Abstract
Overexpression of the yeast Pdr5 ATP-binding cassette transporter leads to pleiotropic drug resistance to a variety of structurally unrelated cytotoxic compounds. To identify Pdr5 residues involved in substrate recognition and/or drug transport, we used a combination of random in vitro mutagenesis and phenotypic screening to isolate novel mutant Pdr5 transporters with altered substrate specificity. A plasmid library containing randomly mutagenized PDR5 genes was transformed into appropriate drug-sensitive yeast cells followed by phenotypic selection of Pdr5 mutants. Selected mutant Pdr5 transporters were analyzed with respect to their expression levels, subcellular localization, drug resistance profiles to cycloheximide, rhodamines, antifungal azoles, steroids, and sensitivity to the inhibitor FK506. DNA sequencing of six PDR5 mutant genes identified amino acids important for substrate recognition, drug transport, and specific inhibition of the Pdr5 transporter. Mutations were found in each nucleotide-binding domain, the transmembrane domain 10, and, most surprisingly, even in predicted extracellular hydrophilic loops. At least some point mutations identified appear to influence folding of Pdr5, suggesting that the folded structure is a major substrate specificity determinant. Surprisingly, a S1360F exchange in transmembrane domain 10 not only caused limited substrate specificity, but also abolished Pdr5 susceptibility to inhibition by the immunosuppressant FK506. This is the first report of a mutation in a yeast ATP-binding cassette transporter that allows for the functional separation of substrate transport and inhibitor susceptibility.
INTRODUCTION
Multiple or pleiotropic drug resistance (MDR/PDR) in the yeast Saccharomyces cerevisiae can result from overexpression of the Pdr5 ATP-binding cassette (ABC) multidrug transporter, leading to MDR/PDR to a variety of structurally unrelated xenobiotics (Balzi and Goffeau, 1994; Egner et al., 1995a). Hence, development of PDR in yeast is similar to an important type of MDR in mammalian cells (Kane, 1996) mediated by the overexpression of the MDR1 (Chen et al., 1986) [P-glycoprotein (Pgp)] or multidrug resistance-associated protein (Cole et al., 1992) gene products. MDR development can also be a major impediment to curative cancer chemotherapy in tumor cells, because Pgp (Ueda et al., 1987) and multidrug resistance-associated protein (Zaman et al., 1994) act as energy-dependent efflux pumps that extrude a broad range of anticancer drugs (Gottesman et al., 1995; Kane, 1996).
The predicted architecture of Pgp comprises two homologous halves, each with six predicted transmembrane domains (TMD) and one cytoplasmic nucleotide-binding domain (NBD). Although it is established that ATP binding and hydrolysis are essential for substrate transport, a suspected functional interaction of the NBDs with certain TMDs of Pgp and the molecular basis for its broad substrate specificity are only poorly understood (Gottesman et al., 1995; Kane, 1996). Several structure-function studies were performed to identify Pgp domains and residues implicated in drug recognition and binding. For instance, photoaffinity labeling, the identification of spontaneously occurring Pgp mutations, and site-directed mutagenesis revealed protein segments throughout the molecule important for substrate specificity and transport (Gottesman et al., 1995; Kane, 1996). Mutations affecting substrate recognition by Pgp were predominantly found in the TMDs, with the most relevant mutations in the TMD5/6 and TMD11/12 (Gottesman et al., 1995; Kane, 1996). For instance, the TMD6 G338A/A339P double mutation in hamster Pgp (Devine et al., 1992) and mutations in human Pgp such as the V338A or G341V (Loo and Clarke, 1996) exchange result in altered drug specificity. Recently, mutagenesis of TMD11 of mouse mdr3 by alanine scanning confirmed a contribution of this TMD to substrate recognition and binding (Hanna et al., 1996).
The mutational analysis of the NBDs of the mouse mdr1 and mdr3 homologues of human Pgp revealed the importance of NBDs for transporter function (Azzaria et al., 1989; Beaudet and Gros, 1995). Interestingly, both the N-terminal and the C-terminal NBDs containing the Walker A, Walker B, and the ABC protein consensus motifs appear to influence substrate specificity (Hoof et al., 1994; Beaudet and Gros, 1995). Furthermore, replacements of the N-terminal K433 and/or the C-terminal K1076 to M in the Walker A motif of human Pgp were shown to abolish the drug-stimulated ATPase activity (Mueller et al., 1996). However, the photoaffinity label 8-azido-ATP retained its ability to bind to the mutant Pgp molecules, albeit with a decreased affinity for the double lysine mutant (Mueller et al., 1996). The results indicate that both NBDs may act in cooperation, and that a distinct step after ATP binding is impaired in these Pgp mutants.
In the yeast S. cerevisiae, a variety of different ABC transporters were identified by molecular cloning approaches or through the yeast genome sequencing project (Egner et al., 1995a; Decottignies and Goffeau, 1997; Kuchler and Egner, 1997). Functional homologues of mammalian Pgps and MRP in S. cerevisiae include plasma membrane ABC efflux pumps such as Pdr5 (Balzi et al., 1994; Bissinger and Kuchler, 1994), Snq2 (Servos et al., 1993), and Yor1 (Katzmann et al., 1995; Zhifeng et al., 1996), respectively. However, despite their high homology, Pdr5 and Snq2 mediate resistance to a distinct subset of compounds with only little overlap in substrate specificity (Balzi et al., 1994; Bissinger and Kuchler, 1994; Hirata et al., 1994). The structural domains of Pdr5 and Snq2 that determine substrate specificity and the substrate recognition and transport mechanisms are unknown. To date, site-directed mutagenesis has been used to analyze structure-function relationships of eukaryotic ABC transporters (Kane, 1996). In this study, we used a combination of random in vitro mutagenesis and a genetic screen to isolate mutant Pdr5 transporters with altered substrate specificity. This approach identified several Pdr5 residues whose mutational change led to a drastic alteration of drug substrate specificity. Strikingly, mutations were not only identified in predicted TMDs, but also in each of the two most highly conserved NBDs and in the predicted extracellular loop 6. Our results also show that it is possible to modulate Pdr5 substrate specificity to a large extent, and they imply that the folded structure of eukaryotic ABC multidrug transporters may represent a primary determinant of their broad drug substrate specificity. Furthermore, our data suggest a novel strategy for generating custom-made resistance efflux pumps with limited substrate specificity and sensitivity to inhibitors and perhaps even with enhanced drug transport capacity. This may be of therapeutic importance for certain mammalian multidrug resistance genes that could be used in gene therapy applications to achieve chemoprotection of certain cells or tissues.
MATERIALS AND METHODS
Media and Yeast Strains
Rich medium (YPD), synthetic complete, and selective (drop-out) media supplemented with auxotrophic components to maintain plasmids were prepared essentially as described elsewhere (Rose et al., 1990). All S. cerevisiae strains used in this study are listed in Table 1. Strain YNK410 was derived from YPH499 in two steps. First, the PDR5 gene was disrupted by a functional LEU2 gene using the DNA insert of the plasmid pTCA/lem1::LEU2 (Kralli et al., 1995). Next, the LEU2 gene at the PDR5 locus was disrupted by the Escherichia coli LacZ gene under the control of the CYC1 minimal promoter linked to three glucocorticoid response elements (GT3Z gene).
Table 1.
Genotypes of S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| YPH499 | MATa ura3-52 his3-Δ200 leu2-Δ1 trp1-Δ63 lys2-801amb ade2-101oc | Sikorski and Hieter (1989) |
| YPH500 | MATα (otherwise isogenic to YPH499) | Sikorski and Hieter (1989) |
| YKKB-13 | YPH500 Δpdr5::TRP1 | Bissinger and Kuchler (1994) |
| YYM5 | YPH500 Δsnq2::hisG | Mahé et al. (1996a) |
| YYM3 | YPH500 Δpdr5::TRP1 Δsnq2::hisG | Mahé et al. (1996a) |
| YYM4 | MATa (otherwise isogenic to YYM3) | This study |
| YB107 | YPH499 fpr1-2::ADE2 | Benton et al. (1994) |
| YB169A | YPH499 fpr2-1::URA3 | Benton et al. (1994) |
| YB105 | YPH500 fpr3-2::HIS3 | Benton et al. (1994) |
| YB171A | YPH499 fpr1-2::ADE2 fpr2-1::URA3 fpr3-2::HIS3 | Benton et al. (1994) |
| YNK410 | MATa Δpdr5::GT3Z (otherwise isogenic to YPH499) | This study |
Drug Susceptibility Assays
Drug susceptibility to different compounds of yeast strains was tested by spotting serial dilutions of exponentially growing cell cultures from liquid YPD or selective medium onto appropriate drug-containing plates (Bissinger and Kuchler, 1994). Agar plates containing different drugs at various concentrations were prepared by adding the substances from stock solutions to 10 ml of YPD or selective agar equilibrated at 50°C before pouring into Petri dishes. Serial dilutions of exponentially growing (A600 of 1) cells were prepared by diluting the cultures to 0.2 A600 and further down to an A600 of 0.02 and 0.002 in sterile water. Five microliters of each dilution were spotted onto the appropriate plates and growth was monitored after incubation of the cells for 2 d at 30°C in the dark. Quantitative determination of the minimal inhibitory concentration (MIC) values for various drugs were performed in microtiter plates with selective medium essentially as described previously (Sanglard et al., 1995). The MIC score was defined as the lowest drug concentration at which a significant inhibition of cell growth could be observed (Sanglard et al., 1995). The drugs fluconazole (Pfitzer U.K., Sandwich, United Kingdom), ketoconazole, and itraconazole (Janssen Pharmaceuticals, Beerse, Belgium), and FK506 (Fujisawa USA, Melrose Park, IL) used in this study were dissolved in dimethyl sulfoxide. The dyes rhodamine 123 and rhodamine 6G (Sigma Chemical, St. Louis, MO) were dissolved in ethanol, whereas the cycloheximide (Sigma) stock solution was prepared with sterile water.
Flow Cytometry and Fluorescence Microscopy of Yeast Cells
For flow cytometry analysis, rhodamine 6G was added at a final concentration of 2.5 μg/ml to yeast cells (1 A600) grown in selective medium lacking leucine. Rhodamine 6G loading of cells was performed by a 60-min incubation at 30°C with or without preincubation for 20 min with 10 μg/ml FK506. Finally, cells were diluted 1:10 in ice-cold water, and data were collected and analyzed with a Becton Dickinson FACScan and Lysis II (Cell Quest) software.
For fluorescence microscopy of rhodamine 123, yeast strains YPH500 (PDR5) and YKKB-13 (Δpdr5) were grown to 2 A600 units in YPD. Equivalents of 3 A600 of cells were harvested, washed once with labeling buffer (2% glucose, 50 mM KPi, pH 7.5) and resuspended in 1.5 ml of labeling buffer containing 50 μM rhodamine 123 from a 10-mM stock solution. Cells were incubated at 30°C and shaken for 10 min, washed quickly twice with 1.5 ml of labeling buffer lacking rhodamine 123, and resuspended again in 1.5 ml of labeling buffer. At indicated time points, 10-μl samples were washed once with cold labeling buffer containing 10 mM NaN3. Intracellular rhodamine 123 fluorescence was monitored microscopically with a Zeiss Axiovert 10 fluorescence microscope equipped with an appropriate filter set. Photomicrographs were taken with a Fuji Provia 1600 film (RSP135) sensitivity-pushed to ASA 1600 using a 1-s exposure time.
Hydroxylamine Mutagenesis and Screening for Mutant Pdr5 Transporters
The PDR5 plasmid pCKSF1 (CEN6 LEU2; Bissinger and Kuchler, 1994) was randomly mutagenized in vitro according to a previously published method (Rose and Fink, 1987). Briefly, 10 μg of plasmid DNA were dissolved in 0.5 ml of hydroxylamine solution (90 mg NaOH, 350 mg hydroxylamine-HCl in 5 ml water, pH about 6.5, freshly made up before use) and incubated for 20 h at 37°C. The reaction was terminated by adding 10 μl of 5 M NaCl, 50 μl bovine serum albumin (1 mg/ml), and 1 ml of ethanol. After precipitation at −70°C, the plasmid DNA was reprecipitated three times with ethanol. Finally, the pool of mutagenized plasmid DNA was dissolved in water and used for yeast transformations.
The strain YYM4 (Δpdr5 Δsnq2) was transformed with the mutagenized plasmid pCKSF1 by the Li-acetate method (Rose et al., 1990). About 3000 LEU+ transformants were tested for their ability to complement the Δpdr5 drug hypersensitivity by replica plating onto solid selective medium lacking leucine and containing 75 ng/ml cycloheximide. Growth was monitored after incubation of the cells for 3 d at 30°C.
