Abstract
The discovery of myostatin and our introduction to the “Mighty Mouse” over a decade ago spurred both basic and applied research and impacted popular culture as well. The myostatin-null genotype produces “double muscling” in mice and livestock and was recently described in a child. The field’s rapid growth is by no means surprising considering the potential benefits of enhancing muscle growth in clinical and agricultural settings. Indeed, several recent studies suggest that blocking myostatin’s inhibitory effects could improve the clinical treatment of several muscle growth disorders, whereas comparative studies suggest that these actions are at least partly conserved. Thus, neutralizing myostatin’s effects could also have agricultural significance. Extrapolating between studies that use different vertebrate models, particularly fish and mammals, is somewhat confusing because whole genome duplication events have resulted in the production and retention of up to four unique myostatin genes in some fish species. Such comparisons, however, suggest that myostatin’s actions may not be limited to skeletal muscle per se, but may additionally influence other tissues including cardiac muscle, adipocytes, and the brain. Thus, therapeutic intervention in the clinic or on the farm must consider the potential of alternative side effects that could impact these or other tissues. In addition, the presence of multiple and actively diversifying myostatin genes in most fish species provides a unique opportunity to study adaptive molecular evolution. It may also provide insight into myostatin’s nonmuscle actions as results from these and other comparative studies gain visibility in biomedical fields.
I. Introduction
- II. Myostatin-Null and Transgenic Phenotypes
- A. Murine models
- B. Domesticated species
- C. Humans
- D. Fish
- III. Cellular Actions of Myostatin
- A. Proteolytic processing and regulated bioactivity
- B. Regulation of myoblast proliferation, differentiation, and quiescence
- C. Receptors and signaling
- D. Conserved action and novel insights from comparative models
- IV. Evolution of the Myostatin/GDF-11 Gene Subfamily
- A. Phylogenetic analysis
- B. Impact of natural and artificial selection
- V. Functional Divergence and Comparative Genomics of Myostatin
- A. Genomic organization
- B. Differential gene expression
- C. Alternative processing
- VI. Novel Actions
- A. Adipose tissue
- B. Cardiac muscle
- C. Brain
VII. Final Thoughts: Implications for Biomedical, Agricultural, and Evolutionary Sciences
I. Introduction
SINCE ITS INITIAL discovery in 1997 by Alexandra McPherron and Se-Jin Lee (1), myostatin and the myostatin-null phenotype have intrigued different scientific and pop culture communities. The now very well known “double muscling” phenotype popularized by the generation of myostatin knockout mice (Fig. 1, B and C) was already well known among animal scientists and other biologists interested in livestock production because similar phenotypes, indeed the term “double muscling” itself, had been characterized in many cattle breeds (Fig. 1A) and in the Texel sheep. The genetic basis for the phenotype in these animals, however, was only revealed with myostatin’s discovery and with studies utilizing cattle that subsequently followed (2,3,4,5). The field has grown considerably over the past decade and has generated over 500 scientific articles while to date, 1137 and 642 core nucleotide and protein records, respectively, have been deposited into GenBank. To put this into perspective, this is approximately twice the number of comparable sequence records for another potent and more readily recognized regulator of skeletal muscle growth, IGF-I.
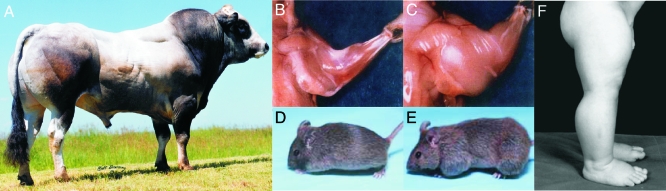
Figure 1.
