Abstract
The alternative splicing code that controls and coordinates the transcriptome in complex multicellular organisms remains poorly understood. It has long been argued that regulation of alternative splicing relies on combinatorial interactions between multiple proteins, and that tissue-specific splicing decisions most likely result from differences in the concentration and/or activity of these proteins. However, large-scale data to systematically address this issue have just recently started to become available. Here we show that splicing factor gene expression signatures can be identified that reflect cell type and tissue-specific patterns of alternative splicing. We used a computational approach to analyze microarray-based gene expression profiles of splicing factors from mouse, chimpanzee and human tissues. Our results show that brain and testis, the two tissues with highest levels of alternative splicing events, have the largest number of splicing factor genes that are most highly differentially expressed. We further identified SR protein kinases and small nuclear ribonucleoprotein particle (snRNP) proteins among the splicing factor genes that are most highly differentially expressed in a particular tissue. These results indicate the power of generating signature-based predictions as an initial computational approach into a global view of tissue-specific alternative splicing regulation.
INTRODUCTION
Alternative splicing generates multiple mRNA products from a single gene, thereby increasing transcriptome and proteome complexity. In contrast to the prokaryotic rule of ‘one gene-one polypeptide’, alternative splicing expands the protein coding potential of eukaryotic genomes by allowing a single gene to produce proteins with different properties and distinct functions. Several studies based on large-scale expressed sequence tag (EST) analysis estimated that >60% of human genes undergo alternative splicing, and this number more recently increased to >80% when microarray data became available (1,2). Alternative splicing is regulated in response to signaling pathways, and is specific to a developmental stage and tissue type.
The removal of introns from precursor mRNAs requires accurate recognition of splice sites by the spliceosome, an assembly of uridine-rich small nuclear RNAs packaged as ribonucleoprotein particles (snRNPs) that function in conjunction with numerous non-snRNP proteins (3,4). The selection between different splice sites on a particular pre-mRNA substrate relies on an intricate interplay involving the cooperative binding of trans-acting splicing proteins to cis-acting sequence elements in the pre-mRNA. In mammals, these cis-elements include short and highly degenerate 5′ and 3′ splice signals, additional regulatory sequences termed splicing enhancers and silencers located in either exons or introns, the sizes of the exons and introns and secondary structures of the pre-mRNA. The trans-acting factors are commonly classified as splicing activators or repressors depending on whether they facilitate or suppress the assembly of snRNPs onto splice sites. However, many of these factors are also essential for ‘constitutive’ splicing, making it unrealistic to distinguish between proteins required for the operation and regulation of the splicing reaction (5,6). Contrasting with the multitude of sequence-specific DNA-binding proteins that control transcription, there are very few known regulatory proteins that selectively control the splicing of specific genes. Although such factors exist and a good example is the brain-specific NOVA1 protein in mammals (7), in the vast majority of cases splicing factors are ubiquitously expressed and modulate splicing of several genes in distinct cell types. Indeed, specificity of splicing regulation is largely achieved with non-specific RNA-binding proteins (8).
According to the current view, regulation of alternative splicing uses combinatorial interactions of many positively and negatively acting proteins. Tissue-specific splicing decisions could therefore result from differences in the concentration and/or activity of these proteins (2,6,8). An immediate prediction from this model is that the relative abundance of multiple splicing proteins should differ in a tissue-specific manner. To explore this idea, we performed a large-scale computational analysis of mRNA expression data obtained from DNA microarray studies of different cell types and tissues derived from human, chimpanzee and mouse. Our results show for the first time that splicing factor gene expression signatures can be identified that correlate with tissue-specific patterns of alternative splicing.
MATERIALS AND METHODS
Selection of splicing-related genes
A list of 254 human splicing-related genes and several murine orthologues was previously described (9). The remaining mouse genes were identified in Ensembl (10) (http://www.ensembl.org), through the Family classification and BLAST (11) search, and by searching SwissProt (12) (http://us.expasy.org/sprot/) with appropriate keywords.
‘Perl’ scripts, relying on Bioperl (13) (http://www.bioperl.org) and modules from the Ensembl PERL API (14) were used for consistent annotation of genes and subsequent cross-linking with the Affymetrix probe set annotation. Annotation for the selected probe sets was validated with a Perl script. The first step of the pipeline consisted in BLASTing (11) and/or BLATing (15) of each probe against both the respective transcriptome [comprising RefSeq (16), GenBank (17) and transcripts from the UCSC Genome Browser database (18)] and genome (Mouse mm8 and Human hg18, NCBI 36). The program subsequently parsed the outcome and extracted the associated transcriptomic and genomic annotations from the tables in the UCSC genome annotation database (18).
Microarray data pre-processing
All the microarray data analysis was done using R and several packages available from CRAN (19) and Bioconductor (20). The raw data (CEL files) were normalized and summarized with the Robust MultiArray Average method from the ‘affy’ package (21). An initial quality assessment was done to remove microarrays with poor quality, using quality diagnostics with probe level models and array quality control metrics for all arrays (average background was <200, scale factors <6, percentage of present calls, RNA degradation for GAPDH and beta-actin - 3′/5′ ratio).
Cell culture and real-time quantitative PCR
C2 mouse myoblasts were cultured at 30% confluence in DMEM supplemented with 20% FCS. For the differentiation experiments the cells were grown in DMEM containing 20% FCS until they reached 90% confluency. At this stage the cells were changed to low serum media (DMEM supplemented with 2% horse serum—differentiation media) and allowed to differentiate for a maximum period of 4 days.
Primary mouse erythroid progenitors were obtained from fetal livers of E12.5 mouse embryos and were subject to differentiation in stem-Pro-34 medium supplemented with Epo and iron-saturated human transferrin as described previously (22).
