Abstract
The Salmonella typhimurium gene flhE is located at the end of a large flagellar locus in at least 10 peritrichously flagellated Gram-negative bacterial genera, but it shares no significant similarity with other genes. This study shows that flhE is transcribed as part of an flhBAE flagellar operon, under the control of the flagellar master regulator FlhD2C2. Deletion of the chromosomal flhE gene did not affect swimming motility, but it abolished swarming motility across solid agar. Swarming was restored to the ΔflhE mutant by the 130 aa putative envelope protein FlhE, but not by a truncated version lacking the N-terminal signal peptidase I recognition sequence. The ΔflhE mutant was indistinguishable from the wild-type parent in number and distribution of flagella, secretion of flagellin subunits, and flagellar gene expression, and there were no obvious differences in cell-surface LPS and extracellular polysaccharide. The ΔflhE mutant was able to swarm when non-ionic surfactant was included in agar medium, and it showed differences to the wild-type in binding calcofluor and Congo red dyes, and in biofilm production. The data show that the flhE gene is part of the flagella regulon but that it has no role in flagella biogenesis. It appears, nevertheless, to act at the cell envelope to influence flagella-dependent swarming.
INTRODUCTION
Motile bacteria like Escherichia coli and Salmonella typhimurium are able to swim through liquid by rotation of peritrichous helical flagella extending from their cell surface (Macnab, 1996). Flagella biogenesis and chemotaxis require over 50 genes transcribed in an ordered programme (Macnab, 1996). At the apex of this regulon is the master regulator flhDC, which activates genes encoding the flagella early structures, the type III flagella subunit secretion apparatus (Kutsukake et al., 1990; Soutourina & Bertin, 2003), and a flagellar sigma-factor (σ28) that activates late genes encoding the multicomponent flagellar filament, motor and chemotaxis apparatus (Chadsey et al., 1998; Karlinsey et al., 2000). Assembly of the flagella is strictly ordered, and coupled to this expression programme. The FlhD2C2 activator also controls non-flagellar genes in an extended motility regulon (Stafford et al., 2005; Soutourina & Bertin, 2003). Multicellular swarming migration over solid surfaces (Allison & Hughes, 1991) requires increased flagella production via upregulation of flhDC (Fraser & Hughes, 1999; Dufour et al., 1998; Hay et al., 1997; Givskov et al., 1995), and other cell surface components that facilitate assembly of swarm cell rafts (Hay et al., 1999; Allison et al., 1994), and surface lubrication by LPS (Toguchi et al., 2000; Belas et al., 1995), exopolysaccharide (EPS) (Gygi et al., 1995) and secreted surfactant (Toguchi et al., 2000; Gygi et al., 1995; Lai et al., 2005).
Motility genes are clustered within three loci around the chromosome of Sal. typhimurium and related bacteria (Macnab, 1996), and their approximate function has, in virtually all cases, been established. In this paper, we re-examine the gene flhE. This gene was given a flagellar nomenclature due to its location at the end of a large flagellar and chemotaxis gene locus, but an early report has indicated that it is not involved in motility (Minamino et al., 1994).
METHODS
Bacterial strains and plasmids
Bacterial strains were grown at 37 °C in LB, unless stated otherwise. Swarm cells were isolated after 6 h incubation on swarm agar (0.6% Bacto agar plus 0.5% glucose; Wang et al., 2004). Wild-type Sal. typhimurium SJW1103 (Yamaguchi et al., 1984) is motile, and the isogenic flhDC mutant SJW1368 is non-motile (Ohnishi et al., 1994). Deletion of flhE was achieved by the method of Datsenko & Wanner (2000) to create the ΔflhE strain, using primers ΔflhEFor (TCCGATAACCGTCATATCCGCATGCACGGCGACCATTGGAGGAAAATAATGGTGTAGGCTGGAGCTGCTTC) and ΔflhERev (TCCGGCAACCTACCTCACTTTATAAAACAGCGTTTCTATTTATTCAAATTCCGGGGATCCGTCGACC), and the pKD4 (KmR) plasmid as a template (Datsenko & Wanner, 2000). Deletion of the flhE gene was verified by PCR. The entire flhE gene was amplified by PCR using primers FlhEfor (TGGAGGAAACATATGCGTAAATGGCTGGCGTTG) and FlheRev (AACCCTCGAGGCGGTAGTTCACAATCACC), and cloned (XbaI-HindIII) 5′ of the arabinose-inducible promoter of expression vector pBAD18, to create pBAD18-FlhE. A derivative gene encoding N-terminally truncated FlhEΔN (lacking aa 1-16) was cloned into pBAD18 after PCR using primers FlhE-T (AATTCTAGAAATAATTTTGTTAACTTTAAGAAGATATACCATGGGCGAAGGCGCGTGGCAG) and FlhERev to make pBAD18-FlhEΔN.
