Abstract
Saccharomyces cerevisiae Srs2 helicase plays at least two distinct functions. One is to prevent recombinational repair through its recruitment by sumoylated Proliferating Cell Nuclear Antigen (PCNA), evidenced in postreplication-repair deficient cells, and a second one is to eliminate potentially lethal intermediates formed by recombination proteins. Both actions are believed to involve the capacity of Srs2 to displace Rad51 upon translocation on single-stranded DNA (ssDNA), though a role of its helicase activity may be important to remove some toxic recombination structures. Here, we described two new mutants, srs2R1 and srs2R3, that have lost the ability to hinder recombinational repair in postreplication-repair mutants, but are still able to remove toxic recombination structures. Although the mutants present very similar phenotypes, the mutated proteins are differently affected in their biochemical activities. Srs2R1 has lost its capacity to interact with sumoylated PCNA while the biochemical activities of Srs2R3 are attenuated (ATPase, helicase, DNA binding and ability to displace Rad51 from ssDNA). In addition, crossover (CO) frequencies are increased in both mutants. The different roles of Srs2, in relation to its eventual recruitment by sumoylated PCNA, are discussed.
INTRODUCTION
DNA helicases are present in all kingdoms and perform an extraordinary variety of functions in cells. They play important roles in DNA metabolic processes such as replication, repair, recombination and transcription. Their DNA unwinding and translocase activities begin to be well characterized at the biochemical and structural levels (1). However, much remains to be understood about their roles in vivo. Since all organisms express multiple DNA helicases, a high degree of functional specialization should exist. This is particularly true for the control of homologous recombination (HR) (2). In Saccharomyces cerevisiae, at least six different helicases, Sgs1, Srs2, Mph1, Mer3, Pif1 and Rrm3 are positively or negatively involved in HR, increasing or decreasing recombination rates. They all perform specific functions that cannot be executed by another one. When and how these helicases act remains unclear. Partial answers to these questions have been recently obtained for Srs2.
Srs2 belongs to the SF1 helicase family and is structurally and functionally related to the bacterial UvrD helicase (3,4). Mutants of SRS2 were first described as partial suppressors of the radiation sensitivity of rad6 and rad18 mutants (5), defective in postreplicative DNA repair (PRR). They were later isolated as hyper-recombination mutants (6) and as suppressors of the methyl methane-sulfonate (MMS) sensitivity of rad18 cells (3). In RAD18 background, the single srs2Δ mutants are sensitive to ultraviolet (UV) light (3), a phenotype dependent on RAD51, RAD52, RAD55 or RAD57 genes [(7) and our unpublished data] that are all involved in the formation/stabilization of presynaptic Rad51 nucleofilaments. This epistasis of rad51 over srs2, together with data showing that Srs2 prevents recombinational repair in cells containing leaky alleles of RAD51 or RAD52 (7–10), led to the proposal that Srs2 eliminates toxic recombination intermediates (11) rather than preventing their formation. This hypothesis was later supported by the ability of Srs2 to dismantle Rad51 nucleoprotein filaments formed in vitro on single-stranded DNA (ssDNA) through its translocase activity (12,13). In the absence of any treatment, toxic recombination intermediates are also formed in double mutants involving srs2Δ and mutations affecting recombination and/or replication such as rad54Δ and sgs1Δ. The synthetic lethality or sickness of the double mutants is suppressed by a third mutation in RAD51, RAD52, RAD55 or RAD57 (10,14,15)). Recently, it was proposed that not only the translocase but also the helicase activity of Srs2 may reverse nonproductive recombination intermediates resulting from strand invasion of the homologous duplex DNA (16).
While the deletion of SRS2 sensitizes wild-type cells to UV, it increases the resistance of the highly UV sensitive rad6 and rad18 mutants. In these PRR-deficient contexts, it has been demonstrated that the high sensitivities of rad6Δ and rad18Δ cells is related to the prevention of recombinational repair by Srs2 which is recruited by a sumoylated form of PCNA (17,18). Indeed, deletion of the Small Ubiquitin-related Modifier (SUMO)-specific ligase gene SIZ1, which is responsible for the sumoylation of PCNA on the K164 lysine or PCNA mutations that prevent its sumoylation suppress DNA damage sensitivities of rad18Δ or rad6Δ cells, as does the deletion of SRS2 (17–19).
Srs2 was also shown to be involved in crossover (CO) control and proposed to unwind the invading strand of recombination intermediates, allowing conversion but preventing formation of COs (20,21). This role of Srs2 would involve the helicase activity, and appears to be dependent on PCNA sumoylation (21).
These genetic data show that Srs2 plays different roles. One is to eliminate toxic recombination intermediates. Importantly, in wild-type cells, a SIZ1 deletion has no significant effect on repair, mutagenesis or recombination (18,19), suggesting that elimination of toxic intermediates does not depend on PCNA sumoylation. Another role of Srs2 is to prevent recombinational repair. This has been evidenced only in PRR-deficient contexts, and the extrapolation of this activity to wild-type cells remains questionable in view of the absence of effects of the single siz1 mutation.
In a hope to obtain mutations that separate the functions, we screened for new Srs2 mutations suppressing the MMS sensitivity of rad18 cells. We describe here two of them which confer UV resistance to rad18 cells but are not by themselves UV or MMS sensitive. However, they do have an effect on the incidence of reciprocal recombination associated to gene conversion events. The biochemical study of the purified mutant proteins indicates that the mutations affect differently the activities of Srs2, although their biological effects are very similar.
MATERIALS AND METHODS
Yeast strains and genetic analysis
Except for CO experiments, the strain used in this study are isogenic derivatives of either FF1852 (MATa leu2-3,112 trp1-289 ura3-52 ade5) or of FF18733 (MATa leu2-3,112 trp1-289 ura3-52 his7-2 lys1-1). The MATa haploids in the FF1852 series are: FF1886, rad18::LEU2 srs2R1; FF1888, rad18::LEU2 srs2R3; FF18672, srs2::LEU2; D84-3B, siz1::KAN; D48-3C, srs2R1; D52-2C, srs2R3; the MATα in the same background is FF18238, rad18::LEU2. The MATa haploids in the FF18733 series are FF182029, pol30-K127R; FF182031, pol30-K127/164R; FF182027, pol30-K164R; D83-5B, rad18::KAN srs2::KAN. In the same background, the MATα strains are FF181496, sgs1::URA3; FF18974, rad54::LEU2.