The C2 cell RNA samples used in the quantitative real-time PCR (qRT-PCR) experiments were collected at days −2 and −1 prior to differentiation and at days 0, 1, 2, 3 and 4 after changing to differentiation media. Primary mouse erythroid RNA was collected at 0, 24, 36, 48 and 60 h after induction of differentiation. The RNA was extracted using the RNeasy extraction kit according to the manufacturer's instructions (Qiagen, Germantown, MD, USA) and treated with RNase-free DNaseI (Roche Diagnostics, Indianapolis, IN, USA) to remove any possible genomic DNA contaminant. The concentration of RNA was determined using the Nanodrop, Wilmington, DE, USA (Nucliber) and RNA quality was assessed by gel electrophoresis. Only samples yielding distinct 28S and 18S bands and A260/A280 ratios between 1.8 and 2.1 were used in this study. Production of cDNA was carried out using Superscript II reverse transcriptase following the manufacturer's protocol (Invitrogen, Carlsbad, CA, USA). About 0.6 μg of total RNA was used in a 20 μl reaction volume. Isolated cDNA from brain, heart, kidney, liver and testes was purchased from Ambion. A total of 30 ng of cDNA was used for each SYBR Green measurement.
The primers used in the qRT-PCR assay (Supplementary Table 1) were designed with the Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The cDNA was amplified in tubes containing 25 μl reaction volume with 50% of SYBR Green PCR master mix (Applied Biosystems, Foster City, CA, USA). Primers were added at a final concentration of 300 nM, which proved to be the best concentration for all the sets of primers tested. All reactions were performed in the ABI7000 Sequence Detector (Applied Biosystems).
The relative quantification of mRNA levels at the various C2 differentiation stages was calculated using 18S as an endogenous reference and the sample at day 0 as the calibrator. For the erythropoiesis experiments we used Rnase Inhibitor as an endogenous reference and the sample at 0 h as the calibrator. For the adult tissues experiments, RNU6A was used as the endogenous reference. The quantities obtained for each gene were extracted from a standard curve of CT versus quantity of mRNA obtained from a serial dilution of either a mix of C2 cell cDNA extracts or a mix of erythropoietic progenitors at the stages of differentiation used for the analysis. For tissue samples, the standard curve was obtained from serial dilutions of a mix of all tissues.
RESULTS
Splicing factor expression during cell differentiation
To study splicing factor expression during differentiation, we first established a list of human and mouse genes associated with splicing and next we compared the corresponding expression profiles from data sets obtained from microarray studies that analyzed cell differentiation. A list containing 254 human genes associated with splicing was previously reported by Barbosa-Morais et al. (9). Here, we searched for the respective orthologues in the mouse genome. Both human and mouse lists contain genes that encode known splicing factors, spliceosome-associated proteins and proteins with a domain structure similar to bona fide splicing factors (9).
We selected transcript profiling studies performed with myotube, adipocyte and erythroid cells differentiated in vitro and whole mouse testis collected from birth to adulthood. In total, we studied four distinct differentiation processes and for each process we analyzed two independent data sets covering a total of 126 arrays (Table 1 and Supplementary Table 2). We identified 181 splicing-related genes (SRGs) for which 240 probe sets are present in the Affymetrix Murine Genome U74v2 platform that was used in all selected microarray studies (Supplementary Table 3).
Table 1.
Microarray data sets used to study mouse differentiation processes
| Description | Data set ID | GEO Acc. number | Reference | Arrays number | Time |
Times for fold-change |
||
|---|---|---|---|---|---|---|---|---|
| Range (h) | Points | T1 (h) | T2 (h) | |||||
| Myogenesis | Myog1 | GSE989 | (24) | 23 | −24 to 240 | 8 | 24 | 48 |
| Myog2 | GSE1984 | 10 | 0 to 48 | 5 | 24 | 48 | ||
| Adipogenesis | Adip1 | GSE2192 | (27) | 15 | 0 to 240 | 4 | 48 | 96 |
| Adip2 | (26) | 13 | 0 to 96 | 7 | 48 | 96 | ||
| Spermatogensis | Sperm1 | GSE640 | (29) | 12 | 24 to 1440 | 9 | 336 | 720 |
| Sperm2 | GSE926 | (30) | 19 | 0 to 1344 | 11 | 336 | 720 | |
| Erythropoiesis | Ery1 | GSE628 | (28) | 17 | 0 to 30 | 6 | 15 | 30 |
| Ery2 | Von Lindern, unpublished data | 17 | 0 to 60 | 5 | 30 | 60 | ||
The GEO accession number, references and number of arrays analyzed are indicated. The time range refers to the total differentiation period. The number of time points studied for each differentiation process is also indicated. For our expression analysis, T1 and T2 correspond to the indicated number of differentiation hours.
All expression values were obtained from Gene Expression Omnibus (23) (http://www.ncbi.nlm.nih.gov/projects/geo). Data for myogenesis were obtained from published studies using the in vitro model of C2C12 myoblasts undergoing differentiation induced by serum restriction (24,25). Adipocyte differentiation in vitro was induced by hormonal treatment on two distinct models: the 3T3-L1 preadipocyte cell line (26), and NIH-3T3 fibroblasts (27). Two distinct cell models were also used to analyze erythroid differentiation in vitro. One model consisted of G1E cells derived from GATA-1-null embryonic stem cells; these cells proliferate in culture as immature erythroblasts and undergo terminal erythroid maturation when GATA-1 function is restored (28). The other model consisted of primary erythroid progenitors from mouse fetal livers; these cells proliferate in serum-free medium under the control of erythropoietin (Epo), stem cell factor (SCF) and dexamethasone (Dex) and undergo terminal differentiation when exposed to Epo in the absence of SCF and Dex (22). Spermatogenesis was examined in vivo (29,30).
