Abstract
DdLim, a multi-domain member of the cysteine-rich family of LIM domain proteins, was isolated from Dictyostelium cells where it localizes in lamellipodia and at sites of membrane ruffling. The transcription and expression of DdLim are developmentally regulated, and the timing of its increased association with the actin cytoskeleton coincides with the acquisition in starved cells of a motile, chemotactic behavior. Vegetative cells that overexpress DdLim contain large lamella and exhibit ruffling at the cortex. The high frequency of large, multinucleated mutant cells found in suspension culture suggests that excess DdLim interferes with cytokinesis. DdLim was also identified as a protein in a Dictyostelium cell lysate that associated indirectly, but in a guanosine triphosphate-dependent manner, with a GST-rac1 fusion protein. The data presented suggest that DdLim acts as an adapter protein at the cytoskeleton-membrane interface where it is involved in a receptor-mediated rac1-signaling pathway that leads to actin polymerization in lamellipodia and ultimately cell motility.
INTRODUCTION
Vegetative cells of Dictyostelium discoideum hunt bacteria by folic acid-based chemotaxis. This type of cell motility is not particularly fast, and at a low cell density with a plentiful supply of bacteria, the cells do not need to change their average position and feed by projecting lamellipodia toward the bacteria, which are subsequently bound and engulfed (Maniak et al., 1995). During starvation, Dictyostelium cells initiate a developmental program that by 6 h endows on preaggregation-competent cells the ability to move at a velocity of up to 10 μm/min in a cAMP-dependent chemotactic response. Lamellipodial protrusion in both growth phase- and preaggregation-competent cells has been shown to require the polymerization of actin at the site of chemoreceptor activation. The polarized, highly motile behavior of preaggregation-competent cells, compared with the random walk of growth phase cells, suggests that subtle differences exist in the regulation and coupling of molecular forces derived from actin polymerization to the protrusion of filopodia and lamellipodia. In this context, Hall and coworkers (Nobes and Hall, 1995) have demonstrated that the regulation of actin cytoskeleton protrusions implicated in cell motility is dependent on the presence of activated forms of the small guanosine triphosphatases (GTPases), rac, rho, and cdc42. In mammalian cells, these proteins trigger the formation of lamellipodia, stress fibers, and filopodia, respectively. Significantly, certain isoforms of rac are induced in the transition of Dictyostelium cells from the growth phase to the highly motile preaggregation-competent stage (Bush et al., 1993).
One of the major objectives in cell motility research is to understand the molecular mechanism of actin polymerization-based protrusion of lamellipodia and filopodia. To address this objective we have developed a new screening strategy designed to identify and characterize cytoskeleton-associated proteins responsible for the change in motile behavior of Dictyostelium cells from growth to the preaggregation-competent stages. Candidates are defined as proteins whose expression level increases in this developmental transition and whose interaction with the cytoskeleton is determined by calcium ions or other second messengers known to play a regulatory role in motility (Brundage et al., 1993). The first protein identified from this screen, named DdLim, is a unique member of the actin cytoskeleton-associated, cysteine-rich protein (CRP) family of the zinc ion-binding LIM domain family of proteins (Sadler et al., 1992; Crawford et al., 1994; Weiskirchen et al., 1995). Although most LIM domain proteins function as transcription factors and have been implicated in the differentiation and development of cells (for review see Sadler et al., 1992; Sanchez-Garcia and Rabbitts, 1994) the function of the CRP family of LIM domain proteins has not been fully established, although their interaction with actin-binding proteins (ABPs) and localization at focal contacts and stress fibers in mammalian cells points to a regulatory or structural role in the actin cytoskeleton (Sadler et al., 1992; Arber et al., 1994). The multiplicity of protein binding domains in the CRP family of LIM proteins may allow them to function as a template or adapter capable of promoting specific protein-protein interactions at the cytoskeleton. The specificity of protein-protein interactions mediated by CRPs, already demonstrated for the CRP1 protein which binds to zyxin (Crawford et al., 1992; Schmeichel and Beckerle, 1994, 1997; Arber and Caroni, 1996), might have evolved via a mix-and-match combination of different LIM genes, in which case the few known members of this family will probably represent a small fraction of the total number of CRPs.
Elucidation of the cellular functions of CRPs will no doubt emerge from comparative biochemical and cell biological studies on the role of these proteins in different cytoskeleton-dependent processes in diverse cell systems. As part of this approach we present a characterization of the DdLim protein isolated from Dictyostelium with the aim of understanding its role in the dramatic developmental transformation that allows slowly moving vegetative cells to acquire the highly motile and chemotactic characteristics of preaggregation cells. A mutant cell that overexpresses the DdLim protein is described, and a characterization of its phenotypes implicates DdLim in the regulation of a number of cell processes that rely on the cytoskeleton including cytokinesis, growth, and the regulation of cytoskeleton protrusions including lamellipodia- and actin-based ruffling at the cell cortex.
MATERIALS AND METHODS
Isolation and Peptide Analysis of DdLim
Triton X-100–insoluble cell fractions of Dictyostelium cells were isolated essentially as described by Prassler et al. (1997) with modified stabilizing and extraction buffers. The lysis buffer contained 10 mM piperazine-N,N′-bis-ethanesulfonic acid (PIPES) pH 6.8, 20 mM KCl, 2 mM MgS04, 5 mM EGTA, and 30% glycerol, and the Ca2+-extraction buffer consisted of 10 mM PIPES, pH 7.0, 2 mM MgS04, 20 mM CaCl2, and a protease inhibitor cocktail; (leupeptin, bestatin A, antipain, pepstatin A, 10 μg/ml each). After extraction of the pellet for 1 h on ice, the sample was centrifuged for 20 min at 130,000 × g, and the supernatant was applied to a DE52-column equilibrated in the extraction buffer. Actin, α-actinin, and DdLim were bound to the column and eluted using a linear NaCl gradient from 0–0.5 M in extraction buffer. DdLim eluted from this column between 220 and 280 mM NaCl, and these fractions were pooled and further fractionated on a Mono Q column equilibrated in 20 mM Tris-HCl, pH 8.0, 0.2 mM dithiothreitol (DTT), 0.1 mM CaCl2, protease inhibitor cocktail (1:1000 vol/vol). Fractions containing DdLim eluted at 480 mM NaCl. The protein was separated from others using SDS-PAGE and eluted from the polyacrylamide gel by electrophoresis. Partial amino acid sequencing of DdLim was achieved by digestion of this purified protein with Lys-C (Boehringer Mannheim, Mannheim, Germany). The peptide fragments were then separated on reverse-phase high-pressure liquid chromatography and microsequenced on an LF 3600 Porton gas phase sequencer (Beckman, Fullerton, CA).
The degenerated oligonucleotide (P1) GCIGTIACIGAYHSIGTIGC was designed from the DdLim peptide sequence, AVTDSV, and the 3′-antisense oligonucleotide (P2) ARIG CRTTIGCIGTIGCCAT was designed from the peptide sequence MATANA (Y = C/T; R = A/G; H = A/C/T: S = G/C). PCR was performed using 1 μg of genomic DNA and 1 nmol of each primer per 100 μl reaction volume. The cycling conditions were one cycle at 96°C for 150 s; 30 cycles at 94°C for 1 min, 53°C for 1 min, 72°C for 90 s; one cycle at 72°C for 5 min.
A cDNA expression library from mRNA obtained from D. discoideum AX3 strain cells starved for 4 h (Clontech, Palo Alto, CA) was screened with a 130-base pair (bp) probe obtained by PCR from genomic Dictyostelium DNA. cDNA inserts from positive phages were amplified by PCR using λgt11-specific oligonucleotide primers. PCR products were directly subcloned into the pGEMT vector (Promega, Madison, WI) and sequenced in both orientations using the dye termination method (Perkin Elmer-Cetus, Norwalk, CT) on an ABI fluorescent sequencer (Applied Biosystems, Foster City, CA) using uni-, reverse, and sequence-specific oligonucleotide primers. The sequence of DdLim was analyzed using the FASTA and BESTFIT programs of the University of Wisconsin Genetics Computer Group software (Devereux et al., 1984) and the MIPSX database (Max Planck Institute for Biochemistry, Martinsried, Germany).