For CO experiments, the strains used are derivatives of W303, as were the strains used in our previous experiments on COs (21). The wild-type SRS2 and the mutants srs2R1 and srs2R3 are tagged by HA. We found no effect of the HA tag on the resistance to genotoxic agents, indicating that the Srs2 proteins are functional. The ‘CY2715’ wild-type HA-SRS2 strain (22) was a gift of Dr Foiani. HA-SRS2 was replaced by HA-srs2R1 or srs2R3 by the pop-in/pop-out method. For this, srs2R1 and srs2R3 were cloned into the URA3 pRS406 vector. The linearized plasmids were integrated into the HA-SRS2 strain, and pop-out events were selected on 5-fluoroorotic acid (5FOA), a drug that allows only the growth of Ura− cells (23). The presence of the srs2R1 or srs2R3 mutation was tested by the suppression of rad18Δ MMS sensitivity in meiotic analysis of diploids obtained by crossing the strains with rad18Δ haploids. It was confirmed by DNA sequencing. The strains were crossed with the srs2Δ haploid bearing the ectopic recombination system (D498-1C) to isolate from the meiotic progeny the wild-type strain ‘D508-10C’ of genotype MATa arg 4-Bg URA3::arg4- RV::ura3-1 ade2-1 trp1-1 leu2-3,112 his3-11,15 can1-100 HA3SRS2::srs2. The corresponding HA-srs2R1 and HA-srs2R3 strains are ‘D511-4D’ and ‘D509-8B’, respectively. Genetic analyses were performed according to published procedures (24).
Determination of CO frequencies
This analysis was performed as previously described (21). Briefly, CO frequencies associated to gene conversion events were determined in haploid cells carrying two arg4 alleles mutated at different sites and located on different chromosomes. One allele is at its endogenous location on chromosome VIII, and the other one is located on chromosome V, between a wild-type and a mutated allele of URA3 in direct orientations. When gene conversion occurs without CO, the two URA3 and ura3-1 genes remain in the parental direct repeat configuration in which loss of URA3 is a frequent event. A CO associated to a conversion of an arg4 allele generates a reciprocal translocation and a separation of the URA3 and ura3-1 alleles on each of the translocated chromosomes. In this situation, loss of the URA3 information is much less frequent. Spontaneous and independent Arg+ convertants are selected on medium lacking arginine and patched on the same medium. To determine the relative frequencies of URA3 losses, the plates are replicated on a medium containing 5-FOA, a drug that allows only the growth of Ura− cells (23). Patches that give rise to numerous Ura− papillae derive from a conversion without CO and patches with no or a very few Ura− papillae reveal a CO event. This was verified by molecular analyses on a sample of convertants (21).
UV treatment
Stationary phase (RAD18 background) or exponentially growing (rad18Δ context) cells were washed in sterile water and resuspended at appropriate dilutions. UV irradiation (254 nm) was applied after plating on YPD and survival was determined after 3 days of incubation at 30°C. For spot assays, exponentially growing cells were resuspended in water at 3.106 cells/ml and 7 µl of 10-fold serial dilutions were spotted onto plates lacking leucine to select for cells that retained the plasmid. The control and UV-irradiated plates were incubated as described above.
Plasmids and proteins
In order to clone the mutant genes into baculoviruses, we first constructed the p14HB-srs2R1 and p14HB-srs2R3 plasmids by the gap repair method (25). The centromeric replicative p14HB plasmid contains the whole wild-type SRS2 coding sequence and, upstream of the ORF, a BamHI site which was introduced by site-directed mutagenesis in p14H (3). A gap was made in the region of either the srs2R1 or srs2R3, and the gapped plasmids were introduced into the corresponding mutant cells. For srs2R1, BbvCI-gapped p14HB (13) was introduced in FF1886 and the repaired plasmid was recovered. For srs2R3, the same procedure was followed using XbaI-gapped p14HB introduced into FF1888. The BamHI-EcoRI fragment of p14HB-srs2R1 or p14HB-srs2R3 was cloned into pBacPAK-His1. The recombinant baculoviruses were constructed as recommended by the manufacturer (Invitrogen, Cergy Pontoise, France). It encodes the Srs2 proteins with 17 additional amino acid residues (MGHHHHHHVVDKLGSQM) fused to its N-terminal methionine. The wild-type fusion protein was found to complement the genotoxic sensitivity of srs2Δ cells (data not shown). Srs2 wild-type and mutant proteins were produced and purified as described previously (13).
To overproduce Srs2 wild-type and mutant proteins in yeast, the HindIII-SphI fragment of p14HB vectors was cloned into the multicopy plasmid YEp13 (26). To eliminate the BamH1 site and restore the genuine sequence, the EagI-Bsu36I fragment that contains the BamH1 site was replaced by the corresponding fragment of p14H.
The K41A mutation in the Walker A site in Srs2 was introduced by using mutagenic DNA primers and QuikChange site-directed mutagenesis kit (Stratagene, Massy, France).
ATPase assay
ATPase activity was measured by linking ATP hydrolysis to the oxidation of NADH as described previously (27). The dependence of the ATPase reaction on ssDNA cofactor (a 56 mer) was examined by the above method at 37°C in a buffer containing 50 mM Tris–HCl pH7.6, 100 mM NaCl, 7 mM MgCl2, 5 mM DTT, 80 µgml−1 BSA and 1 mM ATP. Values for the Michaelis-Menten constants kcat and Km for ATP at saturating amount of ssDNA were derived by fitting data directly to the Michaelis-Menten equation.
Helicase assay
A forked DNA built by the annealing of two oligonucleotides, P1 (5′-AGAAGGTTTCGAATCAGAGGTAGGTGCCCGGCCTCCAACTTGCCGTATTCCTGGT) and Cy5-5′-labeled P2 (5′-Cy5-ACCAGGAATACGGCAAGTTGGAGGCCGGGCTGGATGGAGACTAAGCTTTGGAAGT) was used to assay the helicase activity of Srs2. DNA substrate (20 fmol) was incubated at 25°C with 62 nM Srs2 in buffer A (50 mM Tris–HCl pH 7.6, 7 mM MgCl2, 5 mM DTT, 2 mM ATP and 25 µgml−1 BSA). Reactions were stopped by addition of 150 mM EDTA. The reaction products were resolved by electrophoresis on a 8% nondenaturating polyacrylamide gel in Tris-borate-EDTA (TBE) buffer and quantified by ImageQuant software.