Fluorescence microscopy of cells
Cells scraped from swarm plates were resuspended in saline (to an OD600 of 0.05), and fixed onto glass slides using 4% paraformaldehyde (in 20 mM PIPES, pH 7.4) before blocking with PBS (50 mM NaPO4/Na2PO4, pH 7.4, 150 mM NaCl) plus 3% (w/v) BSA for 1 h at 25 °C. Primary anti-flagellin antibody (1/1000, v/v, in PBS) was added for 2 h before washing (2×10 min, PBS), incubation with AlexaFluor-488/594-conjugated anti-rabbit secondary antibody (1/1000, v/v, in PBS; Molecular Probes) (2 h, 25 °C), and further washing (3×10 min, PBS). Cell membranes were stained for 10 min with SynaptoRed (in the dark), and coverslips were mounted using ProLong Anti-fade reagent (Molecular Probes), and visualized using a fluorescence microscope (Leica DM IRBE). Images were captured by a CCD digital camera (Hamamatsu) and processed using OpenLab software (Improvision).
Cell fractionation
Harvested swarm cells were resuspended in PBS, and diluted to an OD600 of 1.0. Total extracellular FliC protein was prepared by shearing (5 min vortex) of harvested cells, and TCA precipitation (10%, v/v, final concentration) of cell-free supernatant at 4 °C for 1 h. Extracellular protein was centrifuged for 1 h at 300 000 g to separate filament (pellet) from monomeric flagellin (soluble fraction, precipitated with 10% TCA, 4 °C, 1 h). Cells were separated into cytosolic and membrane fractions according to Auvray et al. (2001).
LPS and EPS extraction
Crude LPS was prepared from swarm cells (number of cells equivalent to 1 ml culture at an OD600 of 1), according to Hitchcock & Brown (1983). LPS was also extracted by a hot-phenol method for analysis by urea (high molecular mass) and deoxycholate-SDS (low molecular mass) PAGE, and visualized using silver staining (Guard-Petter et al., 1995). EPS was isolated and visualized according to Gygi et al. (1995).
Biofilm assay
Overnight cultures grown in biofilm LB (10 g tryptone l-1, 5 g yeast extract l-1) were inoculated at a 1 in 10 dilution into 96-well PVC microtitre plate wells (Falcon) containing fresh biofilm LB plus 0.5-2% glucose, and incubated overnight at 30 °C. Biofilm was washed twice with distilled water, air-dried for 30 min, and stained for 15 min with 1% crystal violet before washing with water and air drying. Biofilm was quantified as absorbance at 550 nm, following extraction with 95% ethanol (Mireles et al., 2001).
In vivo assay of transcription
Transcription was assessed as cell β-galactosidase activity (Miller, 1972) of gene fusions created by EcoRI/BamHI cloning of flhB (using primers FlhBPromEco, GAATTCACACGAGACTTTCTTTATC; and FlhBPromBam, GGATCCGCAAACCCTGGATAG) and fliC (primers FliCpromEco, GAATTCTTTTGCAAAAATAATGC; and FliCpromBam, GGATCCTCAATTACAACTTGATG) promoter fragments into the lacZ fusion vector pGPS123 (Stafford et al., 2005), which is identical to pRS551 except that KmR is replaced by GmR (Simons et al., 1987). For RT-PCR, RNA was extracted from swarm cells using hot acidic phenol. After removal of contaminating DNA by using Rq1 DNase (Promega), cDNA specific for the flhB and flhE genes was synthesized using Mu-MLV reverse transcriptase (Promega), and primers flhBRevRT (TTCGGCGTGGCGATATAATG) and flhERevRT (ATTGCTCCGCACTTTTAACG), resulting in cDNA originating within flhB and flhE, respectively. In the final step, primers flhBRevRT/flhBForRT (internal to flhB, ACCGCTCATCGCGGGCGTGG) and flhERevRT/flhEForRT (flhE internal, TGGCGTTGTTGCTCTTTCC) were used to amplify internal fragments of flhB and flhE. To assess transcripts spanning the flhBA and flhAE intergenic regions, primer pairs flhBForRT/flhARevRT (TCGCGAAGTTACCGCCGACCAGG) and flhAForRT (TCCGATAACCGTCATATCC)/flhERTRev were used. All PCR reactions used Taq polymerase, and products were analysed on 1.5% agarose ethidium bromide (EtBr) gels.