Expression of DdLim as an amino-terminal hexa-histidine fusion protein was achieved by introducing restriction sites into the DdLim cDNA sequence using GCGGATCCATGTCTGCTTCAGTTAAATGTGG (P3), and CCCAAGCTTCAG AGAGAGTCAGGTAGTGT (P4), which were cloned in the pQE30 expression vector (Qiagen, Chatsworth, CA). The histidine-tagged DdLim was purified from soluble bacterial extracts after ITPG induction for 3 h using a Ni-NTA-agarose as recommended by the manufacturer (Qiagen). Overexpression of DdLim in Dictyostelium was achieved by modifying the DdLim cDNA by adding an EcoRI restriction site to the 5′-(GGAATTCAAAAATGTCTGCTTCAGTAA; P5), and a HindIII restriction site to 3′-end using the P4 primer, which were then cloned into the pDEXRH expression vector (Faix et al., 1992).
Expression of DdLim as a green flourescent protein (GFP)-fusion protein was achieved by adding EcoRI sites to the 5′- (GCGGAATTCAAAATGTCTGCTTCAGTTA; P6) and 3′- (CGGAATTCTTATTGT TCTTCTTCGTATTGTTG; P7) ends of the DdLim–coding region by PCR. This strategy allowed cloning into the EcoRI site of a red-shifted (S65T) Dictyostelium GFP-expression vector (Westphal et al., 1997).
Dictyostelium cells were transformed by the calcium phosphate method (Nellen et al., 1984), and transformants were selected for resistance to G418 at 20 μg/ml. DdLim-overproducing cells were identified on a protein colony blot using affinity-purified anti-DdLim polyclonal antibodies. Independent clones were isolated as described by Wallraff and Gerisch (1991).
Polyclonal antibodies directed against affinity-purified DdLim were raised in a rabbit by subcutaneous immunization with recombinant DdLim in complete Freund’s adjuvant using a standard immunization protocol. After 6 wk, the immunoglobulin G antibody fraction in serum obtained from the rabbit was affinity-purified using bacterially expressed DdLim covalently coupled to an N-hydroxysuccinimide-activated Sepharose column according to the manufacturers instructions (Pharmacia, Piscataway, NJ). Bound antibodies were eluted with 100 mM glycine, pH 3.0, neutralized, and then dialyzed against phosphate-buffered saline. The titer of these antibodies was established and then employed for iodination, Western blotting, and immunofluorescence labeling.
Growth and Development of Dictyostelium Cells
The Dictyostelium discoideum strain AX2 was cultivated in nutrient medium at 23°C and shaken in suspension at 150 rpm, or cells were cultivated in medium on Petri dishes. For development to aggregation competence, cells were washed twice with 17 mM K-Na phosphate buffer and resuspended in this buffer to a cell density of 1 × 107 cells/ml and shaken as described above for 12 h. For filter-based development, cells were washed as described above and resuspended to a density of 1 × 108 cells/ml and handled as described by Newell et al. (1969). For RNA and protein preparations, cells were scraped from the filter with a razor blade and resuspended in 50 mM HEPES, pH 7.4, for RNA, or lysed in 20 mM Tris-HCl, pH 7.5, 0.1% SDS, 1 mM DTT, 10 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride for determination of protein concentration (Furukawa et al., 1992). Equal amounts of cell lysates, as determined by the method of Bradford (1976), were separated by SDS-PAGE, electrophoretically transferred to nitrocellulose, and probed with 125I-labeled, anti-DdLim antibodies. The DdLim expression level was then quantified using a Fuji phospho-imager (Fuji, Stamford, CT).
Miscellaneous
DNA and RNA were prepared as described by Witke et al. (1986). Northern and Southern blots were hybridized in the presence of 50% or 30% formamide. SDS-PAGE was performed on 8–18% gradient gels or on 12% mini gels using the buffer system described by Laemmli (1970). For immunoblotting, proteins were transferred to nitrocellulose and probed with 125I-labeled antibodies. For Northern blots, total cellular RNA was separated in 1.2% agarose gels in the presence of 6% formaldehyde and probed with 32P-labeled DdLim cDNA.
Electron Microscopy and Immunofluorescence and Live Cell Imaging of GFP-DdLim
Scanning electron microscopy was performed on cells that had settled on acid-washed glass coverslips for 20 min and cells were fixed according to Claviez et al. (1986) with 1% glutaraldehyde and 0.02% osmium tetroxide, dehydrated in alcohol and acetone, and then critical point dried. The specimens were coated with gold to a thickness of 20 nm just before examination with a Joel JSM 35C scanning electron microscope (Joel, Tokyo, Japan) at 25 kV.
For immunofluorescence labeling, cells from suspension cultures were allowed to adhere for 30 min on glass coverslips before fixation. Cells were either fixed for 10 min with cold methanol at −20°C or for 15 min in a solution of 15% saturated picric acid and 2% paraformaldehyde (pH 6.0) and postfixed with 70% ethanol and further processed as described by Faix et al. (1996).
GFP-DdLim was imaged in live cells using a fluorescence microscope optimized for the detection of red-shifted GFP that is described in Westphal et al. (1997).
Expression and Purification of Dictyostelium rac1A and Human rac1 and Affinity-binding Experiments
D. discoideum rac1A (Bush and Cardelli, 1993), human rac1, and human cdc42 (Nobes and Hall, 1995) were expressed in Escherichia coli using pGEX vectors (Pharmacia) as glutathione S-tranferase (GST)-fusion proteins, and purified on Sepharose beads according to the instructions of the manufacturer. For binding assays, purified GST-fusion proteins were bound to GST Sepharose beads and loaded with either GDP or GTPγS essentially as described by Hart et al., (1996).
Vegetative D. discoideum cells (1 × 109) were washed twice in phosphate buffer and lysed with buffer containing 25 mM Tris-HCl, pH 7.4, 50 mM NaCl, 5 mM MgCl2, 2 mM DTT, 1% n-octylpolyoxyethylene (Bachem, Torrance, CA), 2 mM benzamidine, and protease inhibitors as described above. The lysate was centrifuged for 30 min at 10,000 × g, and 1 ml of the supernatant (at approximately 2 mg/ml) was incubated with 250 μg of GST-rac1A, human rac1, cdc42, or GST alone. After 1 h of incubation the beads were washed five times with lysis buffer, and bound proteins eluted with SDS sample buffer were subjected to SDS-PAGE analysis.
RESULTS
DdLim Associates with the Actin Cytoskeleton
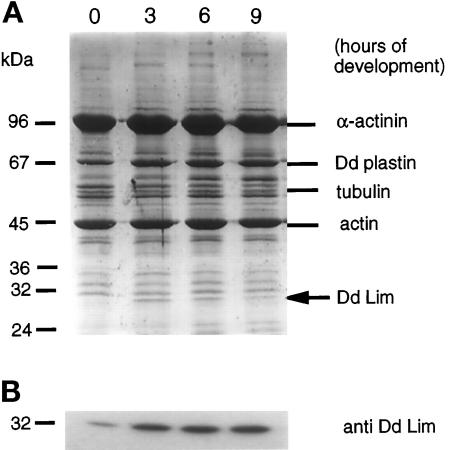
The calcium ion extract of a Triton X-100–insoluble cytoskeleton fraction of Dictyostelium cells has proven to be a rich and valuable source of structural and regulatory actin-binding proteins (Jungbluth et al., 1995; Prassler et al., 1997). This specificity was shown in a Western blot analysis of the calcium ion extract, which identified α-actinin and Ddplastin, two well-known Ca2+-regulated actin-binding proteins (our unpublished results). The relative amount of these cross-linking proteins in the Ca2+-extractable, Triton X-100–insoluble cytoskeleton was invariant during the different developmental stages of Dictyostelium as seen in the Coomassie-stained gel (Figure 1A). This property, therefore, does not satisfy one of our search criteria designed to identify candidate proteins, and this is in accord with an earlier report that showed an α-actinin knock-out mutant exhibits normal cell motility (Witke et al., 1992). On the other hand, a number of protein bands can be seen in this SDS-PAGE whose intensity increases from 0–6 h poststarvation. One of these proteins, which migrates with an apparent molecular weight of 30 kDa, was chosen for further study and identified by sequence analysis (see below) to be a member of the LIM family of proteins.
Figure 1.
(A) Association of cytoskeletal proteins of D. discoideum with detergent-extracted cytoskeleton preparations from different developmental stages. Cells were starved in shaking culture (as indicated) and lysed with 1% Triton X-100 in the presence of cytoskeleton-stabilizing buffer (10 mM PIPES, pH 6.8, 20 mM KCl, 2 mM MgSO4, 30% glycerol). Triton X-100–insoluble material was pelleted and extracted with Ca2+-buffer (10 mM PIPES, pH 7.0, 20 mM KCl, 2 mM MgSO4, 20 mM CaCl2, protease inhibitors). Proteins of the Ca2+ extracts were separated on SDS-PAGE, visualized by Coomassie blue staining, and analyzed for the accumulation of proteins during the transition of vegetative cells (t0) to highly motile preaggregation stage (t3 to t9). The increased association of a minor protein component, a 30 kDa, with the Ca2+-extractable fraction of Triton X-100–insoluble material from aggregation-competent cells is shown in panel A by an arrow and is more clearly visible in Western blots with anti-DdLim–specific antibodies (Figure 1B). The exact values indicated in the text were quantified by phospho-image analysis of Western blots using 125I-labeled anti-DdLim antibodies.