Electrophoretic mobility shift assay
The same forked DNA (20 fmol) used in helicase assay was incubated with increasing amount of Srs2 protein in buffer A without ATP for 30 min at 25°C. Gel shift was detected after electrophoresis on a 6% nondenaturating polyacrylamide gel using a Storm 960 apparatus (GE Healthcare Biosciences, Saclay, France).
Electron microscopy (EM)
Rad51 filaments on ssDNA were formed by incubation at 37°C of 3.5 µM Rad51 protein in 42 mM MOPS pH 7.2, 3 mM Mg(OAc)2, 1 mM DTT, 20 mM NaCl, 2.5 mM ATP with 11 µM (ntd) ΦX174 viral (+) strand for 3 min followed by addition of 330 nM RPA and subsequent incubation for 15 min. Five microliters of this reaction was mixed with 5 µl of various amount of Srs2 diluted in 20 mM phosphate buffer pH 7.8, 200 mM NaCl, 10% glycerol and 1 mM ß-mercaptoethanol and incubated for 10 min at 37°C. The reaction mixtures were then diluted 20-fold in 10 mM Tris–HCl pH 7.5, 50 mM NaCl, 5 mM MgCl2 without any chemical fixation and analyzed by EM as previously described (28).
Immunoprecipitation
For immunoprecipitation assays, yeast native extracts from FF182029 treated with 0.3% MMS were prepared as described previously (18). Extract (2.5 mg of protein) was incubated overnight in buffer B (50 mM Tris–HCl pH 7.4, 150 mM NaCl and supplemented with protease inhibitors) at 4°C with 1.7 µg of purified HisSrs2, HisSrs2R1 or HisSrs2R3 protein and 1 µg of antibody against His-tag (Roche Diagnostics, Meylan, France). Prewashed protein G agarose beads (Roche Diagnostics) were then introduced for 5 hrs at 4°C. Beads were washed six times with buffer B and bound proteins were eluted in SDS loading buffer. Srs2 and PCNA were detected by Western blotting using polyclonal anti-Srs2 antibody (sc-11991, Santa Cruz Biotechnology, Heidelberg, Germany), and a polyclonal rabbit anti-PCNA serum (a generous gift from Martine Heude).
RESULTS
Isolation of srs2 mutations that partially suppress the UV sensitivity of rad18Δ mutant but do not confer UV sensitivity in otherwise wild-type cells
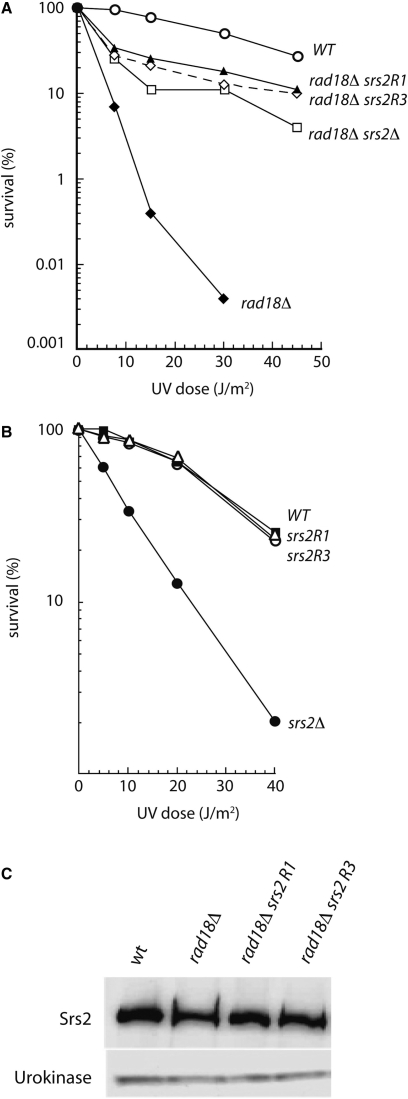
In a search for new suppressors of the sensitivity to the alkylating agent MMS of rad18Δ cells, we isolated several srs2 mutants, which fell into two groups. Mutations belonging to the first one have the same effects as srs2Δ: they partially suppress the MMS and UV sensitivities of rad18Δ cells, and in the RAD18 context, the mutants are UV sensitive and show increased rates of spontaneous and UV-induced intragenic recombination. The mutations of the second group also partially suppress the sensitivities to genotoxic agents of rad18Δ cells but they do not confer UV sensitivity in the RAD18 context. We focused our attention on two mutants of this last class, srs2R1 and srs2R3. Figure 1A shows that they both suppress the UV sensitivity of rad18Δ cells, as efficiently as does srs2Δ. However, unlike srs2Δ cells, the single srs2R1 and srs2R3 mutants are not UV sensitive (Figure 1B). The suppression of UV sensitivity in rad18Δ cells is not due to a down regulation of the mutated proteins, as judged by western blot analysis (Figure 1C). As reminded in the introduction, the UV sensitivity of srs2Δ cells is believed to be due to the binding of recombination proteins to ssDNA, before and/or during replication, generating toxic DNA structures if not processed by Srs2. That srs2R1 and srs2R3 are not UV sensitive suggests that these two mutated proteins are still able to remove the toxic recombination intermediates induced by UV treatment.
Figure 1.
UV survival of srs2R1 and srs2R3 mutants. (A) Like srs2Δ, srs2R1 and srs2R3 mutations partially suppress the UV sensitivity of rad18Δ cells. (B) srs2R1 and srs2R3 mutants are as resistant as SRS2 cells to UV, while srs2Δ cells are sensitive after irradiation. (C) Western blot analysis indicates that Srs2R1 and Srs2R3 are present in the same amount as Srs2 in rad18Δ cells.