A total of 102 transformants that failed to complement the Δpdr5 drug hypersensitivity phenotype were selected and further analyzed for Pdr5 protein expression by immunoblotting using a polyclonal rabbit anti-Pdr5 antiserum (Egner et al., 1996). Out of the 102 selected transformants, 12 showed no expressed Pdr5 protein and 20 transformants expressed a truncated Pdr5 variant. The Pdr5 protein levels were severely reduced in 35 candidates, whereas 35 other transformants expressed wild-type or nearly wild-type amounts of Pdr5. The latter 35 transformants were further characterized by testing their drug sensitivity using the compounds rhodamine 123 (75 μg/ml), fluconazole (2 μg/ml), ketoconazole (250 ng/ml), and itraconazole (100 ng/ml). On the basis of these drug tests, six candidate mutant genes were selected for further characterization. The corresponding plasmids were named pCKSF1–14, -26, -46, -57, -71, and -127, recovered from yeast, and used to transform E. coli for their isolation. Plasmids were then transformed back into yeast strain YYM4 to verify the observed drug-resistance profile. The DNA sequences of mutagenized PDR5 genes in the respective plasmids were determined by the Sanger dideoxy chain termination method (Sanger et al., 1977) using a set of primers covering the entire PDR5 gene.
Site-directed Mutagenesis and Construction of Mutant PDR5 Genes
Mutations within the original PDR5 plasmids pCKSF1–14, -26, -46, -57, -71, and -127 were further analyzed by subcloning to determine the actual mutation causing the altered drug resistance phenotype of each mutant transporter. To do this, segments of the wild-type PDR5 gene in pCKSF1 were replaced by the corresponding fragments of the mutated DNA. Mutations within the N-terminal half of Pdr5 were separated from others by inserting the 1377-bp HindIII–SalI fragment obtained from PDR5-14, -71, -46, -57, -127, and the 649-bp SalI–Bst1107I fragments isolated from PDR5-14 and PDR5-26, into pCKSF1, thereby replacing the corresponding fragments in the wild-type PDR5 gene. To separate mutations within TMD6 of Pdr5–127 and within the C-terminal half of the Pdr5 mutants, nucleotide substitutions were introduced into wild- type PDR5 by site-directed mutagenesis (Chameleon double-stranded site-directed mutagenesis kit, Stratagene, La Jolla, CA). Relevant nucleotide exchanges are depicted in Table 2. Nucleotide exchanges to alter codons and silent exchanges generating novel restriction sites are underlined. Modified endonuclease restriction sites are indicated in bold letters. The successful introduction of mutational changes in the constructs was verified by DNA sequencing.
Table 2.
Oligonucleotides used in site-directed mutagenesis
| Mutation | Sequencea–c |
|---|---|
| MscI | |
| PDR5-26C1427Y | 5′-GGTATGACATATGGCCAGTACATGG-3′ |
| PDR5-46G905S/G908S | 5′-CAGCTTTAATGAGTGCTTCAAGTGCTGG-3′ |
| PDR5-57G1009C | 5′-GTTGCTGGTGAATGTTTAAACG-3′ |
| Eco47III | |
| PDR5-127 S1360F | 5′-GCAGAAAGCGCTGCTAACTTAGCCTTTTTGTTG-3′ |
All oligonucleotides are in the sense orientation.
Underlined, modified nucleotides.
Bold, modified or newly generated endonuclease restriction sites.
Subcellular Fractionation
Subcellular fractionation of yeast cells was performed according to a previously published protocol (Egner et al., 1996) with the following modifications. Cells were grown exponentially (A600 of 1) in selective medium lacking leucine. Cells corresponding to 100 A600 units were harvested by centrifugation at 3700 × g for 10 min. The pellet was washed once with cold 10 mM NaN3 and resuspended in 2 ml of chilled lysis buffer (0.8 M sorbitol, 10 mM 3-(N-morpholino)propanesulfonic acid, pH 7.2, 1 mM EDTA, 1 mM phenylmethane sulfonylfluoride, 5 mM N-ethylmaleimide) and an equal volume of cold glass beads was added. Cell lysis was achieved by vortexing the suspension five times for 1 min, with 1 min on ice in between. The lysate was spun for 10 min at 2000 × g to remove unlysed cells and debris. The resulting supernatant (1.5 ml) was layered onto a step gradient containing 1 ml each of 12, 18, 24, 30, 36, 42, 48, and 54% sucrose, and a final 0.5 ml step of 60% sucrose (wt/vol) in lysis buffer. The gradient was spun for 3 h at 150,000 × g (34,000 rpm) in an SW-40 Ti rotor (Beckman Instruments, Fullerton, CA) at 4°C, and 0.7-ml or 1.0-ml fractions, respectively, were harvested from bottom to top. Immunoblot analysis of the fractions with polyclonal antisera raised against Pdr5, Pma1, DPAP A, and Sec63 was performed as previously described (Egner et al., 1996).
Indirect Immunofluorescence
Immunofluorescence of yeast cells was carried out as described elsewhere (Kuchler et al., 1993) using the following modifications. Cells were grown in selective medium lacking leucine to an A600 of about 1 and fixed by adding formaldehyde to a final concentration of 5% (vol/vol). After an overnight incubation at 30°C in 5% formaldehyde, 50 mM MgCl2, and 100 mM KPi (pH 6.5), about 10 A600 units of cells were harvested by centrifugation and washed in 1 ml of solution of 100 mM Tris-HCl (pH 7.6), and 10 mM EDTA. Cells were then incubated for 10 min at room temperature in 0.6 ml 0.1 M Tris-H2SO4 (pH 9.4) and 2% β-mercaptoethanol and washed with spheroplasting buffer (0.1 M sodium citrate, pH 7.0, 60 mM EDTA, 1 M sorbitol). Cell walls were removed by incubation of the cells in 1 ml of spheroplasting buffer containing 25 mM β-mercaptoethanol and 70 μg/ml Zymolyase-100T (Seikagaku Kogyo Co., Tokyo, Japan) for 45 min at 30°C with gentle shaking. Spheroplasts were washed twice with 1 ml of solution of 50 mM Tris-HCl (pH 7.6), 20 mM EDTA, 1.2 M sorbitol, and twice with 1 ml of phosphate-buffered saline (PBS). Finally, the spheroplasts were suspended to an A600 of 2 in PBS. Aliquots of 30-μl spheroplast suspensions were applied to polyethylenimine-pretreated slides. After a 30-min incubation, the unbound spheroplasts were removed by washing six times with PBS. Slides were blocked for 10 min with blocking buffer (PBS, 0.025% Nonidet-P-40; 5% bovine serum albumin) prior to antibody incubation for 90 min with anti-Pdr5 antibodies diluted in blocking buffer. Slides were washed 15 times with PBS before decoration with the fluorescein isothiocyanate (FITC)-conjugated secondary antibodies (goat anti-rabbit IgG, Oncogene Science, Manhasset, NY) also diluted in blocking buffer. After a 90-min incubation, the slides were washed 15 times with PBS, stained for 5 min with 4,6-diamino-2-phenylindole hydrochloride (DAPI) (1 μg/ml in PBS), and finally washed again six times with PBS. Staining of Pdr5 was visualized with a Zeiss Axiovert 10 fluorescence microscope equipped with an appropriate FITC filter set. Photomicrographs were taken with a Kodak TMY400 black and white film using a 2-s exposure time.
Estradiol Accumulation and Dexamethasone Response Assays
Estradiol accumulation assays in yeast cells were performed as outlined previously using minor modifications (Mahéet al., 1996a). Cells were cultivated to exponential growth phase (1.5 A600) at 30°C in 5 ml of drop-out medium lacking leucine. [3H]-estradiol (151 Ci/mmol, Amersham, Arlington Heights, IL) was added to a final concentration of 2 nM and cells were incubated at 30°C with shaking. At indicated time points, 1-ml samples were harvested by a brief centrifugation (13,000 × g, 30 s), and the cell pellet was washed four times with 1 ml of cold PBS, 2% glucose, and 2 μM unlabeled estradiol and finally resuspended in 50 μl of water. [3H]-estradiol uptake was determined by liquid scintillation counting in a Packard 1600 TR Tri-Carb counter.
Cellular response to dexamethasone was monitored by the production of β-galactosidase (β-gal) from the reporter gene GT3Z genomically integrated in strain YNK410 (Kralli et al., 1995). Yeast cultures (2 ml) of the β-gal reporter strain YNK410 carrying both the glucocorticoid receptor expression plasmid pRS314-GN795 (Lefstin et al., 1994) and Pdr5 mutant plasmids, as indicated, were grown to saturation in selective media at 30°C. Cultures were diluted 1:20 in fresh selective media (200 μl) containing dexamethasone or the carrier ethanol and grown for an additional 12 h in 96-well microtiter plates at 30°C. Dexamethasone-dependent β-galactosidase activity was measured essentially as descibed elsewhere (Iniguez-Lluhi et al., 1997).
RESULTS
Previous structure-function studies on mammalian Pgps and several naturally occurring Pgp mutations identified protein segments and amino acid residues implicated in drug transport (Gottesman et al., 1995; Kane, 1996). To identify such domains in yeast Pdr5, we subjected the entire PDR5 gene to random in vitro mutagenesis and screened for mutants conferring altered drug resistance to selected Pdr5 substrates.
Substrates of the Pdr5 Multidrug Transporter
The Pdr5 ABC transporter can transport a wide variety of substrates including the protein synthesis inhibitor cycloheximide (Balzi et al., 1994), the mycotoxin sporidesmin (Bissinger and Kuchler, 1994), azole antifungals such as fluconazole, ketoconazole, and itraconazole (Sanglard et al., 1995), the fluorescent compounds rhodamine 123 and rhodamine 6G (Kolaczkowski et al., 1996), cationic lipophilic dyes that specifically stain the mitochondria of living cells, and steroids such as dexamethasone (Kralli et al., 1995) and estradiol (Mahéet al., 1996a). Cycloheximide (Balzi et al., 1994; Bissinger and Kuchler, 1994; Hirata et al., 1994; Mahéet al., 1996b), fluconazole, itraconazole, and ketoconazole (Sanglard et al., 1995) were previously shown to be substrates for Pdr5, but not for Snq2.
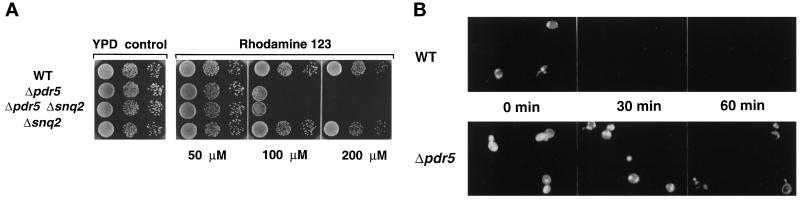
To examine rhodamine 123 toxicity in yeast cells lacking Pdr5, Snq2, or both ABC transporters, cell growth was monitored on agar plates containing increasing concentrations of rhodamine 123 (Figure 1A). The wild-type strain and the Δsnq2 mutant strain, exposed to rhodamine 123 concentrations up to 200 μM, showed no significant growth inhibition when compared with the YPD control. In contrast, growth of the Δpdr5 mutant and the Δpdr5 Δsnq2 double deletion strain was strongly inhibited by 100 μM rhodamine 123, and both strains were unable to grow at 200 μM rhodamine 123.
Figure 1.
(A) Rhodamine 123 drug susceptibility assay of Δpdr5 and Δsnq2 null mutants. Cells of the wild-type strain and of the isogenic Δpdr5, Δsnq2, and Δpdr5 Δsnq2 null mutants were grown to exponential growth phase (A600 of 1) in YPD. The cultures were diluted to an A600 of 0.2, and 5 μl of a serial dilution (100 to 10−2) were spotted onto YPD plates containing the indicated rhodamine 123 concentrations. The plates were incubated at 30°C for 48 h in the dark. (B) Rhodamine 123 efflux in wild-type and Δpdr5 null mutant cells. Exponentially growing cells were labeled with 50 μM rhodamine 123 for 10 min and, after washing of the cells, intracellular rhodamine 123 fluorescence was monitored at the time points indicated.
Fluorescence microscopy was used to directly visualize the enhanced accumulation of rhodamine 123 in the Δpdr5 mutant compared with the isogenic wild-type strain (Figure 1B). Cells were preincubated for 10 min with 50 μM rhodamine 123, and cellular release of the fluorescence dye was monitored after 0, 30, and 60 min. Interestingly, wild-type cells showed a reduced steady state accumulation of the dye compared with the Δpdr5 cells (Figure 1B, 0 min). The rhodamine 123 staining of wild-type cells rapidly decreased with no detectable fluorescence signal after 30 min. In contrast, in Δpdr5 cells rhodamine 123 staining was observed even 60 min after dye loading of the cells. The enhanced accumulation of rhodamine 123 in Δpdr5 cells (Figure 1B) and the growth hypersensitivity of cells lacking Pdr5 on rhodamine 123 containing medium (Figure 1A) clearly showed that rhodamine efflux across the yeast plasma membrane is mediated by Pdr5, but not by the related ABC transporter Snq2, confirming the distinct substrate specificity of Pdr5 and Snq2 (Balzi and Goffeau, 1994; Bissinger and Kuchler, 1994; Hirata et al., 1994). The structurally unrelated Pdr5 substrates cycloheximide, rhodamines, and antifungal azoles were further used in the genetic screen for the selection of mutant Pdr5 transporters after random in vitro mutagenesis of PDR5.