“Double muscling” and the myostatin-null phenotype. A, Muscle hypertrophy in Piedmontese cattle breeds is due to a missense mutation within the third exon of the bovine myostatin gene (4). [Picture reproduced with permission from the North American Piedmontese Cattle Association (NAPA, www.piedmontese.org).] B and C, Forearm musculature of wild-type (B) and myostatin “knock-out” (C) mice. [Reprinted by permission from Macmillan Publishers Ltd: Nature (Ref. 1), copyright 1997.] D and E, Wild-type (D) and follistatin transgenic (E) mice (31). [Copyright 2001 National Academy of Sciences, U.S.A.] F, Leg musculature of a 7-month-old infant boy with a null mutation within the splice donor site on exon 1 of the myostatin gene (26). [Copyright 2004 Massachusetts Medical Society. All rights reserved.]
Such scientific and cultural interest is not surprising when one considers the obvious and potential impact of manipulating myostatin production and/or bioactivity in the clinic or on the farm, which has likely helped spur interest with the general public as well. Indeed, a survey of web sites using the Yahoo search engine and the keyword “myostatin” identifies nearly 270,000 sites. Many of these sites are not associated with scientific journals or institutions, but rather with companies that market nutritional supplements that presumably block or neutralize myostatin. Most of these products—Musclegen (Fareplant, Inc., Westerville, OH), MyoStim (Champion Nutrition, Inc., Concord, CA), Myo-Blast (Cytodyne Technologies, Inc., Manasquan, NJ), Anabol X (Pinnacle, Inc., Central City, NE), etc.—are composed of sulfated polysaccharides isolated from a brown marine plant, Cystoseira canariensis, that have been reported to bind myostatin and other cytokines (6). However, a thorough assessment of their neutralizing activity (7) suggests that they are completely ineffective in vivo even when administered at very high doses (1200 mg/d). Nevertheless, the pursuit of perfect form—the archetypal Vitruvian Man—whether for commercial or biomedical reasons or even for less scrupulous reasons (e.g., performance enhancement) will no doubt continue to fuel interest in myostatin research.
A thorough review of myostatin biology, from a purely biomedical perspective, was published in 2004 by Se-Jin Lee (8). The goals of the current review, however, are to highlight recent advances in the field and to discuss, from a comparative perspective, how studies using a diverse array of vertebrate models have influenced our understanding of muscle biology, basic evolutionary processes, and the pursuit of novel therapies for treating muscle growth disorders.
II. Myostatin-Null and Transgenic Phenotypes
A. Murine models
Myostatin is a highly conserved member of the TGFβ superfamily and possesses all of the structural components common to the family: nine invariant cysteine residues, an “RXXR” furin-type proteolytic processing site, and a bioactive C-terminal domain (8). Its expression in mammals is limited primarily to skeletal muscle, which in mammals appears to be the principal target tissue. The muscle mass of myostatin-null cattle, sheep, mice, and humans is dramatically increased and produces a phenotype often referred to as “double muscling” (Fig. 1C). Indeed, the mass of individual muscles from myostatin knockout mice, appropriately named “Mighty mice,” is often double that of comparable muscles from wild-type mice (1). Enhanced muscle growth in these animals is due to increases in both cell number, or hyperplastic growth, and cell size, or hypertrophic growth, which results in larger and heavier myofibers. Another hypermuscular phenotype was also described at approximately the same time as the mighty mouse: the “Compact” mouse. This line was generated by artificially selecting for high carcass protein content and resembled Mighty mice in many ways (9). Although linkage mapping identified only a single affected locus (Cmpt), maximum likelihood analysis suggested that the phenotype was not due to a single gene despite the fact that a 12-bp deletion in the myostatin gene was eventually proven responsible (10). However, subsequent mapping identified several modifying loci (11,12) including a region that contains myogenin, a myogenic regulatory factor involved in the differentiation and maturation of skeletal muscle, which likely explains the differences between the two murine models. As expected, administering myostatin has the opposite effect and can induce muscle atrophy and a cachectic state as in transgenic mice overexpressing myostatin (13) or in mice receiving Chinese hamster ovary (CHO) cell transplants stably expressing myostatin (14).
