Abstract
Experiments in cultured cells with Ran-binding protein M (RanBPM) suggest that it links cell surface receptors and cell adhesion proteins. In this study, we undertake a genetic study of RanBPM function in the germline stem cell (GSC) niche of Drosophila melanogaster ovaries. We find that two RanBPM isoforms are produced from alternatively spliced transcripts, the longer of which is specifically enriched in the GSC niche, a cluster of somatic cells that physically anchors GSCs and expresses signals that maintain GSC fate. Loss of the long isoform from the niche causes defects in niche organization and cell size and increases the number of GSCs attached to the niche. In genetic mosaics for a null RanBPM allele, we find a strong bias for GSC attachment to mutant cap cells and observe abnormal accumulation of the adherens junction component Armadillo (β-catenin) and the membrane skeletal protein Hu-li tai shao in mutant terminal filament cells. These results implicate RanBPM in the regulation of niche capacity and adhesion.
Introduction
The undifferentiated state of germline stem cells (GSCs) depends on paracrine signals from the stem cell niche, the somatic microenvironment that specifies and maintains stem cell fate (Xie and Spradling, 2000). The core mechanisms that define and maintain GSCs have been described; however, the development and regulation of the niche itself is less well understood (for review see Dansereau and Lasko, 2008). The core of the GSC niche in Drosophila melanogaster ovaries is a cluster of cap cells (CpCs) that physically anchors two to three GSCs to the anterior tip of each ovariole, the egg-producing units of the ovary (Forbes et al., 1996a; Cox et al., 1998; Xie and Spradling, 1998, 2000; Song et al., 2002). CpCs and GSCs are attached by adherens junctions, which maintain the GSCs in close proximity to the regulatory signals from the niche, such as the bone morphogenetic protein homologue Decapentaplegic (Xie and Spradling, 1998; Song et al., 2004). GSCs divide asymmetrically to produce a cystoblast, the daughter cell that is displaced from the niche and differentiates, and a GSC that remains attached to the CpCs and retains its undifferentiated state (Forbes et al., 1996a; Cox et al., 1998; Xie and Spradling, 1998, 2000; Zhu and Xie, 2003). The cystoblasts begin to differentiate by expressing Bag of marbles (Bam), which drives four incomplete mitotic divisions to create an interconnected cyst of 15 accessory nurse cells and one oocyte (McKearin and Ohlstein, 1995; Ohlstein and McKearin, 1997). In the GSCs, however, Decapentaplegic signal transduction leads to the transcriptional repression of bam via the repressor proteins Mad and Medea, which bind directly to a bam transcriptional silencer (Chen and McKearin, 2003a,b, 2005; Song et al., 2004). Close association with the niche is required for GSC fate because mutant GSCs lacking the adherens junction components Armadillo (Arm) or DE-cadherin move away from the maintenance signals expressed by the niche and differentiate (Song et al., 2002).
The number of GSCs in an ovariole is related to the number of CpCs in its niche, suggesting that the niche has a finite capacity to support stem cells. When the number of CpCs is increased by constitutively activating the Notch receptor during niche development, a greater number of GSCs are observed, whereas reducing Notch expression reduces the number of CpCs and reduces the capacity of the niche (Ward et al., 2006; Song et al., 2007). In an underpopulated niche, both daughter cells of a dividing GSC can be maintained as stem cells; the GSC divides perpendicularly to the germarial axis so that both daughter cells remain in contact with the CpCs and retain stem cell identity (Xie and Spradling, 2000; Zhu and Xie, 2003). These results suggest that the close association of an undifferentiated germline cell with the GSC niche and its signals is sufficient to specify and maintain GSC fate. Indeed, when niche morphogenesis is completed during pupal development, GSC fate is specified in an anterior subset of primordial germ cells located in close proximity to the newly formed CpC clusters (Asaoka and Lin, 2004; Gilboa and Lehmann, 2004).
We identified a Gal4 enhancer trap inserted in the Drosophila Ran-binding protein M (RanBPM) gene that reports gene expression in the GSC niche of the ovary. Drosophila RanBPM is related to two vertebrate proteins named RanBPM (or RanBP9) and RanBP10. RanBP10 shares 68% amino acid identity with RanBPM but lacks an N-terminal glutamine-rich region found in RanBPM (Wang et al., 2004). Both of these proteins have been implicated in binding the small GTPase Ran and in regulating microtubule dynamics (Nakamura et al., 1998; Schulze et al., 2008), although the RanBPM–Ran interaction has not been replicated in a subsequent study (Nishitani et al., 2001). RanBPM and RanBP10 contain a SPRY domain, a protein–protein interaction domain found in the SplA and ryanodine receptors (Ponting et al., 1997), a lissencephaly homology (LisH) motif that functions in Lis-1 as a stable dimerization domain (Kim et al., 2004; Gerlitz et al., 2005), an α-helical C terminal to LisH (CTLH) motif of unknown function, and a CRA (CT11-RanBPM) domain, which is also implicated in protein–protein interactions (Menon et al., 2004). These proteins do not contain a consensus Ran-binding domain. Both RanBPM and RanBP10 act as ligand-dependent coactivators of the androgen receptor (Rao et al., 2002; Harada et al., 2008), and both proteins bind to the Met receptor through their conserved SPRY domains (Wang et al., 2002, 2004; Sattler and Salgia, 2007). RanBPM interacts with several other cell surface receptors and cell adhesion molecules, including the integrin LFA-1 (lymphocyte function-associated antigen-1; Denti et al., 2004) and the neural cell adhesion molecule L1 (Cheng et al., 2005). RanBPM was also isolated in a multiprotein CTLH complex that includes the Arm repeat–containing proteins ArmC8α and β and muskelin, a kelch repeat protein that mediates cell spreading responses to the matrix adhesion molecule thrombospondin-1 (Adams et al., 1998; Umeda et al., 2003; Kobayashi et al., 2007). These interactions support the hypothesis that RanBPM acts as an adaptor between the cell adhesion machinery and different cell signaling pathways (Denti et al., 2004; Cheng et al., 2005), but functional data have been limited by the lack of loss of function studies.
Using a range of genetic and cell biology tools, we characterized the Drosophila RanBPM gene and analyzed its expression and function during oogenesis. We found that RanBPM encodes two protein isoforms, and with an isoform-specific antibody we found that the long isoform is specifically enriched in the CpCs and terminal filament (TF) of the GSC niche. Our analyses of a null RanBPM mutant in genetic mosaics and a RanBPM mutant that fails to express the niche-specific long isoform indicate that RanBPM regulates the cell shape, size, and organization of the GSC niche. Interestingly, these changes are accompanied by an increase in the number of GSCs attached to the mutant CpCs, implicating RanBPM in the control of niche capacity for GSCs.
Results
The RanBPM locus and mutagenesis
Vertebrate RanBPM and its paralogue RanBP10 are most similar to the predicted Drosophila gene CG11763. Given the high degree of sequence similarity between the vertebrate and Drosophila genes (Fig. 1 A), and to maintain consistency with a significant literature that uses the term RanBPM, we have adopted the same name for CG11763. Sequence analysis of four cDNAs indicates that through alternative splicing, RanBPM encodes two protein isoforms with predicted masses of 67 and 140 kD (short and long RanBPM; Fig. 1, A–C). As for vertebrate RanBPM, both Drosophila isoforms include a core region with a SPRY domain, a LisH motif, an α-helical CTLH motif, and a CRA domain. In addition, long RanBPM has an unstructured N-terminal extension that is not conserved but is rich in glutamine and may be related to a similar glutamine-rich N-terminal region in vertebrate RanBPM that is not present in RanBP10.
Figure 1.
Summary of RanBPM protein isoforms, transcripts, and alleles. (A) Two RanBPM isoforms share a core region with a SPRY domain, LisH/CTLH motifs, and a CRA domain. Long RanBPM has an unstructured N-terminal extension. Drosophila RanBPM is highly homologous to mouse RanBPM and mouse RanBP10: the percent amino acid identity within the four conserved domains is shown. Protein fragments used to make antibodies are noted. (B) RanBPM is alternatively spliced to regulate the use of alternate start codons (ATG1–2). Exons (1–12) are colored white for UTRs, black for coding regions, and gray for the alternate coding region. Transposable element insertions: a Gal4 P element in the 5′ UTR (NP6392, RanBPM-Gal4), a weak loss of function P element insertion in the first intron (CB-6232-3), and a strong loss of function piggyBac insertion in exon 11 (f06647). RanBPMnull (Δnull) is a deletion of the region between the FRT sequences in CB-6232-3 and f06647, and the N-terminal deletion RanBPMΔN is a P element excision derivative of CB-6232-3. (C) Antibodies raised against the N-terminal fragment of long RanBPM (αN-term) recognize a 140-kD band (L, long RanBPM) in heterozygous extracts that is absent from RanBPMnull extracts. Antibodies raised against the core domains of RanBPM (α-Core) recognize two protein isoforms: long RanBPM (140 kD) and short RanBPM (67 kD). Several cross-reacting bands are marked with asterisks. (D) Summary of complementation tests between various RanBPM alleles. Blue boxes indicate complete lethality of the transheterozygote, whereas each number indicates the percentage of surviving transheterozygotes over the number of survivors expected if the alleles had fully complemented. Relative allele strength at 25°C is null > f06647 > ΔN > CB. ΔN is a temperature-sensitive lethal allele, and these tests were performed at the restrictive temperature of 25°C. (E) RanBPMΔN does not express long RanBPM (L), which is recognized with the N-terminal antibody, but does express short RanBPM (S), which is recognized with the core antibody. RanBPMnull was included as a control to identify bands specific to RanBPM.