srs2R1 and srs2R3 mutations do not show a negative interaction when combined with either rad54Δ or sgs1Δ
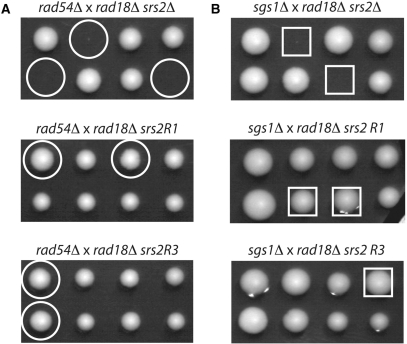
Toxic recombination intermediates are also formed in different double mutants involving srs2Δ, notably srs2Δ rad54Δ cells, which are dead, and srs2Δ sgs1Δ, which are extremely sick. We asked whether srs2R1 or srs2R3 affect the viability of rad54Δ and sgs1Δ mutants, as does srs2Δ. We performed tetrad analysis on diploids heterozygous for srs2R1 and rad54Δ or sgs1Δ, and srs2R3 and rad54Δ or sgs1Δ. In these diploids, rad18 was also heterozygous but this mutation has no effect on the viability of their meiotic progeny. While the negative interaction between srs2Δ and rad54Δ or sgs1Δ were once more evidenced, no interaction was observed between srs2R1 or srs2R3 and rad54Δ or sgs1Δ: the double mutants rad54Δ srs2R1, rad54Δ srs2R3, sgs1Δ srs2R1 and sgs1Δ srs2R3 are viable and have no growth defects, as judged by the size of the colonies (Figure 2). Thus, Srs2R1 and Srs2R3 proteins have retained the capacity to disrupt toxic recombination structures formed not only after UV irradiation but also in the absence of Rad54 or Sgs1.
Figure 2.
srs2R1 and srs2R3 do not affect the viability of rad54Δ or sgs1Δ cells. Two tetrads from crosses indicated above each picture are shown. Circles and squares indicate the rad54Δ srs2 and sgs1Δ srs2 combinations, respectively. (A) While rad54Δ srs2Δ cells are dead, rad54Δ srs2R1 and rad54Δ srs2R3 double mutants are perfectly viable. (B) The severe growth defect seen in sgs1Δ srs2Δ cells is not observed in either sgs1Δ srs2R1 or sgs1Δ srs2R3 double mutants.
Molecular characterization of srs2R1 and srs2R3 mutations
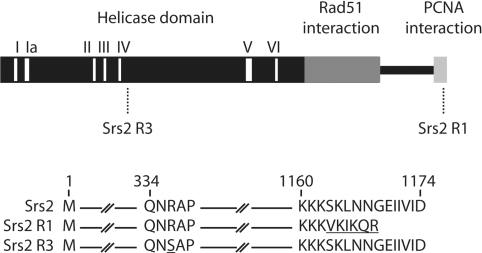
The nature of srs2R1 and srs2R3 mutations was determined by DNA sequencing after PCR amplification of the entire genes (Figure 3). srs2R3 has a G to T transversion leading to the substitution of the arginine in position 337 by a serine. This mutation is close to the helicase domain IV (amino acid 310 to 321). srs2R1 mutation is the addition of one adenine in position 3480. The resulting protein is shorter by six amino acids and five of the last six amino acids of the C-terminal end are changed. This modification occurs in the interacting domain with sumoylated PCNA (18).
Figure 3.
(A) Localization of srs2R1 and srs2R3 mutations. (B) Modifications induced by mutations srs2R1 and srs2R3 at the protein level.
Unexpectedly, although the srs2R1 and srs2R3 mutations were located in a different region of the gene, the mutants have similar phenotypes. In order to determine the defective properties of these mutants, we purified the mutated proteins and compared their biochemical properties to those of the wild-type protein.
Unlike Srs2R1, Srs2R3 is affected for all of its biochemical functions
Srs2 is a ssDNA-dependent ATPase (12,13,29). The ATP hydrolysis is believed to reflect its translocase activity on ssDNA. Therefore, we determined the kcat and the Km for ATP of the wild-type and the two mutated Srs2 proteins, using a 56-mer oligonucleotide as ssDNA cofactor (Table 1). The ATPase activity was determined with saturating amounts of ssDNA (data not shown) and, therefore, represents the true catalytic activity once the protein is bound to the ssDNA. Srs2R1 binds and hydrolyzes ATP as efficiently as Srs2 (Km 94 µM versus 143 µM and kcat 2778 min−1 versus 3316 min−1). These kcat values are in the same range that the one previously reported measured with ΦX viral (+) strand as cofactor (29). Unlike Srs2R1, Srs2R3 is strongly affected for ATP hydrolysis, with only about 10% of the wild-type activity (kcat 373 min−1 versus 3316 min−1). Surprisingly, Srs2R3 binds more efficiently ATP than the wild-type protein (Km 19 µM versus 143 µM). However, the ATP intracellular concentration in yeast is supposed to be superior than 1 mM (30), a much higher value than the calculated Km of Srs2 for ATP. It indicates that the ATP binding parameter can be disregarded for the in vivo function.
Table 1.
Km and kcat values of the enzymes for ATP
| Protein | Km (µM) | kcat (min−1) |
|---|---|---|
| Srs2 | 143 | 3316 |
| Srs2R1 | 94 | 2278 |
| Srs2R3 | 19 | 373 |
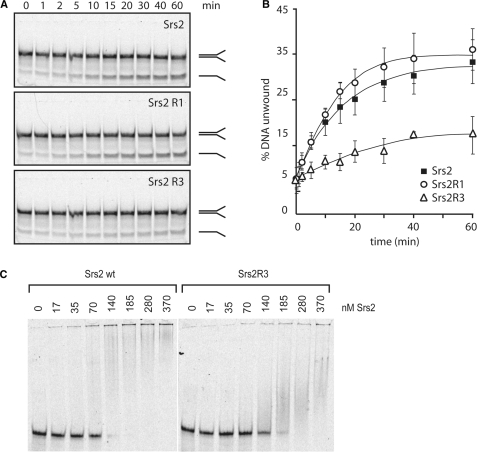
We next studied the DNA helicase activities of the two Srs2 mutant proteins, using a 5′-Cy5-labeled forked DNA containing a 30-bp duplex region flanked by two single-stranded tails of 25 nucleotides. As shown in Figure 4A and B, the wild-type Srs2 and the mutated Srs2R1 proteins have a similar helicase activity, while Srs2R3 unwinds poorly the DNA substrate. The attenuated helicase ability of Srs2R3 may result from an ATPase defect and/or from a default in DNA binding. For this reason, we compared the DNA binding abilities of Srs2R3 and of the wild-type protein with a DNA mobility shift assay. We incubated increasing amounts of wild-type Srs2 and Srs2R3 with the same DNA substrate used in the helicase assay followed by resolution of the reaction mixtures in nondenaturating polyacrylamide gels. As shown in Figure 4C, Srs2R3 binds less efficiently forked DNA than wild-type Srs2. At 140 nM protein, almost all DNA was bound to Srs2 while <50% of it was bound by Srs2R3.
Figure 4.