B. Domesticated species
Similar phenotypes have also been described in some domestic breeds of cattle including the Piedmontese (Fig. 1A), Belgian Blue, and Marchigiana, all of which possess mutant alleles for myostatin (2,3,4). The Piedmontese is marketed as the “Myostatin Breed” because its standard and the North American Piedmontese Cattle Association’s registry require proof of at least one mutant myostatin allele, which may be the first cattle registry based on a particular genotype (www.piedmontese.org). Meat from Piedmontese scores high in palatability studies and is particularly tender, more so than the other breeds (15,16,17). By contrast, meat from the double-muscled Texel sheep (see below) is very tough (18,19). Thus, enhanced musculature itself does not necessarily impact meat palatability because other genetic factors clearly contribute.
An 11-nucleotide deletion in the coding sequence of the C-terminal bioactive domain and the introduction of premature stop codons resulting from the frame-shift are responsible for enhanced muscularity in the Belgian Blue (2). This coincidentally maps to the previously characterized muscular hypertrophy (mh) locus. Double muscling in the other two breeds results from a transversion mutation (G to T) in Marchigiana myostatin (20), which again introduces a stop codon, and a missense mutation in the Piedmontese that converts a critical cysteine to tyrosine (3,4). This particular cysteine is necessary for proper folding of mature myostatin (see Sections III.A and B) and is required for full bioactivity (21). A two-nucleotide deletion in the canine myostatin gene has also been described in whippets, a sight hound used in dog racing (22,23). Dogs heterozygous for the mutation are more muscular than wild-type dogs and even run faster, which the authors suggest is the first indication of a performance-enhancing polymorphism associated with the myostatin gene. Unfortunately, dogs homozygous for the mutation are often referred to as “bully whippets” due to their extreme muscularity and are frequently destroyed by breeders.
Texel sheep are also double muscled due to a mutation in the myostatin gene. Several quantitative trait loci mapping studies for muscle, carcass, and/or fat traits identified markers close to the myostatin gene (18,19,24,25). Fine mapping ultimately identified an interval that included the myostatin gene (25). Unlike all the other mutations discussed, double muscling in the Texel is due to a G to A transition mutation in myostatin’s 3′ untranslated region rather than in the coding sequence. The mutation introduces a target site for specific microRNAs, mir1 and mir206. Both are highly expressed in skeletal muscle, whereas mir1 is also expressed at appreciable levels in cardiac muscle. Binding of these particular microRNAs perturbs myostatin translation and results in lower circulating levels, which in turn contributes to skeletal muscle hypertrophy. A survey of human single nucleotide polymorphism databases identified approximately 2500 polymorphisms that introduce identical microRNA binding sites in the 3′ untranslated regions of other genes. Conversely, another 2500 single nucleotide polymorphisms that destroy existing binding sites were also identified. Thus, identifying the source of the Texel’s muscle hypertrophy presents a unique model system for investigating the contribution of microRNA to phenotypic variation.
C. Humans
A splice site mutation on the first intron-exon boundary of the human myostatin gene was recently described in a young boy (Fig. 1F) (26). Skeletal muscle hypertrophy was immediately evident at birth and was confirmed by ultrasonography. The child’s exceptional strength was also evident because at just 4.5 yr of age, he could suspend two 3-kg dumbbells with both arms extended. Mechanistically, this mutation removes the 5′ small nuclear RNA pairing region used by the spliceosome. The first intron is not removed during pre-mRNA processing, which results in the expression of mature transcripts that include in-frame stop codons. Translation is therefore terminated prematurely and well before the bioactive domain. Several physical exams suggest that the child is developing normally and is healthy and fit. Cardiac muscle expresses activin receptors and is sensitive to myostatin’s negative effects (see Section IV.B). However, all assessments to date indicate normal cardiac performance. Although such claims should be expressed with a modicum of skepticism, this study is the first to suggest that the successful manipulation of myostatin production and/or bioactivity in a clinical setting could have many beneficial outcomes without deleterious side effects.