To test whether the two data sets corresponding to the same differentiation process were temporally synchronized, we performed a time–course analysis of the expression level of the following differentiation marker genes: the muscle-specific troponin C (Tnnc1) (31) and Ca2+ channel ryanodine receptor 1 (Ryr1) (32); the adipogenic complement factor D–adipsin (Cfd) (33) and peroxisome proliferator-activated receptor (Pparγ) (27); the erythroid-specific markers glycophorin A (Gypa) (34) and Slc4a1 (35); the male germ cell lineage markers lactate dehydrogenase C (Ldhc) (36) and phosphoglycerate kinase 2 (Pgk2) (37). For myogenesis, adipogenesis and spermatogenesis the distinct data sets were approximately synchronous and were directly used as biological replicates (Supplementary Figure 1). For erythroid differentiation, maturation of the cell type used in one study (G1E–ER4 cells) occurred significantly faster than that of primary fetal liver progenitors used in the other study. This difference was corrected considering that the last time points of both experiments were biologically equivalent (Supplementary Figure 1).
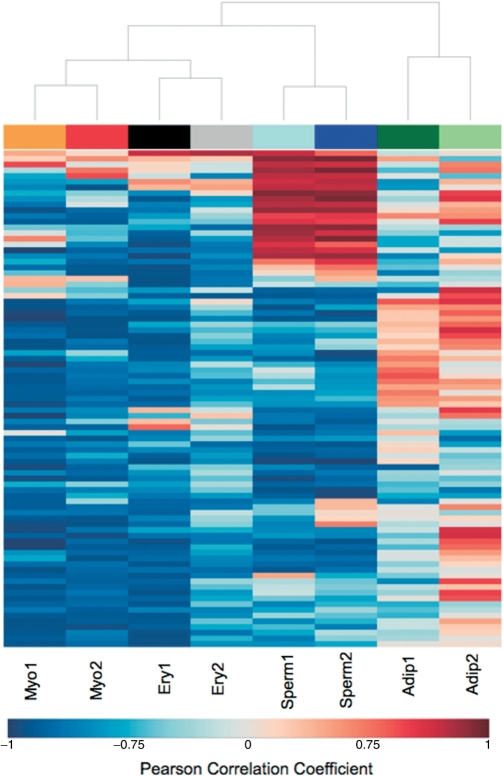
Next, for each differentiation process, we searched for variation in expression of splicing-related genes along time. For each splicing-related gene on each data set, we estimated the Pearson correlation coefficient between expression level and differentiation time point. Only genes with absolute correlation values >0.75 [P-values <0.05, corrected for multiple hypotheses testing using the Benjamini and Hochberg method (38)] in both data sets were selected for further analysis. The Pearson correlation coefficients of this subset of genes were used to cluster the microarray data sets (Figure 1). The hierarchical clustering results revealed consistency between the two data sets for each differentiation process indicating that similar groups of genes were found up- or down-regulated in the two independent experimental studies performed with each cell type. The only exception was found for adipogenesis data sets, where the different expression patterns for some splicing-related genes can be due to the distinct cell lines used in both experiments. As shown in Figure 1, during myotube and erythroid differentiation most splicing-related genes presented a negative correlation, meaning that the expression decreased along time. In contrast, several splicing-related genes increased expression during adipocyte and sperm cell differentiation.
Figure 1.
Variation in expression of splicing-related genes during cell differentiation. Hierarchical clustering display of Pearson correlation values between gene expression and time, for the splicing-related genes with the absolute correlation values >0.75 in both data sets of at least one differentiation process. The negative and positive correlation values are represented by blue and red colors, respectively.
Identification of cell type specific variations in splicing factor expression
To identify cell type specific variations in splicing factor expression, we had to compare microarray data sets derived from different biological systems and experimental assays. To address this issue, we developed a new approach that is based on regression modeling methods. Polynomial models were fitted to the splicing factor expression profiles along each differentiation process, and the best model was selected by the Akaike's Information Theoretic Criterion in a Stepwise Algorithm (39) as implemented in the ‘stats’ package (19). Since the selected regression models were essentially linear or quadratic (meaning that gene expression variations were constant throughout differentiation or showed only one inflexion point), for further analysis we reduced each differentiation process to three time points, T0, T1 and T2 (Table 1). T0 corresponds to the time when cultured cells were switched to differentiation medium or to the first day postpartum for testis. T2 corresponds to terminally differentiated cells or adult testis, and T1 corresponds to an intermediate stage specific to each differentiation process. During myogenesis, the proliferating mononucleate myoblasts withdraw from the cell cycle and subsequently fuse to form multinucleate myotubes; we, therefore, considered that T1 corresponds to the time when irreversible cell cycle withdrawal occurs, ∼24 h after serum restriction (24). Likewise, for adipogenesis T1 corresponds to the time when cells withdraw permanently from the cell cycle at ∼2 days after hormonal stimulation (26,27). In contrast, during erythropoiesis cells undergo three to four rapid cell divisions accompanied by a decrease in cell size and the accumulation of hemoglobin; in this case, we considered that T1 corresponds to the stage of proliferating capacity, which occurs at ∼15 h in GE1 cells and at 30 h in fetal liver erythroid progenitors (28). Based on the observation that >99% of male germ cell-specific transcripts are first expressed during or after the occurrence of meiosis (29), we considered the onset of sperm cell meiosis (taking place at ∼14 days after birth) as T1 for spermatogenesis.