Srs2R3 is defective in both helicase activity and DNA binding. (A) Kinetics of helicase activity of wild-type and mutated Srs2. (B) Quantification of experiments showed in (A). Each point represents the average of three independent assays. Error bars show the standard deviation. (C) Srs2R3 binds forked DNA less efficiently than Srs2 as observed by gel shift assay.
Srs2R3, but not Srs2R1, disrupts less efficiently the Rad51 presynaptic filament
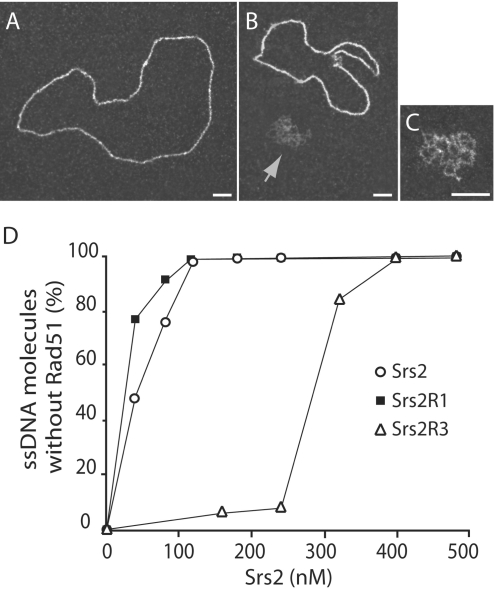
Srs2 is able to disrupt Rad51 presynaptic filaments (12,13). Because srs2R1 and srs2R3 mutants are not UV sensitive (Figure 1), we hypothesized that Srs2R1 and Srs2R3 remove toxic recombination intermediates by releasing Rad51 from the nucleoprotein filament. EM was used to monitor this activity for both mutated and wild-type proteins. Rad51 was first incubated for 3 min with ΦX174 ssDNA followed by addition of RPA and subsequent incubation for another 15 min in order to assemble Rad51 nucleoprotein filaments. As a function of the amount of Srs2 introduced into the reaction, a progressive loss of Rad51 filaments, coupled with a concomitant formation of RPA-ssDNA complexes, was observed (Figure 5). At 120 nM Srs2 or Srs2R1, almost all the observed DNA molecules were covered by RPA while, at the same concentration, <5% of the Rad51 nucleoprotein filaments were destabilized by Srs2R3. 400 nM of Srs2 were required to obtain disruption of all Rad51 filaments. Because disassembly of the presynaptic filament requires the translocase activity of Srs2 on ssDNA, a function that requires ATP hydrolysis, the weak capacity of Srs2R3 protein to remove Rad51 from ssDNA may result from two failing activities: a reduced ability to bind ssDNA and/or a weak ATPase activity.
Figure 5.
Srs2R3 disrupts the Rad51 presynaptic filament less efficiently than Srs2 and Srs2R1. (A) Rad51-ssDNA nucleoprotein filament. (B) Preformed Rad51-ssDNA complexes were incubated for 10 min with Srs2. The arrows point to the ssDNA covered with RPA. (C) Blow-up of ssDNA covered with RPA. (D) The percentage of disrupted Rad51 presynaptic filament was determined for various amounts of helicases. For each concentration, 300–500 molecules were counted. Scale bars, 50 nm.
Taken together, the results from biochemical and EM analyses clearly indicate an attenuated activity for all the tested functions of Srs2R3, while Srs2R1 behaves like the wild-type Srs2 protein.
Srs2R1 does not bind SUMO-modified PCNA
It was previously reported that Srs2 preferentially interacts with the sumoylated forms of PCNA (17,18). This interaction involves the C-terminal 138 residues of Srs2 protein, but a C-terminal truncation of the last six amino acids strongly reduces the two-hybrid interaction with both SUMO and PCNA.
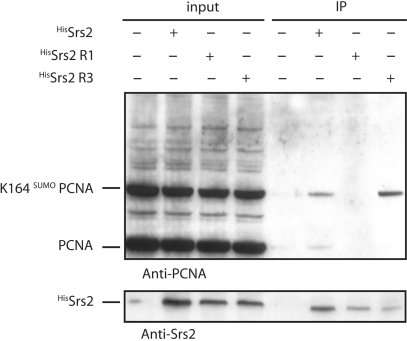
The Srs2R1 is modified in its C-terminal domain. The protein lost its six last residues and 5 of the six last amino acids are changed (Figure 3). Therefore, we asked if the ability of Srs2R1 to interact with sumoylated PCNA is affected. Purified His-tagged Srs2 wild-type or mutated proteins were incubated with total cell extracts in order to examine their binding to PCNA and sumoylated PCNA. Extracts were prepared from a pol30-K127R mutant treated with 0.3% MMS before lysis to induce extensive SUMO conjugation at lysine 164 (K164) (31). The pol30-K127R mutant was used because this lysine was shown to be sumoylated to a small extent in a Siz1-independent manner, a modification that plays a minor biological role (17,18). Under these conditions, the HisSrs2 and HisSrs2R3 recombinant proteins clearly bound the K164-sumoylated PCNA while no interaction could be detected with the HisSrs2R1 recombinant protein (Figure 6). Interestingly, we detected a very faint interaction between unmodified PCNA and Srs2 and no interaction between unmodified PCNA and either Srs2R1 or Srs2R3, even with longer exposure of the blot (data not shown). These data differ from results found previously in two independent studies (17,18) showing an important interaction of Srs2 with non-sumoylated PCNA. The reason for this discrepancy is not clear, it could reflect small differences in experimental procedures. However, as we will show below, the interaction between Srs2 and unmodified PCNA does not seem to play a role in vivo.
Figure 6.
Srs2R1 does not interact with sumoylated PCNA. Immunoprecipitation experiments were performed with total cell lysates prepared from pol30-K127R mutant treated with 0.3% MMS to generate maximal levels of SUMO-modified PCNA. Input and bound fractions were analyzed by western blot as indicated.