D. Fish
Myostatin-null zebrafish have been described in three studies, and although the results are far from conclusive, they suggest that myostatin’s effects may extend beyond skeletal muscle. Injection of antisense morpholinos was reported to enhance the rate of somite formation in developing embryos, the size of individual somites and whole embryos, and the gene expression of some myogenic regulatory factors, but it did not specifically influence myogenesis itself or muscle mass (27). None of these effects were seen in transgenic zebrafish overexpressing a dominant-negative myostatin, although a minor increase in muscle cell number was noted, but only in female fish (28). Changes in somitogenesis were also not noted in the developing Mighty Mouse (1). A third study (29) reported to have generated “giant zebrafish” by injecting double-stranded myostatin RNA from a distant and unrelated species (tilapia). Overall growth enhancement appeared to continue well past the effective half-life of the treatment and even in juvenile fish. Each of these studies was performed before the discovery of a second class of fish myostatin paralogs (see Section IV.A), and it is therefore difficult to determine which if any myostatin transcript was affected. Nevertheless, these studies indicate that the biological actions of myostatin in fish may not be limited to the growth and development of skeletal muscle, but may influence these processes in many tissues. Muscle growth is more limited in zebrafish than in other fish species that possess “indeterminate” growth (30). Thus, this particular model may be better suited for investigating the divergent actions of myostatin in nonmuscle tissues (see Section V.B).
III. Cellular Actions of Myostatin
A. Proteolytic processing and regulated bioactivity
Several myostatin binding proteins have been identified and include follistatin (31), follistatin-like related gene (FLRG; also known as follistatin like-3, FSTL-3, and follistatin-related peptide or FLRP) (32), growth/differentiation factor-associated serum protein (GASP)-1 (33), and titin (T)-cap (34). T-cap is a sarcomeric protein that binds the N-terminal domain of titin where it helps regulate the cytoskeletal organization of muscle cells. T-cap also binds myostatin presumably in the golgi and prevents its secretion. The N-terminal peptide that results from proteolytic processing of pro-myostatin also binds myostatin with high affinity, and like follistatin, FLRG and GASP-1, can prevent receptor binding and activation (31). Such interactions commonly occur between proteolyzed fragments of TGFβ superfamily members (35), and although the exact mechanism of ligand activation has yet to be determined for myostatin, it appears to strongly resemble that of its superfamily siblings. This includes removal of the signal peptide from prepro-myostatin and proteolysis of pro-myostatin at the furin/PACE cleavage site (Fig. 2). This separates the bioactive domain from the N-terminal latency-associated peptide (LAP), which binds to the disulfide-linked myostatin dimer. The two proteins are then secreted as a small latent complex where they enter the circulation or potentially bind to the extracellular matrix forming a large latent complex. Proteolytic cleavage of LAP then releases myostatin from circulating and extracellular complexes (see Ref. 8 for a detailed review of processing events and proteases). Thus, myostatin bioactivity is not mediated per se by increased synthesis or release from skeletal muscle, but by three independent proteolytic events, of which the latter two may be regulated.
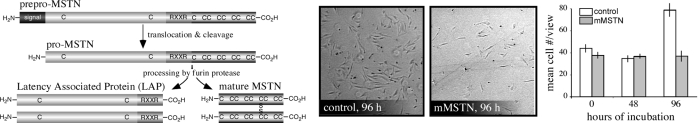
Figure 2.
Proteolytic processing of mature myostatin (MSTN) and conserved bioactivity. Left, Pro-myostatin is cleaved by a furin class protease at a conserved RXXR (R, arginine; X, any amino acid) epitope, and the resulting peptides dimerize via disulfide linkages at the indicated cysteines producing mature myostatin. The dominant-negative LAP sequesters myostatin dimers in a latent complex and can prohibit receptor activation. Right, Primary myosatellite cells from rainbow trout were incubated in 100-mm dishes with 50 ng/ml mouse myostatin (R&D Systems, Minneapolis, MN) or equimolar amounts of bovine serum albumin, and myostatin was added at 0 and 48 h. At each time point, cells were manually counted in each of six views. Mean values for each time point are shown (histogram), as are representative images.