RESULTS AND DISCUSSION
The flhE gene in the flagellar loci of peritrichously flagellated bacteria
The flhE gene in Sal. typhimurium is located immediately downstream of the flhBA genes, and the flhA stop codon overlaps the flhE start codon (Fig. 1). Nevertheless, a transposon insertion in flhE has indicated that the gene is not essential for swimming motility, casting doubt on its flagellar gene nomenclature (Minamino et al., 1994). Our renewed interest in flhE was prompted by its presence in the flagellar gene loci of over 10 genera of peritrichous Gram-negative bacteria (Fig. 1). In the Enterobacteriaceae Esc. coli, Serratia marcescens, Erwinia carotovora, Yersinia pestis, Citrobacter rodentium and Shigella flexneri, flhE is located as in Sal. typhimurium, i.e. immediately distal to flhBA encoding the integral membrane flagellar export proteins FlhB and FlhA (except in Shigella, which contains no flhA gene). The flhE genes from the human pathogens Esc. coli, Y. pestis, Cit. rodentium and Shi. flexneri are similarly located at the end of motility gene locus, with non-flagellar genes downstream, and they are apparently transcribed independently from flhE. In Erwinia and Serratia, the flhBA(E) genes lie immediately adjacent to the chemotaxis genes cheBYZ, and within a still larger flagellar gene cluster containing the divergently transcribed flgAMN and flgBCDEFGHIJKL genes. In the free-living soil microbes Azotobacter vinelandii and Chromohalobacter salexigens, flhE is located downstream of the flhFG genes thought to be involved in flagellar assembly and gene regulation (McCarter 2001), while in Ralstonia metallidurans, flhE is separated from flhG by the fliA gene that encodes the flagella-specific sigma factor σ28. The flhE gene is thus always linked to flagella genes, and has not been located separately from flagellar gene loci. The sequence identity between the deduced amino acid sequence of Sal. typhimurium FlhE, and those of other Enterobacteriaceae, ranges from 37 to 83%, while it is lower (28-38%) for A. vinelandii, R. metallidurans and Chr. salexigens. All the flhE genes are 400±25 bp and encode proteins of approximately 14 kDa, with a predicted N-terminal signal peptidase I leader sequence, and a predicted periplasmic or outer membrane location; the FlhE sequences of the 10 genera in Fig. 1 contain between 7 and 13 apparently randomly distributed proline residues. blast searches revealed no significant similarity of FlhE to any class of proteins.
Fig. 1.
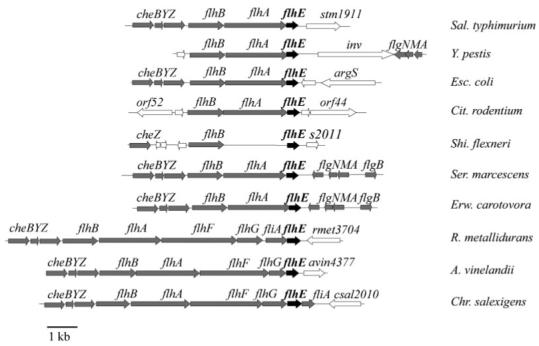
Location of flhE gene in peritrichous flagellated eubacteria. The flhE gene is shown in black, flagellar genes are in grey and non-flagellar genes are in white. Using the predicted amino acid sequence of the Sal. typhimurium LT2 flhE gene (AAL20828.1) with the blastp algorithm (Altschul et al., 1997) and the colibase website (Chaudhuri et al., 2004), flhE was identified in Y. pestis (GenBank accession no. NP_669822), Esc. coli (AAC74948), Cit. rodentium (colibase GL096266), Shi. flexneri (AAN43476), Ser. marcescens (colibase GL076148), Erw. carotovora (colibase CAG74604), R. metallidurans (YP_585844), A. vinelandii (ZP_00417385) and Chr. salexigens (ABE59363).
flhE is transcribed in an flhBAE operon activated by FlhD2C2
The flhBA operon is transcribed from a class II (early) flagellar promoter upstream of flhB, and is therefore activated by the flagellar master regulator FlhD2C2 (Fig. 2). To assess whether expression of flhE is flagellar-like, we purified RNA from wild-type and flhDC− strains, and performed RT-PCR using primers targeted within the flhE gene and the class II flhB gene. The results (Fig. 2) show that transcription of flhE was dependent on FlhD2C2, i.e. it mirrored that of flhB. We assessed whether flhE was transcribed as part of a contiguous polycistronic messenger RNA molecule by measuring transcription across the flhBA and flhAE intergenic regions in the wild-type and flhDC− strains. Fig. 2 shows that flhE is transcribed as part of a polycistronic messenger RNA in an flhDC-dependent manner, and it indicates that there is no post-transcriptional processing of the messenger RNA in vivo. These expression data establish flhE as part of the FlhD2C2 regulon, and indeed as part of an flhBAE operon. Together with the conserved location of flhE in the flagellar loci of peritrichously flagellated bacteria, this gives renewed validity to its designation as a flagellar gene. We investigated the possible function of flhE in motility.
Fig. 2.
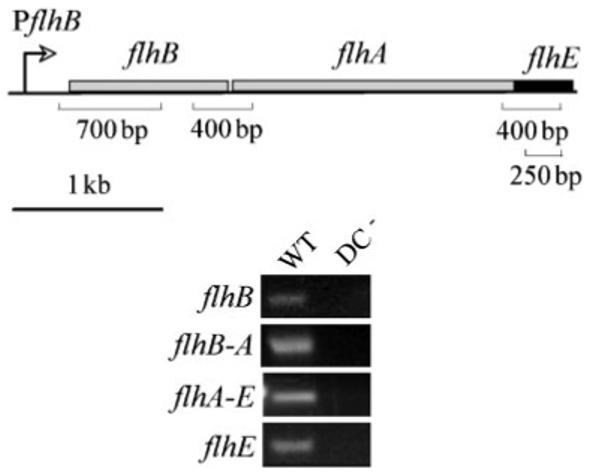
Transcription of flhE as part of the Sal. typhimurium flhBAE operon. RT-PCR reactions were performed on RNA extracted from swarm cells of wild-type (WT) and flhDC− strains, and products (sizes and corresponding locations are indicated in the top panel) within the flhB and flhE coding sequences, and spanning junctions between flhB-A and flhA-E were visualized by UV illumination of 1.5% agarose EtBr gels.