Random In Vitro Mutagenesis of PDR5 and Mutant Screen
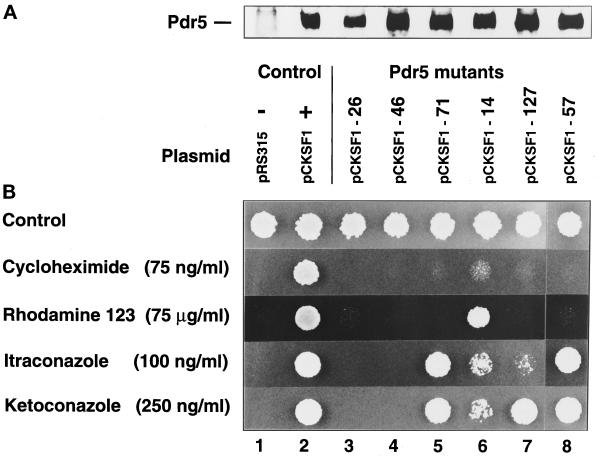
A library of hydroxylamine-mutagenized CEN-based plasmids derived from the PDR5 plasmid pCKSF1 (Bissinger and Kuchler, 1994) was transformed into the drug-sensitive Δpdr5 Δsnq2 strain YYM4, followed by phenotypic selection of Pdr5 mutants conferring altered drug resistance patterns. About 3000 LEU+ transformants were screened for their ability to grow in the presence of 75 ng/ml cycloheximide, a drug concentration that normally allowed growth of cells expressing wild-type Pdr5. Thirty-five transformants that failed to grow, but still expressed nearly wild-type Pdr5 protein levels, were tested for their sensitivity to other structurally unrelated Pdr5 substrates, including rhodamine 123, and the antifungal azoles fluconazole, ketoconazole, and itraconazole (Sanglard et al., 1995). Plasmids were recovered from six individual transformants that exhibited altered drug resistance profiles and named pCKSF1–14, -26, -46, -57, -71, and -127. To verify the corresponding drug resistance phenotype, these plasmids were transformed back into the drug- sensitive yeast strain YYM4. The resulting transformants were tested for levels of Pdr5 expression and their pattern of resistance to different drugs (Figure 2).
Figure 2.
(A) Protein expression levels of wild-type and mutant Pdr5 transporters. Yeast strain YYM4 (Δpdr5 Δsnq2) was transformed with either wild-type (pCKSF1, +) PDR5 plasmid, control plasmid (pRS315, −), or individual PDR5 mutant plasmids. Protein extract of each transformant equivalent to 0.4 A600 units of cells was prepared, separated by SDS-PAGE through a 7.5% gel, and transferred to nitrocellulose membranes. Immunoblots were incubated with anti-Pdr5 antiserum to visualize the relative amounts of wild-type versus mutant Pdr5 transporters. (B) Drug specificity of wild-type and mutant Pdr5 transporters. Cells of strain YYM4, bearing wild-type or different mutant Pdr5 transporters as indicated, were grown to exponential growth phase (A600 of 1) in selective medium lacking leucine. The cultures were diluted to an A600 of 0.02 and 5 μl of the cell suspensions were spotted on drop-out minus leucine plates containing the desired antifungal agents and incubated at 30°C for 48 h.
As can be seen in Figure 2A, all other Pdr5 mutants except Pdr5–26 were expressed at nearly identical levels, when compared with the expression of the wild-type control. As expected, no Pdr5 protein was detectable in the empty vector control. The spot test on drug plates in Figure 2B shows the drug resistance profile of cells expressing Pdr5 mutant variants. All cells expressing mutant Pdr5 proteins grew on the synthetic complete medium at the same rate as the wild-type control. In contrast, cells with the control vector (pRS315) were unable to grow on plates containing the drugs at the indicated concentrations, whereas the Pdr5 wild-type cells (pCKSF1) grew as on the control medium (Figure 2B). The resistance phenotypes of cells expressing Pdr5 mutants could be grouped according to their different phenotypes. First, growth of cells carrying the mutant PDR5 plasmids pCKSF1–26 and -46 was strongly inhibited on all drugs tested. Second, cells expressing the mutants Pdr5–71 and Pdr5–57 were only able to grow in the presence of itraconazole and ketoconazoles, but no longer on other drugs such as cycloheximide or rhodamine 123. Third, cells with the mutant Pdr5–14 grew on rhodamine 123 only, whereas growth in the presence of antifungal azoles at the indicated concentrations was clearly reduced and nearly abolished for cycloheximide. Finally, cells with mutant Pdr5–127 were able to grow on ketoconazole medium but not on media containing the related compound itraconazole, or rhodamine 123 and cycloheximide. Taken together, the isolated Pdr5 mutants comprised apparently nonfunctional Pdr5 transporters (Pdr5–26 and -46) and mutant Pdr5 transporters with restricted drug transport properties (Pdr5–14, Pdr5–57, Pdr5–71, and Pdr5–127). Interestingly, the relative resistance conferred by Pdr5–14 and Pdr5–71 for rhodamine 123 and azoles (itraconazole and ketoconazole) was reversed, indicating a switch in substrate utilization between the mutants Pdr5–14 and Pdr5–71.
Determination of the Minimal Inhibitory Concentrations of the Pdr5 Mutants
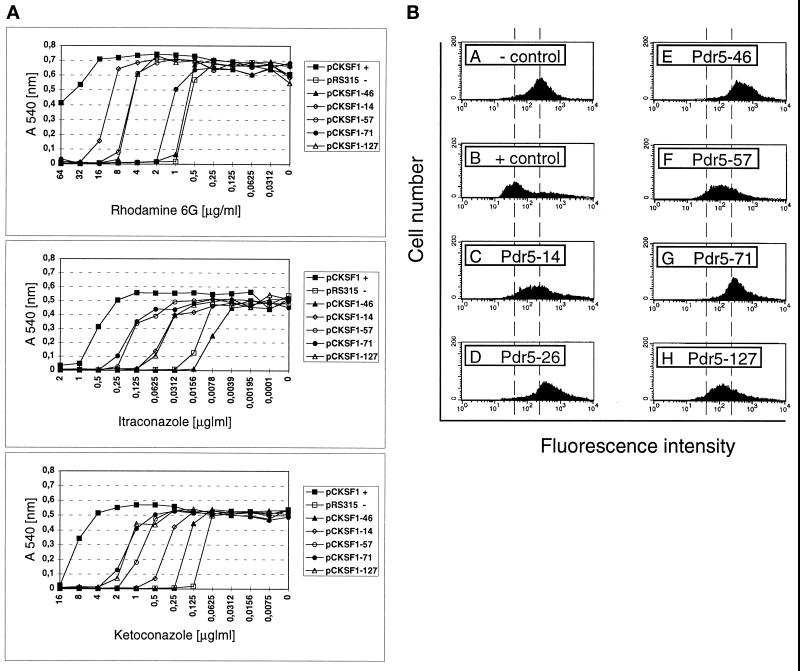
The drug susceptibility was further quantified by determining the MIC for Pdr5 mutants using a microtiter plate assay (Sanglard et al., 1995). The MIC values were determined for itraconazole, ketoconazole, and rhodamine 6G (Figure 3A and Table 3). The latter compound displayed a similar resistance profile for the Pdr5 mutants as rhodamine 123 at a concentration of 10 μg/ml (our unpublished results), indicating that substrate recognition and transport by Pdr5 is quite similar for both rhodamines. First, presumably because the initial screen was performed for loss of cycloheximide resistance, all isolated Pdr5 mutants conferred reduced resistance to the tested compounds when compared with cells expressing wild-type Pdr5. The MIC values obtained for the cells carrying mutant PDR5 plasmids pCKSF1–46, -14, -57, -71, and -127 were consistent with the results obtained by the spot tests (see Figure 2B), although cells expressing Pdr5–57 and Pdr5–127 showed a slightly higher resistance to rhodamines than observed in the spot test. Moreover, the MIC values clearly demonstrated a switch in the substrate specificity for rhodamines and azoles between the mutant transporters Pdr5–14 and Pdr5–71, respectively. Furthermore, mutant Pdr5–57 exhibited a similar resistance pattern as Pdr5–71 for the azole compounds but not for rhodamine 6G (Figure 3A and Table 3).
Figure 3.
(A) Susceptibilities of wild-type Pdr5 cells and Pdr5 mutants to rhodamine 6G and azole derivatives. Cells of strain YYM4, transformed with the wild-type PDR5 plasmid pCKSF1, the control plasmid pRS315, or the mutant plasmids as indicated, were grown overnight in selective medium and subjected to the MIC assay by the microdilution methods described in MATERIALS AND METHODS. The optical density (A540) of each well on the microtiter plate was determined after a 48-h incubation at 30°C. Each value was plotted against its corresponding drug concentration. (B) Rhodamine 6G accumulation in cells expressing Pdr5 or mutant Pdr5 transporters. Rhodamine 6G accumulation was determined by flow cytometry after preloading of Pdr5 wild-type cells (+ control, pCKSF1) and cells bearing the mutant transporters Pdr5–14, -26, -46, -57, -71, and -127 and control cells (− control, pRS315) with rhodamine 6G at a concentration of 2.5 μg/ml.
Table 3.
MIC values for cells carrying wild-type Pdr5 or mutant Pdr5 transporters
| Plasmid | Rhodamine 6G | Itraconazole | Ketoconazole |
|---|---|---|---|
| pCKSF1 (+) | >32 (>32×) | 1 (64×) | 16 (128×) |
| pRS315 (−) | 1(1×) | 0.0156(1×) | 0.125 (1×) |
| pCKSF1-46 | 1(1×) | 0.0156(1×) | 0.25(2×) |
| pCKSF1-14 | 16(16×) | 0.0625(4×) | 0.50(4×) |
| pCKSF1-57 | 8(8×) | 0.25(16×) | 1(8×) |
| pCKSF1-71 | 2(2×) | 0.25(16×) | 2(16×) |
| pCKSF1-127 | 8(8×) | 0.0625(4×) | 2(16×) |
MIC values (μg/ml) were determined from the drug susceptibilities of the individual transformants as indicated in Figure 3A. The relative drug resistance is expressed as fold resistance over background level [pRS315 (−)].
Pdr5-mediated Rhodamine 6G Efflux in Pdr5 Mutant Cells
The intracellular accumulation of rhodamine 6G (Figure 3B) was also determined by flow cytometry analysis of dye preloaded cells expressing wild-type Pdr5 (+ control, pCKSF1), the different Pdr5 mutants (-26, -46, -14, -57, -71, and -127), and cells lacking Pdr5 (− control, pRS315). Cells expressing mutant Pdr5–14, -57, and -127 yielded an increased fluorescence intensity when compared with the rhodamine 6G efflux mediated by wild-type Pdr5, demonstrating their reduced rhodamine 6G efflux capacity. In contrast, the increase in the fluorescence signal of cells expressing mutant Pdr5–26, -46, and -71 was similar or even slightly enhanced when compared with control cells lacking Pdr5, suggesting a lack of rhodamine export capacity in these mutant Pdr5 transporters (Figure 3B). The data for rhodamine 6G efflux obtained from the flow cytometry experiments are in good agreement with the measured MIC values (Figure 3A).