For most TGFβ superfamily ligands, the formation of large latent complexes requires the binding of TGFβ LAP peptides to latent TGFβ binding proteins (LTBP) (35). Anderson et al. (36) recently identified a large extracellular pool of unprocessed myostatin in skeletal muscle that may be mediated in part by the association of myostatin with LTBP-3. Coexpression of LTBP-3 and myostatin in a human kidney epithelial cell line (293T) sequestered myostatin within the extracellular matrix and attenuated its bioactivity, whereas the ectopic expression of LTBP-3 in mouse skeletal muscle increased fiber cross-sectional area. Another matrix-associated protein, decorin, also binds myostatin at a 1:1 ratio and with relatively high affinity (∼10–20 nm Kd), but not LAP, and prevents receptor activation (37). The spatiotemporal expression patterns of both myostatin and decorin in rat skeletal muscle are similar and are consistent with decorin’s ability to sequester myostatin outside the cell (38). This small leucine-rich proteoglycan binds TGFβ as well and regulates collagen fibrillogenesis and myoblast proliferation (39). Myostatin and TGFβ may help to maintain fibrosis in some forms of muscular dystrophy because both factors induce myogenic cells to differentiate into myofibroblasts (40,41,42). Myostatin also stimulates fibroblast proliferation in vitro as well as the secretion of TGFβ1 from mouse C2C12 myoblasts (42). Stably overexpressing decorin in myoblasts accelerates the differentiation rate, whereas gene transfer of decorin in vivo has similar effects on skeletal muscle regeneration (43), actions that are mediated by decorin sequestration of myostatin and TGFβ as well as the up-regulation of follistatin (42). Crossing mdx mice, models for Duchenne muscular dystrophy, with Mighty mice ameliorates much of the dystrophic phenotype including fibrosis (44). Furthermore, skeletal muscle from Mighty mice regenerates more readily than wild-type muscle and has reduced fibrosis after notexin- or laceration-induced muscle injury (42,45). These studies indicate that myostatin may associate with the extracellular matrix independent of LAP and suggest that neutralizing the coregulatory relationship between myostatin and TGFβ with decorin could aid in the clinical treatment of some muscular dystrophies.
The nullifying effects of LAP and follistatin on myostatin bioactivity have been demonstrated quite conclusively using in vitro and/or in vivo systems, with LAP receiving the most attention. Both proteins bind myostatin and prevent receptor binding and activation in vitro, whereas transgenic mice overexpressing LAP or follistatin develop hypermuscularity (Fig. 1) similar to that seen in the Mighty Mouse (31,46,47,48,49). Conversely, muscles of follistatin knockout mice are smaller (50), which is consistent with increased myostatin activity and/or muscle growth inhibition. The muscle mass of follistatin transgenic mice (Fig. 1, D and E), however, is bigger than that of LAP transgenics (31), suggesting that ligands in addition to myostatin may serve to inhibit skeletal muscle development. Potential candidates include activin and growth/differentiating factor (GDF)-11, both of which bind follistatin (51,52), as well as activin type 2 receptors (Acvr2 and Acvr2b, also known as ActRIIa and ActRIIb), which also mediate myostatin’s effects (31,48). These results suggest that double muscling alone does not maximize the tissue’s growth potential. Indeed, transgenic Mighty mice overexpressing follistatin have even greater muscle mass than follistatin transgenics alone (53), which produces muscles four times larger than those in wild-type mice (i.e., “quadruple muscling”). The effects were dose dependent because muscles of follistatin transgenic mice with only a single myostatin gene were smaller than follistatin transgenics lacking both myostatin genes, but larger than those with two functional copies (i.e., FS-Tg/mstn−/− > FS-Tg/mstn−/+ > FS-Tg/mstn+/+). The biggest differences were seen in offspring from myostatin-null mothers, which the authors suggest could be due to the lack of maternal transfer of myostatin or a myostatin-regulated factor into the fetal circulation. Activin receptors are expressed throughout the placenta (54,55), so these results could just as likely be due to the lack of placenta stimulation or even to the repartitioning of energy because the myostatin-null phenotype is also associated with reduced adiposity (1,56,57,58). The results are nevertheless intriguing and suggest that normal skeletal muscle is even more plastic than originally presumed and once again that multiple factors contribute to the negative regulation of muscle development.