Loss of flhE attenuates swarming but not swimming
We set out to re-examine a possible role for flhE in motility by constructing an flhE deletion strain using the method of Datsenko & Wanner (2000). We confirmed the report by Minamino et al. (1994) that the swimming phenotype of such a mutant is at most only marginally reduced from the wild-type (Fig. 3). However, in common with several Gram-negative species, Salmonella is also capable of swarming motility, which is a form of flagella-dependent mass migration that is assayed as movement across the surface of denser 0.6% agar, rather than the standard 0.35% agar. The ability of Sal. typhimurium to swarm was severely attenuated by flhE loss, and was restored by a plasmid expressing the flhE gene in trans from the pBAD18 arabinose-inducible promoter (Fig. 3). In contrast, a truncated version of FlhE lacking the putative N-terminal 16 aa leader signal peptidase I sequence (MRKWLALLLFPLTVQA), and representing the mature form of the protein (aa 17-130), did not complement the swarming defect, even at high induction levels, indicating the importance of its secretion (Fig. 3). These data suggest that FlhE is a cell envelope protein that does have a role in flagellar-dependent motility, i.e. not cell swimming motility, but swarming migration.
Fig. 3.
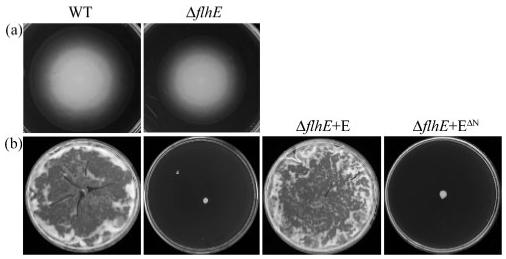
Effect of flhE deletion on Sal. typhimurium motility. Wild-type (WT), the ΔflhE mutant and the ΔflhE mutant complemented with pBAD18-FlhE (E) and FlhEΔN pBAD18-FlhEΔN (EΔN) were incubated on media for (a) swimming (0.35% agar, 6 h) and (b) swarming (0.6% agar+0.5% glucose, overnight).
Loss of flhE does not impair flagellar gene expression or assembly
To test if transcription of flagella genes is altered in the ΔflhE mutant, plasmid-borne transcriptional lacZ fusions were constructed to the flagellar class II promoter controlling the FlhD2C2-dependent flhBAE operon, and to the flagellar class III (σ28-dependent) fliC promoter. The activity of these promoter fusions during growth in liquid culture revealed that while transcription of the class II flhB(AE) and class III fliC promoters was reduced 122- and 421-fold, respectively, in an flhDC mutant compared with the wild-type (Fig. 4a), transcription of both genes was unaltered in the ΔflhE strain.
Fig. 4.
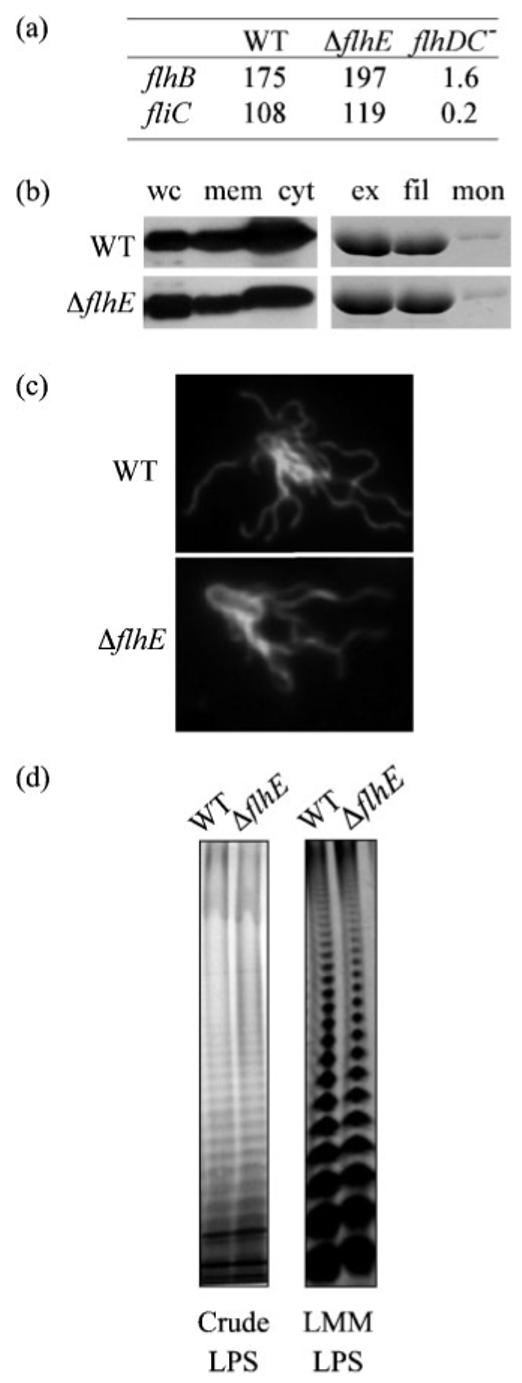
Flagellar and LPS production in the wild-type and ΔflhE mutant. (a) Activity (10-2×Miller units) of plasmid lacZ gene fusions to the promoter regions of flhB (class II) and fliC (class III) in Sal. typhimurium wild-type (WT) and ΔflhE and flhDC− mutants. (b) Flagellin (FliC) in whole swarm cells (wc), and derived membrane (mem) and cytosolic (cyt) fractions. Total extracellular flagellin (ex) was separated into filament (fil) and soluble monomeric (mon), and analysed by SDS-PAGE and immunoblotting. (c) Swarm cells were stained using SynaptoRed and rabbit-anti-flagellin fluorescent antibodies, and viewed at 540 and 366 nm, respectively, by fluorescence microscopy. (d) Crude and low-molecular-mass LPS (LMM) from wild-type and ΔflhE strains was visualized by SDS-PAGE (12%) and silver staining.