Cellular Localization of Mutant Pdr5 Transporters
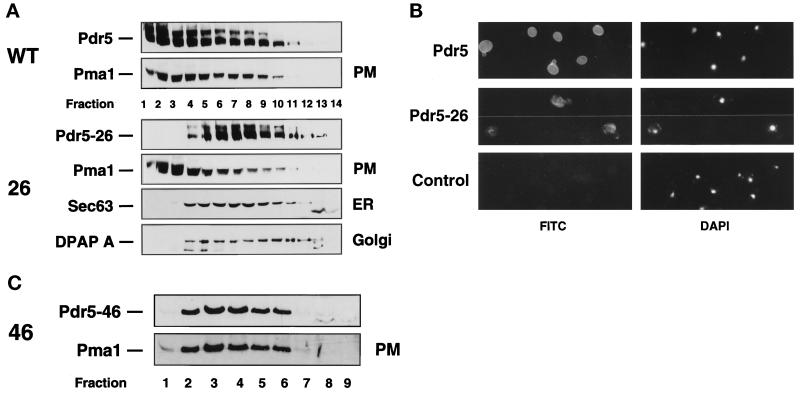
To determine the subcellular localization of the mutant Pdr5 transporters, sucrose gradient fractionation of cell lysates was performed (Egner et al., 1995b). The mutant transporters with limited substrate specificity such as Pdr5–14, -57, -71, and -127 cofractionated in the sucrose gradient with the Pma1 plasma membrane H+-ATPase. Their sedimentation was indistinguishable from that of wild-type Pdr5 (our unpublished results). Conversely, Pdr5–26, which conferred no resistance to any of the drugs tested (Figure 2B), did not cofractionate with the plasma membrane markers Pma1, wild-type Pdr5, or the Golgi membrane marker dipeptidylaminopeptidase DPAP A (Figure 4A). Instead, Pdr5–26 mainly peaked in fractions 5–9, the fractions where the endoplasmic reticulum (ER) marker Sec63 was also predominantly found. The results indicate that mutant transporters Pdr5–14, -57, -71, and -127 were correctly localized to the plasma membrane, whereas cell surface targeting of Pdr5–26 through the secretory pathway was blocked, leading to the accumulation of Pdr5–26 in the ER membrane. The ER localization of Pdr5–26 was also shown by indirect immunofluorescence (Figure 4B). All cells expressing Pdr5–26 clearly showed an intracelluar perinuclear fluorescence staining typical for yeast ER membrane proteins. In contrast and as shown before (Egner et al., 1995b), wild-type Pdr5 was present predominantly at the cell surface (Figure 4B). However, the second nonfunctional mutant transporter isolated, Pdr5–46, was correctly localized to the plasma membrane. Sucrose gradient fractionation revealed a step by step cofractionation of Pdr5–46 with the Pma1 plasma membrane H+-ATPase (Figure 4C). Hence, the apparent lack of drug resistance in cells expressing Pdr5–26 or Pdr5–46 is most likely caused by impaired plasma membrane targeting or loss of transporter function, respectively.
Figure 4.
(A) Subcellular distribution of wild- type Pdr5 and the mutant transporters Pdr5–26 and Pdr5–46. The supernatant of a 2000 × g centrifugation step obtained from a cell-free cell lysate, prepared as described in MATERIALS AND METHODS, was layered on a 12–60% (wt/vol) sucrose gradient. Membranes of subcellular organelles were separated during a 3-h spin at 150,000 × g. Aliquots of the gradient fractions were separated by SDS-PAGE on 7.5% gels. The distribution of Pdr5, mutant Pdr5–26, the plasma membrane marker Pma1, Golgi membrane marker DPAP A, and ER membrane marker Sec63 was analyzed by immunoblotting. Fractions (0.7 ml) were harvested from bottom (number 1) to top (number 14). (B) Indirect immunofluorescence of wild-type Pdr5 and the mutant Pdr5–26. YYM4 cells carrying wild-type PDR5, the mutant PDR5-26, or the control plasmid were prepared for indirect immunofluorescence as described in MATERIALS AND METHODS. Visualization of Pdr5 and Pdr5–26 was performed with polyclonal rabbit anti-Pdr5 antibodies and FITC-conjugated goat anti-rabbit IgG. Nuclear DNA was visualized by staining with DAPI. (C) Subcellular localization of Pdr5–46. The subcellular distribution of Pdr5–46 and the plasma membrane marker Pma1 was analyzed by immunoblotting of aliquots (1 ml fractions; bottom to top numbered 1–9) harvested after sucrose density gradient centrifugation of a cell lysate as described in A.
Mutant Pdr5–127 Is Insensitive to Inhibition by FK506
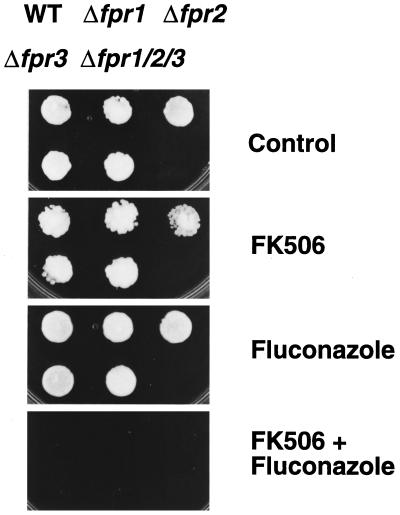
It was recently shown that the macrocyclic lactone and immunosuppressive agent FK506 is a inhibitor of Pdr5, since it blocks Pdr5-mediated dexamethasone transport (Kralli and Yamamoto, 1996). We therefore tested the inhibitory effect of FK506 on the transport activity of wild-type and mutant Pdr5 transporters. Since the mechanism of inhibition is unknown, we first determined whether the inhibitory FK506 effect on wild-type Pdr5 function is mediated by known FK506-binding proteins (FKBPs), the primary cellular targets of FK506 (Kunz and Hall, 1993). Thus, we tested Pdr5 activity and the reversing effect of FK506 on Pdr5 in yeast cells lacking the genes FPR1, FPR2, and FPR3 encoding FKBPs (Benton et al., 1994). First, resistance to fluconazole (Figure 5) or other azoles (our unpublished results) of wild-type, isogenic single Δfpr1, Δfpr2, and Δfpr3 and the isogenic Δfpr1 Δfpr2 Δfpr3 triple deletion strain was identical, showing that Pdr5-mediated resistance to the antifungals does not require tested FKBPs (Figure 5). Second, in the absence of any other drugs, FK506 alone was not toxic to these cells (Figure 5) or to Δpdr5 cells (Figure 6A). In contrast, FK506 rendered wild-type cells hypersensitive to fluconazole and ketoconazole, a phenotype normally caused by the lack of Pdr5 (Figure 5 and Figure 6). Notably, FK506 rendered all wild-type and single Δfpr1, Δfpr2, and Δfpr3, as well as the isogenic Δfpr1 Δfpr2 Δfpr3 triple mutant sensitive to fluconazole (Figure 5), ketoconazole, and itraconazole (our unpublished results), demonstrating that these FKBPs are not required for inhibition of Pdr5-mediated drug efflux. These data suggest that the immunosuppressive agent FK506 may exert its inhibitory function by direct interaction with Pdr5.
Figure 5.
Inhibition of Pdr5 by FK506 does not require the presence of the FKBPs Fpr1, Fpr2, and Fpr3. Cells of wild-type strain YPH499 (WT) and the isogenic null mutants YB107 (Δfpr1), YB169A (Δfpr2), YB105 (Δfpr3), and YB171A (Δfpr1 Δfpr2 Δfpr3) were spotted on YPD plates containing no drugs (control), 10 μg/ml FK506, 1.5 μg/ml fluconazole, or a combination of both compounds as indicated. Cell growth was monitored after 48 h of incubation at 30°C.
Figure 6.
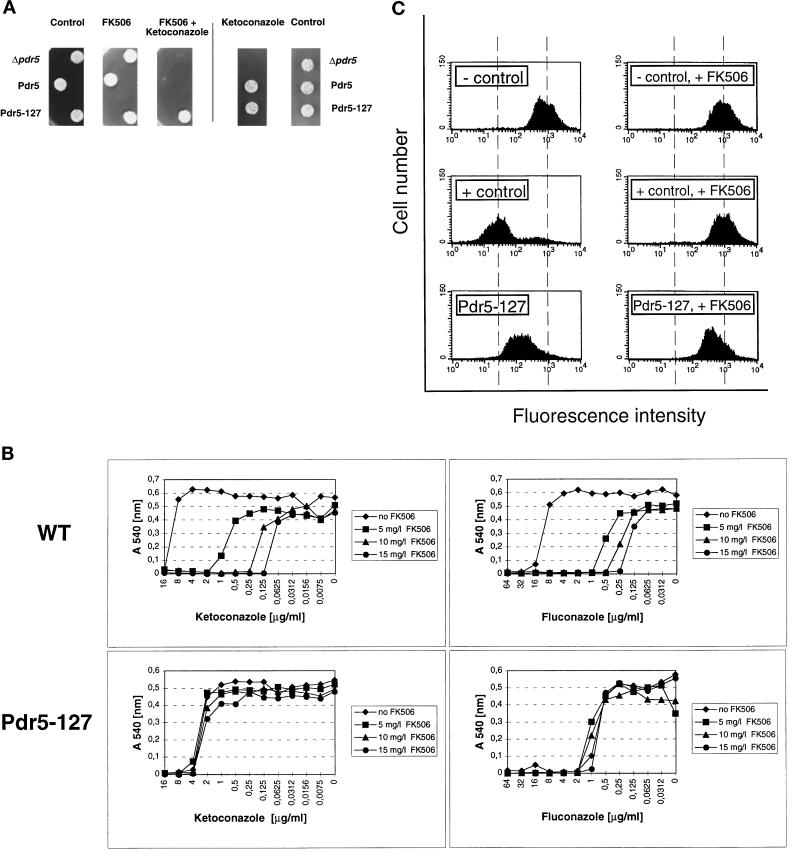
(A) Pdr5 drug transport activity is inhibited by FK506, but not in the mutant Pdr5–127. Cells of strain YYM4 (Δpdr5 Δsnq2) and YYM4 containing wild-type Pdr5 or mutant Pdr5-127 transporters were grown to exponential growth phase (A600 of 1) in selective medium lacking leucine. The cultures were diluted to an A600 of 0.02, and 5 μl were spotted onto appropriate plates containing 10 μg/ml FK506, 0.250 μg/ml ketoconazole, or a combination of both compounds, as indicated. Cell growth was monitored after a 48-h incubation at 30°C. (B) Susceptibilities of cells carrying wild-type Pdr5 or mutant transporter Pdr5–127 to FK506 inhibition. Cells of strain YYM4 with wild-type Pdr5 or the mutant Pdr5–127 were grown overnight in selective medium lacking leucine. The MIC assay was done by the microdilution method described in MATERIALS AND METHODS. The optical density (A540) of each well on the microtiter plate was determined after a 48-h incubation at 30°C. Each A540 value was plotted against the corresponding drug concentration. (C) Rhodamine 6G accumulation in cells expressing Pdr5 and Pdr5–127 in the presence of FK506. Wild-type Pdr5 cells (+ control, pCKSF1), Pdr5–127 expressing cells, and control cells lacking Pdr5 (− control, pRS315) were loaded for 60 min with rhodamine 6G (2.5 μg/ml) with or without FK506 (10 μg/ml) preincubation for 20 min. Relative intracellular rhodamine 6G accumulation was determined by flow cytometry.
Next, we determined the inhibitory effect of FK506 on cells with mutant Pdr5 transporters compared with wild-type Pdr5 and cells lacking Pdr5. From all mutants tested (our unpublished results), only mutant Pdr5–127 showed a lack of FK506 inhibition. As can be seen in Figure 6A, cells lacking Pdr5 were unable to grow in the presence of 0.250 μg/ml ketoconazole, whereas cells harboring wild-type Pdr5 exhibited no growth inhibition when compared with the control plate lacking drugs. Simultaneous exposure of wild-type Pdr5 cells to both ketoconazole and FK506 totally abolished growth, demonstrating inhibition of Pdr5-mediated ketoconazole transport by FK506. In addition, FK506 also reversed wild-type Pdr5-mediated drug resistance to other substrates such as fluconazole (see Figures 5 and 6B), rhodamines (see Figure 6C), itraconazole, and cycloheximide (our unpublished results). Interestingly, however, the mutant transporter Pdr5–127, which confers resistance to ketoconazole in the spot test (see Figure 2B), was not sensitive to inhibition by FK506 (Figure 6A). We also compared the ketoconazole and fluconazole MIC values of wild-type Pdr5 and mutant Pdr5–127 at different FK506 concentrations ranging from 0 to 15 μg/ml. Wild-type Pdr5-mediated ketoconazole and fluconazole resistance was strongly inhibited at 5 μg/ml FK506 (Figure 6B). The MIC values for cells carrying wild-type Pdr5 further decreased at FK506 concentrations of 10 and 15 μg/ml. By contrast, the MIC values of Pdr5–127 for ketoconazole were unchanged at all FK506 concentrations tested. Interestingly, the insensitivity of Pdr5–127 to FK506 inhibition was not exclusively restricted to ketoconazole transport, since the residual transport activity of Pdr5–127 for fluconazole could also not be blocked by FK506 (Figure 6B). Similarly, rhodamine 6G transport activity of Pdr5–127 showed a severely decreased susceptibly to FK506 inhibition when compared with wild-type Pdr5 (Figure 6C). Flow cytometry analysis showed that 10 μg/ml FK506 blocked rhodamine 6G efflux in wild-type Pdr5 cells, resulting in a dye accumulation as in cells lacking Pdr5. In contrast, the intracellular rhodamine 6G concentration in cells carrying Pdr5–127 was decreased in the presence of FK506 when compared with Δpdr5 cells, demonstrating a lack of Pdr5–127 inhihition by FK506. Taken together, our results imply that structural changes in the Pdr5–127 transporter could deteriorate or even prevent FK506–Pdr5 interaction.