B. Regulation of myoblast proliferation, differentiation, and quiescence
Myostatin appears to prevent myoblast hyperplasia in mammals by inhibiting cell cycle progression past the G1 and G2 stages (59,60,61). These actions are mediated in part by reduced Cdk 2 levels and activity, a concomitant increase in p21 Cdk-inhibitor, and consequently, the hypophosphorylation of Rb. Myostatin also inhibits myoblast differentiation (62,63,64), although the teleological significance of this particular effect may appear controversial because conflicting data suggest that myostatin initiates cell cycle withdrawal, which is a necessary prerequisite for differentiation (59,60,64,65). However, studies with primary myosatellite cells (also known as “skeletal muscle stem cells” located below the sarcolemma and basal lamina) from myostatin-null mice suggest that myostatin-stimulated cell cycle withdrawal is accompanied by cellular quiescence (65,66) rather than differentiation. This explains the apparent discrepancy and supports earlier studies indicating that myostatin is a myoblast survival factor (60). A model for myostatin action in mammals suggests that in the absence of other myogenic regulators, myostatin inhibits myoblast hyperplasia by stimulating cell cycle withdrawal and delays differentiation by inducing cellular quiescence. Recent studies further suggest that myostatin-induced cellular quiescence is reversible and is associated with reduced expression of the myogenic regulatory factors Pax-3, Myf-5, and MyoD (67). It is therefore inaccurate to describe myostatin’s actions as solely inhibitory. Indeed, myostatin initiates the first and necessary step in the differentiation process, cell cycle withdrawal, and prevents apoptosis of the quiescent cells. Its negative effects on myofiber hypertrophy are due in part to the inhibition of myosatellite cell activation, proliferation, and/or renewal because the fusion of these cells with existing myofibers is largely responsible for postnatal muscle growth (68). These cells are more abundant in skeletal muscle of myostatin-null mice, which proliferate more rapidly than those isolated from wild-type mice (65). Myostatin also inhibits protein synthesis in differentiated C2C12 myotubes (61). These studies together suggest that myostatin ultimately limits skeletal muscle size by inhibiting the hyperplastic growth of developing myoblasts and thus, the number of cells that eventually differentiate into mature myofibers, and by reducing myofiber protein synthesis and myosatellite cell renewal, both of which inhibit the hypertrophic growth of mature muscle.
Cell culture studies have used recombinant myostatin generated in both prokaryotic and eukaryotic expression systems and have generated similar, but not identical results. Recombinants from both systems inhibit myoblast proliferation in vitro; however, those generated in bacteria are functionally active only at very high concentrations (2–10 μg/ml, 100–500 μm) (59,61,63,65,69,70). By contrast, myostatin generated in CHO cells is biologically active at concentrations 100- to 1000-fold lower (14,46,71). The production of biologically active myostatin in bacteria is actually quite surprising because the formation of disulfide-linked dimers (Fig. 2) requires an oxidizing environment that does not exist in bacterial cytoplasm. Myostatin recombinants generated in prokaryotic expression systems are therefore monomeric unless denatured and refolded in the presence of an appropriate oxidizing agent, which itself does not guarantee proper folding. Indeed, most of the studies using recombinant myostatin generated in bacteria attempted to refold the peptides. Proper folding is critical to biological activity because double muscling in Piedmontese cattle results from the production of misfolded peptides (3,4) that lack a residue critical to the formation of the cysteine knot structure common to all TGFβ superfamily proteins (72,73,74,75). It appears, therefore, that only a very small fraction of myostatin generated in bacteria is biologically active because such preparations are primarily composed of either nonfunctional monomers or misfolded dimers. Whether or not the growth inhibitory effects of bacterial recombinants are mediated through Acvr2 or Acvr2b, by preventing endogenous myostatin from binding to other proteins (e.g., LAP) or by cross-reacting with receptors for other TGFβ superfamily ligands is not known. The use of myostatin generated in bacteria, which is commercially available from many different sources, has helped to define basic cellular responses and has been validated to some degree by studies with recombinant myostatin generated in eukaryotic cells, which is also commercially available. However, the continued use of bacterial recombinants could ultimately prove misleading especially when used in vivo, with tissues other than skeletal muscle (see Section VI) or when trying to define mechanisms of myostatin action.