Loss of swarming motility could be due to attenuated post-transcriptional expression, or assembly of flagellar structural subunits. To examine this, levels of FliC protein were analysed by immunoblotting whole-cell, cytosolic, membrane-associated, extracellular and filament-incorporated fractions in the ΔflhE strain, and compared with the wild-type (Fig. 4b). These assays showed that the intracellular level of FliC was unaltered, as was external flagellin in the filaments. The stability of the flagella to shearing in the ΔflhE mutant was also unchanged (data not shown). It remained possible that the number or distribution of flagella on the cell surface was changed by the ΔflhE mutation, so we examined wild-type and ΔflhE mutant cells harvested from swarm agar, and fixed to glass slides. Fig. 4(c) shows representative merged fluorescence microscopy images highlighting flagella (visualized using anti-flagellin primary antibody and FITC-labelled secondary antibody) and cell membranes (stained with SynaptoRed). The images indicate no obvious change in flagellar number (approximately 15 per cell) or distribution. The combined data establish that the ΔflhE mutation does not reduce flagella gene expression, assembly or stability, or differentiation into swarm cells. The attenuation of flagellar-dependent swarming must have a non-flagellar cause.
Altered surface and biofilm properties of the ΔflhE strain
Transposon mutations attenuating swarming motility of flagellated bacteria have been mapped to genes involved in the biosynthesis, not only of cell-free surfactants (Nakano et al., 1992; Eberl et al., 1999), but also of LPS (Toguchi et al., 2000; Belas et al., 1995) and EPS. Fig. 4(d) shows that representative samples of crude LPS from the wild-type and ΔflhE strains, extracted according to Hitchcock & Brown (1983), failed to highlight any obvious differences. Furthermore, low-molecular-mass (Fig. 4d) and high-molecular-mass LPS (data not shown) were analysed by silver staining (Guard-Petter et al., 1995), and, again, no changes between the two strains were evident. In common with LPS, some components of the EPS are thought to reduce surface resistance, and aid in swarming migration; for example, a mutation in the cmfA gene of the strongly swarming Proteus mirabilis abolished swarming migration due to loss of an EPS rich in galacturonic acid and galactosamine (Gygi et al., 1995). However, when crude acid hydrolysable EPS was assessed according to Gygi et al. (1995), again no differences were observed between the mutant and the wild-type (data not shown). This is unsurprising, since the biosynthetic pathways for LPS and several types of EPS are well characterized, and FlhE shares no motifs with their enzymes.
Such transposon mutations attenuating swarming motility commonly reduce the ‘wettability’ of the bacterial cell surface (Toguchi et al., 2000; Gygi et al., 1995; Belas et al., 1995; Lai et al., 2005), and swarming by such mutants, and of the weakly swarming Esc. coli K-12, can be recovered by the addition of external surfactants such as Tween 80 (Niu et al., 2005; Toguchi et al., 2000). The Sal. typhimurium ΔflhE strain was incubated on 0.6% agar plates containing the non-ionic detergent Tween 80 to increase wetting and reduce the surface tension of the agar. As shown in Fig. 5(a), swarming was recovered to almost the wild-type level. However, this could not be restored by addition of spent medium from a wild-type culture, indicating that the swarming defect of the ΔflhE strain was not due to the absence of a secreted surfactant, such as serawettin from Ser. marcescens (Matsuyama et al., 1992).
Fig. 5.