To study the effect of loss of RanBPM function, we identified in public stock collections transposable elements that map to the RanBPM gene. These include RanBPM-Gal4, a viable and fertile P element insertion in the 5′ untranslated region (UTR), RanBPMCB, a semilethal and semisterile P element insertion in the first intron, and RanBPMf06647, a homozygous lethal piggyBac element insertion in exon 11 (Fig. 1 B). The RanBPMCB and RanBPMf06647 insertions fail to complement one another and Df(2R)12, a large deletion that removes the RanBPM genomic region (Fig. 1 D), and their excision led to a high frequency of reversion to fertility and viability (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200711046/DC1). RanBPMCB and RanBPMf06647 are hypomorphic alleles, and Western analysis showed that these alleles express both long and short RanBPM (unpublished data).
To generate stronger alleles, we induced imprecise excision of the RanBPMCB P element and isolated a deletion that precludes expression of the long RanBPM isoform (RanBPMΔN; see Materials and methods; Fig. 1, B and E). RanBPMΔN bears a 1.5-kb deletion that extends from the first intron into the second exon, deleting both of the alternative splice acceptor sites for exon 2 (Fig. 1 B). RanBPMΔN is a temperature-sensitive allele: it is viable at 22°C and lethal at 25°C, allowing us to collect homozygous mutants at low temperatures. Western analysis of RanBPMΔN homozygotes showed loss of long RanBPM expression and retention of short RanBPM expression (Fig. 1 E). Although RanBPMΔN mutants express reduced levels of short RanBPM, they express sufficient quantities for viability and fertility at low temperatures.
We also generated a null allele by targeted deletion of the RanBPM coding region (see Materials and methods; Fig. 1 B). Western analysis of RanBPMnull homozygous larvae confirmed the absence of both RanBPM isoforms (Fig. 1 C). Complementation tests showed that RanBPMnull is lethal in trans with Df(2R)12 and the strong hypomorph RanBPMf06647, is semilethal with the weaker allele RanBPMCB, and is temperature-sensitive lethal with RanBPMΔN (Fig. 1 D). When short RanBPM expression was induced with a ubiquitous promoter in a RanBPMnull background (see Materials and methods), the lethality of RanBPMnull was rescued at 22°C. Short RanBPM expression was not sufficient to rescue the lethality of RanBPMnull at 25°C, mimicking the temperature sensitivity of the RanBPMΔN allele, which also expresses only short RanBPM. Collectively, these data indicate that our mutations comprise a single complementation group corresponding to RanBPM and show that RanBPM is an essential gene in Drosophila.
RanBPM expression pattern
RanBPM-Gal4, a P{GawB } enhancer-trap insertion in the 5′ UTR of RanBPM (Fig. 1 B), reports gene expression in a small subset of somatic cells in the ovary (Fig. 2, A–C). The P{GawB } element contains the GAL4 coding region under the control of a minimal promoter that can respond to enhancers near the P element chromosomal insertion point (Hayashi et al., 2002). RanBPM-Gal4 drives UAS-LacZ or UAS-GFP expression at the anterior tip of the germarium, specifically in the CpCs and TF cells of the GSC niche (Fig. 2, B and C).
Figure 2.
Long RanBPM is expressed in the stem cell niche. (A and A′) The germline stem cell (GSC) niche in Drosophila ovaries is made up of two somatic cell types: eight to nine cells in a single-filed array make up the terminal filament (TF), and a tight cluster of five or six cap cells (CpC) directly contacts the GSCs. In A′, a hedgehog-LacZ (hh-LacZ) reporter marks the TF and CpC cluster. The positions of two GSCs are noted with dotted circles. (B and C) The RanBPM-Gal4 enhancer trap expresses GAL4 in the CpC and TF of the stem cell niche (green). In B, RanBPM-Gal4; UASp-LacZ ovaries were stained for β-galactosidase (green) and Vasa (red), a germline marker. (C) A RanBPM-Gal4; UAS-GFP germarium counterstained with DAPI (red). Not all of the niche cells show the same level of RanBPM-Gal4 expression, regardless of the marker used. Because the RanBPM protein is expressed in all niche cells (see D), we believe this is an artifact of the enhancer trap or the UAS-Gal4 system. (D) The long RanBPM isoform, which is visualized with an isoform-specific antibody (RanBPM-N, green), is specifically enriched in the CpC and TF. A patched-LacZ (ptc-LacZ) reporter marks the CpC cluster (red). (E) In pupal ovaries, RanBPM-N staining (green) is enriched at the TF cell membranes. Hts (red) marks the fusomes of germline cysts and cell membranes. (F) As a negative control, pupal ovaries from RanBPMnull were stained with the N-terminal antibody. The absence of signal shows the specificity of the RanBPM-N signal in the stem cell niche. Bars: (A′, C, and D) 10 μm; (B) 30 μm; (E and F) 50 μm.
To study the endogenous RanBPM protein expression pattern, we raised antibodies to a core RanBPM fragment including the SPRY domain and LisH motif (Fig. 1 A). This anticore antiserum recognizes both RanBPM isoforms by Western analysis (Fig. 1 C). We immunostained ovaries with the anticore antiserum and observed staining at cell membranes and throughout the cytoplasm and nuclei of somatic follicle cells and germline cells of the ovary (Fig. 3). To confirm the specificity of the anticore antiserum, we stained genetic mosaics of RanBPMnull and observed that germline and follicle cells mutant for RanBPM were no longer recognized (Fig. 3, A and B).
Figure 3.
Expression pattern of RanBPM in the Drosophila ovary and embryo. (A and B) In RanBPMnull mutant cells marked by lack of GFP expression (A and B, green), labeling with anti-RanBPM is highly reduced (A′ and B′), confirming the specificity of the antiserum. Ovaries were counterstained with antibodies to α-spectrin (α-Spec, red) to outline the fusomes of dividing cysts and cell membranes. The arrows in A and A′ indicate mutant GSCs, and the arrowheads indicate wild-type GSCs. (C) RanBPM (C″, green) is expressed in all germline cells at the anterior of the germarium (the arrows indicate a GSC) and in the somatic follicle cells (arrowheads). Hts (C′, red) marks cell membranes and the spectrosomes and fusomes of the germline. (D) RanBPM expression (D″, green) is reduced in the germline after completion of cyst mitosis. Antibodies to Bag of marbles (D′, Bam, red) were used to specifically mark the mitotic cells of the germarium. The reduction of RanBPM expression coincides with the disappearance of Bam expression (arrowheads). (E) At later stages of development, RanBPM (E′, red) becomes enriched in the nurse cell (NC) and follicle cell nuclei and at nurse cell membranes (arrows). Membranes are labeled with fluorescent phalloidin (E″, F-actin, green), and DNA is labeled with DAPI (E‴, blue). Bars: (A–D) 20 μm; (E) 75 μm.
We observed that RanBPM localizes to the cytoplasm, nucleus, and at the membrane and that its distribution varies depending on cell type and developmental stage. At the anterior tip of the germarium, RanBPM is expressed in the GSCs and in dividing cysts, and its expression is reduced in the germline as individual egg chambers are formed (Fig. 3, C and D). We colabeled wild-type germaria with antibodies recognizing RanBPM and Bam, which is expressed only during cyst mitoses (McKearin and Ohlstein, 1995), and found that the reduction in RanBPM expression coincides with the completion of cyst mitoses (Fig. 3 D). At later stages, RanBPM becomes enriched in the nuclei of follicle cells and germline cells and at the nurse cell membranes (Fig. 3 E). Our results are consistent with the different localization patterns reported for vertebrate RanBPM in various cell types: some studies report cytoplasmic and membrane localization (Zou et al., 2003; Denti et al., 2004; Kramer et al., 2005), whereas others report nuclear (Nishitani et al., 2001; Rao et al., 2002) or perinuclear staining (Shibata et al., 2004).
The anticore antiserum staining pattern contrasts with the RanBPM-Gal4 expression pattern, suggesting that the two RanBPM isoforms might be differentially expressed. To test this possibility, we raised an isoform-specific antiserum against the N-terminal portion of long RanBPM that is not present in short RanBPM (Fig. 1 A). By Western analysis, the anti–N-terminal antiserum recognizes the long isoform but not the short isoform (Fig. 1 C). We immunostained ovaries from adult females using this anti–N-terminal antiserum and observed staining in the nuclei and cytoplasm of the CpCs and TF cells of the niche, coincident with the patched-LacZ niche cell marker (Fig. 2 D; Forbes et al., 1996a). In pupal ovaries, long RanBPM is also enriched in the niche cells, but in contrast to its localization in adult ovaries, it is localized to the cell periphery, where it colocalizes with the membrane skeletal protein α-spectrin (Fig. 2 E). We did not observe this staining pattern in homozygous RanBPMnull pupal ovaries (Fig. 2 F) or in RanBPMΔN mutant ovaries (not depicted), confirming the specificity of this antiserum for the long isoform of RanBPM. These results indicate that RanBPM-Gal4 reports expression of the long RanBPM isoform in the CpCs and TF. The specific expression of long RanBPM in the GSC niche strongly suggests a specific role for this isoform in the niche.