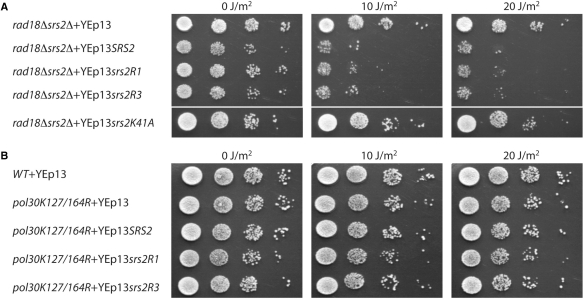
Overexpression of Srs2R1 and Srs2R3 sensitizes rad18Δ srs2Δ but not pol30-K127/164R cells to UV radiation
Although the Srs2R3 protein still interacts with K164-sumoylated PCNA, the mutation suppresses the UV sensitivity of rad18Δ cells to the same extent as does the deletion of SRS2 (Figure 1A). Since the different activities of Srs2 tested are present but less efficient in srs2R3, we predicted that overexpressing the mutant protein would sensitize the double mutant rad18Δ srs2Δ cells. To test this idea, we introduced SRS2 or srs2R3 under their own promoter into the multicopy yeast vector YEp13. The rad18Δ srs2Δ double mutant cells were transformed and UV survival assays performed. As predicted, overexpression of srs2R3 strongly reduces UV survival of these cells, and furthermore, to a similar extent as does the wild-type SRS2 overexpression (Figure 7A). In the same conditions, overexpression of the Srs2K41A helicase- and ATPase-dead mutant protein (32) has no effect (Figure 7A), indicating that indeed the srs2R3 overexpression effect relates to an increased activity and not simply to the abnormally high protein concentration. These results support the view that a single copy of srs2R3 does not produce enough protein to prevent recombinational repair in UV-treated rad18Δ cells, and consequently that the Srs2R3 protein suffers from a decreased specific activity rather than from a complete deficiency in a particular activity.
Figure 7.
Overexpression of Srs2R1 and Srs2R3 sensitizes rad18Δ srs2Δ cells (A) but not pol30-K127/164R cells (B) to UV radiation. Ten-fold serial dilutions of an equal number of exponentially phase growing cells were spotted onto plates and irradiated with ultraviolet light at indicated doses.
When we did the same experiment with srs2R1, we also observed that its overexpression sensitizes rad18Δ srs2Δ cells (Figure 7A). One possible explanation for this result is that a residual interaction (although not detected in our experiments) between the overproduced Srs2R1 proteins and K164-sumoylated PCNA allows enough helicase recruitment to prevent recombinational repair. A second explanation is that overproduction of Srs2 prevents recombinational repair by a recruitment through unmodified PCNA or directly through its interaction with Rad51 (12). In order to differentiate between these possibilities, srs2R1 was overexpressed in the pol30-K127/164R mutant unable to be sumoylated on lysine 127 or sumoylated or ubiquitinated on lysine 164. Overproduction of Srs2R1 does not sensitize the pol30-K127/164R mutant to UV (Figure 7B). Similar results are obtained when wild-type SRS2 or srs2R3 were overexpressed. Altogether, these data indicate that, at least in a context where PCNA cannot be ubiquitinated, the recruitment of Srs2 to prevent recombinational repair is strictly dependent upon its interaction with sumoylated PCNA.
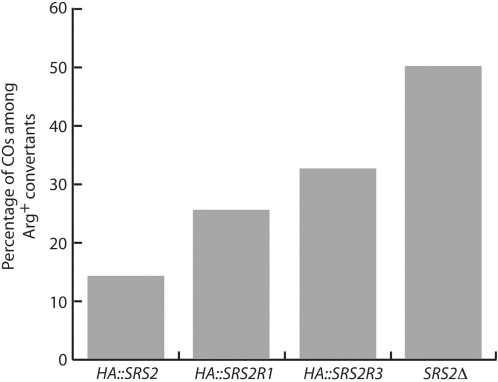
Srs2R1 and Srs2R3 mutants show increased CO frequencies
The absence of Srs2 was shown to enhance both DSB-induced and spontaneous CO frequencies (20,21). The proposed interpretation is that Srs2 acts on mitotic recombination intermediates at a postsynaptic stage. It unwinds the invading and elongated strand from its template, promoting synthesis-dependent strand annealing (SDSA), a mechanism that allows gene conversion but not associated COs. A partially unanswered question concerns the recruitment of Srs2 to the D-loop. Using a genetic system allowing to determine the frequency of COs associated to a selectable conversion event involving ectopic heteroalleles (see Materials and methods section), we previously showed (21) that CO frequencies rise from 11% in wild-type cells to 50% in both the srs2Δ mutant and in the pol30-K127/164R mutant that can neither be sumoylated nor ubiquitinated at the lysine 164 of PCNA (31). This result strongly suggested that SUMO-modified PCNA recruits Srs2 into the D-loop. However, because the double PCNA mutant is also deficient in PRR, the high incidence of COs observed in this strain is possibly also due to the PRR deficiency, and not only to the lack of Srs2 recruitment by PCNA. To address this question, we used the srs2R1 mutation, which prevents the interaction of the helicase with PCNA. We introduced at their native locus a HA-tagged version of SRS2 or of srs2R1 into the strain used by Robert et al. (21). The percentage of COs associated with spontaneous gene conversion events was found to be increased 1.8-fold in srs2R1 as compared with SRS2 cells (25 and 14%, respectively) (Figure 8). This result shows that the interaction between SUMO-modified PCNA and Srs2 is indeed involved in CO control. However, the level of COs measured in srs2R1 (25%) is half that in srs2Δ or in the double PCNA mutants (50%). It suggests that Srs2 controls CO both in a PCNA-dependent and independent fashion. We cannot formally exclude that a possible residual interaction between PCNA and Srs2R1 could account for this result. The same experiment was carried out with srs2R3. Interestingly, this mutant leads to a 2.3-fold increase in COs over the wild-type level (33 and 14%, respectively). The percentage is still lower than that observed in srs2Δ mutants. The effect of this mutation likely results from the decreased helicase activity of Srs2R3, and thus from less efficient repair through the SDSA pathway.
Figure 8.
CO bias observed in srs2 mutants. The percentage of CO was determined using three individual segregants for each genotype.
DISCUSSION
Early studies on srs2 mutations revealed two opposite effects with respect to radiations. In the highly sensitive rad6 or rad18 haploid mutants, deficient for PRR, srs2 suppresses the sensitivity of a subpopulation of cells (5), while in PRR-proficient cells, it sensitizes a subpopulation of cells (3). It is now understood that these effects reflect two roles of Srs2. One is the elimination of potentially toxic structures formed by recombination proteins (11), and a second role is the prevention of recombinational repair through its recruitment by sumoylated PCNA, evidenced only in PRR-deficient cells (17,18). The ability of Srs2 to disrupt in vitro Rad51 nucleofilaments on ssDNA would be sufficient to account for the elimination of both the toxic and the recombination intermediates. It is likely that toxic structures are also formed after UV treatment of rad18 srs2 cells, but this cannot be directly tested because rad51, as well as other mutants affected in the formation of Rad51 nucleofilaments, has a synergistic effect with rad18 on UV survival, regardless of the status of Srs2.