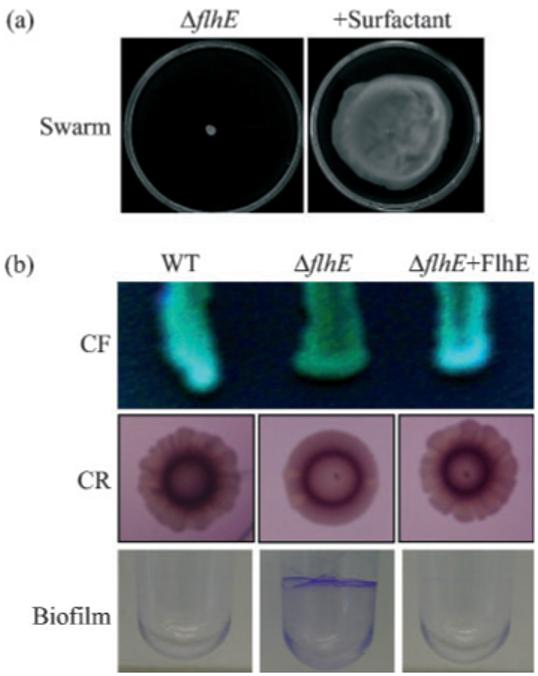
Recovery of ΔflhE swarming and changes in ΔflhE mutant colony morphology. (a) The ΔflhE mutant incubated (14 h) on swarm agar with and without Tween 80 (0.02% w/v) surfactant. (b) Wild-type (WT), the ΔflhE mutant and the ΔflhE mutant carrying pBAD18-FlhE were grown on agar containing calcoflour (CF; 200 μg ml-1) for 72 h, agar containing Congo red (40 μg ml-1) and Coomassie blue (20 μg ml-1) (CR) for 96 h, and in LB in PVC wells for 16 h before visualization of the biofilm with crystal violet (1%).
It therefore seemed possible that other unknown FlhE-related surface properties were influencing the ability to swarm. We incubated the wild-type, the ΔflhE mutant, and the ΔflhE mutant complemented with FlhE, on LB agar containing calcofluor, an LB agar containing Congo red and Coomassie blue, which have been used to highlight altered sugar composition (binding to β-glucans, particularly cellulose) and expression of thin aggregative filaments (curli) in the Sal. typhimurium extracellular matrix (Solano et al., 1998; Römling et al., 1998). Fig. 5(b) shows that the ΔflhE mutant colonies have altered colony morphology on both media, and that this phenotype reverted to wild-type when FlhE was provided in trans. Such changes in calcofluor-binding properties of colonies have been shown to correlate with mutations in the bcs operons responsible for biosynthesis of cellulose (Solano et al., 2002). This change is concomitant with defects in biofilm formation on a glass surface (Solano et al., 1998). However, the ΔflhE mutant colonies were still able to make biofilm under similar conditions (i.e. glass in adherence test medium) (data not shown), suggesting that the ΔflhE change in calcofluor binding was not due to alteration in cellulose production. Despite the Salmonella wild-type SJW1103 not displaying an rdar phenotype on Congo-red-containing medium, it did display a lacy edged colony morphology, while the ΔflhE colonies did not (Fig. 5b); this is another indicator of altered extracellular matrix composition. Altered colony morphology on Congo red plates can be associated with loss of thin aggregative filaments (tafi, also known as curli), encoded in Salmonella by the agf/csg operon (Römling et al., 1998; Solano et al., 2002; Guard-Petter, 2001). Nevertheless, there are examples in the literature of many variations in Congo red colony morphology, depending not only on curli expression, but also on expression of other factors, such as LPS and polysaccharide biosynthesis genes (e.g. wzxE and wcaI) (Solano et al., 2002).
The extracellular matrices of Salmonella and Esc. coli are also involved in biofilm formation on other inert surfaces, such as PVC and polystyrene (Mireles et al., 2001; Römling et al., 1998), and reduced swarming and increased adherence to PVC have been reported in a ddhC mutant (defective in O antigen synthesis) (Mireles et al., 2001). We assessed biofilm formation by wild-type and ΔflhE strains growing on the PVC surface of microtitre wells. After crystal violet staining (Fig. 5b), quantification according to Mireles et al. (2001) confirmed the visual impression that the ΔflhE mutant formed approximately fivefold more biofilm than wild-type under all conditions tested (0.5-2% glucose). Altered biofilm formation on PVC surfaces can also be associated with altered curli expression levels, but this is not the case for the ΔflhE strain, since assessment of curli levels using anti-CsgA antisera indicated unchanged curli expression (data not shown). Nonetheless, the extracellular matrix is complex, and new components continue to come to light (Wang et al., 2004; Branda et al., 2005).
Conclusion
The data suggest that flhE belongs to the flagellar regulon, but is not required for individual cell motility, or any aspect of flagellar production. The data suggest that it nevertheless has a role in the swarming motility of peritrichously flagellated Gram-negative bacteria, possibly influencing the composition of the extracellular matrix, and increasing surface lubrication or wettability. The protein sequences deduced from the flhE genes cited in Fig. 1 are short sequences of 138-158 aa that have no significant similarity with any protein in the current sequence databases. All FlhE proteins have a putative signal peptidase I leader sequence, indicative of a periplasmic or outer-membrane location, and removal of this N-terminal sequence (aa 1-16) apparently results in a loss of function. FlhE proteins have 7-13 proline residues, and proline-rich regions are often involved in protein-protein interactions (Seifert et al., 2004; Larsen et al., 1993). The flhE gene is not associated in the genome with other unknown genes, suggesting that it is not part of a pathway, but rather that it may encode a structural protein that acts alone on the surface, or contributes to a matrix-specific biofilm; for example, a protein that influences interaction with other cells in raft formation, or lubrication for surface movement. These possibilities are as yet unsupported by data, and it remains to be seen what this motility protein does.