Ability of Pdr5 Mutants to Transport Steroid Substrates
Pdr5 can also transport specific steroid substrates in vivo (Kralli et al., 1995; Kolaczkowski et al., 1996; Mahéet al., 1996a), thereby reducing the intracellular levels of such steroids and their ability to induce a cellular response (Kralli et al., 1995; Mahéet al., 1996a). Thus, we tested the ability of Pdr5 mutants to transport steroids using two different approaches. First, we measured the intracellular accumulation of estradiol, and second, the ability of dexamethasone to modulate the response of an appropriate reporter gene.
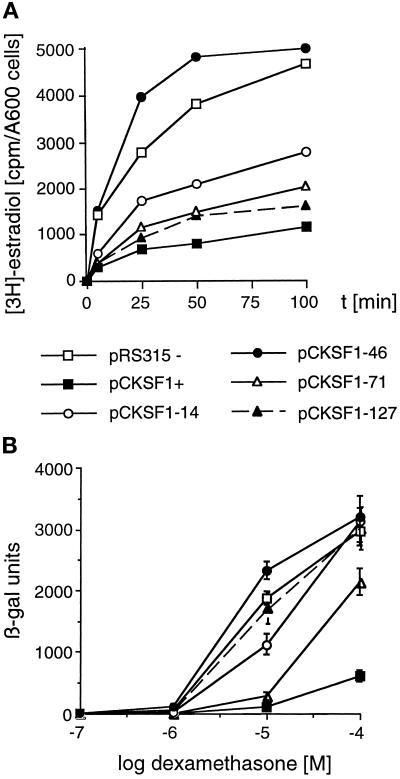
Incubation of YYM4 cells expressing wild-type Pdr5, the Pdr5 mutants 14, 46, 71, 127 or no Pdr5 with radiolabeled estradiol showed that cells carrying mutant 46 accumulated estradiol at levels compared with the ones in cells lacking Pdr5, indicating that Pdr5–46 is unable to transport estradiol. However, Pdr5–14, -71, and -127 accumulated intermediate to low levels of estradiol, suggesting they all retained their ability, albeit somewhat reduced when compared with wild-type Pdr5, to transport estradiol (Figure 7A). Estradiol accumulation in these cells was also measured in the presence of 10 μg/ml FK506. Similar to the inhibition of Pdr5-mediated azole transport, estradiol efflux was blocked by FK506 for wild-type Pdr5 and all Pdr5 mutants tested with the only exception of Pdr5–127 (our unpublished results).
Figure 7.
(A) Estradiol accumulation in the Δpdr5 Δsnq2 strain YYM4 expressing mutant Pdr5 transporters. Exponentially growing cells of YYM4 transformed with control plasmid pRS315 (−), wild-type PDR5 plasmid pCKSF1 (+), and individual plasmids expressing Pdr5 mutants (46, 14, 71, 127) were incubated with [3H]-estradiol. Samples were analyzed for intracellular steroid levels at the indicated time points. (B) Response of wild-type or mutant Pdr5 expressing cells to dexamethasone. Cells of strain YNK410 with the human glucocorticoid receptor expression plasmid pRS314-GN795 and either the control plasmid pRS315, the wild-type Pdr5 expressing plasmid pCKSF1, or the Pdr5 mutant plasmids as indicated, were treated with the indicated concentrations of dexamethasone for 12 h and assayed for β-galactosidase activity [β-galactosidase units are defined as described previously (Iniguez-Lluhi et al., 1997)]
The intracellular levels of dexamethasone were assessed by measuring the ability of dexamethasone to induce expression of a β-galactosidase reporter gene, which is under the control of glucocorticoid response elements. In strain YNK410 that expresses the mammalian glucocorticoid receptor, dexamethasone can induce β-galactosidase expression in a dose-dependent manner (Figure 7B). Introduction of wild-type Pdr5 renders cells resistant to dexamethasone; more than 20-fold higher levels of dexamethasone are required to induce a response. As seen in Figure 7B, Pdr5–46 confers no dexamethasone resistance, as the response was the same as in cells lacking Pdr5. Mutant 14 caused a small decrease in response, suggesting a very reduced ability to transport dexamethasone. Mutant 71, although not as efficient as wild-type Pdr5, conferred more than sevenfold increased resistance to dexamethasone. Surprisingly, mutant 127 showed a complete lack of dexamethasone transport, allowing as good a response as in cells lacking Pdr5.
On the basis of the different abilities of the Pdr5 mutants to lower estradiol levels or to decrease dexamethasone response, we conclude that Pdr5–46 does not transport either estradiol or dexamethasone. Mutant Pdr5–14 shows a severely reduced ability for steroid transport, especially for dexamethasone, whereas Pdr5–71 can transport both steroids. Interestingly, Pdr5–127 suggested that it retains its ability to transport estradiol, but has lost the ability to mediate transport of dexamethasone.
Amino Acid Substitutions in the Mutant Pdr5 Transporters
To identify the actual mutations responsible for the drug resistance phenotype (Table 4, compare with Figure 8), the mutated PDR5 genes were subjected to DNA sequencing. In most cases where multiple mutations were detected, they were separated by subcloning and/or replacement of corresponding DNA fragments in wild-type PDR5 or by site-directed mutagenesis of PDR5 (Table 2). The contribution of the respective single residue changes or clusters of mutations to the modulation of Pdr5 substrate specificity was examined by spot tests after transformation of strain YYM4 with the corresponding plasmids (our unpublished results).
Table 4.
Amino acid substitutions found in mutant Pdr5 transporters after random in vitro mutagenesis
| Mutant | Amino Acid Substitutiona | Domainb |
|---|---|---|
| Pdr5-14 | C199Y | NBD1, Walker A |
| A676V | TMD 5 | |
| T1460I | Extracellular loop 6 | |
| V1467I | Extracellular loop 6 | |
| Pdr5-26 | G557D | Extracellular loop 1 |
| A1398T | TMD 11 | |
| P1421S | Extracellular loop 6 | |
| C1427Y | Extracellular loop 6 | |
| Pdr5-46 | V149M | N-terminal cytoplasmic domain |
| G905S | NBD2, Walker A | |
| G908S | NBD2, Walker A | |
| Pdr5-57 | G138D | N-terminal cytoplasmic domain |
| G1009C | NBD2, C-motif | |
| Pdr5-71 | G302D | NBD1, C-motif |
| Pdr5-127 | S140N | N-terminal cytoplasmic domain |
| V150L | N-terminal cytoplasmic domain | |
| T360I | N-terminal cytoplasmic domain | |
| V782I | TMD 6 | |
| V783I | TMD 6 | |
| S1360F | TMD 10 |
Bold, amino acid substitutions that influence Pdr5 function.
Compare with Figure 8, showing the predicted membrane topology of Pdr5.
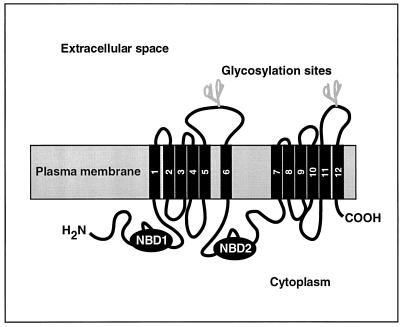
Figure 8.
Predicted membrane topology of Pdr5. The cartoon is based on the structural model for yeast ABC transporters such as Pdr5 and Snq2 (Servos et al., 1993; Bissinger and Kuchler, 1994). NBD, nucleotide-binding domain.
Amino Acid Substitutions in the Predicted Loop 6 that Lead to Pdr5 Mislocalization
In cells carrying the mutant transporter Pdr5–26, drug efflux was totally abolished (Figure 2B). Cell fractionation and indirect immunofluorescence experiments indicated that Pdr5–26 was not properly targeted to the plasma membrane, as it was found in the ER membrane (Figure 4). Four mutations were detected in the original Pdr5–26 mutant (Table 4), namely the exchange G557D in the extracellular loop 1 between the TMD1 and 2, an A1398T substitution within TMD11, and a double mutation at the positions P1421S and C1427Y in the predicted extracellular loop 6 between TMD11 and 12. Strikingly, only the C1427Y mutation in predicted loop 6 caused an ER retention of Pdr5–26 (Figure 4). This result clearly showed that a cysteine residue in extracellular loop 6 (C1427) is required for proper protein assembly in the ER membrane and/or the cell surface targeting of Pdr5.
Amino Acid Substitutions in Predicted Loop 6 that Alter Substrate Specificity
In the original Pdr5–14 mutant transporter, four amino acid substitutions were detected (Table 4). The A676V substitution in TMD5 and the C199Y exchange in NBD1 did not cause the Pdr5–14 phenotype. Interestingly, the C199Y mutant also retained function (our unpublished results), although the amino acid exchange was found in the Walker A motif of NBD1 (GRPGSGCTT), which is highly conserved among various ABC transporters (Bissinger and Kuchler, 1994). It was the double mutation T1460I/V1467I in the predicted extracellular loop 6 between TMD11 and 12 that severely altered the substrate specificity of Pdr5–14 (Figures 2 and 3 and Table 3). The functional importance of the last extracellular loop for substrate specificity was previously also shown for human Pgp (Zhang et al., 1995). Replacements of the extracellular loop between TMD11 and 12 of human Pgp (MDR1) by the corresponding regions of its homologue MDR2/MDR3 (Van der Bliek et al., 1987), an ABC transporter normally not involved in MDR development (van Helvoort et al., 1996), appeared to create an even more efficient drug pump for certain Pgp substrates such as actinomycin D, colchicine, and doxorubicin but not for vincristine (Zhang et al., 1995). Although yeast Pdr5 and human Pgp have a reversed TMD organization with respect to the localization of their NBDs, an involvement of extracellular loop 6 of Pdr5 in substrate specificity is evident from our results.
Amino Acid Substitutions in the Nucleotide-binding Domains
Three amino acid exchanges were detected in the original Pdr5–46 variant (Table 4). First, a single V149 M substitution at the N terminus of Pdr5 did not influence Pdr5 transporter activity. However, the double G905S/G908S mutation in the Walker A motif (GASGAGKTT) of NBD2 totally abolished transport for all Pdr5 substrates tested (Figures 3 and 7 and Table 3). These results showed that a functional N-terminal NBD1 alone is not sufficient for proper Pdr5 function, indicating that substrate transport requires the presence of both functional NBDs in one Pdr5 transporter molecule.
In the mutant transporter Pdr5–57, we found a silent G138D mutation near the N-terminal end of Pdr5, whereas a G1009C exchange close to the C-motif of NBD2 (Table 4) clearly reduced resistance to the tested drugs (Figure 3 and Table 3).
In Pdr5–71 the single substitution G302D near the C-motif of NBD1 was detected (Table 4). Based on homology comparisons, G302 in the NBD1 of Pdr5 corresponds precisely to G1009 in NBD2, the amino acid mutated in Pdr5–57. Notably, both glycines are absolutely conserved among NBD1 and NBD2 of ABC transporters from various different species (Figure 9). Although the mutations were found at equivalent glycine residues of NBD1 and NBD2, Pdr5–71 and Pdr5–57 show distinct differences in substrate specificity [see also resistance to rhodamine 6G (Figure 3 and Table 3)]. This could be caused by the difference in the substituted amino acids in the mutant transporters, namely, D and C in Pdr5–71 and Pdr5–57, respectively.
Figure 9.
Alignment of amino acid sequences around the C-motif of NDBs. NBD1 (A) and NBD2 (B) from Pdr5 and other yeast ABC transporters [Pdr10, Pdr15, Cdr1, Cdr2, Snq2, Pdr12 (LPE14c)], human MDR1 (h-MDR1), mouse mdr3 (m-mdr3), and other members of the family of ABC transporters [Plasmodium falciparum (pf-mdr1) and bacterial hemolysin B (Hlyb)].