Long RanBPM affects niche morphology and niche capacity
The unique properties of RanBPMΔN provided a source of adult females at 20°C that only expressed short RanBPM. To study the effects of loss of long RanBPM, we first compared RanBPMΔN mutant niches to heterozygous controls using an antibody to lamin C (LamC), which is expressed at the highest levels in the CpCs and TF (Xie and Spradling, 2000; Ward et al., 2006). Consistent with the proper specification of TF and CpC fates, RanBPM mutant niche cells maintained normal expression of the niche cell markers Engrailed (not depicted) and LamC, and, as in wild type, LamC expression was higher in mutant TF cells than in mutant CpCs (Fig. 4, A and B).
Figure 4.
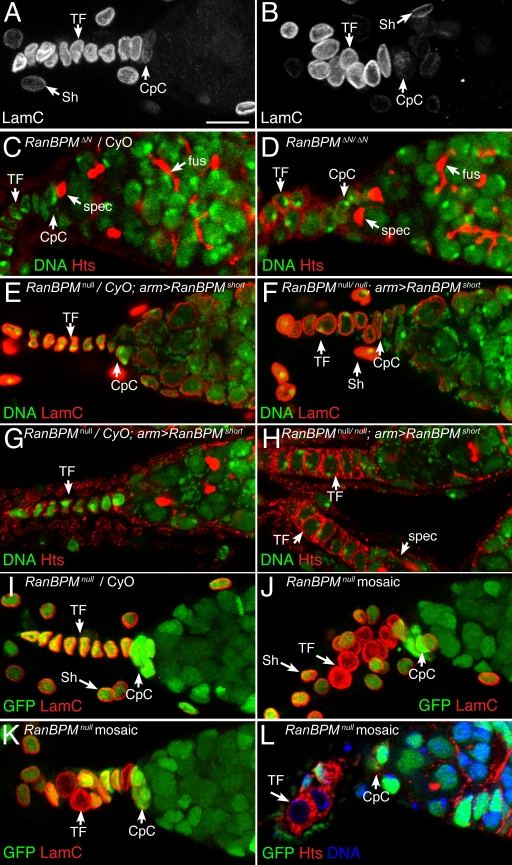
Long RanBPM affects niche cell shape and TF morphology. Loss of long RanBPM causes an increase in niche cell size and abnormal terminal filament (TF) organization. (A–D) Comparisons between heterozygous RanBPMΔN controls (A and C) with homozygous RanBPMΔN mutants (B and D). (E–H) RanBPMnull homozygotes with ubiquitous expression of short RanBPM (F and H) compared with heterozygous controls expressing short RanBPM (E and G). (I–L) A control RanBPMnull heterozygous niche (I) compared with RanBPMnull mosaic niches (J–L). In mosaics, RanBPM mutant cells are marked by lack of GFP expression (green). Antibodies to lamin C (LamC) strongly label the single-filed stack of TF cells, more weakly mark the cap cells (CpC), and also label cells from the sheath that surrounds each ovariole (Sh). Hts antibodies (red) label niche cell membranes, GSC spectrosomes (spec), and cystoblast fusomes (fus). Cell nuclei are labeled with DAPI (A–D and I–L) or PicoGreen (E–H). Each panel with LamC staining is a maximum projection of three optical sections. Bar, 10 μm.
Normal TF cells assume the shape of a short cylinder and stack together in a single-filed array to form a filament (Fig. 4, A, E, and I). We observed a cell-autonomous defect in cell shape and size that was most visible in the TF, wherein RanBPMΔN mutant niche cells are larger and more spherical than normal (Fig. 4, B and D). In addition, the organization of the filament was affected, with mutant cells often misaligned and not forming a normal single-filed array of cells (Fig. 4, B and D).
Loss of long RanBPM from the ovary does not affect the ability of the stem cells to produce functional cystoblasts, and RanBPMΔN homozygotes do not show obvious morphological defects in egg chamber development. To test whether long RanBPM affects stem cell regulation, we scored the number of GSCs attached to the CpC cluster and the rate of stem cell division in RanBPMΔN mutants (Table I). CpCs were identified by their position and morphology, and GSCs were identified by the presence of a spectrosome, a germline-specific cytoplasmic organelle that in GSCs is invariably attached to the site of contact between GSCs and their niche (Lin et al., 1994; Deng and Lin, 1997; de Cuevas and Spradling, 1998). Viewed with antibodies recognizing α-spectrin or an adducin-like protein (Hu-li tai shao [Hts]), the spectrosome appears as an apical sphere in the GSC cytoplasm that contacts the CpCs (Fig. 4, C and D). Mitotic GSCs are characterized by an elongated spectrosome that spreads from the CpC attachment point into the cystoblast (de Cuevas and Spradling, 1998). We did not observe altered stem cell division rates in RanBPMΔN homozygotes (0.43 ± 0.2 divisions per GSC, n = 189) compared with heterozygous controls (0.39 ± 0.2 divisions per GSC, n = 101; P = 0.46). We also measured the maintenance of GSCs in RanBPMΔN homozygotes and did not observe stem cell loss over extended incubations at 20 or 25°C. Instead, we observed a significant increase in the total number of GSCs attached to mutant CpC clusters (3.57 ± 0.84, n = 53 niches) as compared with heterozygous controls (2.73 ± 0.61, n = 37; P = 4.3E-7).
Table I. The effect of RanBPM loss on cell number and size in the ovarian stem cell niche.
| Description/genotype | Isoform
|
Ratio mutant/control |
t test p-value |
Mean | SD | n | |
|---|---|---|---|---|---|---|---|
| Long | Short | ||||||
| GSCs per germarium | |||||||
| Mosaics | − | − | 1.34a | 1.7E-4 | 4.01 | 0.91 | 23 |
| Nonmosaics | + | + | 3.00 | 0.71 | 21 | ||
| ΔN/ΔN | − | + | 1.31a | 4.3E-7 | 3.57 | 0.84 | 53 |
| ΔN/CyO | + | + | 2.73 | 0.61 | 37 | ||
| null/null; arm > RanBPMshort | − | + | 1.06 | 0.39 | 2.22 | 0.72 | 36 |
| null/CyO; arm > RanBPMshort | + | + | 2.09 | 0.52 | 33 | ||
| CpCs per germarium | |||||||
| Mosaics | − | − | 1.00 | 0.96 | 5.95 | 1.4 | 42 |
| Nonmosaics | + | + | 5.94 | 1.2 | 32 | ||
| ΔN/ΔN | − | + | 1.01 | 0.86 | 5.39 | 0.73 | 49 |
| ΔN/CyO | + | + | 5.36 | 0.64 | 36 | ||
| null/ null; arm > RanBPMshort | − | + | 0.81a | 3.9E-7 | 4.08 | 0.71 | 38 |
| null/CyO; arm > RanBPMshort | + | + | 5.00 | 0.66 | 39 | ||
| GSCs per CpC | |||||||
| null/null from mosaics | − | − | 2.27ab | 2.4E-5 | 1.09 | 0.52 | 58 |
| All CpCs from mosaics | mixed | mixed | 1.26ab | 6.7E-3 | 0.63 | 0.17 | 152 |
| +/+ from mosaics | + | + | 0.96 | 0.80 | 0.48 | 0.35 | 94 |
| null/CyO from nonmosaics | + | + | 0.50 | 0.12 | 128 | ||
| ΔN/ΔN | − | + | 1.31a | 1.1E-7 | 0.68 | 0.14 | 261 |
| ΔN/CyO | + | + | 0.52 | 0.11 | 193 | ||
| null/null; arm > RanBPMshort | − | + | 1.32a | 5.2E-5 | 0.56 | 0.14 | 133 |
| null/CyO; arm > RanBPMshort | + | + | 0.42 | 0.11 | 164 | ||
| CpC surface area (μm2) | |||||||
| null/null from mosaics | − | − | 1.28a | 4.0E-3 | 90.2 | 21.7 | 17 |
| +/+ from mosaics | + | + | 70.3 | 19.0 | 27 | ||
| ΔN/ΔN | − | + | 1.35a | 1.4E-11 | 90.1 | 12.8 | 38 |
| ΔN/CyO | + | + | 66.5 | 12.0 | 34 | ||
| null/null; arm > RanBPMshort | − | + | 1.32a | 6.4E-6 | 82.0 | 20.4 | 34 |
| null/CyO; arm > RanBPMshort | + | + | 62.2 | 7.3 | 19 | ||
| Total niche cells | |||||||
| Mosaics | − | − | 1.03 | 0.37 | 14.5 | 1.2 | 19 |
| Nonmosaics | + | + | 14.1 | 1.0 | 11 | ||
| null/null; arm > RanBPMshort | − | + | 0.94 | 1.3E-4 | 11.7 | 0.62 | 38 |
| null/CyO; arm > RanBPMshort | + | + | 12.4 | 0.88 | 39 | ||
| Divisions per GSC | |||||||
| null/null from mosaics | − | − | 0.93 | 0.78 | 0.26 | 0.26 | 92 |
| +/+ from mosaics | + | + | 0.28 | 0.27 | 63 | ||
| ΔN/ΔN | − | + | 1.10 | 0.46 | 0.43 | 0.21 | 189 |
| ΔN/CyO | + | + | 0.39 | 0.23 | 101 | ||
| null/null; arm > RanBPMshort | − | + | 1.47 | 0.22 | 0.33 | 0.38 | 80 |
| null/CyO; arm > RanBPMshort | + | + | 0.22 | 0.33 | 69 | ||
| GSC surface area (μm2) | |||||||
| GSC attached to null/null CpC | − | − | 1.04 | 0.31 | 356 | 41 | 21 |
| GSC attached to wild-type CpC | + | + | 341 | 53 | 22 | ||
Significant difference.