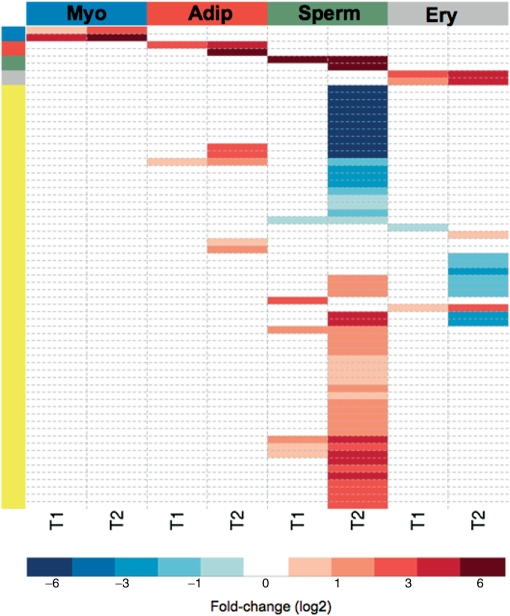
For each differentiation process, the fitted models were used to predict the splicing factor expression levels at time points T0, T1 and T2. Then, to normalize the data, we estimated the fold-changes observed at T1 and T2, relative to T0. We also transformed the residual standard errors from each fitted regression model and used as weights (weights = exponential − residual standard error) to include confidence levels of each prediction (biological variability). Finally, the differentially expressed splicing-related genes for each T1 and T2 differentiation stage were selected using linear models and empirical Bayes methods (40) as implemented in ‘limma’ package (41). The B-statistics gives the log odds of differential expression and it requires an ‘a priori’ value for the estimated proportion of differentially expressed genes. To determine this value, we visually inspected the volcano plot, which compares biological significance (represented by fold-changes) with statistical significance (B-values) (42), finding the value which enabled genes to be distinguished from the majority (43). Additionally, we verified the P-values corresponding to moderated F-statistics. Using the Benjamini and Hochberg method (38), all genes selected as differentially expressed had adjusted P-values <0.01. To validate our approach we included in the analysis the specific differentiation marker genes used for synchronization. Up-regulation of each differentiation marker gene was specifically detected in the respective differentiation process (Figure 3).
Figure 3.
Splicing-related gene expression signatures during cell differentiation. Heatmap with the fold-changes (log2) observed for each gene that is most highly differentially expressed during myotube (Myo), adipocyte (Adip), sperm cell (Sperm) and erythrocyte (Ery) differentiation. The genes and respective fold-changes are presented in detail in Supplementary Table 5. The side colors represent the splicing-related genes (yellow) and the specific differentiation marker genes for myogenesis (Ryr1, Tnnc1 in blue), adipogenesis (Pparg and Cfd in red), spermatogenesis (Ldhc, Pgk2 in green) and erythropoiesis (Gypa and Slc4a1 in gray).
Our analysis revealed that major variations in splicing factor expression occurred at T2. The highest variation was found in spermatogenesis: 47% of total splicing-related genes were up- or down-regulated at T2 relative to T0. The genes that were statistically selected as up- or down-regulated in the different processes included members of the hnRNP and SR protein families, SR protein kinases, DEAD-box RNA helicases, snRNP proteins and several additional spliceosomal proteins (Supplementary Table 4).
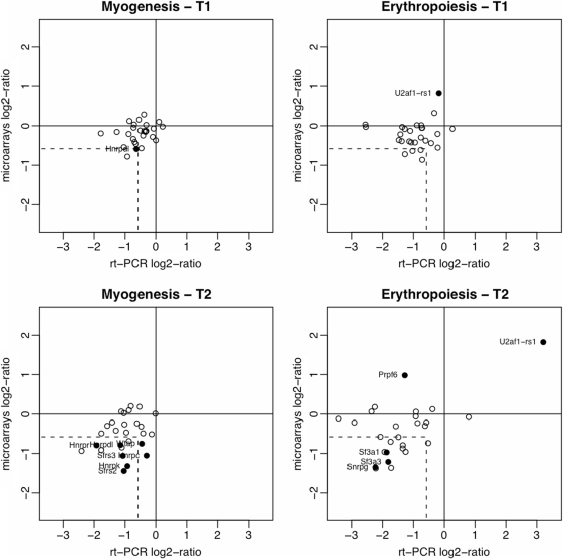
In order to validate the microarray data analysis we determined mRNA expression levels using a more sensitive method. RNA samples were obtained from C2 myoblasts and fetal liver erythroid progenitors and analyzed by qRT-PCR. We started by selecting 12 genes that the microarray data analysis identified as up- or down-regulated during erythroid and myotube differentiation. As shown in Figure 2 (closed circles), expression changes were confirmed for nine genes (75%). Thus, we obtained a validation rate of 75% among independent biological samples for genes identified as differentially expressed in our statistical analysis of microarray fold-changes. We then selected 15 other genes that were not identified as differentially expressed during myogenesis or erythropoiesis. From these, we found four genes down-regulated in myogenesis (Rod1, Hnrpa1, Sfrs10 and Hnrpa2b1) and 10 genes down-regulated in erythropoiesis (Cugbp1, Cugbp2, Ddx17, Snrpb2, U2af1, Sfrs2, Ptbp1, Hnrpdl, Hnrpr and Wtap; open circles in Figure 2). This reveals that the microarray analysis is missing several genes the expression of which is less obviously altered.
Figure 2.
Validation of microarray data analysis by quantitative real-time PCR. The fold-changes in expression of 27 splicing factors at T1 and T2 relative to T0 are indicated. For qRT-PCR analysis, RNA samples were obtained from C2 myoblasts and fetal liver erythroid progenitors. Results are presented as means for at least three independent experiments. Results from microarray data sets are presented as the fold-changes estimated from the linear models. The dashed lines indicate the 1.5-fold-change values (in logarithm scale) for microarray data and qRT-PCR. The differentially expressed genes selected by microarray data analysis for each differentiation stage are indicated with solid circles.
We next asked whether robust differences could be found that distinguish one differentiation process from the others. To identify genes that are most highly differentially expressed in a particular differentiation process we used linear models and empirical Bayes methods (40) as described previously. Following the statistical analysis, a filter was applied to eliminate genes that were similarly differentially expressed in more than one differentiation process. A gene is considered to be part of a ‘signature’ when its expression changes at least 1.5-fold (log2 = 0.58) more than in any other process. As shown in Figure 3 and Supplementary Table 5, we identified gene expression signatures associated with three of the four differentiation processes. The list of genes in each signature included members of the several splicing-related protein families. The gene expression signature associated with spermatogenesis contained the highest number of genes. The signature associated with erythroid differentiation consisted of two genes (U2af1-rs1 and Prpf6), and the adipogenesis signature comprised three genes (Hnrpab, Hnrpdl, Sfrs1). No signature was associated with myotube differentiation, as the genes that were differentially expressed during myogenesis were also found differentially expressed in at least one of the other processes analyzed. This may be related to the finding that splicing factors in muscle are predominantly regulated at the post-transcriptional level (44).