C. Receptors and signaling
All TGFβ superfamily ligands signal through membrane-bound heteromeric serine-threonine kinase receptor complexes composed of two type 1 and two type 2 receptors (76). Ligand binding to type 2 receptors recruits type 1 and both autophosphorylate via transinteractions with one another’s intracellular kinase domains. The signaling pathway to the nucleus is short and quick because specific receptor (R)-Smads are recruited to the complex and are phosphorylated by type 1 receptors. The R-Smads then oligomerize with appropriate co-Smads and translocate into the nucleus. This complex directly binds promoter elements and initiates or represses gene transcription. Myostatin bioactivity is mediated by activin receptors, specifically Acvr2 and Acvr2b. Cross-linking studies and radioreceptor assays indicate that although myostatin binds both, it binds the latter with slightly higher affinity (31). Skeletal muscle mass was increased by 125% in transgenic mice overexpressing dominant-negative Acvr2b (31) and by 60% just 2 weeks after three ip injections of a soluble form of Acvr2b’s extracellular domain (48). Mice homozygous for deactivating mutations in Acvr2 have pectoralis and triceps muscles that are 27–40% larger than the same muscles from wild-type mice (48). These muscles are just 20–26% larger in mice with mutant Acvr2b receptors, suggesting that Acvr2 may play a more important role in regulating myostatin’s actions, at least in these muscles. The relative distribution of each receptor in different skeletal muscles or even within a specific muscle is not known. Thus, the contribution of each receptor may be equally relative and may differ between individual muscles or even fiber types. Myostatin activation of either receptor recruits the type I receptors activin like kinase-4 or -5 (77), which in turn phosphorylate Smads 2 and 3. These transcription factors oligomerize with Smad4 and eventually regulate gene transcription, including the expression of Smad7 and c-ski. This particular Smad is an inhibitory Smad because it sequesters Smad4 in the cytoplasm and prevents it from binding to the Smad2/3 complex (77,78). C-ski is a corepressor that stabilizes the inactive Smad2/3/4 complex on Smad/ski binding elements (79,80). It also appears to inhibit Smad2 and Smad3 signaling in part by directly blocking histone deacetylase activity as well as their association with a transcriptional coactivator (81,82). Nuclear localization of c-ski is required for the differentiation of myoblasts because it enhances myogenin transactivation through direct interactions with MyoD/MEF2 heterodimers (82), which is in direct opposition to myostatin’s negative effects on differentiation. These studies together suggest that in skeletal muscle, Smad7 and c-ski serve as intracellular negative feedback mechanisms for myostatin or other TGFβ superfamily ligands that signal via Smads 2 and 3.
Myostatin signaling is not limited to canonical Smad pathways because it curiously interacts with mitogenic pathways as well. Myoblast proliferation and cell cycle progression are stimulated by IGF-I, a potent mitogen for many different cell types including myoblasts. IGF-I also stimulates myoblast differentiation and the associated cell cycle arrest (83). The mechanisms of IGF-I’s ability to stimulate these normally diametrically opposed cellular activities is currently under dispute and may include the local production of IGF binding protein (IGFBP)-3 (see Section III.D) (84,85,86,87). Nevertheless, IGF-stimulated myoblast differentiation is mediated by increased activity of the Cdk2 inhibitor p21 (88,89,90). Myostatin also stimulates myoblast cell cycle withdrawal and activates p21 (59,60,65), but unlike IGF-I it inhibits rather than stimulates differentiation. Myostatin also activates Akt (58,91,92) and the MAPKs extracellular signal-regulated kinase (Erk)-1/2 (93) and c-Jun N-terminal kinase (JNK) (94). The significance of cross-talk between myostatin and IGF signaling is not known, nor is it known how myostatin activates these pathways while simultaneously arresting the cell cycle. However, myostatin and IGF-I are both survival factors, and thus myostatin activation of these particular aspects of a mitogenic pathway is likely related to its ability to maintain cellular quiescence in growth-arrested cells. Indeed, blocking Erk-1/2 and JNK activation similarly blocks myostatin’s ability to inhibit differentiation (93,94).