As compared with GSCs per CpC from the nonmosaic null/CyO control.
A possible explanation for the observed increase in GSC number is that loss of long RanBPM causes an increase in the number of CpCs in the niche. However, the number of CpCs in control niches (5.36 ± 0.64, n = 36 niches) was essentially the same as in RanBPMΔN mutant niches (5.39 ± 0.73, n = 49; P = 0.86). In normal niches, the CpC cluster's ability to support stem cells is limited so that each cluster of five CpCs supports two or three stem cells, giving a mean of 0.5 stem cells per CpC (0.52 ± 0.11, n = 193). This ratio is consistent with published mean numbers of CpCs and GSCs per ovariole (Xie and Spradling, 2000; Ward et al., 2006; Song et al., 2007). In the RanBPMΔN mutant, each CpC supports on average a 30% increase in total GSCs (0.68 ± 0.14, n = 261, P = 1.1E-7).
Another possible explanation for the observed increase in GSC number is that the mutant CpCs are larger than normal and can therefore support more GSCs. We measured the size of wild-type and RanBPM mutant CpCs and compared their surface areas (see Materials and methods and Table I). RanBPM mutant CpCs were significantly larger than normal (mutant, 90.1 ± 12.8 μm2; control, 66.5 ± 12.0 μm2; P = 1.4E-11). This increase in niche size correlates well with the increased number of GSCs that associate with CpC clusters in the RanBPMΔN mutant.
As an independent test for the niche function of long RanBPM, we ubiquitously expressed short RanBPM with a combination of arm-Gal4 and UASp-RanBPMshort transgenes (null/null; arm > RanBPMshort; Table I), which rescued the lethality of RanBPMnull homozygotes at 18°C. Based on immunostaining, this genotype expresses short RanBPM throughout the germarium and at higher levels than normal (unpublished data). Similar to RanBPMΔN, we observed an increase in cell size and TF misalignment (Fig. 4, E–H), although niche organization was closer to normal than in RanBPMΔN or RanBPMnull mosaics. We observed a significant decrease in the number of CpCs in this genotype (4.08 ± 0.71, n = 38; control, 5.0 ± 0.66, n = 39; P = 3.9E-7), possibly as a result of higher than normal expression of short RanBPM in the GSC niche. As in RanBPMΔN mutants, we observed a 30% increase in the mean number of GSCs per CpC in females expressing only short RanBPM (0.56 ± 0.14, n = 133) as compared with heterozygous controls (0.42 ± 0.11, n = 164; P = 5.2E-5). As a measure of the approximate GSC binding capacity of CpCs expressing only short RanBPM, we measured CpC surface area and observed an increase of ∼30% (82.0 ± 20.4 μm2, n = 34; control, 62.2 ± 7.3 μm2; P = 6.4E-6). Together with the niche-specific expression pattern of long RanBPM and the RanBPMΔN mutant phenotype, these observations indicate that the long isoform of RanBPM functions in the GSC niche to regulate its organization and cell size and that CpC size is an important determinant of niche capacity.
A bias for stem cell attachment to RanBPM-null CpCs in mosaic niches
To investigate the possibility of a niche function for the ubiquitously expressed short RanBPM isoform, we continued our analyses using genetic mosaics of a RanBPM-null allele. To study the complete loss of RanBPM from the GSC niche, we induced mitotic recombination in the gonadal mesoderm during niche morphogenesis to generate niche cells that were homozygous mutant for RanBPMnull (see Materials and methods). RanBPMnull cells in mosaic niches, which were identified by lack of GFP expression, have a very similar niche phenotype to RanBPMΔN mutant cells: mutant TF cells are larger, and both mutant and wild-type cells in mosaic TFs are disorganized (Fig. 4, I–L).
We analyzed the ability of individual RanBPMnull CpCs in mosaic niches to maintain GSCs. Similar to our observations with RanBPMΔN, the number of GSCs per niche is significantly higher in mosaics (4.01 ± 0.91, n = 23 niches) compared with nonmosaic controls (3.00 ± 0.71, n = 21; P = 1.7E-4; Table I). This was not caused by a decrease in the size of GSCs attached to mutant CpCs (attached to wild-type CpC, 341 ± 53 μm2, n = 22; attached to null CpC, 356 ± 41 μm2, n = 21; P = 0.31) or to an increase in CpC number, as the number of CpCs in nonmosaic niches (5.94 ± 1.2, n = 32 niches) was not significantly different from mosaic niches (5.95 ± 1.4, n = 42; P = 0.96). Similar to RanBPMΔN mutant CpCs, the mean surface area of homozygous-null CpCs was increased by 30% relative to controls (wild type, 70.3 ± 19.0 μm2, n = 27; null, 90.2 ± 21.7 μm2, n = 17; P = 4.0E-3). The defects in niche morphology caused by removing long RanBPM are therefore not enhanced by additional removal of short RanBPM, indicating a specific function for the long isoform in these cells.
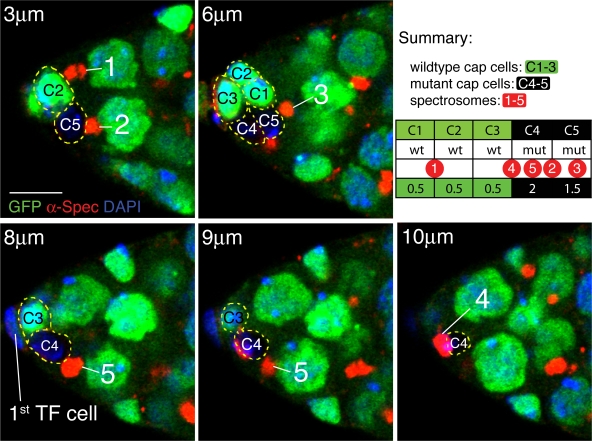
To test whether loss of RanBPM from CpCs affects stem cell regulation, we scored the frequency of cell division for GSCs attached to mutant or wild-type CpCs and the number of GSCs attached to individual CpCs in null mosaics (Table I). We did not observe a correlation between CpC genotype and the rate of GSC division (wild type, 0.28 ± 0.27, n = 63; mutant, 0.26 ± 0.26, n = 92; P = 0.78) or any obvious defects in the development of egg chambers, although it should be noted that, with the exception of the GSCs themselves, we are aware of no method to identify germline cells with respect to initial adhesion to specific CpCs. We next scored the attachment of individual CpCs to individual GSCs by noting the position of the GSC spectrosomes in relation to the CpC membranes (Fig. 5 and Table I). In nonmosaic niches, an individual CpC was associated with 0.5 ± 0.12 spectrosomes (n = 128). Within mosaic niches, RanBPMnull mutant CpCs were attached to twice as many spectrosomes as normal (1.09 ± 0.52, n = 58 CpCs, P = 2.4E-5), whereas the wild-type CpCs in the same niches were attached to normal numbers of spectrosomes (0.48 ± 0.35, n = 94 CpCs, P = 0.80 compared with nonmosaic controls). Individual mutant CpCs in mosaic niches were observed to contact as many as three spectrosomes, allowing the attachment of up to five spectrosomes to the mosaic CpC cluster (Fig. 5). The 2.2-fold increase in the number of GSCs attached to RanBPMnull CpCs is greater than the 30% increase in mutant CpC surface area, indicating a relative increase in the adhesive properties of RanBPMnull CpCs as compared with wild-type CpCs. The increased number of GSCs attached to RanBPMnull CpCs, beyond that observed when only the long isoform was removed from the niche, strongly suggests that short RanBPM functions in the niche to suppress adhesion between the CpCs and GSCs.
Figure 5.