Tissue-specific differences in splicing factor expression
Having identified splicing factor signatures associated with cell differentiation, we next explored variations in splicing factor expression across tissues from human, chimpanzee and mouse. Available mRNA expression data were obtained from a microarray study covering five different tissues in six humans and five chimpanzees using a total of 48 hybridizations (45). This study used the Affymetrix Human Genome hgu133plus2 platform containing 738 probe sets for 208 human splicing-related genes (Supplementary Table 3). Gene expression profiles from adult mouse was obtained from a study that analyzed 24 brain regions and 10 body tissues using a total of 150 array hybridization measurements with the Affymetrix Murine mgu74av2 platform (46) (Supplementary Table 6).
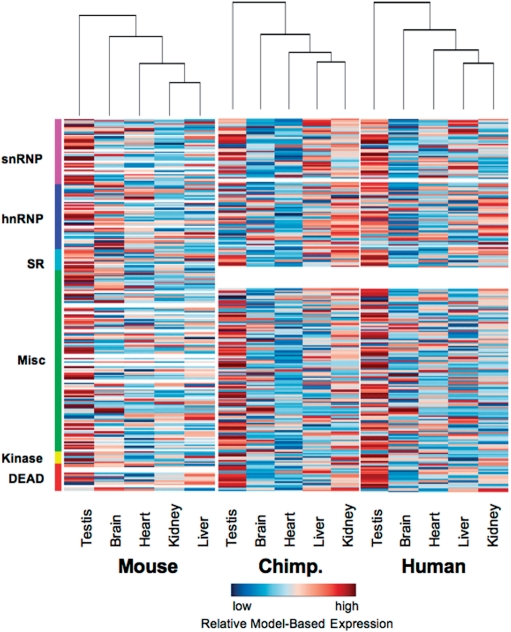
To compare the splicing-related gene expression profiles from human, chimpanzee and mouse datasets, a linear model (40) was fitted for each gene using the expression values from all microarrays and with one regression coefficient for each tissue. Thus, each regression coefficient from the model represents the expression level of the gene in a different tissue. The tissues relatedness was studied performing a hierarchical clustering analysis of the tissues expression profiles using only the splicing-related genes and the non-splicing related genes. We estimated the Euclidean distance among the tissues and used hierarchical clustering with different agglomeration methods (complete, single, average, centroid and ward) as implemented in ‘stats’ package (19). The best hierarchical tree was chosen using the cophenetic correlation value. The results revealed very similar expression profiles of splicing-related genes in human and chimpanzee tissues (Figure 4). For these two organisms, the testis was clearly an outlier, with low concordance in expression of splicing-related genes relative to the other tissues examined. Analysis of mouse tissues also indicated the testis as the main out-group (Figure 4). Most of the 24 mouse brain regions revealed high similarity in expression profiles and were mostly grouped together for both splicing-related genes and all remaining genes (Supplementary Figure 2). Pituitary and retina appeared as an out-group of the brain cluster, and corpus plexus of the fourth ventricle (Cp4v) did not group with the remaining brain regions but rather clustered with the body tissues. Hierarchical clustering of splicing-related gene expression profiles in the 10 body tissues revealed the testis, spleen and thymus as the main out-group (Supplementary Figure 2).
Figure 4.
Tissue expression profiles of splicing-related genes are similar in human, chimpanzee and mouse. Heatmap of adult mouse, chimpanzee and human tissues using microarray-derived expression profiles of splicing-related genes. The expression value for each gene is normalized across the samples to zero mean and 1SD for visualization purposes. Genes with expression levels greater than the mean are colored in red and those below the mean are colored in blue. The expression values for genes that are not present in one of the microarray platforms are represented by white.
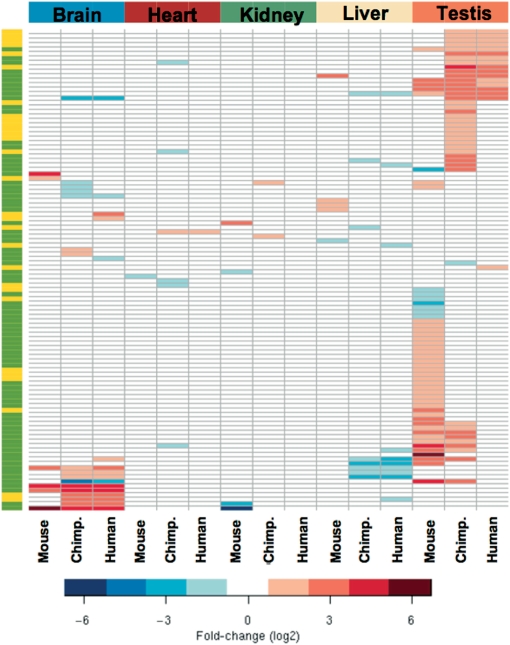
From the human, chimpanzee and mouse microarray data, we identified 154 genes that were differentially expressed between brain, testis, heart, liver and kidney (Supplementary Table 7 and Supplementary Figure 3). From these, seven genes were selected and five of them (71%) were found differentially expressed by qRT-PCR (Supplementary Figure 4). Similarly to the results observed during cell differentiation, the differentially expressed genes code for hnRNP and SR proteins, SR protein kinases, DEAD-box RNA helicases, snRNP proteins and several other splicing-related proteins. From the selected 154 genes, 104 showed tissue-specific expression variation >1.5-fold in at least one of the three organisms (Figure 5 and Supplementary Table 8). Analysis of all mouse data sets further revealed 74 genes with highest expression variation in the 24 brain regions and 10 body tissues (Supplementary Table 9). As shown in Figure 5, testis and brain contain the highest number of splicing-related genes that are more than 1.5-fold differentially expressed. From the human and chimpanzee microarray data sets, we identified 43 genes included in the testis-specific signature and 20 in the brain signature. From the mouse studies our results reveal 49 genes in the testis signature and 6 in the brain signature. Out of the 48 genes included in the signature for spermatogenesis (Supplementary Table 5), 27 appeared also in the adult mouse testis signature (Supplementary Table 8 and Supplementary Figure 5).