Amino Acid Substitutions in Transmembrane Domain 10
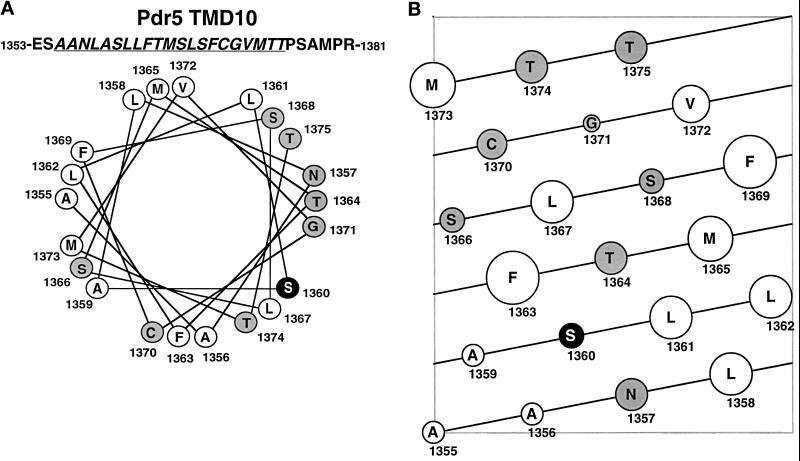
The original Pdr5–127 mutant contained six mutations (Table 4). Five of them, S140N, V150L, and T360I, located in the N-terminal cytoplasmic part, and V782I and V783I in TMD6, had no effect on Pdr5 function. However, the single amino acid exchange S1360F in TMD10 caused the alteration in drug and steroid transport specificity and FK506 inhibition. The secondary structure of TMD10 of Pdr5 was predicted using the PHDsec algorithm (Rost and Sander, 1993, 1994). This algorithm predicts S1360 into the core of an α-helix. Individual residues of the TMD10 α-helix were also classified according to their hydrophobic moment (Eisenberg et al., 1984). Interestingly, a helical wheel projection of TMD10 hints that the S1360F exchange severely disturbs the distribution of hydrophobic and hydrophilic amino acids in the amphipatic helix (Figure 10A). The model indicates that a distinct relative arrangement of polar and nonpolar residues and/or the residue volume size requirement (see Figure 10B; Geneworks Program 2.5.1, Helical Net) in TMD10 is somehow important for Pdr5 substrate specificity. The alteration in TMD10 structure may either directly influence drug and steroid binding or could change the overall structure of the folded transporter, thereby creating transport channels or pores of variable architecture stretching through the phospholipid bilayer (Rosenberg et al., 1997).
Figure 10.
Helical wheel projection of the Pdr5 TMD10. (A) The helical wheel projection of TMD10 (underlined) is viewed from its C terminus. Residues with polar side chains are shaded in gray and hydrophobic amino acids are indicated with open circles. The amino acid S1360, mutated to F in Pdr5–127, is shown by a white letter on a black background. (B) Two-dimentional illustration of the TMD10 α-helix viewed from its sideface. Each diagonal line represents a 360-degree turn of the α-helix. Hydrophilic amino acids (gray) and hydrophobic residues (open circles) are depicted as circles of varying size dependent on their relative volume.
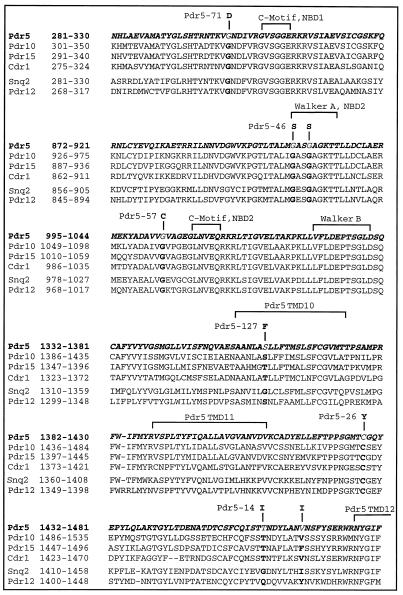
The analysis of mutant Pdr5 transporters obtained through random in vitro mutagenesis revealed that mutations in both NBDs, the extracellular loop 6 between TMD11 and 12, as well as TMD10 alterations can modulate drug substrate specificity. Figure 11 shows an alignment of amino acid sequences from members of the Pdr5 and Snq2 family (Parle-McDermott et al., 1996) of yeast ABC transporters. The alignment includes parts of the consensus ATP-binding motifs Walker A, Walker B, and the C-motif of NBD1 and NBD2, TMD10, and the extracellular loop between TMDs 11 and 12 (Figure 11). Interestingly, five of eight mutated amino acids were perfectly conserved among all members of the yeast Pdr5 and Snq2 subfamilies, implying an important functional role for these residues in other ABC proteins closely related to Pdr5. Furthermore, amino acid substitutions in the conserved G302 of NBD1 and three other amino acids, namely S1360, T1460/V1467, which are not conserved between Pdr5 and Snq2, were found in the mutant transporters Pdr5–71, Pdr5–127, and Pdr5–14, respectively. These mutations clearly altered drug substrate specificity of Pdr5 or as in the case of Pdr5–127, also prevented inhibition of Pdr5 activity by FK506. Interestingly, Snq2-mediated transport of 1,10-phenanthroline, a specific Snq2 substrate, could not be blocked by FK506 (our unpublished results). Since wild-type Pdr5 and Snq2 exhibit quite distinct substrate specificities (Balzi et al., 1994; Bissinger and Kuchler, 1994; Hirata et al., 1994; Mahéet al., 1996b), one can speculate that these nonconserved amino acid residues are involved in determination of substrate specificity of Pdr5 and presumably Snq2.
Figure 11.
Alignment of amino acid sequences from members of the Pdr5 family. Selected regions of the Pdr5 subfamily [Pdr5, Pdr10, Pdr15, Cdr1 (Candida)] and the Snq2 subfamily (Snq2, Pdr12) of yeast ABC transporters were compared. Amino acids whose substitutions alter substrate specificity of Pdr5 are indicated in bold letters.
DISCUSSION
The knowledge about the molecular determinants of the broad substrate specificity of many ABC transporters is particularly important to understand the mechanisms of multidrug resistance efflux pumps. In the present study, we performed a mutational structure-function analysis of the yeast Pdr5 ABC transporter. We have used a random in vitro mutagenesis approach and in vivo phenotypic screening that allowed for the isolation of Pdr5 variants conferring altered drug resistance phenotypes (Figure 1–3 and Table 3). DNA sequencing of six PDR5 alleles identified residues important for Pdr5 function and altered substrate specificities (Table 4).
Mutants Pdr5–26 and Pdr5–46 Are Nonfunctional
In the first group, both Pdr5–26 and Pdr5–46 mutants turned out to be nonfunctional regarding all Pdr5 substrates used in this study (Figures 2, 3, and 7). Cell fractionation and indirect immunofluorescence experiments localized Pdr5–26 to the ER (Figure 4). Interestingly, the single amino acid substitution C1427Y in the predicted extracellular loop 6 between the TMD11 and 12 of Pdr5 caused the efficient retention of Pdr5–26 in the ER. In contrast, a human Pgp variant that lacks all cysteines is both properly targeted to the plasma membrane and retains the ability to confer drug resistance (Loo and Clarke, 1995b).
The ER lumen contains high levels of chaperones and folding enzymes such as Kar2 (BiP), calnexin, and protein disulfide isomerases (Gething and Sambrook, 1992). The oxidizing environment in the ER lumen facilitates the formation of disulfide bonds, a prerequisite for the proper folding and subsequent transport of disulfide bonded proteins (Jämsäet al., 1994). Likewise, retention of Pdr5–26 in the ER could be the consequence of a lack of disulfide bond formation between C1427 and other cysteines of Pdr5 that face the ER lumen. For instance, C1427 could stabilize Pdr5 structure via a disulfide bond with one of the three other cysteine residues present in loop 6 (C1411, C1452, and C1455), all of which are highly conserved among various yeast ABC transporters (Figure 11). Alternatively, C1427 could also build a disulfide bond with one of the highly conserved cysteines within the predicted extracellular loop 3 (C722 and C742) between TMD5 and 6, the only other predicted extracellular loop of Pdr5 with cysteines (Bissinger and Kuchler, 1994). At present, it is not clear whether the inability of Pdr5–26 to confer drug resistance is caused by a mislocalized and inactive protein or by the mislocalization of an otherwise functional transporter, as is the case with ER-mislocalized ΔF508 CFTR mutant (Denning et al., 1992; Jensen et al., 1995; Ward et al., 1995).
Site-directed mutagenesis of several glycines (G251, G268, G269, or G781) in predicted cytoplasmic loops of human Pgp to valine also yielded loss-of-function mutants located in the ER (Loo and Clarke, 1994a). Interestingly, a defective protein kinesis of the temperature-sensitive G269V Pgp mutant could be corrected by a temperature decrease (Loo and Clarke, 1994a), whereas a targeting defect of the G268V Pgp mutant can be overcome by incubation of cells with certain substrates and modulators (Loo and Clarke, 1997). However, incubation of Pdr5–26 cells in the presence of Pdr5 substrates or temperature shift did not render cells more drug resistant (Figure 2B and our unpublished results).
In contrast to Pdr5–26, the Pdr5–46 variant was correctly targeted to the plasma membrane but it was inactive in drug transport (Figures 2–4). The double mutation G905S/G908S in the Walker A motif of NBD2 was shown to inactivate Pdr5–46 for all substrates tested. Both glycines of the Walker A motif are highly conserved among the members of the ABC family (Higgins, 1992), indicating their importance in ABC transporter function. For instance, amino acid exchanges such as G431A and/or G1073A, but also K432R and/or K1074R in the Walker A motif of NBD1 and NBD2 of mouse mdr1, resulted in loss of function (Azzaria et al., 1989). Furthermore, the K433 M and/or the K1076 M replacements in the Walker A motifs of human Pgp abolished drug-stimulated ATPase activity, although ATP binding was only poorly affected (Mueller et al., 1996). Likewise, our results clearly demonstrated the requirement of NBD2 for Pdr5-mediated substrate transport, because a functional NBD1 alone was not sufficient to maintain transport activity of Pdr5–46. Currently it is not known whether ATP binding or another step is impaired in the yeast Pdr5–46 mutant. However, our results demonstrate that both NBDs of Pdr5 are essential for proper transporter function.
Pdr5 Mutants with Altered Substrate Specificity
The second group of mutations were found to either reduce drug resistance (Pdr5–57) and/or alter drug substrate specificity (Pdr5–14, Pdr5–71, and Pdr5–127). Pdr5–14 conferred considerable resistance to rhodamines, but was severely impaired in its ability to confer azole and cycloheximide resistance or to decrease response to dexamethasone (Figure 2, Table 3, and Figure 7). The double mutation T1460I/V1467I in the predicted extracellular loop 6 between TMD11 and 12 (Table 4) was shown to mediate the altered substrate specificity of Pdr5–14. The contribution of an extracellular loop in substrate specificity was previously also documented for loop 6 in human Pgp (Zhang et al., 1995). The results suggest that extracellular loops could form part of the drug-binding domain in Pgp or in its functional yeast homologue Pdr5. However, these regions could also contribute to drug efflux specificity by influencing the folded structure of the transporter. This idea is supported by the observation that the misfolded, loss-of-function Pdr5–26 mutant was also found in the last extracellular loop in close proximity to the mutations in Pdr5–14.
Interestingly, the C199Y amino acid substitution found in the N-terminal Walker A motif in the original Pdr5–14 isolate (GRPGSGCTT) did not cause a mutant Pdr5 phenotype with our test substrates (our unpublished results). The alignment of the Pdr5 Walker A motif with several ABC transporters from yeast and other organisms matched C199 of Pdr5 to a highly conserved lysine found in all other ABC transporters at this position (Bissinger and Kuchler, 1994). The replacement of the corresponding K433 of human Pgp (GNSGCGKST) to M (Mueller et al., 1996) abolished drug-stimulated ATPase activity of human Pgp, indicating the importance of K433 for Pgp function. Chemical modification of C431 with N-ethylmaleimide suggested the highly conserved cysteine residue in the Walker A motif of hamster Pgp (GNSGCGKST) critical for transporter function (Al-Shawi et al., 1994). Likewise, chemical modification of the C431 and C1074 in human Pgp proved critical for human Pgp function (Loo and Clarke, 1995a). In contrast, our results showed that Pdr5 with the C199Y mutation in the Walker A motif of NBD1 still retained wild-type function. However, we cannot exclude that a substitution of C199 by an amino acid other than tyrosine could affect Pdr5 activity or substrate specificity.