RanBPM mutant CpCs are attached to more stem cell spectrosomes than normal. Five optical sections from a single RanBPMnull mosaic niche are shown. Antibodies to α-spectrin (red) mark the spectrosome of individual GSCs and cell membranes. In this example, the CpC cluster, which is composed of five CpCs (C1–5), is directly apposed to five spectrosomes (1–5). The spectrosome numbers also indicate the positions of the GSC nuclei. Two RanBPM mutant CpCs are marked by lack of nuclear GFP expression (C4 and C5). By studying the original optical stack, the attachment of each spectrosome to each CpC was noted (summary), and individual slices were chosen to best show the spectrosomes. Each panel is labeled with a distance in micrometers from the top of the optical stack as determined by LSM510 software. In this example, four of the five spectrosomes contact the two mutant CpCs. The number of spectrosomes per CpC is noted at the bottom of the summary table. When a spectrosome appeared to contact two CpCs, a score of 0.5 was given to each CpC. In this example, the mean number of spectrosomes per wild-type CpC is 0.5, whereas the mean for RanBPMnull CpCs is 1.75. Bar, 5 μm.
RanBPM affects Arm and Hts localization
There are several possible explanations for the defective morphology of RanBPM mosaic niches, such as abnormal cell polarity, cytoskeletal organization, or cell adhesion. The increase in the number of spectrosomes attached to mutant CpCs together with the vertebrate RanBPM literature suggested a role for RanBPM in regulating cell adhesion in the GSC niche. Consistent with this hypothesis, we observed altered localization of Arm (β-catenin), a structural component of adherens junctions and a key component of the Wnt signal transduction pathway (for review see Harris and Peifer, 2005). In a wild-type GSC niche, Arm normally accumulates at the membranes of all CpCs, whereas its expression is reduced in the TF (Fig. 6 A). In mosaic niches, however, Arm accumulates to abnormally high levels in mutant TF cells, where it localizes to cell surfaces especially at the boundary between mutant and wild-type cells (Fig. 6, B and C). By immunostaining, we did not observe changes in Arm localization or expression level in RanBPM mutant CpCs, possibly because the normal staining at the surfaces of all CpCs obscured the increase in mutant CpCs. The alteration in Arm localization in the TF suggests an increase in the stability or expression of Arm in RanBPM mutant niche cells or an increase in the formation of adherens junctions between mutant niche cells and their neighbors.
Figure 6.
Arm localization in the niche is affected by loss of RanBPM. (A) In a RanBPMnull heterozygous control niche, the periphery of each cap cell (CpC) is labeled strongly with antibodies to Armadillo (Arm, red), whereas the terminal filament (TF) is only weakly labeled. (B and C) In RanBPMnull mosaic niches, Arm staining is increased between RanBPM mutant cells. This increase is especially pronounced at the interface between mutant and wild-type TF cells (arrows). Bar, 10 μm.
We also observed altered localization patterns of the actin-binding protein Hts. Hts is related to adducin, a cytoskeletal protein that binds to the barbed ends of actin filaments and promotes their association with spectrin (Li et al., 1998). Normally, Hts colocalizes with filamentous actin and spectrin at the cell cortex of the Drosophila oocyte, consistent with its predicted adducin-like function (Zaccai and Lipshitz, 1996). In RanBPM mutant niche cells, we observed an increase in the cytoplasmic level of Hts (Fig. 4, C, D, G, H, and L). This defect in Hts localization suggests a defect in cytoskeletal organization in RanBPM mutant cells either as a primary effect of RanBPM loss or as a consequence of cell adhesion defects in mutant cells. Although it is possible that the observed increase in Hts staining in RanBPM mutant cells is related to the cell shape defect of mutant cells, loss of RanBPM did not affect the normal localization of F-actin to the cell periphery of niche cells (unpublished data).
RanBPM affects epithelial and dorsal appendage morphology
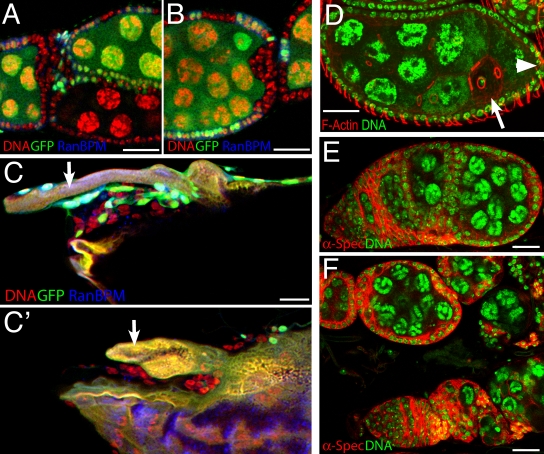
To further characterize the function of RanBPM in cell adhesion, we analyzed RanBPMnull genetic mosaics in the epithelial follicle cell layer that surrounds individual egg chambers. We found that loss of RanBPM from follicle cells can disrupt epithelial organization, causing the single-layered epithelium to become multilayered, which is sometimes associated with fusion of adjacent egg chambers (Fig. 7, A and B). This phenotype is incompletely penetrant and preferentially affects follicle cells at the anterior and posterior poles of an egg chamber.
Figure 7.
RanBPM affects dorsal appendage morphogenesis and follicle cell organization. (A and B) In mosaics, loss of RanBPM causes incompletely penetrant defects in epithelial structure, causing egg chamber fusion (A) and follicle cell multilayering, which mainly affects clones positioned at the anterior or posterior poles of the egg chamber (B). (C and C′) Two partial projections from a single optical stack though a mosaic RanBPMnull late stage oocyte. RanBPM mutant follicle cells are marked by lack of GFP (green) and lack of RanBPM (blue), and nuclei are labeled with DAPI (red). An extended dorsal appendage (arrow) is surrounded by wild-type cells in C, whereas on the other side of the embryo (C′), RanBPM mutant cells surround a short, paddle-like dorsal appendage (arrow). (D) Homozygotes for the hypomorphic RanBPMCB allele are fertile and have normal egg chamber morphology except for an incompletely penetrant defect in oocyte nuclear positioning. The normal posterior position of the oocyte is marked with an arrowhead, and the ectopic position is marked with an arrow. (E and F) The phenotype of RanBPMCB/RanBPMnull is normal except for an oocyte localization defect, but a dominant enhancement after removing one copy of shotgun causes egg chamber fusion (E) and loss of epithelial integrity (F). Bars, 25 μm.
Loss of RanBPM was also observed to cause defects in the morphogenesis of dorsal appendages, the prominent extended chorion structures at the anterior dorsal surface of the eggshell. Dorsal appendages are formed by coordinated epithelial movements and cell shape changes that remodel the dorsal follicle cell epithelium into a pair of elongated tubes (Dorman et al., 2004; Ward and Berg, 2005). We observed the morphology of dorsal appendages during late stages of oogenesis, before follicle cell apoptosis, so the genotype of individual follicle cells could still be determined. We concentrated our analysis on those egg chambers in which one dorsal appendage was surrounded by wild-type GFP-expressing cells, whereas the second was surrounded by RanBPMnull follicle cells, allowing us to compare dorsal appendages of different genotypes at the same stage of development. In these egg chambers, the dorsal appendage surrounded by wild-type cells formed an extended tube (Fig. 7 C), whereas the dorsal appendage surrounded by RanBPM mutant cells was much shorter and paddle-like (Fig. 7 C′). This indicates a role for RanBPM in dorsal appendage morphogenesis, a process known to involve differential cell adhesion (Ward and Berg, 2005; Laplante and Nilson, 2006).
In further support of an adhesive role for RanBPM, we observed a genetic interaction between the hypomorphic allele RanBPMCB and shotgun, which encodes the homophylic adhesion molecule DE-cadherin. Homozygous mutant RanBPMCB females are fertile, and the majority of their egg chambers develop normally, each individually surrounded by a monolayer of follicle cells. However, in 19% of midstage RanBPMCB egg chambers, we observed clear mislocalization of the oocyte away from the posterior (Fig. 7 D). Because DE-cadherin is required for the germline–soma interactions that position the oocyte within the egg chamber (Godt and Tepass, 1998), we tested for a genetic enhancement of RanBPMCB phenotypes by reducing the shotgun gene dose by half.
RanBPMCB is a hypomorphic allele and is semilethal in trans with RanBPMnull (67 survivors from an expected 713 [9.4%]). This reduction in viability was dominantly enhanced in an shg heterozygous background (RanBPMCB shg2/RanBPMnull; five survivors from an expected 412 [1.2%]). In ovaries of the RanBPMCB shg2/RanBPMnull survivors, we observed defects that were not seen in either RanBPMCB/RanBPMnull or shg2 heterozygotes alone. These defects included frequent follicle cell multilayering at the poles of egg chambers, severe egg chamber fusion, and loss of epithelial integrity so that many germline cells were no longer surrounded by a somatic cell layer (Fig. 7, E and F). This genetic enhancement suggests that RanBPM positively regulates cell adhesion in the follicular epithelium.
Discussion
Drosophila RanBPM differentially expresses two protein isoforms: although short RanBPM appears to be expressed in all somatic and germline cells of the ovary, the long isoform is specifically expressed in the GSC niche. We studied the niche function of long RanBPM by analyzing two types of flies expressing only short RanBPM: homozygotes for RanBPMΔN, a hypomorphic allele that specifically disrupts long RanBPM expression, and RanBPMnull females expressing only short RanBPM from a transgene. These studies revealed that long RanBPM is required for the proper arrangement of niche cells and is autonomously required for proper niche cell size. We found that CpCs not expressing long RanBPM are in contact with ∼30% more GSC spectrosomes than wild-type CpCs. This correlates well with our observation that the mean surface area of CpCs lacking long RanBPM is 30% higher than normal. We conclude that loss of long RanBPM increases niche capacity by increasing the potential contact surface between CpCs and stem cells.