Figure 5.
Tissue-specific splicing-related gene expression signatures. Heatmap indicating the fold-changes (log2) observed for each gene that is most highly differentially expressed in the five tissues examined. The left bar highlights genes that are present in both human and mouse Affymetrix platforms (green) or only in one of the two platforms (yellow). The genes and respective fold-changes are presented in detail in Supplementary Table 8.
Concerning the brain-specific splicing factor gene expression signature, the gene list includes the previously reported brain-splicing regulators PTB1, NOVA1, A2bp1/FOX1, and members of the CELF/BRUNOL and ELAVL families. Additionally, we identified the non-SR splicing regulator Y-box protein 1 (47) highly down-regulated and the core snRNP protein SmN (48) highly up-regulated. We detected many genes that were highly differentially expressed in chimpanzee but not in human brain, and we found two genes (TNRC4, encoding for the CELF3/BRUNOL1 protein, and LSM8, encoding for U6 snRNA-associated Sm-like protein LSm8) that were, respectively, highly up- and down-regulated in human but not in chimpanzee brain.
The testis-specific signature included the splicing factor 3a subunit 2 (SF3A2) and the SR protein kinases 1 and 2 (SRPK1 and SRPK2). The genes that were common to the testis-specific signatures from all three organisms (human, chimpanzee and mouse) encode SF3A2, SRPK2, protein phosphatase 1G (PPM1G), the RNA binding protein RDBP and the heterogeneous nuclear ribonucleoprotein HNRPLL. Remarkably, 10 (37%) of the mouse genes included in the testis-signature corresponded to up-regulated snRNPs (Lsm2, Lsm4, Sf3a3, Snrpa, Snrpa1, Snrpc, Snrpd2, Snrpg, Usp39 and U5-40d).
DISCUSSION
In this study, we applied computational methods to identify tissue-specific splicing factor gene expression signatures from published microarray data sets. By using this approach we have identified over 100 splicing-related genes that are most highly differentially expressed in a particular tissue or differentiation process.
Recently, several microarray-based methods have been reported for genome-wide monitoring of splicing events in mammalian tissues (49,50). The increasing availability of splicing-microarray data sets will make it possible to extend our approach and systematically search for differential expression of alternatively spliced isoforms of splicing regulators. Importantly, however, changes in splicing factor mRNA levels may not necessarily reflect on protein expression due to post-transcriptional regulation (51,52). Therefore, further experimental investigation on the candidate tissue-specific splicing regulators identified in this study is required to determine whether specific changes in the protein concentration and/or activity do occur.
Splicing factor signatures correlate with tissue-specific alternative splicing patterns
By using a method that normalizes the number of observed alternative splicing events to the EST coverage in each tissue, Yeo and colleagues (53) found that the brain has the highest proportion (>40%) of alternatively spliced genes, followed by the liver and testis. The brain and testis showed the highest levels of exon skipping, while the liver had the highest frequency of alternative 3′ and 5′ splice site usage. Using a microarray platform with probes that span exon–exon junctions, Pan et al. (54) detected the largest number of tissue-dependent alternative splicing events associated with brain. A more recent analysis performed with human exon microarrays revealed that testis and brain express the largest number of probe sets that are not expressed in any other tissue (55). In that study, tissue-specific probe sets may be from genes that are only expressed in a single tissue, or individual exons that are included in a tissue-specific manner via alternative splicing (55).
Our analysis revealed that the highest number of highly differentially expressed splicing-related genes occurred in the testis and in the brain, whereas the liver showed higher concordance in expression of splicing-related genes relative to other tissues, namely the kidney. Thus, our results specifically distinguish the two tissues with highest abundance of alternatively spliced mRNA isoforms that differ by inclusion or exclusion of an exon, as those with a highest variation in splicing factor expression. Yeo and coauthors (53) have also analyzed microarray expression data for 20 splicing factors of the SR, SR-related and hnRNP protein families across several human tissues and identified liver as an outlier, suggesting an involvement of this group of factors in regulation of liver-specific alternative 3′ and 5′ splice decisions. However, our analysis revealed that variation in expression levels of these factors is not unique to the liver.
SR protein kinases as tissue-specific signatures
According to a current model, small differences in concentration or activity of SR proteins may influence the choice of competing splice sites and therefore control alternative splicing (6). SR proteins form multi-protein complexes that bind to splicing enhancer sequences in the pre-mRNA and stabilize the assembly of the spliceosome at splice sites. One possible mechanism to affect SR protein activity is differential phosphorylation. Indeed, the phosphorylation status of Ser residues within the RS domain of SR proteins has been shown to alter protein–protein interactions and splicing activity (56–58). Several SR-protein kinases have been identified, including SRPK and CLK/STY (59,60). Here, we detected members of both the SRPK and CLK gene families being differently expressed in distinct cell types and tissues. In particular, the SRPK1 and SRPK2 genes were highly up-regulated during mouse spermatogenesis. Moreover, SRPK1 and SRPK2 were included in the testis-specific signature for chimpanzee and mouse (SRPK2 also found for human), whereas SRPK3 was included in the heart signature for human and chimpanzee. We therefore predict that SR protein kinases are likely to play an important role in tissue-specific alternative splicing.