The mutations in Pdr5–57 (G1009C) and Pdr5–71 (G302D) are present at corresponding positions in the C-motif of NBD2 and NBD1, respectively (Figure 9). Pdr5–57 exhibited partial activity for all substrates, conferring considerable drug resistance, albeit to lower extents than wild-type Pdr5. In contrast, Pdr5–71 showed an interesting novel pattern of drug resistance. Like Pdr5–57, it conferred significant resistance to azoles (Figure 3) and it efficiently reduced intracellular levels of estradiol and dexamethasone (Figure 7 and our unpublished results). Unlike Pdr5–57, however, Pdr5–71 had a pronounced defect in rhodamine transport. The distinct phenotypes of Pdr5–57 and -71 may be due to the fact that the functionally equivalent glycines of NBD1 and NBD2 are not substituted by the same amino acid. Alternatively, the results could be explained by distinct functional roles of individual NBDs in their contribution to Pdr5 substrate specificity. Both glycines are highly conserved among NBD1 and NBD2 of ABC transporters from several different species (Figure 9), indicating the importance of these residues in transporter function. Notably, sequence comparison (Figure 9) revealed that G521 of mouse mdr3 represents the equivalent of G302 in Pdr5. The residues, ERGA, present near the C-motif in NBD1 of mouse Pgp (TLVGERGAQLSGGQ) are essential for proper mdr3 function (Beaudet and Gros, 1995). The replacement of ERGA by the homologous DKGT peptide of NBD2 and a T578C substitution in mdr3 caused a reduction in doxorubicin, actinomycin D, and colchicine resistance (Beaudet and Gros, 1995). In summary, the results suggest that the highly conserved glycine residues and/or the peptide segments neighboring the glycines may directly participate in substrate transport of Pdr5, mammalian Pgps, and possibly other ABC transporters. Alternatively, the glycines could also be involved in a functional cross-talk of the NBDs with certain TMDs involved in drug specificity and/or transport.
A comparison of the substrate specificity profile of Pdr5–71 (G302D in NBD1) and Pdr5–46 (G905S/G908S in NBD2) revealed that both mutants no longer conferred resistance to rhodamines when compared with wild-type Pdr5 (Figures 2 and 3 and Table 3). These results indicate that a functional intramolecular substitution of NBDs is unlikely, suggesting that NBD1 and NBD2 are both essential for proper Pdr5 function. Furthermore, when Pdr5–71 and Pdr5–46 were simultaneously coexpressed in one cell, we could not detect elevated rhodamine resistance when compared with transformants expressing only Pdr5–71 or Pdr5–46 (our unpublished results). The apparent failure of cross-complementation between Pdr5–71 and Pdr5–46 also argues against a functional complementation of NBDs in trans.
The mutant Pdr5–127 with the S1360F exchange in TMD10 (Table 4) showed an interesting pattern of substrate recognition. It confers resistance to ketoconazole at levels comparable to Pdr5–71 but better than Pdr5–14 (Figure 3 and Table 3). Moreover, Pdr5–127 could, like Pdr5–71, significantly decrease intracellular estradiol accumulation (Figure 7). However, Pdr5–127 showed a reduced ability to confer resistance to itraconazole and cycloheximide comparable to that of Pdr5–14 (Figures 2 and 3 and Table 3). Furthermore, Pdr5–127 was completely deficient in reducing dexamethasone response, being less active than Pdr5–14 and comparable to the complete lack of Pdr5 (Figure 7).
Many mutations that alter substrate specificity of mammalian Pgps lie within predicted TMDs (Gottesman et al., 1995; Kane, 1996). The helix packing of predicted TMD helices and the aromatic amino acid distribution within individual helices was previously suggested to determine substrate specificity and transport properties of Pgps (Pawagi et al., 1994). In addition, it was shown that the relative distribution of hydrophilic and hydrophobic amino acids in TMD11 of mouse mdr3 influences substrate specificity (Hanna et al., 1996). Alanine-scanning of TMD11 of mouse mdr3 proved the more hydrophilic face of the amphipatic helix mutation sensitive, whereas the nonpolar face appeared mutation insensitive. For instance, substitutions such as F953A and Y949A in TMD11 drastically induced changes in mdr3 substrate specificity. Similarly, a substitution such as S1360F in the predicted TMD10 of Pdr5, as present in Pdr5–127 (Table 4), drastically changed substrate specificity (Table 3). Hence, this mutation may severely disturb the amphiphilic helix arrangement or the amino acid size requirement of TMD10 (Figure 10), thereby directly influencing the interaction of TMD10 with Pdr5 substrates or with other TMDs in the folded Pdr5 transporter, all of which together could form the putative substrate binding pocket. Strikingly, a physical interaction of TMDs in human Pgp was recently demonstrated by chemical cross-linking of engineered cysteines in TMD6 (C332) and TMD12 (C975), which could be inhibited by Pgp substrates such as vinblastine, but not colchicine (Loo and Clarke, 1996). Our data are in good agreement with previous findings for mdr3 (Hanna et al., 1996), suggesting that the distribution of polar and nonpolar residues in TMD10 of Pdr5 may be an important prerequisite for transport specificity. Likewise, serine to phenylalanine substitutions influencing substrate specificity were also found in TMD11 of mouse mdr1 (S941F) and mdr3 (S943F), respectively (Gros et al., 1991). In turn, the F335A substitution in TMD6 of human Pgp affected vinblastine transport (Loo and Clarke, 1994b), whereas the substitution of F978S or F978A in TMD12 resulted in a lack of resistance to colchicine and doxorubicin, and in reduced resistance to vinblastine and actinomycin D (Loo and Clarke, 1993). Taken together, the polarity distribution and/or the volume required for individual sidues in TMD helices of eukaryotic multidrug transporters may play a key role in substrate recognition and drug transport. However, the precise role of TMDs in drug transport and the molecular mechanism(s) of substrate recognition is still ill-defined. The molecular understanding of these processes will likely require three-dimensional structural information. A first three-dimension structure was indeed recently reported for human Pgp, although the resolution was still too low to yield definitive structural information (Rosenberg et al., 1997).
Serine 1360 of Pdr5 TMD10 Is Required for FK506 Inhibition
The identification of residues implicated in protein-substrate interaction and the mechanism(s) of action are of particular interest in the molecular modeling of new drugs or modulators that could overcome drug resistance by specifically blocking the transporter. The observation that FK506 modulates dexamethasone transport activity of Pdr5 (Kralli and Yamamoto, 1996) prompted us to test the Pdr5 mutants for their ability to confer drug resistance in the presence of FK506. We focused on azole antifungals (Sanglard et al., 1995), which are widely used in the clinic to treat infections with pathogenic fungi such as Candida albicans. The azoles fluconazole, ketoconazole, and itraconazole were proven substrates for Pdr5 and for the related ABC drug efflux pumps Cdr1 (Sanglard et al., 1996) and Cdr2 (Sanglard et al., 1997), both of which are highly homologous ABC transporters from C. albicans (Prasad et al., 1995; Sanglard et al., 1995). We found that Pdr5 activity could be blocked by FK506 in both wild-type and all Pdr5 mutants, except in mutant Pdr5–127, which has severely reduced its susceptibility to FK506 inhibition (Figure 6). Interestingly, a sequence alignment of TMD10 from Pdr5, Cdr1 (Figure 11), and Cdr2 (our unpublished results) matched a threonine residue of Cdr1 and Cdr2 to S1360, the amino acid substituted by phenylalanine in Pdr5–127. Furthermore, a helical wheel projection of the predicted TMD10 of Cdr1 and Cdr2 indicates an amphiphilic helix comparable to the one of Pdr5 (our unpublished results). Notably, the impaired sensitivity of Pdr5–127 to FK506 inhibition was not restricted to ketoconazole transport, because even the residual transport activity of Pdr5–127 for other Pdr5–127 substrates failed to be blocked by FK506 (Figure 6).
Recently, the mouse mdr3 Pgp was expressed in yeast Δfpr1 cells lacking FKBP12 (Hemenway and Heitman, 1996). The authors suggested that FK506 could reverse Pgp-mediated multidrug resistance in yeast by inhibiting the drug transporter. Moreover, an inhibition of FKBP12-dependent Pgp function may contribute to reversal of multidrug resistance by FK506. In the case of yeast Pdr5, our results showed that both transport activity and inhibition by FK506 is not dependent on the presence of FKBPs such as Fpr1 (FKBP12), Fpr2, and Fpr3 (Benton et al., 1994). Although we cannot exclude a possible role for at least two other putative FKBPs present in the yeast genome database, our findings suggest that FK506 may interact directly with Pdr5 and that the S1360F mutation somehow prevents Pdr5-FK506 interaction. Such a mechanism would be in agreement with earlier studies on mammalian Pgp that suggest a direct high-affinity interaction of Pgp and FK506 (Rao and Scarborough, 1994).
We have shown that the mutational change of several Pdr5 residues in the most highly conserved cytoplasmic NBDs, in TMD10, and even in predicted extracellular loop 6 can lead to drastic changes in substrate specificity. Our studies imply that the overall structure of the folded ABC transporter in the membrane may represent a primary determinant of their broad substrate specificity. Moreover, it is also conceivable that the membrane lipid composition (Zinser et al., 1991) might somehow influence transporter structure and/or the substrate specificity. It will be of interest to test this idea for Pdr5 and its mutants isolated in this study by their functional expression in mammalian cells, whose membrane lipid compositions are quite different from yeast (Zinser et al., 1991).
The results from our studies have also important general implications, since they indicate that one can now apply such a strategy of mutant isolation for multidrug transporters such as mammalian Pgp or MRP. Hence, it should be possible to isolate drug resistance efflux pumps with limited substrate specificity or enhanced drug transport capacity. For instance, one can envision that gene therapy applications aimed at the in vivo chemoprotection of hematopoietic stem cells (for review, see Kane, 1996) would be greatly facilitated, once Pgp transporters become available that can confer high-level resistance to drugs frequently used in high-dose chemotherapeutic regimens. Although this strategy would not be feasible in mammalian cells for technical reasons, our work demonstrates that it is possible to identify mammalian ABC transporter mutants by random mutagenesis and phenotypic selection of mutant Pgps functionally expressed in yeast.
ACKNOWLEDGMENTS
We thank Jeremy Thorner for sharing research materials, yeast strains, and reagents. We thank Dr. Ihor Bekersky from Fujisawa for the generous gift of FK506. In addition, we are greatly indebted to Keith Yamamoto because part of the initial experiments by A.K. were performed in his laboratory under National Science Foundation grant support. Many critical and helpful comments and suggestions on the manuscript by Susan Kane, Helga Edelmann, and Rudy Pandjaitan are thankfully acknowledged. Special thanks to Helga Edelmann for the advice with the flow cytometry analysis. This work was supported by grants from the Austrian Science Foundation (FWF MOB-10123 and P-AL661-BIO) and in part by National Institutes of Health grant #RO1-CA64645–01A1 to K.K. R.E. was a recipient of a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft and is currently under support of the Herzfelder’schen Familienstiftung. D.S. was supported by a grant from the Swiss Research National Foundation (31–45716.95). A.K. was supported in part by a generous Herb Boyer Fellowship.