To study additional functions of short RanBPM in the niche, we removed both isoforms from the niche by generating genetic mosaics of a null RanBPM allele. We observed that RanBPMnull niche cells share the same cell size and niche morphology defects as those we observed after removing only long RanBPM. In addition, we observed a strong bias for GSC attachment to RanBPMnull CpCs in mosaic niches (2.27 times higher than wild-type CpCs), a greater effect than would be predicted by the loss of long RanBPM alone (1.3 times higher; Table I). The bias for GSC attachment to mutant CpCs supports a role for the short isoform in negatively regulating the adhesive properties of the niche in addition to the requirement for long RanBPM for proper niche cell size and organization. Collectively, our results imply that regulating the size and adhesive properties of the CpCs is an important component of the mechanism that controls their capacity to support stem cells and that the two RanBPM isoforms are important factors in mediating this regulation.
In contrast to the niche, our analysis of RanBPMnull mosaics in the follicle cell epithelium suggests that short RanBPM acts as a positive regulator of cell adhesion. In RanBPMnull mosaics, we observed defects in the organization of the follicle cell layer and defects in dorsal appendage morphogenesis, which are consistent with a decrease in adhesion between mutant cells in the follicle cell layer. This is further supported by a strong genetic enhancement of a weak RanBPM allele when the gene copy number of shotgun (DE-cadherin) is decreased. Why might short RanBPM function as a negative regulator of cell adhesion in the GSC niche but as a positive regulator of cell adhesion in the follicle cell epithelium? We propose that long RanBPM, which is specific to the GSC niche, has additional or altered functions conferred by its N-terminal domain that affect niche cell size and morphogenesis and that long RanBPM or another protein expressed in the niche alters the cell adhesive function of short RanBPM. In this context, it is noteworthy that vertebrate RanBP10 operates antagonistically to RanBPM in the Met pathway, where it competes with RanBPM for binding to the Met receptor but lacks the ability to activate the Ras–extracellular regulated kinase pathway (Wang et al., 2002, 2004). In this case, the functional difference between RanBP10 and RanBPM depends on their different N-terminal sequences.
Although we cannot presently distinguish between direct or indirect effects, our loss of function analyses of Drosophila RanBPM indicate a role for its long isoform in niche morphogenesis and a role for short RanBPM in regulating cell adhesion. Further characterization of RanBPM function in Drosophila awaits the identification of its interacting partners.
Materials and methods
Drosophila stocks and cDNA clones
P{GawB}NP6392 (RanBPM-Gal4; GenBank/EMBL/DDBJ accession no. AG218064), a P element insertion in the 5′ UTR of RanBPM, was obtained from the GetDB enhancer trap collection (Hayashi et al., 2002). The insertion site of P{GawB}NP6392 was confirmed by inverse PCR. P{RS3}RanBPMCB-6232-3 (RanBPMCB; GenBank/EMBL/DDBJ accession no. AJ547101) was obtained from the DrosDel collection through the Szeged Drosophila Stock Center (Ryder et al., 2004), and Pbac{WH}RanBPMf06647 (GenBank/EMBL/DDBJ accession no. CZ489352) was obtained from the Exelixis stock collection at Harvard University (Artavanis-Tsakonas, 2004). The P{UASp} transformation vector and flies carrying the P{UASp-LacZ} reporter were provided by P. Rørth (Temasek Life Sciences Laboratory, National University of Singapore, Singapore). The hedgehog-LacZ and patched-LacZ lines were provided by T. Xie (Stowers Insitute for Medical Research, Kansas City, MO) and A. Spradling (Carnegie Institution, Baltimore, MD). Df(2R)12 and other lines used for mosaic analyses and mutagenesis were obtained from the Bloomington Drosophila Stock Center at Indiana University. Homozygous mutant RanBPM larvae or adults were identified by the absence of a CyO, Krüppel-GAL4, UAS-GFP balancer chromosome (Casso et al., 1999).
RanBPM cDNA clones RH61511, RH58540, RE27154, and RE54051 were obtained from the Drosophila Gene Collection libraries (Stapleton et al., 2002) and were sequenced at the McGill Sheldon Biotechnology Center (GenBank/EMBL/DDBJ accession nos. AY973544, AY973545, and AY973546).
RanBPM mutagenesis and P element reversion
P{RS3}CB-6232-3 was mobilized with the P{Δ2-3}99B transposase source, and Pbac{WH}f06647 was mobilized with a Hsp70-piggyBac transposase source (Thibault et al., 2004). White-eyed excision chromosomes were individually stocked, tested for viability and lethality, and tested for complementation with the RanBPM deletion, Df(2R)12. Excision of P{RS3}CB6232-3 or Pbac{WH}f06647 led to a high frequency of reversion to fertility and viability (Table S1), indicating a strong correlation between each transposable element insertion and its lethal phenotype. RanBPMΔN was isolated at 25°C as a lethal excision derivative of P{RS3}CB-6232-3. A genomic fragment spanning the 1,448-bp RanBPMΔN deletion was isolated by PCR and sequenced (GenBank/EMBL/DDBJ accession no. EU120026).
The RanBPMnull deletion allele (P+Pbac{RS3.WH}RanBPMnull) is an FLP-out derivative of two Flip recombinase target (FRT) sites from the transposable element insertions P{RS3}CB-6232-3 and Pbac{WH}f06647. In brief, genomic DNA between the directly repeated FRT sequences from P{RS3}CB-6232-3 and Pbac{WH}f06647 was removed by transrecombination in the presence of a heat shock–inducible FLP recombinase (Parks et al., 2004; Thibault et al., 2004). The relative organization of the FRT and white+ sequences at the RanBPM locus allowed the screening of recombinant chromosomes for two copies of the white+ marker. The RanBPMnull deletion was confirmed by PCR and Western analysis (Fig. 1 C).
UAS-RanBPM transgene and RanBPM rescue
A genomic DNA fragment containing the coding region for short RanBPM was isolated with primers 5′-GAAGACGACGACGAGGAGGTGG-3′ and 5′-GGGCATCGGTATTGCTGGACAGG-3′. The PCR fragment was cloned into pCR4-Topo (Invitrogen), restricted with NotI and SpeI, and ligated into the NotI and XbaI sites of P{UASp}. The lethality of RanBPMnull was rescued at 22°C in flies of genotype w; RanBPMnull/RanBPMnull; P{UASp-RanBPMshort}/arm-GAL4. As in the RanBPMΔN temperature-sensitive allele, only the short RanBPM isoform was produced in this genotype, and rescue was not successful at 25°C. The insufficiency of the short isoform to confer viability at 25°C indicates an essential function of long RanBPM at higher temperatures.
Genetic mosaic analyses
To study the role of RanBPM in the niche, we used bab-GAL4 and UAS-FLP transgenes to target mitotic recombination to the gonadal mesoderm early in niche development (Bolívar et al., 2006). Mosaic stem cell niches were generated at 25°C in flies of genotype P{FRT}G13 RanBPMnull/P{FRT}G13 P{Ubi-GFP.nls}; bab-Gal4 P{UAST-FLP}. Flies with the bab-Gal4 P{UAST-FLP} chromosome were provided by A. González-Reyes (Spanish Research Council, Universidad Pablo de Olavide, Sevilla, Spain; Bolívar et al., 2006). To generate germline clones and somatic follicle cell mosaics, flies of genotype y w P{hsFLP}1; P{neoFRT}42D pwn1 RanBPMnull/P{neoFRT}42D P{Ubi-GFP.nls} were heat treated (37°C for 1 h) three times on consecutive days in vials, beginning with the first appearance of pupae.
To mark GSCs and dividing cysts, we used mouse monoclonal antibodies recognizing Hts or α-spectrin, which stain the membrane skeleton of somatic and germline cells, the fusomes of dividing cysts, and the spectrosomes of stem cells. LamC antibodies and hedgehog-LacZ and patched-LacZ reporter constructs were used to mark the stem cell niche (Forbes et al., 1996b; Xie and Spradling, 2000).
To compare the number of GSCs per CpC in mosaic niches, an unpaired two-tailed t test assuming unequal variances was performed with Excel (Microsoft). To estimate the surface area of CpCs or GSCs, two measurements were taken from individual cells with the measure tool in Photoshop (Adobe): across and perpendicular to the longest dimension of the cell in an optical section. These measurements were converted to micrometers (73.1 μm per 1,024 pixels), and the surface area was estimated assuming each cell approximated an oblate spheroid using the following formula:
 |
where the angular eccentricity (e) is
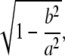 |
a is the semimajor axis, and b is the semiminor axis (Kumar and Mathew, 2003). Calculated surface areas were compared with a two-tailed t test assuming unequal variances.