The increased association of DdLim with the actin cytoskeleton during development can be seen more clearly in a Western blot analysis using specific antibodies raised against this protein (Figure 1B). The acquisition of the motile response in wild-type cells between 3 and 6 h poststarvation is mirrored by an increased accumulation of DdLim in the Ca2+-extracted cytoskeleton to 58% of the total cellular DdLim (Figure 1B).
Sequence Analysis of DdLim
The DdLim protein was purified from the Ca2+-extracted cytoskeleton using two chromatographic steps, concentrated, and subjected to Lys-C proteolytic fragmentation for peptide sequencing. Degenerated oligonucleotides derived from two of these peptide sequences were used to perform a PCR on genomic DNA, which led to the amplification of a 160-bp DNA fragment whose sequence coded for a peptide with the expected amino acids at each of its ends. This DNA fragment was used to screen a cDNA expression library obtained from cells 4 h after starvation (CLONTECH). The longest cDNA clone that hybridized to this probe was 715 bp long with an open reading frame from base pairs 1–634. The first methionine in this frame was located at 34 bp and most likely serves as the initiation codon since the sequence preceding this ATG is unusually rich in A and T bases, which are normally only found in nontranslated regions of the Dictyostelium genome. The coding region is terminated by a stop-codon that forms part of the polyadenylation signal AATAA. The open reading frame of the cDNA clone codes for a polypeptide of 199 amino acid residues with a molecular weight of 22.1 kDa, and the predicted isoelectric point of 4.7 reflects a high content of acidic residues. To establish that this polypeptide represents the full coding length of the cDNA clone, and that the first methionine in the open reading frame is the initiation codon, we expressed the cDNA from base pair +34 to position 642 in Dictyostelium and E. coli. The proteins expressed from this cDNA in both cell systems migrated in an SDS-PAGE gel with a molecular weight of 30 kDa and 32 kDa, respectively (due to the 6 histidine residue tag). Correspondingly, the difference between the calculated and experimentally determined molecular weight must be due to an unusual migratory behavior of this protein during electrophoresis, as is often seen for proteins with a high proportion of acidic residues.
Domain Structure of DdLim
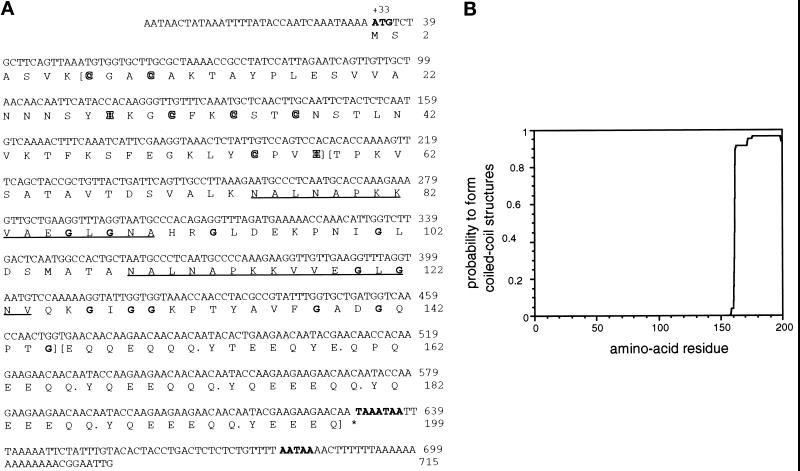
Analysis of the deduced polypeptide sequence of DdLim (Figure 2A) reveals the existence of three distinct structural domains. The amino-terminal region, which spans residues 7–57, exhibits a high degree of homology to family of LIM domain proteins (reviewed by Sadler et al., 1992; Crawford et al., 1994). The essential elements of this protein family are seven cysteine and histidine residues separated by a defined number of amino acids to form a double zinc ion-binding finger structure called the LIM domain (Michelsen et al., 1993, 1994; Kosa et al., 1994). The LIM motif is characterized by the amino acid sequence CX2CX16–23HX2C-X2-CX2CX16–21CX2(C,H,D) (Freyd et al., 1990; Karlsson et al. 1990; Sadler et al., 1992). Whereas the positions of the cysteine and histidine residues are invariant in this sequence, the length of the loops may span from 16 to 23 amino acids. The sequence and size of the loop region probably define the protein or nucleic acid-binding specificity of each LIM domain (Schmeichel and Beckerle, 1997). The amino acid sequence of the LIM domain of DdLim, which can be represented as (CX2CX17HX2C)-X2-(CX2CX17CX2H), has an intervening sequence of 17 amino acids in both loops and conforms exactly to the organization of the LIM domain in members of the CRP family (Stronach et al., 1996). Furthermore, DdLim, like other members of the CRP family, was found to associate with the actin cytoskeleton.
Figure 2.
(A) Nucleotide sequence and derived amino acid sequence of DdLim. The start codon, the stop codon, and two potential polyadenylation signals are indicated in bold letters in the nucleotide sequence. The protein sequence suggests the presence of three different structural domains; the borders of these domains are indicated by parentheses. The N-terminal LIM domain (amino residues 7–57), shows the defined spacing of cysteine and histidine residues (CX2CX17HX2 CX2CX2CX17CX2H) of the LIM domain (shown in outlined letters). The LIM domain is followed by a domain with a high proportion of glycine residues (indicated in bold). Within this domain a 16-amino acid motif is repeated twice (underlined) with two conserved alanine-to-valine exchanges. The high number of glycine residues in this domain is typical of the CRP/MLP group of LIM proteins. The C-terminal domain consists of a hepta-amino acid motif repeated eight times with minor deviations (indicated by a dot after each heptad). The DdLim sequence has been deposited in the GenBank with accession number U97699. (B) Analysis of DdLim sequence by the COILS algorithm of Lupas et al. (1991) predicts, with a high probability, that at least four hepta-repeats (repeat 4 to 7) will form a coiled-coil structure.
A third characteristic that DdLim shares with members of the CRP family is the presence of a glycine-rich domain that normally follows the LIM domain in the protein sequence (Figure 2A). In DdLim, 12 of the 15 glycine residues reside in this second, central domain. A putative nuclear targeting signal (KKYGPK) located within the glycine-rich domain of other CRPs (Stronach et al., 1996) is not present in the DdLim protein. In fact, other than the high content of glycine residues in this domain, there is no sequence homology to the glycine-rich domains of other CRP members. Interestingly though, the glycine-rich domain of DdLim contains two nearly perfect repeat motifs of 16 amino acids with the minor difference that two alanine residues are replaced by valine (Figure 2A, underlined).
DdLim contains a third, C-terminal domain not found in members of the CRP family. This domain spans amino acid residues 146 to the C terminus and shows a periodic arrangement of one α-helix–breaking amino acid followed by 6-helix–forming amino acids. This heptad amino acid arrangement has the motif variation YQEEQQQ and occurs eight times in this domain. The sequence of this domain was analyzed using the coiled-coil algorithm of Lupas et al. (1991) and the pair-coils algorithm (Berger et al., 1995). Employing a MTIDK matrix with a window size of 21 amino acids, and an increased weight of 2.5 at positions a and d, the program predicted, with a high probability (>0.9), that the four perfect heptad repeats between residues 167 and 194 will form a coiled coil (Figure 2B). The same region was predicted by the pair-coil algorithm as a sequence having the capacity to form a dimer. Consistent with this prediction, purified bacterially expressed DdLim eluted on a gel filtration column between ovalbumin (Mr 43,000) and albumin (Mr 67,000; our unpublished results).
