SW, Mizuuchi K, Nash HA. Crystal structure of IHF-DNA complex: a protein induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 39.Grove A, Saavedra TC. The role of surface-exposed lysines in wrapping DNA about the bacterial histone-like protein HU. Biochemistry. 2002;41:7597–7603. doi: 10.1021/bi016095e. [DOI] [PubMed] [Google Scholar]
- 40.White SW, Appelt K, Wilson KS, Tanaka I. A protein structural motif that bends DNA. Proteins. 1989;5:281–288. doi: 10.1002/prot.340050405. [DOI] [PubMed] [Google Scholar]
- 41.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implication for structural proteomics. Structure (Camb) 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 42.Swinger KK, Lemberg KM, Zhang Y, Rice PA. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003;22:3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jia X, Grove A, Ivancic M, Hsu VL, Geiduscheck EP, Kearns DR. Structure of the Bacillus subtilis phage SPO1-encoded type II DNA-binding protein TF1 in solution. J. Mol. Biol. 1996;263:259–268. doi: 10.1006/jmbi.1996.0573. [DOI] [PubMed] [Google Scholar]
- 44.Boelens R, Vis H, Vorgias CE, Wilson KS, Kaptein R. Structure and dynamics of the DNA-binding protein HU from Bacillus stearothermophilus by NMR spectroscopy. Biopolymers. 1996;40:553–559. doi: 10.1002/(sici)1097-0282(1996)40:5<553::aid-bip13>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 45.Williamson DH, Preiser PR, Wilson, Wilson RJM. Organelle DNAs: the bit players in malaria parasite DNA replication. Parasitol. Today. 1996;12:357–362. doi: 10.1016/0169-4758(96)10053-3. (Iain) [DOI] [PubMed] [Google Scholar]
- 46.Mazumdar J, Wilson EH, Masek K, Hunter CA, Striepen B. Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc. Natl Acad. Sci. USA. 2006;103:13192–13197. doi: 10.1073/pnas.0603391103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghosh S, Grove A. Histone-like protein HU from Deinococcus radiodurans binds preferentially to four-way DNA junctions. J. Mol. Biol. 2004;337:561–571. doi: 10.1016/j.jmb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 48.Hodges-Garcia Y, Hagerman P, Pettijohn D. DNA ring closure mediated by protein HU. J. Biol. Chem. 1989;264:14621–14623. [PubMed] [Google Scholar]
- 49.Kamau E, Tsihlis ND, Simmons LA, Grove A. Surface salt bridges modulate the DNA site size of bacterial histone-like HU proteins. Biochem. J. 2005;390:49–55. doi: 10.1042/BJ20050274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miao J, Fan Q, Cui L, Li J, Li J, Cui L. The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation. Gene. 2006;369:53–65. doi: 10.1016/j.gene.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 51.Chandra BR, Olivieri A, Silvestrini F, Alano P, Shrama A. Biochemical characterization of the two nucleosome assembly proteins from Plasmodium falciparum. Mol. Biochem. Parasitol. 2005;142:237–247. doi: 10.1016/j.molbiopara.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 52.Diffley JFX, Stillman B. a close relative of the nuclear, chromosomal high mobility group protein HMG1 in yeast mitochondria. Proc. Natl Acad. Sci. USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grove A, Lim L. High-affinity DNA binding of HU protein from the hyperthermophile Thermotoga maritima. J. Mol. Biol. 2001;311:491–502. doi: 10.1006/jmbi.2001.4763. [DOI] [PubMed] [Google Scholar]
- 54.Lee EC, Hales LM, Gumport RI, Gardner JF. The isolation and characterization of mutants of the integration host factor (IHF) of Escherichia coli with altered, expanded DNA-binding specificities. EMBO J. 1992;11:305–313. doi: 10.1002/j.1460-2075.1992.tb05053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: a protein induced DNA U turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 56.Swinger KK, Lemberg KM, Zhang Y, Rice PA. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003;22:3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swinger KK, Rice PA. IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 58.Grove A. Surface salt bridges modulate DNA wrapping by the type II DNA-binding protein TF1. Biochemistry. 2003;42:8739–8747. doi: 10.1021/bi034551o. [DOI] [PubMed] [Google Scholar]
- 59.Feng ZP, Zhang X, Han P, Arora N, Anders RF, Norton RS. Abundance of intrinsically unstructured proteins in Plasmodium falciparum and other apicomplexan parasite proteomes. Mol. Biochem. Parasitol. 2006;150:256–267. doi: 10.1016/j.molbiopara.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 60.Shi W-X, Larson RG. Atomic Force Microscopic study of aggregation of RecA-DNA nucleoprotein filaments into left-handed supercoiled bundles. Nano Lett. 2005;5:2476–2481. doi: 10.1021/nl051783v. [DOI] [PubMed] [Google Scholar]
- 61.Dame RT, Wyman C, Goosen N. H-NS mediated compaction of DNA visualized by atomic force microscopy. Nucleic Acids Res. 2000;28:3504–3510. doi: 10.1093/nar/28.18.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waller RF, Keeling PJ, vanDooren GG, McFadden GI. Comment on ‘A green algal apicoplast ancestor’. Science. 2003;301:49. doi: 10.1126/science.1084684. [DOI] [PubMed] [Google Scholar]
- 63.Funes S, Reyer-Prieto A, Perez-Martinez X, Gonzalez-Halphen D. On the evolutionary origins of apicoplasts: revsisiting the rhodophyte vs. chlorophyte controversy. Microbes Infect. 2004;6:305–311. doi: 10.1016/j.micinf.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 64.Sato N, Terasawa K, Miyajima K, Kabeya Y. Organization, developmental dynamics and evolution of plastid nucleoids. Int. Rev. Cytol. 2003;232:217–262. doi: 10.1016/s0074-7696(03)32006-6. [DOI] [PubMed] [Google Scholar]
- 65.Arenas AF, Escobar AJG, Gomez-Marin JE. Evolutionary origin of the protozoan parasites histone-like proteins (HU) In Silico Biol. 2007;8:0002. [PubMed] [Google Scholar]
- 66.Chodavarapu S, Felczak MM, Yaniv JR, Kaguni JM. Escherichia coli DNA interacts with HU in initiation at the E.coli replication origin. Mol. Microbiol. 2008;67:781–792. doi: 10.1111/j.1365-2958.2007.06094.x. [DOI] [PubMed] [Google Scholar]
- 67.Vaishnava S, Striepen B. The cell biology of secondary endosymbiosis- how parasites build, divide and segregate the apicoplast. Mol. Microbiol. 2006;61:1380–1387. doi: 10.1111/j.1365-2958.2006.05343.x. [DOI] [PubMed] [Google Scholar]