Generation of RanBPM antibodies, immunostaining, and Western analysis
Rabbit antisera recognizing RanBPM were generated at the McGill Animal Resources Centre against GST fusion proteins containing an N-terminal fragment of RanBPM (αN-term) or a central fragment (αCore). Fragments were amplified from clone RE27154 with primers 5′-GGATCCCTACGTCTCGGTCACACTCTC-3′ and 5′-CTCGAGGACTGCCGTTGTTGCTGTAGA-3′ (N-term) or 5′-GAATTCAGCAGCACCCAGACATCATCGC-3′ and 5′-AGCGGCCGCCGGGGAACTGGGTCAATTTTGCG-3′ (Core). The restriction sites built into the primers were used to clone the PCR fragments into pGEX-5X-1. Antibodies were tested by Western analysis on control and RanBPMnull tissue and by staining RanBPMnull mosaic tissue.
For Western blots, homozygous mutant larvae were identified by the absence of a CyO, Krüppel-GAL4, UAS-GFP balancer chromosome. Roaming third instar larvae from each genotype were selected, washed in PBS, and homogenized directly in 50 μl of loading buffer (5 M urea, 0.125 M Tris, pH 6.8, 4% SDS, 10% β-mercaptoethanol, 20% glycerol, and 0.1% bromophenol blue).
Antibody stainings were performed as in Liu et al. (2003). RanBPM serum was preadsorbed overnight at 4°C with an equal volume of fixed and washed RanBPMnull tissue to decrease background staining and was used at 1:2,500 for immunostaining or 1:10,000 on Western blots. Mouse anti-LamC (ADL67; 1:1) was provided by P. Fischer (University of Texas at Austin, Austin, TX), and mouse anti-Hts (1B1; 1:5), anti–α-spectrin (3A9; 1:15), and anti-Arm (N2 7A1; 1:100) were obtained from the Developmental Studies Hybridoma Bank. Rabbit anti–β-galactosidase (ab616; 1:2,500) was purchased from Abcam. Cell nuclei were stained with PicoGreen (1:1,000) for 30 min after treating egg chambers with 20 μg/ml RNase A for 30 min or with 0.5 μg/ml DAPI for 10 min. AlexaFluor secondary antibodies (488, 555, and 647 nm used at 1:500) and Slowfade Light Antifade reagents were obtained from Invitrogen. Images were acquired on a laser-scanning microscope (LSM510; Carl Zeiss, Inc.) with C-Apochromat 40×/NA 1.2 and 63×/NA 1.2 water-immersion objectives (Carl Zeiss, Inc.) or a 63×/NA 1.4 oil immersion objective (Carl Zeiss, Inc.). Optical projections were performed with the LSM Image Browser (Carl Zeiss, Inc.) set for maximum transparency. Channel levels were manually adjusted with Photoshop to optimize histogram width.
Online supplemental material
Table S1 shows reversion analysis of RanBPM transposon insertions. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200711046/DC1.
Supplementary Material
Acknowledgments
We would like to acknowledge A. Wilde, A. DiAntonio, N. Scantlebury, S. Johnson, A.R. Campos, and J. Jiang for exchanging information before publication. We thank D. Godt for generously providing temporary laboratory space and equipment in Toronto. We are grateful for flies and antibodies provided by P. Rørth, A. González-Reyes, P.A. Fisher, A.C. Spradling, T. Xie, H. Lin, N. Liu, and the Developmental Studies Hybridoma Bank. We thank H. Han for purifying GST-RanBPM fusion proteins for antibody production, B. Hu for transgene injections, and L.A. Nilson, D. Godt, F. Fagotto, and L. Cáceres for helpful comments on the manuscript.
This work was supported by grant RO1-HD036631 to P. Lasko from the National Institute of Child Health and Development.
Abbreviations used in this paper: Arm, Armadillo; Bam, Bag of marbles; CpC, cap cell; CTLH, C terminal to LisH; FRT, Flip recombinase target; GSC, germline stem cell; Hts, Hu-li tai shao; LamC, lamin C; LisH, lissencephaly homology; RanBPM, Ran-binding protein M; TF, terminal filament; UTR, untranslated region.
References
- Adams, J.C., B. Seed, and J. Lawler. 1998. Muskelin, a novel intracellular mediator of cell adhesive and cytoskeletal responses to thrombospondin-1. EMBO J. 17:4964–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas, S. 2004. Accessing the Exelixis collection. Nat. Genet. 36:207. [DOI] [PubMed] [Google Scholar]
- Asaoka, M., and H. Lin. 2004. Germline stem cells in the Drosophila ovary descend from pole cells in the anterior region of the embryonic gonad. Development. 131:5079–5089. [DOI] [PubMed] [Google Scholar]
- Bolívar, J., J. Pearson, L. López-Onieva, and A. González-Reyes. 2006. Genetic dissection of a stem cell niche: the case of the Drosophila ovary. Dev. Dyn. 235:2969–2979. [DOI] [PubMed] [Google Scholar]
- Casso, D., F.A. Ramirez-Weber, and T.B. Kornberg. 1999. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech. Dev. 88:229–232. [DOI] [PubMed] [Google Scholar]
- Chen, D., and D. McKearin. 2003. a. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr. Biol. 13:1786–1791. [DOI] [PubMed] [Google Scholar]
- Chen, D., and D.M. McKearin. 2003. b. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development. 130:1159–1170. [DOI] [PubMed] [Google Scholar]
- Chen, D., and D. McKearin. 2005. Gene circuitry controlling a stem cell niche. Curr. Biol. 15:179–184. [DOI] [PubMed] [Google Scholar]
- Cheng, L., S. Lemmon, and V. Lemmon. 2005. RanBPM is an L1-interacting protein that regulates L1-mediated mitogen-activated protein kinase activation. J. Neurochem. 94:1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, D.N., A. Chao, J. Baker, L. Chang, D. Qiao, and H. Lin. 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12:3715–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansereau, D.A., and P. Lasko. 2008. The development of germline stem cells in Drosophila. Methods Mol. Biol. 450:3–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cuevas, M., and A.C. Spradling. 1998. Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development. 125:2781–2789. [DOI] [PubMed] [Google Scholar]
- Deng, W., and H. Lin. 1997. Spectrosomes and fusomes anchor mitotic spindles during asymmetric germ cell divisions and facilitate the formation of a polarized microtubule array for oocyte specification in Drosophila. Dev. Biol. 189:79–94. [DOI] [PubMed] [Google Scholar]
- Denti, S., A. Sirri, A. Cheli, L. Rogge, G. Innamorati, S. Putignano, M. Fabbri, R. Pardi, and E. Bianchi. 2004. RanBPM is a phosphoprotein that associates with the plasma membrane and interacts with the integrin LFA-1. J. Biol. Chem. 279:13027–13034. [DOI] [PubMed] [Google Scholar]
- Dorman, J.B., K.E. James, S.E. Fraser, D.P. Kiehart, and C.A. Berg. 2004. bullwinkle is required for epithelial morphogenesis during Drosophila oogenesis. Dev. Biol. 267:320–341. [DOI] [PubMed] [Google Scholar]
- Forbes, A.J., H. Lin, P.W. Ingham, and A.C. Spradling. 1996. a. hedgehog is required for the proliferation and specification of ovarian somatic cells prior to egg chamber formation in Drosophila. Development. 122:1125–1135. [DOI] [PubMed] [Google Scholar]
- Forbes, A.J., A.C. Spradling, P.W. Ingham, and H. Lin. 1996. b. The role of segment polarity genes during early oogenesis in Drosophila. Development. 122:3283–3294. [DOI] [PubMed] [Google Scholar]
- Gerlitz, G., E. Darhin, G. Giorgio, B. Franco, and O. Reiner. 2005. Novel functional features of the Lis-H domain: role in protein dimerization, half-life and cellular localization. Cell Cycle. 4:1632–1640. [DOI] [PubMed] [Google Scholar]
- Gilboa, L., and R. Lehmann. 2004. Repression of primordial germ cell differentiation parallels germline stem cell maintenance. Curr. Biol. 14:981–986. [DOI] [PubMed] [Google Scholar]
- Godt, D., and U. Tepass. 1998. Drosophila oocyte localization is mediated by differential cadherin-based adhesion. Nature. 395:387–391. [DOI] [PubMed] [Google Scholar]
- Harada, N., T. Yokoyama, R. Yamaji, Y. Nakano, and H. Inui. 2008. RanBP10 acts as a novel coactivator for the androgen receptor. Biochem. Biophys. Res. Commun. 368:121–125. [DOI] [PubMed] [Google Scholar]
- Harris, T.J., and M. Peifer. 2005. Decisions, decisions: β-catenin chooses between adhesion and transcription. Trends Cell Biol. 15:234–237. [DOI] [PubMed] [Google Scholar]