Gene Expression of DdLim Is Developmentally Regulated
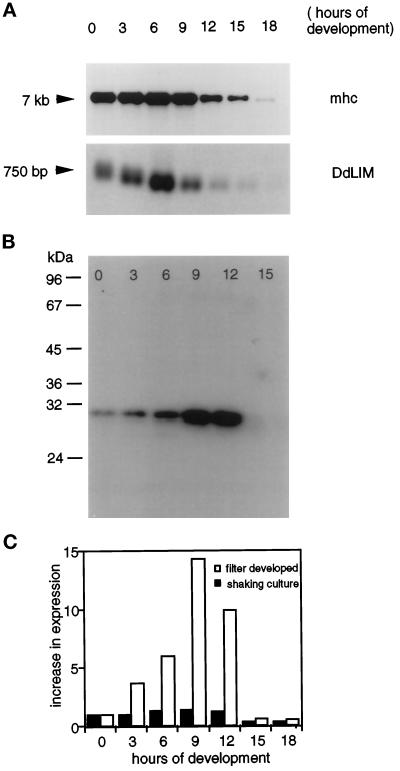
Northern blot analysis performed under highly stringent hybridization conditions identified the DdLim mRNA as a single species of about 750 bp. The transition from vegetative to preaggregation-competent cells was accompanied by a significant increase in the level of DdLim mRNA, and thereafter the message was rapidly degraded such that after 12 h of starvation it was barely detectable (Figure 3A, lower panel).
Figure 3.
(A) Developmental regulation of the transcription and expression of DdLim during the life-cycle of D. discoideum. Five micrograms of total RNA were electrophoretically separated on denaturing agarose-formaldehyde gel and transferred to nitrocellulose paper. Equal loading was confirmed by EtBr staining of the agarose gel. The Northern blot was probed with DdLim cDNA (lower panel) and as loading control with myosin heavy chain cDNA (mhc; upper panel). The DdLim transcript of about 750 bp accumulates between 3 and 6 h of development. (B) The expression of the protein, monitored by Western blotting, appears at a maximum level between 9 and 12 h. Equal concentrations of cellular proteins, as determined by the method of Bradford, were loaded on each lane. Both total RNA and protein were isolated from filter-developed cell at the indicated time points. (C) Influence of the culture condition on the expression level of DdLim. Equal amounts of cellular proteins determined by the method of Bradford were separated by SDS-PAGE, blotted on nitrocellulose, and labeled with [125I]anti-DdLim antibodies. The DdLim signal was quantified by phospho-image analysis. Cells were either starved in phosphate buffer at a cell density of 1 × 107 cells per ml in suspension culture (filled bars), or 5 × 107 cells were plated on filters (empty bars). Only the filter-starved cells showed development to fruiting bodies. The expression level of DdLim greatly increased during this developmental differentiation process, reaching a maximum at 9 h poststarvation.
The expression of the DdLim protein during development was examined in parallel to the mRNA measurements using filter-developed cells. The level of DdLim protein, like the mRNA transcript, accumulated about sixfold in the transition to preaggregation cells, while in cells starved for 9 h, further accumulation resulted in a 14-fold increase compared with vegetative cells (Figure 3, B and C, open bars). Interestingly, the same number of cells starved in phosphate buffer in shaking cultures only exhibited a 1.3-fold increase in DdLim expression in the transition from vegetative cells to preaggregation-competent cells (Figure 3C, solid bars). Cells plated on a surface in nutrient medium at a low cell density did not show any increase in the level of DdLim after prolonged contact with the substratum (our unpublished results), and together these results show that the elevated expression of the DdLim is part of the developmental process.
Southern Blot Analysis Suggests the Presence of a Second Highly Homologous LIM Protein in Dictyostelium
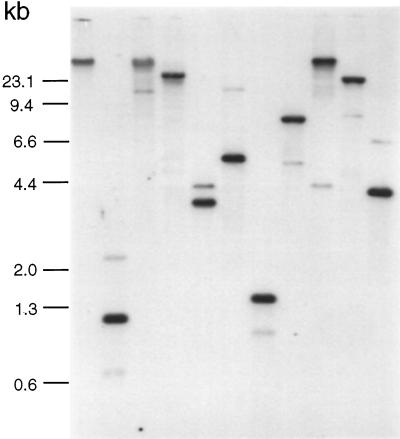
To search for other LIM domain-related genes in Dictyostelium, the full-length DdLim cDNA was used as a probe in a high-stringency hybridization of restricted genomic DNA. The restriction enzymes (KpnI, AsnI, BamHI, HpaI, SspI, SwaI, EcoRI, ClaI, BglII, and BcII) used for the digestion of genomic DNA do not have any internal restriction sites in the DdLim cDNA. In addition to the strong hybridization signal belonging to the DdLim gene (Figure 4), a second band with a weaker intensity was apparent in nearly all lanes and is consistent with the idea that a second DdLim gene or a closely related gene exists in the Dictyostelium genome.
Figure 4.
Southern blot labeled with DdLim DNA probe under stringent conditions (50% formamide) indicates the presence of a second DdLim gene or at least a highly homologous gene. For lanes 1–11, wild- type genomic DNA was digested with KpnI, AsnI, BamHI, HpaI, AluI, SspI, SwaI, EcoRI, ClaI, BglII, and BclI. In all lanes the DdLim gene appears as a strong signal while a second weaker band is also visible.
Cellular Distribution of DdLim
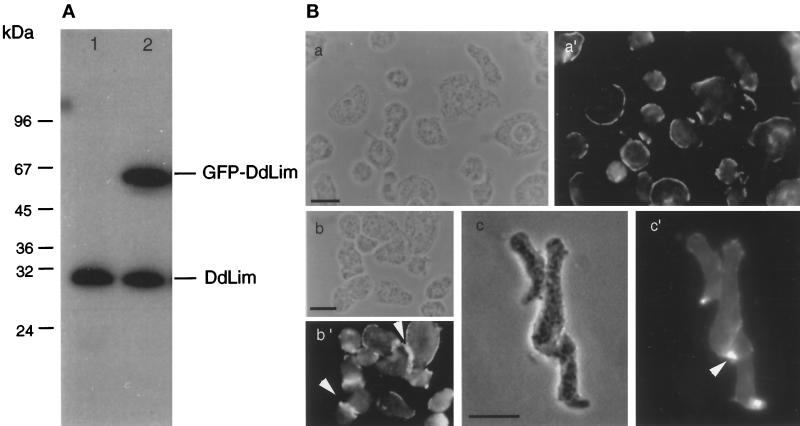
Affinity-purified polyclonal antibodies raised against the bacterially expressed DdLim protein were shown to specifically bind DdLim in Western blots of total protein lysates of Dictyostelium cells (Figure 5A). These antibodies were also used to determine the localization of DdLim within vegetative and aggregating cells by indirect immunofluorescence. In vegetative cells, DdLim was found to localize mainly in the cell cortex (Figure 5B, a′) whereas a weaker signal was seen in the cytoplasm (Figure 5B, a′, b′, and c′). In preaggregation-competent, motile cells, DdLim was primarily localized at the pioneer front (Figure 5B, c′). A strong accumulation of DdLim was also found at the site of cell–cell contacts (Figure 5B, b′ and c′, indicated by arrows).
Figure 5.
(A) Western blot of total cell lysate of wild-type cells (lane 1) and GFP-DdLim cells (lane 2) probed with affinity-purified anti-DdLim antibodies shows the monospecificity of these antibodies used for immunolocalization. GFP-DdLim fusion protein (lane 2) is expressed in these cells to an amount equal to that of the endogenous level of DdLim. (B) Immunolocalization of DdLim in wild-type cells using affinty-purified anti-DdLim antibodies. The phase contrast images are indicated by a, b, and c, the corresponding fluorescence images by a′, b′, and c′. Vegetative (a′ and b′), and preaggregation-competent cells (c′), were fixed with cold methanol and stained with 10 μg/ml of the anti-DdLim antibodies as the first antibody and TRITC-conjugated antirabbit as the second antibody. Scale bar, 10 μm.
To correlate the cellular dynamics of DdLim with the organization of the actin cytoskeleton in lammellipodial protrusions, we expressed a GFP-DdLim fusion protein in WT-cells. In live vegetative cells, GFP-DdLim accumulated at the extreme membrane rim of newly formed protrusions where it remained until the protrusion retracted. In aggregation-competent cells, DdLim transiently accumulated either at the extreme tip of a filopodia or at the front of a newly established region of the cell (our unpublished results). DdLim rapidly dissociated from the cell cortex when the mass of the cell moved in the direction of the leading edge.