ACKNOWLEDGEMENTS
We thank Jean Guard-Bouldin and Gillian Fraser for useful discussions, Johnathan Green and Lyndsey Brawn for assistance with fluorescence microscopy, and Ute Römling for anti-CsgA antisera. This work was supported by a Wellcome Trust Programme grant (C. H.).
Abbreviations
- EPS
exopolysaccharide
- EtBr
ethidium bromide
REFERENCES
- Allison C, Hughes C. Closely linked genetic loci required for swarm cell differentiation and multicellular migration by Proteus mirabilis. Mol Microbiol. 1991;5:1975–1982. doi: 10.1111/j.1365-2958.1991.tb00819.x. [DOI] [PubMed] [Google Scholar]
- Allison C, Emody L, Coleman N, Hughes C. The role of swarm cell differentiation and multicellular migration in the uropathogenicity of Proteus mirabilis. J Infect Dis. 1994;169:1155–1158. doi: 10.1093/infdis/169.5.1155. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auvray F, Thomas J, Fraser GM, Hughes C. Flagellin polymerisation control by a cytosolic export chaperone. J Mol Biol. 2001;308:221–229. doi: 10.1006/jmbi.2001.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belas R, Goldman M, Ashliman K. Genetic analysis of Proteus mirabilis mutants defective in swarmer cell elongation. J Bacteriol. 1995;177:823–828. doi: 10.1128/jb.177.3.823-828.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Chadsey MS, Karlinsey JE, Hughes KT. The flagellar anti-sigma factor FlgM actively dissociates Salmonella typhimurium sigma28 RNA polymerase holoenzyme. Genes Dev. 1998;12:3123–3136. doi: 10.1101/gad.12.19.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri RR, Khan AM, Pallen MJ. coliBASE: an online database for Escherichia coli, Shigella and Salmonella comparative genomics. Nucleic Acids Res. 2004;32:296–299. doi: 10.1093/nar/gkh031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour A, Furness RB, Hughes C. Novel genes that upregulate the Proteus mirabilis flhDC master operon controlling flagellar biogenesis and swarming. Mol Microbiol. 1998;29:741–751. doi: 10.1046/j.1365-2958.1998.00967.x. [DOI] [PubMed] [Google Scholar]
- Eberl L, Soren-Molin I, Givskov M. Surface motility of Serratia liquefaciens MG1. J Bacteriol. 1999;181:1703–1712. doi: 10.1128/jb.181.6.1703-1712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser GM, Hughes C. Swarming motility. Curr Opin Microbiol. 1999;2:630–635. doi: 10.1016/s1369-5274(99)00033-8. [DOI] [PubMed] [Google Scholar]
- Givskov M, Eberl L, Christiansen G, Benedik MJ, Molin S. Induction of phospholipase and flagellar synthesis in Serratia liquefaciens is controlled by expression of the flagellar master operon flhD. Mol Microbiol. 1995;15:445–454. doi: 10.1111/j.1365-2958.1995.tb02258.x. [DOI] [PubMed] [Google Scholar]
- Guard-Petter J. The chicken, the egg and Salmonella enteritidis. Environ Microbiol. 2001;3:421–430. doi: 10.1046/j.1462-2920.2001.00213.x. [DOI] [PubMed] [Google Scholar]
- Guard-Petter J, Lakshmi B, Carlson R, Ingram K. Characterization of lipopolysaccharide heterogeneity in Salmonella enteritidis by an improved gel electrophoresis method. Appl Environ Microbiol. 1995;61:2845–2851. doi: 10.1128/aem.61.8.2845-2851.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi D, Rahman MM, Lai HC, Carlson R, Guard-Petter J, Hughes C. A cell-surface polysaccharide that facilitates rapid population migration by differentiated swarm cells of Proteus mirabilis. Mol Microbiol. 1995;17:1167–1175. doi: 10.1111/j.1365-2958.1995.mmi_17061167.x. [DOI] [PubMed] [Google Scholar]
- Hay NA, Tipper DJ, Gygi D, Hughes C. A nonswarming mutant of Proteus mirabilis lacks the Lrp global transcriptional regulator. J Bacteriol. 1997;179:4741–4746. doi: 10.1128/jb.179.15.4741-4746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay NA, Tipper DJ, Gygi D, Hughes C. A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J Bacteriol. 1999;181:2008–2016. doi: 10.1128/jb.181.7.2008-2016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock PJ, Brown TM. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlinsey JE, Tanaka S, Bettenworth V, Yamaguchi S, Boos W, Aizawa SI, Hughes KT. Completion of the hook-basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol Microbiol. 2000;37:1220–1231. doi: 10.1046/j.1365-2958.2000.02081.x. [DOI] [PubMed] [Google Scholar]