Tissue-specific signatures include several snRNP proteins
It is generally assumed that splicing is regulated by non-snRNP proteins that modulate the association of core components of the spliceosome with the pre-mRNA. This view was for the first time questioned by an RNAi screen in Drosophila cells that unexpectedly detected changes in alternative splicing of endogenous genes after reducing the levels of core spliceosomal proteins (61). These included components of the U1, U2 and U4/U6 snRNPs, and both subunits of the U2 snRNP auxiliary factor, U2AF. More recently, we used RNAi to down-regulate expression of the small subunit of U2AF in human cells and we also observed changes in alternative splicing of transcripts derived from both endogenous genes and exogenous reporter minigenes (62,63). In another study, Massiello and coauthors (64) reported that RNAi-mediated down-regulation of SAP155 (a subunit of splicing factor SF3B, which associates with the U2 snRNP) affected alternative splicing of Bcl-x transcripts. Although some of the effects on alternative splicing induced by RNAi may be indirect, it was also shown that in Saccharomyces cerevisiae substrate selectivity can be modulated by altering the kinetics of spliceosome rearrangement (65). Further support to the idea that fluctuations in the concentration of core spliceosomal proteins may contribute to regulate splicing is provided by the differential cell type and tissue-specific expression profiles presented in this study. Variations in expression of genes that code for Lsm, Sm and snRNP-specific proteins were detected in the course of myotube, erythroid and sperm cell differentiation. Consistent with our results, down-regulation of snRNP synthesis during myogenesis was previously demonstrated by pulse-labeling experiments (66). A decrease in expression of genes that encode snRNP proteins was not observed during adipogenesis, arguing that the variations detected in myogenesis are not related to the cell cycle arrest, which is common to both myotube and adipocyte differentiation. In addition to core snRNP proteins, the U2af1-rs1 gene, which encodes a protein with a high degree of homology to the small subunit of U2AF (67), was found specifically up-regulated during erythroid differentiation. Another U2AF-related gene, U2af1-rs2, was highly up-regulated in the mouse brain. SF3A2 was further identified as part of the testis-signature for human, chimpanzee and mouse, while the snRNP protein SmN appeared in the brain-signature for the three organisms. Clearly, a major task for the future will be to determine whether tissue-specific alternative splicing events are regulated by the differential expression of these snRNP and snSNP-related proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Juan Valcárcel, Ben Blencowe and Doug Black for stimulating discussions and insightful advice. We are also grateful to our colleague Margarida Gama-Carvalho for help and support. This work was supported by grants from Fundação para a Ciência e Tecnologia, Portugal (PTDC/SAU-GMG/69739/2006), and the European Commission (MCRTN Eurythron 005499 and EURASNET, LSHG-CT-2005-518238). A.R.G. is supported by a fellowship from Fundação para a Ciência e Tecnologia (SFRH/BD/22825/2005). Funding to pay the Open Access publication charges for this article was provided by EURASNET.
Conflict of interest statement. None declared.
REFERENCES
- 1.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 3.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 4.Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- 5.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 6.Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nat. Rev. Mol. Cell Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- 7.Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, Darnell RB. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 8.Singh R, Valcarcel J. Building specificity with nonspecific RNA-binding proteins. Nat. Struct. Mol. Biol. 2005;12:645–653. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- 9.Barbosa-Morais NL, Carmo-Fonseca M, Aparicio S. Systematic genome-wide annotation of spliceosomal proteins reveals differential gene family expansion. Genome Res. 2006;16:66–77. doi: 10.1101/gr.3936206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 12.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stabenau A, McVicker G, Melsopp C, Proctor G, Clamp M, Birney E. The Ensembl core software libraries. Genome Res. 2004;14:929–933. doi: 10.1101/gr.1857204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2007;35:D21–25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, Siepel A, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.R Development Core Team. Vienna, Austria: R Foundation for Statistical Computing; 2008. R: A language and environment for statistical computing. ISBN 3-900051-07-0, URL http://www.R-project.org. [Google Scholar]
- 20.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 22.Drissen R, von Lindern M, Kolbus A, Driegen S, Steinlein P, Beug H, Grosveld F, Philipsen S. The erythroid phenotype of EKLF-null mice: defects in hemoglobin metabolism and membrane stability. Mol. Cell. Biol. 2005;25:5205–5214. doi: 10.1128/MCB.25.12.5205-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, Rudnev D, Lash AE, Fujibuchi W, Edgar R. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005;33:D562–566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomczak KK, Marinescu VD, Ramoni MF, Sanoudou D, Montanaro F, Han M, Kunkel LM, Kohane IS, Beggs AH. Expression profiling and identification of novel genes involved in myogenic differentiation. FASEB J. 2004;18:403–405. doi: 10.1096/fj.03-0568fje. [DOI] [PubMed] [Google Scholar]
- 25.Zhao P, Caretti G, Mitchell S, McKeehan WL, Boskey AL, Pachman LM, Sartorelli V, Hoffman EP. Fgfr4 is required for effective muscle regeneration in vivo. Delineation of a MyoD-Tead2-Fgfr4 transcriptional pathway. J. Biol. Chem. 2006;281:429–438. doi: 10.1074/jbc.M507440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burton GR, Nagarajan R, Peterson CA, McGehee RE., Jr Microarray analysis of differentiation-specific gene expression during 3T3-L1 adipogenesis. Gene. 2004;329:167–185. doi: 10.1016/j.gene.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 27.Akerblad P, Månsson R, Lagergren A, Westerlund S, Basta B, Lind U, Thelin A, Gisler R, Liberg D, Nelander S, et al. Gene expression analysis suggests that EBF-1 and PPARgamma2 induce adipogenesis of NIH-3T3 cells with similar efficiency and kinetics. Physiol. Genomics. 2005;23:206–216. doi: 10.1152/physiolgenomics.00015.2005. [DOI] [PubMed] [Google Scholar]