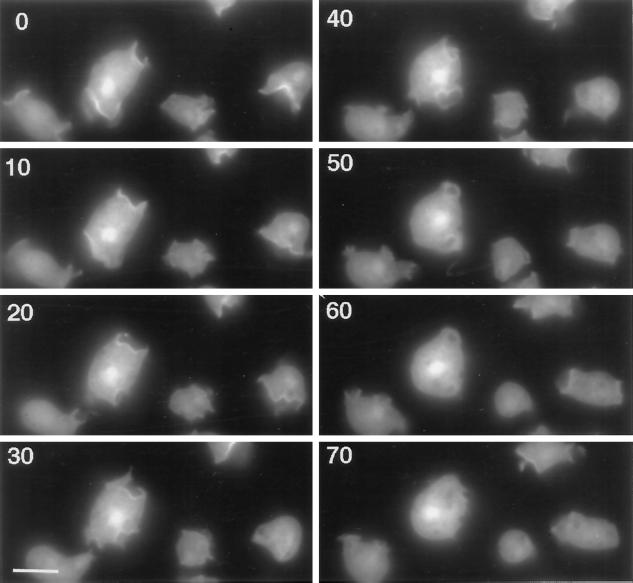
The image sequence shown in Figure 6 highlights several interesting features of the dynamics and localization of the DdLim in growth-phase cells. The largest cell in the image sequence contains three lamellipodia that evolved and dissipated within a 60-s period. Dorsal lamellipodia were first identified as a narrow ring or cup of elevated GFP-DdLim fluorescence at the membrane, which then expanded to a variable range of size and shapes. The closure of these cups was concomitant with the dissipation of GFP-DdLim fluorescence. The fact that GFP-DdLim did not increase in other areas of the cortex during these events would suggest that the recruitment of DdLim to the membrane was specific for lamellipodial protrusions. Together, these localization data define DdLim as a cortical actin cytoskeleton-associated protein where accumulation at the membrane of cytoskeleton protrusions suggests it is involved in the regulation of cell motility. GFP-DdLim also accumulated to different levels in the nucleus of the four cells shown in the image sequence. The intensity of the label was not influenced by protrusive activities at the cell surface, and control cells expressing GFP alone do not show any nuclear labeling (Gerisch et al., 1995). Although the functional significance of the nuclear localization of DdLim is not clear at this point, we note that in a recent report zyxin was shown to shuttle between focal contacts and the nucleus, where it may have a function in the regulation of gene expression (Nix and Beckerle, 1997).
Figure 6.
Image sequence of the distribution of GFP-DdLim in live cells. GFP-DdLim localizes to the extreme rim of newly formed dorsal lamellipodia and dissipates from the cortex upon retraction of the lamellipodia. Scale bar, 10 μm.
Overexpression of DdLim in Dictyostelium discoideum
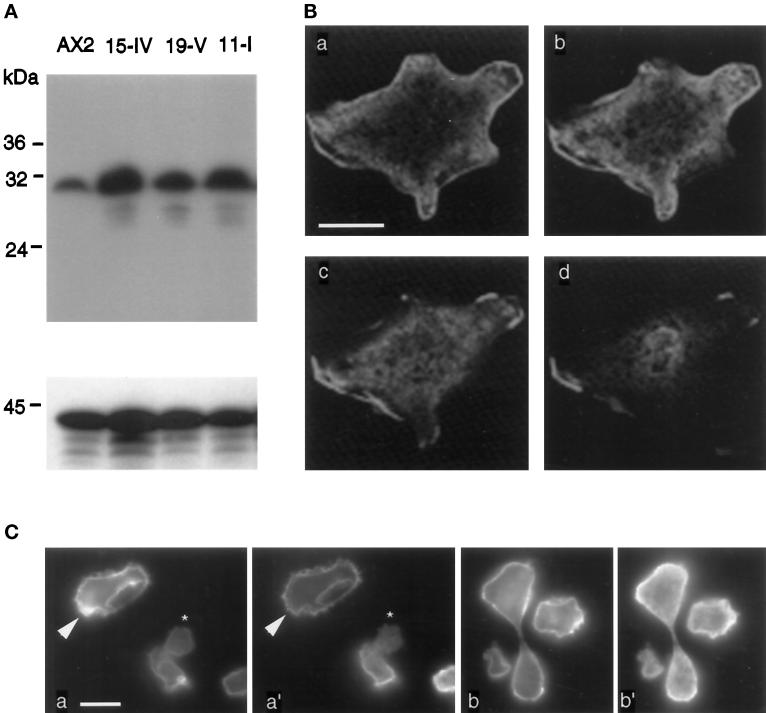
To better understand the role of DdLim in motility, growth, and cytokinesis, the full-length DdLim protein was overexpressed in Dictyostelium cells under control of the constitutively active actin-15 promotor. Wild type cells were transfected with a vector carrying the cDNA from the first methionine. Transfected cells capable of growing in the presence of geneticin (G418) were selected as candidates that overexpressed the DdLim protein. Several independent clones from this selection procedure were isolated, and Western blot analysis of their total cell lysates showed the level of DdLim was five- to tenfold higher than that of wild-type cells (Figure 7A). All of the isolated independent clones proved to have the same morphological defects. In the studies reported hereafter, mutant cells were cultured from spores and used within 3 wk, since prolonged maintenance of these cells in culture resulted in a reduced expression of DdLim (our unpublished results).
Figure 7.
(A) Western blot analysis of wild-type AX2 cells and three independent isolated DdLim-overproducing cells. Equal number of cells were loaded on each lane. Expression levels of DdLim were monitored using anti-DdLim antibodies (upper panel). Equal loading and transfer to nitrocellulose are confirmed by staining with an anti-actin antibody (lower panel). (B) Immunolocalization of DdLim in overexpressing cells using confocal fluorescence microscope. Vegetative cells were fixed with picric acid and paraformaldehyde and labeled as described for Figure 5B. Sections of a cell overproducing DdLim are shown from the substrate contact area (a) to the top of the cell (d). The distance between these sections is 0.5 μm. Scale bar, 10 μm. (C) Double labeling of DdLim-overproducing cells with rhodamine-phalloidin (a and b), anti-DdLim as first antibody, and fluorescein-conjugated antirabbit as the second antibody (a′ and b′).
Staining of DdLim in Overexpressing DdLim+ Cells
The intracellular localization of DdLim in overexpressing mutant cells was investigated by indirect immunofluorescence. The interpretation of immunofluorescent images of DdLim in overexpressing mutant cells using conventional fluorescence microscopy was made difficult because of the intense fluorescence signal associated with numerous crowns and lamellipodia that form on their dorsal surface. Using confocal fluorescence microscopy, it was possible to obtain optical sections of the cell to reject the intense out-of-focus signal associated with these dorsal protrusions. The confocal fluorescence images presented in Figure 7B, a real measure of the distribution of DdLim in the cortex and cytoplasm, clearly showed a preferential localization of the protein at sites of membrane ruffling at the cortex and in lamellipodia on the dorsal surface of mutant cells.
To learn whether DdLim localizes only to F-actin–rich structures at the cell cortex, we performed immunofluorescence in vegetative cells. Figure 7C shows two examples of fixed cells labeled for F-actin (a and b) and for DdLim (a′ and b′). For most areas of these cells the pattern of F-actin staining coincided with that of DdLim antibody label, although some minor areas (indicated by arrows) stained for F-actin but not for DdLim. The small cell in the center of Figure 7C (a and a′, marked by an asterisk) represents a extreme example of this effect. In cells with protrusive activity there was an excellent correlation between the distribution of F-actin and DdLim. These data confirmed the conclusions of earlier data (Figure 6) that demonstrated the association of DdLim at the cortical actin membrane was a prerequisite for the formation and persistence of a lamellipodia. In the dividing cell shown in Figure 7C (b and b′), DdLim localized at the poles of the cell, an area of intensive membrane ruffling and pseudopodia protrusion, and this property might implicate a role for DdLim in cytokinesis.
DdLim+ Cells Exhibit Defects in Growth
The growth of mutant cells was investigated using two different culture conditions. The generation time of DdLim+ cells maintained in suspension culture was 12 h compared with a generation time of 8 h for wild-type cells (our unpublished results). In addition, mutant cells could only attain a maximum cell density of about 3–5 × 106 cells/ml before they reached a stationary growth phase, compared with a value of 1.4 × 107 cells/ml in wild-type cells. On the other hand, DdLim-overexpressing cells grown on bacteria on the surface of an agar plate were indistinguishable from wild-type cells in their growth rate, measured by the increase of the colony size on bacterial plates. Ultimately, overexpression of DdLim in mutant cells grown on an agar surface did not interfere with the ability of these cells to form aggregates or to develop into slugs and fruiting bodies (our unpublished results).