In this study we describe two new srs2 mutants that have acquired the ability to perform recombinational repair in a rad18 context, but have retained the ability to reverse toxic structures in a RAD18 context. The biochemical activities of the two mutant proteins are differently affected. The only known defect of Srs2R1 is the loss of interaction with sumoylated PCNA. This explains its suppressive effect on the UV sensitivity of rad18 cells. The mutation is located in the C-terminal domain of the protein, known to interact with sumoylated PCNA, and deletions in this domain were reported to suppress the sensitivity of PRR mutants (18). Differently, Srs2R3 interacts as efficiently as the wild-type protein with sumoylated PCNA and its cellular amount in the cells is not decreased. However, the ATPase and helicase activities of the protein, as well as its ability to disrupt Rad51 nucleoprotein filaments, are decreased. We suggest that the inability of Srs2R3 to prevent recombinational repair in rad18 cells may be related to the size of the Rad51 presynaptic filament that must be removed. Indeed, EM analyses revealed that UV lesions, if not repaired, induce uncoupling of nascent strands synthesis at replication forks leading to up to 3 kb long ssDNA regions (33). In rad18 cells, only recombination could save the cells, but Srs2 counteracts this repair. Because of its weak translocase activity, Srs2R3 would be unable to fully destroy the Rad51 nucleofilaments, allowing recombinational repair to occur. This interpretation fits with our data showing that overexpression of Srs2R3 in rad18Δ srs2Δ cells largely sensitizes the cells. It fits also with the semi dominance of srs2 mutants in rad18 homozygous diploids: the cells are more sensitive with two copies of SRS2 than with a single one, indicating a gene dosage effect (34).
Removal of toxic recombination structures by Srs2 does not require an interaction with sumoylated PCNA
Contrarily to srs2Δ cells, the two mutants srs2R1 and srs2R3 are not UV sensitive. This indicates that the two mutated proteins have retained the ability to reverse toxic recombination structures. The Srs2R3 protein is therefore in sufficient amount to perform this activity. It suggests that in PRR proficient cells, these structures are limited in size and/or number. The only deficient activity that we observed for Srs2R1 is its binding to sumoylated PCNA. This interaction is therefore not required to remove the dead-end structures. In agreement with this conclusion, the single siz1 mutant, deficient in sumoylation of the lysine 164 of PCNA, shows no UV sensitivity (18). We asked if the potentially toxic intermediates formed in the rad54 or sgs1 mutants, and revealed by their Rad51-dependent synthetic lethality with srs2Δ, depends on PCNA sumoylation. The srs2R1 mutation has no effect on the growth of rad54 or sgs1 cells. Thus, the toxic structures formed in these mutants, as those formed after UV treatment, are reversed by Srs2 independently of an interaction with sumoylated PCNA. In these cases, the recruitment of Srs2 could be mediated through its interaction with Rad51 (12).
Control of COs involves the helicase activity of Srs2 and is partially dependent of its interaction with sumoylated PCNA
It has been proposed that Srs2 prevents COs by promoting SDSA, a recombination pathway non-associated with COs (20,21). This would imply the unwinding activity of Srs2 in order to displace the elongated invading strand from the D-loop. We recently showed that Srs2 has the biochemical activities that are required to perform SDSA (16). However, in vivo evidence for an implication of the helicase function of Srs2 in this process is missing. We hypothesized that the srs2R3 mutation which decreases the helicase activity of Srs2 would increase the percentage of CO associated to conversion events. We indeed found that in this mutant CO frequency is increased from 14% in wild-type cells to 33%, while it reaches 50% in the absence of Srs2.
The approximate correlation between the level of COs and that of the helicase activity supports the idea that Srs2 controls the level of COs through its helicase activity. However, we cannot exclude the possibility that the increased CO frequency observed in srs2R3 cells relates to its decreased DNA binding ability and translocase activity. An srs2 mutant affected specifically in its unwinding activity would be required to answer this question.
The Srs2R1 mutation increases the frequency of COs associated with conversions from 14% in wild-type cells to 25%. This indicates that the interaction with sumoylated PCNA is involved in CO control. However, since the percentage of COs in srs2R1 mutant is half that in srs2Δ cells, it suggests that the enrollment of the Srs2 helicase to disrupt D-loops is only partially dependent on sumoylated PCNA.
It is important to note that the effect on CO incidence of srs2R1 and of srs2R3 is the only phenotype of these mutants in a RAD18 context. In both cases, it is likely due to a decreased efficiency in the processing of recombination intermediates into the SDSA pathway. This would be related to a limited Srs2 helicase activity, for different reasons in each of the mutants.
What is the role of PCNA sumoylation?
The absence of genetic effects of the single siz1 or srs2R1 mutation raises the question of the role of PCNA sumoylation at K164 in wild-type cells. Our favorite model is the following one. Since PCNA is sumoylated even in the absence of any treatment during S phase (31) and since the same lysine is the ubiquitin target of Rad18/Rad6, it is reasonable to believe that the sumoylated PCNA drives replication and avoids unintended mutation and recombination events to take place. On one hand, sumoylation of PCNA on the lysine 164 would protect against unnecessary ubiquitination and, consequently, hinder the error-prone and error-free Rad18-dependent pathways from acting. Notably, it would prevent a recruitment of an error prone polymerase that could introduce untargeted mutations. On the other hand, the sumoylated PCNA would recruit Srs2 that counteracts recombinational repair.
When the replicative polymerase encounters a blocking lesion, a single-stranded region upstream of the synthesis block is formed, due to the uncoupling of the two nascent strands synthesis upon fork progression (33). Desumoylation of PCNA through a specific isopeptidase would occur, or new unmodified PCNA monomers would replace the sumoylated PCNA trimers. The absence of sumoylated PCNA opens two nonexclusive possibilities: ubiquitination of PCNA and formation of a Rad51 nucleoprotein filament on the ssDNA region. Fork restoration could then be mediated through the Rad18 pathways or by recombination. It is important to note that the choice of one or the other pathway is likely not determined by the ubiquitination state of PCNA: it is known that recombinational repair plays an important role in cells unable to ubiquitinate PCNA (e.g. rad18 or more evidently rad18 srs2 mutants) and in cells where PCNA is presumably ubiquitinated (e.g. rev3 or rad5 cells). Therefore, a possibility is that both PCNA ubiquitination and formation of a Rad51 nucleoprotein filament occur concomitantly. If fork restoration occurs by translesion synthesis, the Rad51 nucleoprotein filament might be eliminated upon progression of the polymerase.