- Hayashi, S., K. Ito, Y. Sado, M. Taniguchi, A. Akimoto, H. Takeuchi, T. Aigaki, F. Matsuzaki, H. Nakagoshi, T. Tanimura, et al. 2002. GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis. 34:58–61. [DOI] [PubMed] [Google Scholar]
- Kim, M.H., D.R. Cooper, A. Oleksy, Y. Devedjiev, U. Derewenda, O. Reiner, J. Otlewski, and Z.S. Derewenda. 2004. The structure of the N-terminal domain of the product of the lissencephaly gene Lis1 and its functional implications. Structure. 12:987–998. [DOI] [PubMed] [Google Scholar]
- Kobayashi, N., J. Yang, A. Ueda, T. Suzuki, K. Tomaru, M. Takeno, K. Okuda, and Y. Ishigatsubo. 2007. RanBPM, Muskelin, p48EMLP, p44CTLH, and the armadillo-repeat proteins ARMC8alpha and ARMC8beta are components of the CTLH complex. Gene. 396:236–247. [DOI] [PubMed] [Google Scholar]
- Kramer, S., T. Ozaki, K. Miyazaki, C. Kato, T. Hanamoto, and A. Nakagawara. 2005. Protein stability and function of p73 are modulated by a physical interaction with RanBPM in mammalian cultured cells. Oncogene. 24:938–944. [DOI] [PubMed] [Google Scholar]
- Kumar, V.A., and S. Mathew. 2003. A method for estimating the surface area of ellipsoidal food materials. Biosystems Engineering. 85:1–5. [Google Scholar]
- Laplante, C., and L.A. Nilson. 2006. Differential expression of the adhesion molecule Echinoid drives epithelial morphogenesis in Drosophila. Development. 133:3255–3264. [DOI] [PubMed] [Google Scholar]
- Li, X., Y. Matsuoka, and V. Bennett. 1998. Adducin preferentially recruits spectrin to the fast growing ends of actin filaments in a complex requiring the MARCKS-related domain and a newly defined oligomerization domain. J. Biol. Chem. 273:19329–19338. [DOI] [PubMed] [Google Scholar]
- Lin, H., L. Yue, and A.C. Spradling. 1994. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development. 120:947–956. [DOI] [PubMed] [Google Scholar]
- Liu, N., D.A. Dansereau, and P. Lasko. 2003. Fat Facets interacts with Vasa in the Drosophila pole plasm and protects it from degradation. Curr. Biol. 13:1905–1909. [DOI] [PubMed] [Google Scholar]
- McKearin, D., and B. Ohlstein. 1995. A role for the Drosophila Bag-of-Marbles protein in the differentiation of cystoblasts from germline stem cells. Development. 121:2937–2947. [DOI] [PubMed] [Google Scholar]
- Menon, R.P., T.J. Gibson, and A. Pastore. 2004. The C terminus of fragile X mental retardation protein interacts with the multi-domain Ran-binding protein in the microtubule-organising centre. J. Mol. Biol. 343:43–53. [DOI] [PubMed] [Google Scholar]
- Nakamura, M., H. Masuda, J. Horii, K. Kuma, N. Yokoyama, T. Ohba, H. Nishitani, T. Miyata, M. Tanaka, and T. Nishimoto. 1998. When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to gamma-tubulin. J. Cell Biol. 143:1041–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani, H., E. Hirose, Y. Uchimura, M. Nakamura, M. Umeda, K. Nishii, N. Mori, and T. Nishimoto. 2001. Full-sized RanBPM cDNA encodes a protein possessing a long stretch of proline and glutamine within the N-terminal region, comprising a large protein complex. Gene. 272:25–33. [DOI] [PubMed] [Google Scholar]
- Ohlstein, B., and D. McKearin. 1997. Ectopic expression of the Drosophila Bam protein eliminates oogenic germline stem cells. Development. 124:3651–3662. [DOI] [PubMed] [Google Scholar]
- Parks, A.L., K.R. Cook, M. Belvin, N.A. Dompe, R. Fawcett, K. Huppert, L.R. Tan, C.G. Winter, K.P. Bogart, J.E. Deal, et al. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36:288–292. [DOI] [PubMed] [Google Scholar]
- Ponting, C., J. Schultz, and P. Bork. 1997. SPRY domains in ryanodine receptors (Ca(2+)-release channels). Trends Biochem. Sci. 22:193–194. [DOI] [PubMed] [Google Scholar]
- Rao, M.A., H. Cheng, A.N. Quayle, H. Nishitani, C.C. Nelson, and P.S. Rennie. 2002. RanBPM, a nuclear protein that interacts with and regulates transcriptional activity of androgen receptor and glucocorticoid receptor. J. Biol. Chem. 277:48020–48027. [DOI] [PubMed] [Google Scholar]
- Ryder, E., F. Blows, M. Ashburner, R. Bautista-Llacer, D. Coulson, J. Drummond, J. Webster, D. Gubb, N. Gunton, G. Johnson, et al. 2004. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics. 167:797–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler, M., and R. Salgia. 2007. c-Met and hepatocyte growth factor: potential as novel targets in cancer therapy. Curr. Oncol. Rep. 9:102–108. [DOI] [PubMed] [Google Scholar]
- Schulze, H., M. Dose, M. Korpal, I. Meyer, J.E. Italiano Jr., and R.A. Shivdasani. 2008. RanBP10 is a cytoplasmic guanine nucleotide exchange factor that modulates noncentrosomal microtubules. J. Biol. Chem. 283:14109–14119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, N., N. Tsunekawa, S. Okamoto-Ito, R. Akasu, A. Tokumasu, and T. Noce. 2004. Mouse RanBPM is a partner gene to a germline specific RNA helicase, mouse vasa homolog protein. Mol. Reprod. Dev. 67:1–7. [DOI] [PubMed] [Google Scholar]
- Song, X., C.H. Zhu, C. Doan, and T. Xie. 2002. Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science. 296:1855–1857. [DOI] [PubMed] [Google Scholar]
- Song, X., M.D. Wong, E. Kawase, R. Xi, B.C. Ding, J.J. McCarthy, and T. Xie. 2004. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag-of-marbles, in germline stem cells in the Drosophila ovary. Development. 131:1353–1364. [DOI] [PubMed] [Google Scholar]
- Song, X., G.B. Call, D. Kirilly, and T. Xie. 2007. Notch signaling controls germline stem cell niche formation in the Drosophila ovary. Development. 134:1071–1080. [DOI] [PubMed] [Google Scholar]
- Stapleton, M., G. Liao, P. Brokstein, L. Hong, P. Carninci, T. Shiraki, Y. Hayashizaki, M. Champe, J. Pacleb, K. Wan, et al. 2002. The Drosophila gene collection: identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 12:1294–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault, S.T., M.A. Singer, W.Y. Miyazaki, B. Milash, N.A. Dompe, C.M. Singh, R. Buchholz, M. Demsky, R. Fawcett, H.L. Francis-Lang, et al. 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36:283–287. [DOI] [PubMed] [Google Scholar]
- Umeda, M., H. Nishitani, and T. Nishimoto. 2003. A novel nuclear protein, Twa1, and Muskelin comprise a complex with RanBPM. Gene. 303:47–54. [DOI] [PubMed] [Google Scholar]
- Wang, D., Z. Li, E.M. Messing, and G. Wu. 2002. Activation of Ras/Erk pathway by a novel MET-interacting protein RanBPM. J. Biol. Chem. 277:36216–36222. [DOI] [PubMed] [Google Scholar]
- Wang, D., Z. Li, S.R. Schoen, E.M. Messing, and G. Wu. 2004. A novel MET-interacting protein shares high sequence similarity with RanBPM, but fails to stimulate MET-induced Ras/Erk signaling. Biochem. Biophys. Res. Commun. 313:320–326. [DOI] [PubMed] [Google Scholar]
- Ward, E.J., and C.A. Berg. 2005. Juxtaposition between two cell types is necessary for dorsal appendage tube formation. Mech. Dev. 122:241–255. [DOI] [PubMed] [Google Scholar]
- Ward, E.J., H.R. Shcherbata, S.H. Reynolds, K.A. Fischer, S.D. Hatfield, and H. Ruohola-Baker. 2006. Stem cells signal to the niche through the Notch pathway in the Drosophila ovary. Curr. Biol. 16:2352–2358. [DOI] [PubMed] [Google Scholar]
- Xie, T., and A.C. Spradling. 1998. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 94:251–260. [DOI] [PubMed] [Google Scholar]
- Xie, T., and A.C. Spradling. 2000. A niche maintaining germ line stem cells in the Drosophila ovary. Science. 290:328–330. [DOI] [PubMed] [Google Scholar]
- Zaccai, M., and H.D. Lipshitz. 1996. Role of Adducin-like (hu-li tai shao) mRNA and protein localization in regulating cytoskeletal structure and function during Drosophila oogenesis and early embryogenesis. Dev. Genet. 19:249–257. [DOI] [PubMed] [Google Scholar]
- Zhu, C.H., and T. Xie. 2003. Clonal expansion of ovarian germline stem cells during niche formation in Drosophila. Development. 130:2579–2588. [DOI] [PubMed] [Google Scholar]
- Zou, Y., S. Lim, K. Lee, X. Deng, and E. Friedman. 2003. Serine/threonine kinase Mirk/Dyrk1B is an inhibitor of epithelial cell migration and is negatively regulated by the Met adaptor Ran-binding protein M. J. Biol. Chem. 278:49573–49581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.