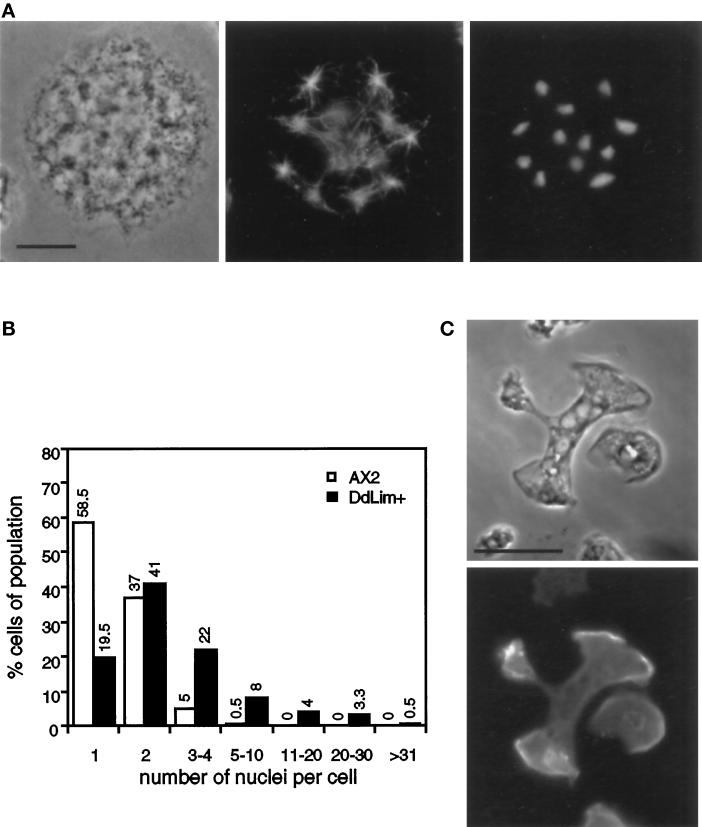
Cytokinesis Defect of DdLim+ Cells
Mutant cells that had been grown in suspension were plated onto coverslips, fixed, and stained with the DNA stain, DAPI. As mentioned earlier, these cells were large and contained many nuclei (Figure 8A), which might suggest that the excess DdLim interferes with cytokinesis. This idea was supported by an analysis of the nuclei count per cell for wild-type and DdLim+ cells, shown in Figure 8B. In wild-type cells grown in shaking culture, 58% of all cells were mononucleated whereas only 20% of mutant cells contained a single nuclei, while the majority contained between two and four nuclei. Furthermore, since 16% of all DdLim+ cells contained more than five nuclei and one cell was found to harbor 40 nuclei, it would appear that although mutant cells proceed to an advanced stage in mitosis they do not complete cytokinesis (Figure 8A). This defect in cytokinesis was apparently corrected for when large cells grown in suspension culture were transferred to a surface, after which their average size and number of nuclei decreased within a relatively short period of time. This effect was probably related to the large number of lamellipodia simultaneously projected by these cells onto the substratum, which increased the chance that two or more lamellipodia would pull against each other and lead to a traction-mediated cyto-fission event (Figure 8C; DeLozanne and Spudich, 1987). We suspect that this effect is responsible for the finding that mutant cells grown on a surface exhibited a normal development.
Figure 8.
(A) Localization of microtubule and nuclei in a dividing DdLim+ cell. Cells were fixed with cold methanol and stained with anti-α-tubulin (middle) and DAPI (right). Cells were grown in shaking culture for six generations and plated on glass cover slips for 30 min before fixation. Scale bar, 10 μm. (B) Quantitative analysis of the number of nuclei in wild-type Dictyostelium cells (open bars) and DdLim-overproducing cells (solid bars). Cells grown for 12 generations in shaking culture were plated on glass cover slips for 10 min before methanol fixation and nuclei staining with DAPI. The nuclei of 400 cells were counted. (C) Large DdLim-overexpressing cells were allowed to settle for 30 min on a glass coverslip upon which they immediately started to divide by traction-mediated fission. DdLim is located at the leading edge of the separating cell parts.
Morphology of DdLim+ Cells
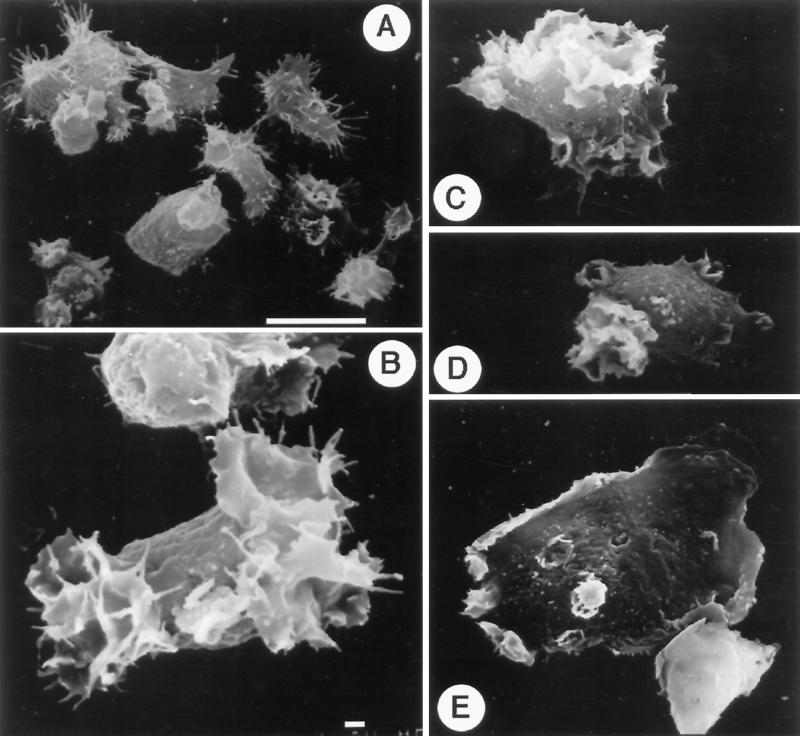
Phase-contrast image microscopy revealed that most DdLim+ cells grown in suspension were at least threefold to fivefold larger than wild-type cells. Mutant cells were also found to harbor many large vacuoles (Figure 8C). To investigate the morphology of DdLim+ cells in more detail, cells were imaged using scanning electron microscopy. High-resolution images of these cells revealed the presence of many thin cytoplasmic protrusions that appeared as either circular lamellae on the dorsal surface, often containing multiple layers of lamellae, or as a very large lamella in contact with the substratum (Figure 9, B, C, and D). Although crowns and lamella were also found in wild-type cells, they never occurred at the same frequency or attained the large size of those found in mutant cells (Figure 9A). Furthermore the filopodia of mutant cells were reduced to very short spikes, or were visible only as part of a lamella scaffold, whereas the larger number of filopodia seen in wild-type cells were normally long and straight (Figure 9, A and B). The largest of the mutant cells were usually well spread, had a smooth surface appearance interspersed by dorsal lamella, and showed extensive membrane ruffling at the cell rim (Figure 9E).
Figure 9.
Scanning electron microscope micrographs of wild-type Dictyostelium cells (A), and DdLim+ cells (B–E). Panels A and C–E were recorded at the same magnification (2000×; the scale bar in panel A is 10 μm. Micrograph B was recorded at 4000× magnification; scale bar, 1 μm.
Evidence that DdLim Associates with the GTP-bound Form of rac1
The enhanced protrusion of lamellipodia- and membrane-ruffling activity observed in DdLim+ cells is similar to the behavior of mammalian cells microinjected with activated forms of small GTPase-signaling proteins (Nobes and Hall, 1995). This analogy prompted us to test whether the excess DdLim in mutant cells might be involved in regulating the protrusion of these actin filament assemblies through an association with an activated isoform of rac or cdc42.
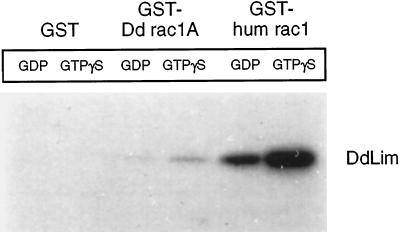
GST-fusion proteins of rac1 and cdc42 were separately incubated in their inactive (GDP-bound) or activated (GTPγS-bound) form with proteins present in total cell lysates of Dictyostelium cells. Cytosolic proteins capable of interacting with the GTPase fusion proteins were isolated by recovering the GST-fusion complex with glutathione-Sepharose beads. The beads were then washed, and specifically bound proteins were eluted, separated by SDS-PAGE, and electrophoretically transferred to nitrocellulose, which was then probed for the presence of DdLim using anti-DdLim antibodies. DdLim was identified as one of a number of proteins that associated with human rac1 and, to a lesser extent, with Dictyostelium rac1A, while no association was detected with human cdc42 (our unpublished results) or the control GST-beads (Figure 10). The amount of DdLim that associated with the activated (GTPγS bound) form of human rac1 and Dictyostelium rac1A was more than fivefold higher than with the corresponding GDP-bound form of rac1, which suggests the association of DdLim with rac1 is specific. Since it was not possible to show that bacterially expressed DdLim was able to bind to human rac1 directly (our unpublished results), other additional protein components of the cytosolic fraction of Dictyostelium must be involved in the formation of the DdLim-rac1 complex. Work is in progress to solidify these studies and to scale up the experiment in an effort to identify and characterize other components of the GST–rac1(GTPγS) complex.