- 28.Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- 29.Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc. Natl Acad. Sci. USA. 2003;100:12201–12206. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol. Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- 31.Hastings KE, Emerson CP., Jr cDNA clone analysis of six co-regulated mRNAs encoding skeletal muscle contractile proteins. Proc. Natl Acad. Sci. USA. 1982;79:1553–1557. doi: 10.1073/pnas.79.5.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacLennan DH, Duff C, Zorzato F, Fujii J, Phillips M, Korneluk RG, Frodis W, Britt BA, Wortont RG. Ryanodine receptor gene is a candidate for predisposition to malignant hyperthermia. Nature. 1990;343:559–561. doi: 10.1038/343559a0. [DOI] [PubMed] [Google Scholar]
- 33.Djian P, Phillips M, Green H. The activation of specific gene transcription in the adipose conversion of 3T3 cells. J. Cell Physiol. 1985;124:554–556. doi: 10.1002/jcp.1041240327. [DOI] [PubMed] [Google Scholar]
- 34.Lahlil R, Lecuyer E, Herblot S, Hoang T. SCL assembles a multifactorial complex that determines glycophorin A expression. Mol. Cell Biol. 2004;24:1439–1452. doi: 10.1128/MCB.24.4.1439-1452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paw BH, Davidson AJ, Zhou Y, Li R, Pratt SJ, Lee C, Trede NS, Brownlie A, Donovan A, Liao EC, et al. Cell-specific mitotic defect and dyserythropoiesis associated with erythroid band 3 deficiency. Nat. Genet. 2003;34:59–64. doi: 10.1038/ng1137. [DOI] [PubMed] [Google Scholar]
- 36.Bonny C, Cooker LA, Goldberg E. Deoxyribonucleic acid-protein interactions and expression of the human testis-specific lactate dehydrogenase promoter: transcription factor Sp1 plays a major role. Biol. Reprod. 1998;58:754–759. doi: 10.1095/biolreprod58.3.754. [DOI] [PubMed] [Google Scholar]
- 37.McCarrey JR, Kumari M, Aivaliotis MJ, Wang Z, Zhang P, Marshall F, Vandeberg JL. Analysis of the cDNA and encoded protein of the human testis-specific PGK-2 gene. Dev. Genet. 1996;19:321–332. doi: 10.1002/(SICI)1520-6408(1996)19:4<321::AID-DVG5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 39.Hastie TJ, Pregibon D. Generalized linear models. In: Chambers JM, Hastie TJ, editors. Chapter 6 of Statistical Models. Wadsworth & Brooks/Cole: 1992. [Google Scholar]
- 40.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 41.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 42.Jin W, Riley RM, Wolfinger RD, White KP, Passador-Gurgel G, Gibson G. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat. Genet. 2001;29:389–395. doi: 10.1038/ng766. [DOI] [PubMed] [Google Scholar]
- 43.Conboy CM, Spyrou C, Thorne NP, Wade EJ, Barbosa-Morais NL, Wilson MD, Bhattacharjee A, Young RA, Tavare S, Lees JA, et al. Cell cycle genes are the evolutionarily conserved targets of the E2F4 transcription factor. PLoS ONE. 2007;2:e1061. doi: 10.1371/journal.pone.0001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Pāābo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- 46.Zapala MA, Hovatta I, Ellison JA, Wodicka L, Del Rio JA, Tennant R, Tynan W, Broide RS, Helton R, Stoveken BS, et al. Adult mouse brain gene expression patterns bear an embryologic imprint. Proc. Natl Acad. Sci. USA. 2005;102:10357–10362. doi: 10.1073/pnas.0503357102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stickeler E, Fraser SD, Honig A, Chen AL, Berget SM, Cooper TA. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J. 2001;20:3821–3830. doi: 10.1093/emboj/20.14.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grimaldi K, Horn DA, Hudson LD, Terenghi G, Barton P, Polak JM, Latchman DS. Expression of the SmN splicing protein is developmentally regulated in the rodent brain but not in the rodent heart. Dev. Biol. 1993;156:319–323. doi: 10.1006/dbio.1993.1080. [DOI] [PubMed] [Google Scholar]
- 49.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 50.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 51.Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 55.Clark TA, Schweitzer AC, Chen TX, Staples MK, Lu G, Wang H, Williams A, Blume JE. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prasad J, Colwill K, Pawson T, Manley JL. The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol. Cell Biol. 1999;19:6991–7000. doi: 10.1128/mcb.19.10.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prasad J, Manley JL. Regulation and substrate specificity of the SR protein kinase Clk/Sty. Mol. Cell Biol. 2003;23:4139–4149. doi: 10.1128/MCB.23.12.4139-4149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 59.Gui JF, Lane WS, Fu XD. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- 60.Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- 61.Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc. Natl Acad. Sci. USA. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pacheco TR, Coelho MB, Desterro JM, Mollet I, Carmo-Fonseca M. In vivo requirement of the small subunit of U2AF for recognition of a weak 3′ splice site. Mol. Cell Biol. 2006;26:8183–8190. doi: 10.1128/MCB.00350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pacheco TR, Moita LF, Gomes AQ, Hacohen N, Carmo-Fonseca M. RNA interference knockdown of hU2AF35 impairs cell cycle progression and modulates alternative splicing of Cdc25 transcripts. Mol. Biol. Cell. 2006;17:4187–4199. doi: 10.1091/mbc.E06-01-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Massiello A, Roesser JR, Chalfant CE. SAP155 Binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. FASEB J. 2006;20:1680–1682. doi: 10.1096/fj.05-5021fje. [DOI] [PubMed] [Google Scholar]
- 65.Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol. Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- 66.Gabanella F, Carissimi C, Usiello A, Pellizzoni L. The activity of the spinal muscular atrophy protein is regulated during development and cellular differentiation. Hum. Mol. Genet. 2005;14:3629–3642. doi: 10.1093/hmg/ddi390. [DOI] [PubMed] [Google Scholar]
- 67.Mollet I, Barbosa-Morais NL, Andrade J, Carmo-Fonseca M. Diversity of human U2AF splicing factors. FEBS J. 2006;273:4807–4816. doi: 10.1111/j.1742-4658.2006.05502.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.