- Kutsukake K, Ohya Y, Iino T. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J Bacteriol. 1990;172:741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai HC, Soo PC, Wei JR, Yi WC, Liaw SJ, Horng YT, Lin SM, Ho SW, Swift S, Williams P. The RssAB two-component signal transduction system in Serratia marcescens regulates swarming motility and cell envelope architecture in response to exogenous saturated fatty acids. J Bacteriol. 2005;187:3407–3414. doi: 10.1128/JB.187.10.3407-3414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen RA, Wood GE, Postle K. The conserved proline-rich motif is not essential for energy transduction by Escherichia coli TonB protein. Mol Microbiol. 1993;10:943–953. doi: 10.1111/j.1365-2958.1993.tb00966.x. [DOI] [PubMed] [Google Scholar]
- Macnab RM. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt FC, others, editors. Washington, DC: American Society for Microbiology; 1996. pp. 123–145. [Google Scholar]
- Matsuyama T, Kaneda K, Nakagawa Y, Isa K, Hara-Hotta H, Yano I. A novel extracellular cyclic lipopeptide which promotes flagellum-dependent and -independent spreading growth of Serratia marcescens. J Bacteriol. 1992;174:1769–1776. doi: 10.1128/jb.174.6.1769-1776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter LL. Polar flagellar motility of the Vibrionaceae. Microbiol Mol Biol Rev. 2001;65:445–462. doi: 10.1128/MMBR.65.3.445-462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbour, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Minamino T, Iino T, Kutuskake K. Molecular characterization of the Salmonella typhimurium flhB operon and its protein products. J Bacteriol. 1994;176:7630–7637. doi: 10.1128/jb.176.24.7630-7637.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mireles JR, II, Toguchi A, Harshey RM. Salmonella enterica serovar typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J Bacteriol. 2001;183:5848–5854. doi: 10.1128/JB.183.20.5848-5854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano MM, Corbell N, Besson J, Zuber P. Isolation and characterization of sfp: a gene that functions in the production of the lipopeptide biosurfactant, surfactin, in Bacillus subtilis. Mol Gen Genet. 1992;232:313–321. doi: 10.1007/BF00280011. [DOI] [PubMed] [Google Scholar]
- Niu C, Graves JD, Mokuolu FO, Gilbert SE, Gilbert ES. Enhanced swarming of bacteria on agar plates containing the surfactant Tween 80. J Microbiol Methods. 2005;62:129–132. doi: 10.1016/j.mimet.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Ohnishi K, Ohto Y, Aizawa S, Macnab RM, Iino T. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J Bacteriol. 1994;176:2272–2281. doi: 10.1128/jb.176.8.2272-2281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Bian Z, Hammar M, Sierralta WD, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998;180:722–731. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert TB, Bleiweis AS, Brady LJ. Contribution of the alanine-rich region of Streptococcus mutans P1 to antigenicity, surface expression, and interaction with the proline-rich repeat domain. Infect Immun. 2004;72:4699–4706. doi: 10.1128/IAI.72.8.4699-4706.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Solano C, Sesma B, Alvarez M, Humphrey TJ, Thorns CJ, Gamazo C. Discrimination of strains of Salmonella enteritidis with differing levels of virulence by an in vitro glass adherence test. J Clin Microbiol. 1998;36:674–678. doi: 10.1128/jcm.36.3.674-678.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano C, Garcia B, Valle J, Berasain C, Ghigo JM, Gamazo C, Lasa I. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol Microbiol. 2002;43:793–808. doi: 10.1046/j.1365-2958.2002.02802.x. [DOI] [PubMed] [Google Scholar]
- Soutourina OA, Bertin PN. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev. 2003;27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- Stafford GP, Ogi T, Hughes C. Binding and transcriptional activation of non-flagellar genes by the Escherichia coli flagellar master regulator FlhD2C2. Microbiology. 2005;151:1779–1788. doi: 10.1099/mic.0.27879-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toguchi A, Siano M, Burkart M, Harshey RM. Genetics of swarming motility in Salmonella enterica serovar typhimurium, critical role for lipopolysaccharide. J Bacteriol. 2000;182:6308–6321. doi: 10.1128/jb.182.22.6308-6321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Frye JG, McClelland M, Harshey RM. Gene expression patterns during swarming in Salmonella typhimurium, genes specific to surface growth and putative new motility and pathogenicity genes. Mol Microbiol. 2004;52:169–187. doi: 10.1111/j.1365-2958.2003.03977.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Preston JF, III, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Fujita H, Taira T, Kutsukake K, Homma M, Iino T. Genetic analysis of three additional fla genes in Salmonella typhimurium. J Gen Microbiol. 1984;130:3339–3342. doi: 10.1099/00221287-130-12-3339. [DOI] [PubMed] [Google Scholar]