Figure 10.
Interaction of DdLim with rac1-GST fusion proteins. Autoradiogram of a Western blot of Dictyostelium proteins that associate with a GST-, Dictyostelium rac1A-, and human rac1-affinity resin probed with 125I-labeled anti-Lim antibodies. DdLim present in the cell lysate was recovered mainly with activated human rac1, and to a lesser extent with Dictyostelium rac1A. No interaction of DdLim was found with a GST-control-resin. The binding of DdLim to both rac1-proteins was about 5-fold more in the prescence of rac1 charged with GTPγS compared with GDP.
DISCUSSION
Chemotactic motility in preaggregation-competent cells of Dictyostelium is achieved in part by the regulation of actin polymerization and organization of actin filament assemblies into filopodia and lamellipodia at the site of cAMP receptor activation. This is clearly a complicated process that demands the coordination of local activities of membrane-bound receptors, signaling molecules, and cytoskeleton effector complexes that can both promote actin polymerization and couple the molecular forces generated during this reaction to the protrusion of filopodia or lamellipodia and ultimately cell motility. To identify cytoskeleton-associated proteins involved in these processes we developed a search strategy based on the assumption that the expression level of candidate proteins should increase in the transition of cells from the slowly motile vegetative stage to the highly motile and chemotactic preaggregation stage. Furthermore, the association of these proteins with the cytoskeleton should be regulated by second messengers, e.g., Ca2+, known to play a role in regulating motility (Brundage et al., 1993).
Analysis of a Ca2+-extract of the Triton X-100–insoluble cytoskeleton of Dictyostelium at different times during starvation identified a number of proteins that satisfied these conditions. The first protein characterized from this pool of candidates, DdLim, a small, multidomain member of the CRP family of proteins, contains a single LIM domain that harbors two loop regions capable of promoting protein–protein interactions. Although DdLim does not bind to F-actin directly (our unpublished results), it is extracted from the cytoskeleton by Ca2+ along with actin, α-actinin, and a host of other ABPs (Figure 1A). This coelution behavior suggests that DdLim, like CRP1, which binds to zyxin, associates indirectly with F-actin through its interaction with ABPs.
Evidence that DdLim plays an important role in the regulation of the organization and dynamics of the actin cytoskeleton at the cell cortex and plasma membrane was obtained from a number of independent experiments that included the similarity in the organization of the polypeptide domains of DdLim to members of the CRP family, the immunocytochemical localization of DdLim, the induced expression of DdLim in motile, preaggregation-competent cells, and by the fact that a mutant overexpressing DdLim exhibited a number of defects associated with the regulation and organization of the actin cytoskeleton that included growth, cytokinesis, and an increased number of lamellipodia that projected from their dorsal surface. Interestingly an apparent correction of the growth and cytokinesis defects was observed by plating mutant cells onto a surface, whereupon the increased protrusive activity of lamellipodia in these cells evidently enhanced traction-mediated cytofission. One caveat of these experiments is that the high level of DdLim in mutant cells may lead to dominant negative effects on certain cell functions. For example, the cytokinesis defect observed in suspension-grown DdLim+ cells could result from a DdLim-mediated sequesterization of activated rac1 generated during cytokinesis or, alternatively, an ABP required for cytokinesis might be sequestered by the excess DdLim.
Western blot analysis of an incubation mixture of a Dictyostelium cell lysate with GTPγS-activated human rac1-GST fusion protein coupled to Sepharose beads identified DdLim as one of the proteins in a GTPγS–rac1 complex. Rac1 is a small GTP-binding protein implicated in the regulation of lamellipodia protrusion in mammalian cells (Ridley et al., 1992). A similar experiment conducted with Dictyostelium rac1A (Bush et al., 1993) beads showed a much weaker association to DdLim. We speculate therefore that Dictyostelium should contain a rac isoform more similar to the human rac1, in terms of its binding preference and perhaps function, and this might be a member of the large family of Dictyostelium rac genes identified by Bush et al. (1993). The inability to demonstrate a direct interaction between purified activated human rac1 and DdLim suggests that rac1 associates with DdLim as part of a multiprotein complex.
Based on the findings that 1) DdLim is a component of the actin cytoskeleton; 2) DdLim localizes at the leading edge of lamellipodia in cells undergoing a chemotactic response; 3) DdLim was identified as a component of a GTP-rac1 protein complex; 4) both DdLim and rac-like proteins in Dictyostelium are induced in preaggregation-competent cells; 5) overexpression of DdLim in Dictyostelium cells increased the frequency of lamellipodial protrusions in vegetative cells compared with wild-type cells; and 6), the DdLim+ mutant cells exhibited a number of phenotypes associated with an unusual regulation of the actin cytoskeleton, we speculate that rac1 and DdLim are part of a signaling-effector pathway that links activation of the cAMP receptor to cell motility. In a working model consistent with these findings, we envision that cAMP binding to the cAMP receptor would indirectly activate a small GTPase- signaling protein with close homology to human rac1. We anticipate the signaling protein would trigger the formation of a complex with DdLim and ABPs at the plasma membrane. This would then lead to an explosive polymerization of actin with concomitant production of molecular forces that lead to the protrusion of a lamellipodium at the site of receptor activation. Hydrolysis of the GTP bound to rac1 would inactivate its signaling function and result in the dissociation of the DdLim-ABPs complex and thereby uncouple the receptor- mediated signaling pathway from actin filament dynamics at the leading edge. Polarized cell motility mediated by a chemotactic response will therefore only occur if there is a sustained, localized stimulation of the cAMP receptor with the subsequent generation of activated rac1. Consistent with this model, image analysis of GFP-DdLim in live cells showed recruitment of the protein to the membrane preceded the formation of filopodia and lamellipodia. Furthermore, the persistence of lamellipodia was dependent on the presence of GFP-DdLim at the membrane. Loss of GFP-DdLim from the membrane invariably heralded the collapse of a lamellipodium. An elevated level of DdLim was also found at the two opposing poles of cells during cytokinesis. These regions exhibit intensive membrane ruffling and pseudopodia protrusion activity (Neujahr et al., 1997).
If the cAMP receptor is the only target for activated rac1, how does overexpression of DdLim result in the protrusion of an unusually large number of lamellipodia in vegetative cells, which have only 10% of the cAMP receptor level of preaggregation-competent cells (Gerisch, 1987). To address this question, vegetative DdLim+ cells were plated at low density to ensure the concentration of cAMP under these conditions was below the threshold value for cAMP receptor activation. The mutant cells were still capable of protruding numerous lamellipodia under these conditions, which would suggest these protrusions were formed by activation of receptors other than the cAMP receptor. The folic acid receptor might be a potential candidate since it has been implicated in regulation of lamellipodia formation in vegetative cells (Gerisch, 1987). We suggest, therefore, that DdLim, and probably rac1, regulate the formation of lamellipodia in both growth phase and preaggregation-competent Dictyostelium cells through a common mechanism that involves membrane receptor activation and small GTPase signaling.
In summary, we have used a new subtractive screening approach to identify cytoskeleton proteins that are responsible for the acquisition of a highly motile behavior in preaggregation-competent Dictyostelium cells. DdLim, the first protein characterized from a pool of candidate proteins, is a LIM domain-containing protein that shares significant structural and sequence similarities to LIM domain proteins found in the cytoskeleton of mammalian cells. A number of experiments described in this work suggest that DdLim is involved in the regulation of the actin cytoskeleton and plays an important role in cell motility and cytokinesis.
ACKNOWLEDGMENTS
The authors thank G. Rahn for iodination of DdLim antibodies. We are grateful to Dr. J. Kuhlmann and Dr. Robert Cool for providing human rac1- and cdc42-GST-fusion constructs. This work was supported by a grant from the Max Planck Society (MA215) awarded to G.M. The authors thank Dr. G. Gerisch for useful discussions.