In this scheme where PCNA is ubiquitinated upon replication arrest, the single siz1 mutation is not expected to have any effects on survival to genotoxic treatments, nor on induced mutation or recombination frequencies. The only siz1 effects that may be predicted in otherwise wild-type cells is an increase of untargeted mutations or recombination events, due to inadvertent switch of polymerases or/and Rad51 binding. However, these effects could be too small to overcome the other sources of spontaneous mutation or recombination and to be easily detectable.
ACKNOWLEDGEMENTS
We are grateful to Marco Foiani and Martine Heude for providing strains and antibodies. We also thank Pablo Radicella, Stéphanie Marsin, Serge Boiteux and Eric Coïc for helpful discussions and technical advices. This study was supported by grants of the Institut National du Cancer (PL003), the European Community (C.L.B., S.G., F.F., X.V., LSHG-CT-2003-503303), the Agence Nationale de la Recherche (ANR-07-BLAN-0350-01) and fellowships of La Ligue contre le Cancer (P.D.) and the Association pour la Recherche sur le Cancer (C.L.B.), the Centre National de la Recherche Scientifique and the Commissariat à l'Energie Atomique. Funding to pay the Open Access publication charges for this article was provided by Commissariat à l'Energie Atomique.
Conflict of interest statement. None declared.
REFERENCES
- 1.Singleton MR, Dillingham MS, Wigley DB. Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 2.Wu L, Hickson ID. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- 3.Aboussekhra A, Chanet R, Zgaga Z, Cassier-Chauvat C, Heude M, Fabre F. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 1989;17:7211–7219. doi: 10.1093/nar/17.18.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veaute X, Delmas S, Selva M, Jeusset J, Le Cam E, Matic I, Fabre F, Petit MA. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 2005;24:180–189. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence CW, Christensen RB. Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J. Bacteriol. 1979;139:866–876. doi: 10.1128/jb.139.3.866-876.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aguilera A, Klein HL. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119:779–790. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aboussekhra A, Chanet R, Adjiri A, Fabre F. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaytor MD, Nguyen M, Livingston DM. The complexity of the interaction between RAD52 and SRS2. Genetics. 1995;140:1441–1442. doi: 10.1093/genetics/140.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milne GT, Ho T, Weaver DT. Modulation of Saccharomyces cerevisiae DNA double-strand break repair by SRS2 and RAD51. Genetics. 1995;139:1189–1199. doi: 10.1093/genetics/139.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schild D. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics. 1995;140:115–127. doi: 10.1093/genetics/140.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chanet R, Heude M, Adjiri A, Maloisel L, Fabre F. Semidominant mutations in the yeast Rad51 protein and their relationships with the Srs2 helicase. Mol. Cell Biol. 1996;16:4782–4789. doi: 10.1128/mcb.16.9.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 13.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 14.Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 15.Heude M, Chanet R, Fabre F. Regulation of the Saccharomyces cerevisiae Srs2 helicase during the mitotic cell cycle, meiosis and after irradiation. Mol. Gen. Genet. 1995;248:59–68. doi: 10.1007/BF02456614. [DOI] [PubMed] [Google Scholar]
- 16.Dupaigne P, Le Breton C, Fabre F, Gangloff S, Le Cam E, Veaute X. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: implications for crossover incidence during mitotic recombination. Mol. Cell. 2008;29:243–254. doi: 10.1016/j.molcel.2007.11.033. [DOI] [PubMed] [Google Scholar]
- 17.Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 19.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 20.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robert T, Dervins D, Fabre F, Gangloff S. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 2006;25:2837–2846. doi: 10.1038/sj.emboj.7601158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liberi G, Chiolo I, Pellicioli A, Lopes M, Plevani P, Muzi-Falconi M, Foiani M. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J. 2000;19:5027–5038. doi: 10.1093/emboj/19.18.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 24.Sherman F, Hicks J. Micromanipulation and dissection of asci. Methods Enzymol. 1991;194:21–37. doi: 10.1016/0076-6879(91)94005-w. [DOI] [PubMed] [Google Scholar]
- 25.Orr-Weaver TL, Szostak JW. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc. Natl. Acad. Sci. USA. 1983;80:4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broach JR, Strathern JN, Hicks JB. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979;8:121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- 27.Pullman ME, Penefsky HS, Datta A, Racker E. Partial resolution of the enzymes catalyzing oxidative phosphorylation. I. Purification and properties of soluble dinitrophenol-stimulated adenosine triphosphatase. J. Biol. Chem. 1960;235:3322–3329. [PubMed] [Google Scholar]
- 28.Beloin C, Jeusset J, Revet B, Mirambeau G, Le Hegarat F, Le Cam E. Contribution of DNA conformation and topology in right-handed DNA wrapping by the Bacillus subtilis LrpC protein. J. Biol. Chem. 2003;278:5333–5342. doi: 10.1074/jbc.M207489200. [DOI] [PubMed] [Google Scholar]
- 29.Van Komen S, Reddy MS, Krejci L, Klein H, Sung P. ATPase and DNA helicase activities of the Saccharomyces cerevisiae anti-recombinase Srs2. J. Biol. Chem. 2003;278:44331–44337. doi: 10.1074/jbc.M307256200. [DOI] [PubMed] [Google Scholar]
- 30.Somsen OJ, Hoeben MA, Esgalhado E, Snoep JL, Visser D, van der Heijden RT, Heijnen JJ, Westerhoff HV. Glucose and the ATP paradox in yeast. Biochem. J. 2000;352(Pt 2):593–599. [PMC free article] [PubMed] [Google Scholar]
- 31.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 32.Krejci L, Macris M, Li Y, Van Komen S, Villemain J, Ellenberger T, Klein H, Sung P. Role of ATP hydrolysis in the antirecombinase function of Saccharomyces cerevisiae Srs2 protein. J. Biol. Chem. 2004;279:23193–23199. doi: 10.1074/jbc.M402586200. [DOI] [PubMed] [Google Scholar]
- 33.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 34.Palladino F, Klein HL. Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics. 1992;132:23–37. doi: 10.1093/genetics/132.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]