REFERENCES
- Al-Shawi MK, Urbatsch IL, Senior AE. Covalent inhibitors of P-glycoprotein ATPase activity. J Biol Chem. 1994;269:8986–8992. [PubMed] [Google Scholar]
- Azzaria M, Schurr E, Gros P. Discrete mutations introduced in the predicted nucleotide-binding sites of the mdr1 gene abolish its ability to confer multidrug resistance. Mol Cell Biol. 1989;9:5289–5297. doi: 10.1128/mcb.9.12.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzi E, Goffeau A. Genetics and biochemistry of yeast multidrug resistance. Biochim Biophys Acta. 1994;1187:152–162. doi: 10.1016/0005-2728(94)90102-3. [DOI] [PubMed] [Google Scholar]
- Balzi E, Wang M, Leterme S, Van DL, Goffeau A. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J Biol Chem. 1994;269:2206–2214. [PubMed] [Google Scholar]
- Beaudet L, Gros P. Functional dissection of P-glycoprotein nucleotide-binding domains in chimeric and mutant proteins. J Biol Chem. 1995;270:17159–17170. doi: 10.1074/jbc.270.29.17159. [DOI] [PubMed] [Google Scholar]
- Benton BM, Zang J-H, Thorner J. A novel FK506- and rapamycin-binding protein (FPR3 gene product) in the yeast Saccharomyces cerevisiae is a proline rotamase localized to the nucleolus. J Cell Biol. 1994;127:623–639. doi: 10.1083/jcb.127.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissinger PH, Kuchler K. Molecular cloning and expression of the Saccharomyces cerevisiae STS1 gene product. A yeast ABC transporter conferring mycotoxin resistance. J Biol Chem. 1994;269:4180–4186. [PubMed] [Google Scholar]
- Chen CJ, Chin JE, Ueda K, Clark DP, Pastan I, Gottesman MM, Roninson IB. Internal duplication and homology with bacterial transport proteins in the MDR1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell. 1986;47:381–9. doi: 10.1016/0092-8674(86)90595-7. [DOI] [PubMed] [Google Scholar]
- Cole SP, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, Stewart AJ, Kurz EU, Duncan AM, Deeley RG. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- Decottignies A, Goffeau A. Complete inventory of the yeast ABC proteins. Nature Genet. 1997;15:137–145. doi: 10.1038/ng0297-137. [DOI] [PubMed] [Google Scholar]
- Denning GM, Anderson MP, Amara JF, Marshall J, Smith AE, Welsh MJ. Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature. 1992;358:761–764. doi: 10.1038/358761a0. [DOI] [PubMed] [Google Scholar]
- Devine SE, Ling V, Melera PW. Amino acid substitutions in the 6th transmembrane domain of P-glycoprotein alter multidrug resistance. Proc Natl Acad Sci USA. 1992;89:4564–4568. doi: 10.1073/pnas.89.10.4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner R, Mahé Y, Pandjaitan R, Huter V, Lamprecht A, Kuchler K. ATP binding cassette transporters in yeast: from mating to multidrug resistance. In: Rothman S, editor. Membrane Protein Transport. Greenwich: JAI Press Inc.; 1995a. pp. 57–96. [Google Scholar]
- Egner R, Mahé Y, Pandjaitan R, Kuchler K. Endocytosis and vacuolar degradation of the plasma membrane localized Pdr5 ATP binding cassette multidrug transporter in Saccharomyces cerevisiae. Mol Cell Biol. 1995b;15:5879–5887. doi: 10.1128/mcb.15.11.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Schwartz E, Komarony M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Gething M-J, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Gottesman MM, Hrycyna CA, Schoenlein PV, Germann UA, Pastan I. Genetic analysis of the multidrug transporter. Annu Rev Genet. 1995;29:607–649. doi: 10.1146/annurev.ge.29.120195.003135. [DOI] [PubMed] [Google Scholar]
- Gros P, Dhir R, Talbot F. A single amino acid substitution strongly modulates the activity and substrate specificity of the mouse mdr1 and mdr3 drug efflux pumps. Proc Natl Acad Sci USA. 1991;88:7289–93. doi: 10.1073/pnas.88.16.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna M, Brault M, Kwan T, Kast C, Gros P. Mutagenesis of transmembrane domain 11 of P-glycoprotein by alanine scanning. Biochemistry. 1996;35:3625–3635. doi: 10.1021/bi951333p. [DOI] [PubMed] [Google Scholar]
- Hemenway CS, Heitman J. Immunosuppressant target protein FKBP12 is required for P-glycoprotein function in yeast. J Biol Chem. 1996;271:18527–18534. doi: 10.1074/jbc.271.31.18527. [DOI] [PubMed] [Google Scholar]
- Higgins CF. ABC transporters: from microrgansims to man. Ann Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- Hirata D, Yano K, Miyahara K, Miyakawa T. Saccharomyces cerevisiae YDR1, which encodes a member of the ATP-binding cassette (ABC) superfamily, is required for multidrug resistance. Curr Genet. 1994;26:285–294. doi: 10.1007/BF00310491. [DOI] [PubMed] [Google Scholar]
- Hoof T, Demmer A, Hadam MR, Riordan JR, Tummler B. Cystic fibrosis-type mutational analysis in the ATP-binding cassette transporter signature of human P-glycoprotein MDR1. J Biol Chem. 1994;269:20575–20583. [PubMed] [Google Scholar]
- Iniguez-Lluhi JA, Lou DY, Yamamoto KR. Three amino acid substitutions selectively disrupt the activation but not the repression function of the glucocorticoid receptor N-terminus. J Biol Chem. 1997;272:4149–4156. doi: 10.1074/jbc.272.7.4149. [DOI] [PubMed] [Google Scholar]
- Jämsä E, Simonen M, Makarow M. Selective retention of secretory proteins in the yeast endoplasmic reticulum by treatment of cells with a reducing agent. Yeast. 1994;10:355–370. doi: 10.1002/yea.320100308. [DOI] [PubMed] [Google Scholar]
- Jensen TJ, Loo MA, Pind S, Williams DB, Goldberg AL, Riordan JR. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- Kane SE. Multidrug resistance of cancer cells. In: Testa B, Meyer UA, editors. Advances in Drug Research. Vol. 28. San Diego: Academic Press; 1996. pp. 181–252. [Google Scholar]
- Katzmann DJ, Hallstrom TC, Voet M, Wysock W, Golin J, Volckaert G, Moye-Rowley WS. Expression of an ATP-binding cassette transporter-encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6875–6883. doi: 10.1128/mcb.15.12.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski M, van der Rest M, Cybularz-Kolaczkowska A, Soumillion JP, Konings WN, Goffeau A. Anticancer drugs, ionophoric peptides, and steroids as substrates of the yeast multidrug transporter Pdr5p. J Biol Chem. 1996;271:31543–31548. doi: 10.1074/jbc.271.49.31543. [DOI] [PubMed] [Google Scholar]
- Kralli A, Bohen SP, Yamamoto KR. LEM1, an ATP-binding cassette transporter, selectively modulates the biological potency of steroid hormones. Proc Natl Acad Sci (USA) 1995;92:4701–4705. doi: 10.1073/pnas.92.10.4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralli A, Yamamoto KR. An FK506-sensitive transporter selectively decreases intracellular levels and potency of steroid hormones. J Biol Chem. 1996;271:17152–17156. doi: 10.1074/jbc.271.29.17152. [DOI] [PubMed] [Google Scholar]
- Kuchler K, Dohlman H, Thorner J. The a-factor transporter (STE6 gene product) and cell polarity in Saccharomyces cerevisiae. J Cell Biol. 1993;120:1203–1215. doi: 10.1083/jcb.120.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler K, Egner R. Unusual protein secretion and translocation pathways in yeast: implication of ABC transporters. In: Rubartelli A, Kuchler K, editors. Unusual Secretory Pathways: From Bacteria to Man. B. Holland: Landes Bioscience Publishers; 1997. pp. 49–85. [Google Scholar]
- Kunz J, Hall MN. Cyclosporin A, FK506 and rapamycin: more than just immunosupression. Trends Biochem Sci. 1993;18:334–338. doi: 10.1016/0968-0004(93)90069-y. [DOI] [PubMed] [Google Scholar]
- Lefstin JA, Thomas JR, Yamamoto KR. Influence of a steroid receptor DNA-binding domain on transcriptional regulatory functions. Genes Dev. 1994;8:2842–2856. doi: 10.1101/gad.8.23.2842. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Functional consequences of phenylalanine mutations in the predicted transmembrane domain of P-glycoprotein. J Biol Chem. 1993;268:19965–19972. [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Functional consequences of glycine mutations in the predicted cytoplasmic loops of P-glycoprotein. J Biol Chem. 1994a;269:7243–7248. [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Mutations to amino acids located in predicted transmembrane segment 6 (TM6) modulate the activity and substrate specificity of human P-glycoprotein. Biochemistry. 1994b;33:14049–14057. doi: 10.1021/bi00251a013. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Covalent modification of human P-glycoprotein mutants containing a single cysteine in either nucleotide-binding fold abolishes drug-stimulated ATPase activity. J Biol Chem. 1995a;270:22957–22961. doi: 10.1074/jbc.270.39.22957. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Membrane topology of a cysteine-less mutant of human P-glycoprotein. J Biol Chem. 1995b;270:843–848. doi: 10.1074/jbc.270.2.843. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Inhibition of oxidative cross-linking between engineered cysteine residues at positions 332 in predicted transmembrane segments (TM) 6 and 975 in predicted TM12 of human P-glycoprotein by drug substrates. J Biol Chem. 1996;271:27482–27487. doi: 10.1074/jbc.271.44.27482. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Correction of defective protein kinesis of human P-glycoprotein mutants by substrates and modulators. J Biol Chem. 1997;272:709–712. doi: 10.1074/jbc.272.2.709. [DOI] [PubMed] [Google Scholar]
- Mahé Y, Lemoine Y, Kuchler K. The ATP binding cassette transporters Pdr5 and Snq2 of Saccharomyces cerevisiae can mediate transport of steroids in vivo. J Biol Chem. 1996a;271:25167–25172. doi: 10.1074/jbc.271.41.25167. [DOI] [PubMed] [Google Scholar]
- Mahé Y, Parle-McDermott AG, Nourani A, Delahodde A, Lamprecht A, Kuchler K. The ATP-binding cassette multidrug transporter Snq2 of Saccharomyces cerevisiae: a novel target for the transcription factors Pdr1 and Pdr3. Mol Microbiol. 1996b;20:109–117. doi: 10.1111/j.1365-2958.1996.tb02493.x. [DOI] [PubMed] [Google Scholar]
- Mueller M, Bakos E, Welker E, Varadi A, Germann UA, Gottesman MM, Morse BS, Roninson IB, Sarkadi B. Altered drug-stimulated ATPase activity in mutants of the human multidrug resistance protein. J Biol Chem. 1996;271:1877–1883. doi: 10.1074/jbc.271.4.1877. [DOI] [PubMed] [Google Scholar]
- Parle-McDermott AG, Hand NJ, Goulding SE, Wolfe KH. Sequence of 29 kb around the PDR10 locus on the right arm of Saccharomyces cerevisiae chromosome XV: similarity to part of chromosome I. Yeast. 1996;12:999–1004. doi: 10.1002/(SICI)1097-0061(199609)12:10B%3C999::AID-YEA976%3E3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Pawagi AB, Wang J, Silverman M, Reithmeier RA F, Deber CM. Transmembrane aromatic amino acid distribution in P-glycoprotein: a functional role in broad substrate specificity. J Mol Biol. 1994;235:554–564. doi: 10.1006/jmbi.1994.1013. [DOI] [PubMed] [Google Scholar]
- Prasad R, Dewergifosse P, Goffeau A, Balzi E. Molecular cloning and characterization of a novel gene of Candida albicans, Cdr1, conferring multiple resistance to drugs and antifungals. Curr Genet. 1995;27:320–329. doi: 10.1007/BF00352101. [DOI] [PubMed] [Google Scholar]
- Rao US, Scarborough GA. Direct demonstration of high affinity interactions of immunosuppressant drugs with the drug binding site of the human P-glycoprotein. Mol Pharmacol. 1994;45:773–776. [PubMed] [Google Scholar]
- Rose MD, Fink GR. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987;48:1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Rosenberg MF, Callaghan R, Ford RC, Higgins C. Structure of the multidrug resistance P-glycoprotein to 2.5 nm resolution determined by electron microscopy and image analysis. J Biol Chem. 1997;272:10685–10694. doi: 10.1074/jbc.272.16.10685. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–77. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen GG, Coulson AR. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci (USA) 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- Sanglard D, Ischer F, Monod M, Bille J. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother. 1996;40:2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D, Kuchler K, Ischer F, Pagani JL, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servos J, Haase E, Brendel M. Gene SNQ2 of Saccharomyces cerevisiae, which confers resistance to 4-nitroquinoline-N-oxide and other chemicals, encodes a 169 kDa protein homologous to ATP-dependent permeases. Mol Gen Genet. 1993;236:214–218. doi: 10.1007/BF00277115. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K, Cardarelli C, Gottesman MM, Pastan I. Expression of a full-length cDNA for the human MDR1 gene confers resistance to colchicine, doxorubicin, and vinblastine. Proc Natl Acad Sci (USA) 1987;84:3004–3008. doi: 10.1073/pnas.84.9.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Bliek AM, Baas F, Ten H dL T, Kooiman PM, Van dV KT, Borst P. The human mdr3 gene encodes a novel P-glycoprotein homologue and gives rise to alternatively spliced mRNAs in liver. EMBO J. 1987;6:3325–3331. doi: 10.1002/j.1460-2075.1987.tb02653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helvoort A, Smith A J, Sprong H, Fritzsche I, Schinkel A H, Borst P, van Meer G. MDR1 P-glycoprotein is a lipid translocase of broad specificity, while MDR3 P-glycoprotein specifically translocates phosphatidylcholine. Cell. 1996;87:507–517. doi: 10.1016/s0092-8674(00)81370-7. [DOI] [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Zaman GJ, Flens MJ, van Leusden MR, de Haas M, Mulder HS, Lankelma J, Pinedo HM, Scheper RJ, Baas F, Broxterman HJ, Borst P. The human multidrug resistance-associated protein MRP is a plasma membrane drug-efflux pump. Proc Natl Acad Sci (USA) 1994;91:8822–8826. doi: 10.1073/pnas.91.19.8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Collins KI, Greenberger LM. Functional evidence that transmembrane 12 and the loop between transmembrane 11 and 12 form part of the drug-binding domain in P-glycoprotein encoded by MDR1. J Biol Chem. 1995;270:5441–5448. doi: 10.1074/jbc.270.10.5441. [DOI] [PubMed] [Google Scholar]
- Zhifeng C, Hirata D, Tsuchiya E, Osada H, Miyakawa T. The multidrug resistance-associated protein (MRP) subfamiliy (Yrs1/Yor1) of Saccharomyces cerevisiae is important for the tolerance to a broad range of organic anions. J Biol Chem. 1996;271:14712–14716. doi: 10.1074/jbc.271.25.14712. [DOI] [PubMed] [Google Scholar]
- Zinser E, Sperka-Gottlieb CD M, Fasch E, Kohlwein FP, Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]