REFERENCES
- Arber S, Caroni P. Specificity of single LIM motifs in targeting and LIM/LIM interactions in situ. Genes Dev. 1996;10:289–300. doi: 10.1101/gad.10.3.289. [DOI] [PubMed] [Google Scholar]
- Arber S, Halder G, Caroni P. Muscle LIM protein a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell. 1994;79:221–231. doi: 10.1016/0092-8674(94)90192-9. [DOI] [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein dye-binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brundage RA, Fogarty KE, Tuft RA, Fay FS. Chemotaxis of newt eosinophils: calcium regulation of chemotactic response. Am J Physiol. 1993;265:1527–1543. doi: 10.1152/ajpcell.1993.265.6.C1527. [DOI] [PubMed] [Google Scholar]
- Bush J, Franek K, Cardelli J. Cloning and characterization of seven novel Dictyostelium discoideum rac-related genes belonging to the rho family of GTPases. Gene. 1993;136:61–68. doi: 10.1016/0378-1119(93)90448-c. [DOI] [PubMed] [Google Scholar]
- Claviez M, Brink M, Gerisch G. Cytoskeletons from a mutant of Dictyostelium discoideum with flattened cells. J Cell Sci. 1986;86:69–82. doi: 10.1242/jcs.86.1.69. [DOI] [PubMed] [Google Scholar]
- Crawford AW, Michelsen JW, Beckerle MC. An interaction between zyxin and α-actinin. J Cell Biol. 1992;116:1381–1393. doi: 10.1083/jcb.116.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford AW, Pino JD, Beckerle MC. Biochemical and molecular characterization of the chicken cysteine-rich protein, a developmentally regulated LIM-domain protein that is associated with the actin cytoskeleton. J Cell Biol. 1994;124:117–127. doi: 10.1083/jcb.124.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLozanne A, Spudich JA. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–385. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Gerisch G, Noegel AA. Overexpression of the csA cell adhesion molecule under its own cAMP-regulated promoter impairs morphogenesis in Dictyostelium. J Cell Sci. 1992;102:203–214. doi: 10.1242/jcs.102.2.203. [DOI] [PubMed] [Google Scholar]
- Faix J, Steinmetz M, Boves H, Kammerer RA, Lottspeich F, Mintert U, Murphy J, Stock A, Aebi U, Gerisch G. Cortexillins, major determinants of cell shape and size, are actin-bundling proteins with a parallel coiled-coil tail. Cell. 1996;86:631–642. doi: 10.1016/s0092-8674(00)80136-1. [DOI] [PubMed] [Google Scholar]
- Freyd G, Kim SK, Horvitz HR. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature. 1990;344:876–879. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- Furukawa R, Butz S, Fleischmann E, Fechheimer M. The Dictyostelium discoideum 30,000 dalton protein contributes to phagocytosis. Protoplasma. 1992;169:18–27. [Google Scholar]
- Gerisch G. Cyclic AMP and other signals controlling cell development and differentiation in Dictyostelium. Annu Rev Biochem. 1987;56:853–79. doi: 10.1146/annurev.bi.56.070187.004225. [DOI] [PubMed] [Google Scholar]
- Gerisch G, Albrecht R, Heizer C, Hodgkinson S, Maniak M. Chemoattractant-controlled accumulation of coronin at the leading edge of Dictyostelium cells monitored using a green fluorescent protein-coronin fusion protein. Curr Biol. 1995;5:1280–1285. doi: 10.1016/s0960-9822(95)00254-5. [DOI] [PubMed] [Google Scholar]
- Hart M, Callow MG, Souza B, Polakis P. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO J. 1996;15:2997–3005. [PMC free article] [PubMed] [Google Scholar]
- Jungbluth A, Eckerskorn C, Gerisch G, Lottspeich F, Stocker S, Schweiger A. Stress-induced tyrosine phosphorylation of actin in Dictyostelium cells and localization of the phosphorylation site to tyrosine-53 adjacent to the DNase I binding loop. FEBS Lett. 1995;375:87–90. doi: 10.1016/0014-5793(95)01165-b. [DOI] [PubMed] [Google Scholar]
- Karlsson O, Thor S, Norberg T, Ohlsson H, Edlund T. Insulin enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature. 1990;344:879–882. doi: 10.1038/344879a0. [DOI] [PubMed] [Google Scholar]
- Kosa JL, Michelsen JW, Louis HA, Olsen JI, Davis DR, Beckerle MC, Winge DR. Common metal ion coordination in LIM domain proteins. Biochemistry. 1994;33:468–477. doi: 10.1021/bi00168a011. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequence. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Maniak M, Rauchenberger R, Albrecht R, Murphy J, Gerisch G. Coronin involved in phagocytosis: dynamics of particle-induced relocalization visualized by a green fluorescent protein tag. Cell. 1995;83:915–924. doi: 10.1016/0092-8674(95)90207-4. [DOI] [PubMed] [Google Scholar]
- Michelsen JW, Schmeichel K, Beckerle MC, Winge DR. The LIM motif defines a specific zinc-binding protein domain. Proc Natl Acad Sci USA. 1993;90:4404–4408. doi: 10.1073/pnas.90.10.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelsen JW, Sewell AK, Louis HA, Olsen JI, Davis DR, Winge DR, Beckerle MC. Mutational analysis of the metal sites in a LIM domain. J Biol Chem. 1994;269:11108–11113. [PubMed] [Google Scholar]
- Nellen W, Silan C, Firtel RA. DNA-mediated transformation in Dictyostelium discoideum regulated expression of an actin gene fusion. Mol Cell Biol. 1984;4:2890–2898. doi: 10.1128/mcb.4.12.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neujahr R, Heizer C, Gerisch G. Myosin-II-independent processes in mitotic cells of Dictyostelium discoideum: redistribution of the nuclei, re-arrangement of the actin system and formation of the cleavage furrow. J Cell Sci. 1997;110:127–137. doi: 10.1242/jcs.110.2.123. [DOI] [PubMed] [Google Scholar]
- Newell PC, Telser A, Sussman M. Alternative developmental pathways determined by environmental conditions in the cellular slime mold Dictyostelium discoideum. J Bacteriol. 1969;100:763–768. doi: 10.1128/jb.100.2.763-768.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix DA, Beckerle MC. Nuclear-cytoplasmic shuttling of the focal contact protein, Zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Prassler J, Stocker S, Marriott G, Heidecker M, Kellermann J, Gerisch G. Interaction of a Dictyostelium member of the plastin/fimbrin family with actin filaments and actin-myosin complexes. Mol Biol Cell. 1997;8:83–95. doi: 10.1091/mbc.8.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Sadler I, Crawford AW, Michelsen JW, Beckerle MC. Zyxin and cCRP: two interactive LIM domain proteins associated with the cytoskeleton. J Cell Biol. 1992;119:1573–1587. doi: 10.1083/jcb.119.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Garcia I, Rabbitts TH. The LIM domain: a new structural motif found in zinc-finger-like proteins. Trends Genet. 1994;10:315–320. doi: 10.1016/0168-9525(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Schmeichel KL, Beckerle MC. The LIM domain is a modular protein binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Schmeichel KL, Beckerle MC. Molecular dissection of a LIM domain. Mol Biol Cell. 1997;8:219–230. doi: 10.1091/mbc.8.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stronach BE, Siegrist SE, Beckerle MC. Two muscle-specific LIM proteins in Drosophila. J Cell Biol. 1996;134:1179–1195. doi: 10.1083/jcb.134.5.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallraff E, Gerisch G. Screening for Dictyostelium mutants defective in cytoskeletal proteins by colony immunoblotting. Methods Enzymol. 1991;196:334–348. doi: 10.1016/0076-6879(91)96030-u. [DOI] [PubMed] [Google Scholar]
- Weiskirchen R, Pino JD, Macalama T, Bister K, Beckerle M. The cysteine-rich protein family of highly related LIM domain proteins. J Biol Chem. 1995;270:28946–28954. doi: 10.1074/jbc.270.48.28946. [DOI] [PubMed] [Google Scholar]
- Westphal M, Jungbluth A, Heidecker M, Muehlbauer B, Heizer C, Schwartz J-M, Marriott G, Gerisch G. Microfilament dynamics during cell movement and chemotaxis monitored using a GFP-actin fusion protein. Curr Biol. 1997;7:176–183. doi: 10.1016/s0960-9822(97)70088-5. [DOI] [PubMed] [Google Scholar]
- Witke W, Schleicher M, Lottspeich F, Noegel A. Studies on the transcription, translation, and structure of α-actinin in Dictyostelium discoideum. J Cell Biol. 1986;103:969–975. doi: 10.1083/jcb.103.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witke W, Schleicher M, Noegel AA. Redundancy in the microfilament system-abnormal development of Dictyostelium cells lacking two F-actin cross-linking proteins. Cell. 1992;68:53–62. doi: 10.1016/0092-8674(92)90205-q. [DOI] [PubMed] [Google Scholar]