D. Conserved action and novel insights from comparative models
Most in vitro studies of myostatin’s actions were performed using C2C12 myoblasts. This immortalized mouse cell line was established from myosatellite cells originally isolated from dystrophic skeletal muscle (95). Although the cells maintain full myogenic potential and are capable of differentiating into contractile myotubes, they can also differentiate into osteogenic cells when exposed to bone morphogenic protein-2, another TGFβ superfamily member (96). Nevertheless, results from these studies are consistent with those using primary porcine embryonic myogenic cells (PEMC). They suggest that the C2C12 model is physiologically relevant and that myostatin’s growth inhibitory actions, which have been described in vivo with mice, cattle, sheep, dogs, and humans (see Section II), are likely conserved in eutherian mammals. Myostatin and TGFβ are both expressed in PEMC (97), and both inhibit proliferation in a dose-dependent manner (98). Their expression is reduced in differentiated cells (97), whereas the cytokines also stimulate the gene expression and secretion of IGFBP-3 and -5 (98). As their names imply, the IGFBPs bind the IGFs with high affinity and in doing so can either attenuate or potentiate their action depending on the tissue, the differentiation status of the specific cell type, and the location of binding (i.e., circulation or extracellular) (99). IGFBP-3 and -5, however, inhibit myoblast proliferation in both IGF-dependent and -independent manners (85,100,101,102,103), the latter effects being potentially mediated by the nuclear localization and functional interaction with a subunit of RNA polymerase II (103). Nuclear localization of IGFBP-3 also occurs in PEMCs and increases in response to TGFβ stimulation (myostatin was not tested) (104). Furthermore, immunoneutralization of IGFBP-3 blocks myostatin’s antiproliferative effects in these cells (85). An emerging model of myostatin action therefore suggests that IGFBP-3 and -5 partly mediate the cytokine’s effects and that the extracellular sequestration of IGF-I and the nuclear localization of IGFBP-3 and possibly IGFBP-5 are involved.
Myostatin genes, putative promoters, and/or cDNA have been cloned in several avian species including turkey (Meleagris gallopavo), chicken (Gallus gallus), pigeon (Columba livia), duck (Anas platyrhynchos), goose (Anser anser), and quail (Coturnix chirensi) (4,105). Initial studies with avians, for that matter with most nonmammalian vertebrates, attempted to correlate changes in myostatin expression to key periods of skeletal muscle development or to physiological responses to catabolic insult (see Section V.B). Although primarily descriptive, such studies were the first to suggest that myostatin’s growth inhibitory actions may be conserved in skeletal muscle of avians and fish. Recent studies, however, are beginning to assess not only myostatin’s actions in different vertebrates, but the myostatin attenuating actions of LAP as well. Polymorphisms in the chicken myostatin gene are associated with differences in body weight (106), whereas the immunoneutralization of myostatin in 3-d-old embryos has a small, but significant and positive effect on both body weight and muscle mass (107). Administration of polyclonal antibodies against LAP in ovo decreased thigh and leg weights of post-hatch chickens, providing the first functional evidence that LAP inhibits the biological activity of myostatin in a nonmammalian vertebrate (108). Chicken embryonic myoblasts and primary myosatellite cells are both sensitive to myostatin’s suppressive effects (109,110), although myosatellite cells isolated from chicken pectoralis major are more responsive to myostatin than those from biceps femoris (110). It is unknown whether this differential response is due to tissue sensitivity per se (i.e., receptor binding) or to tissue-specific expression of myostatin binding proteins that could potentially influence bioavailability. Nevertheless, these results suggest that muscle sensitivity to myostatin may differ between fiber types and/or muscle groups. Indeed, the degree of muscle growth enhancement in myostatin-null animals can vary considerably between muscle groups (1). The use of primary chicken embryonic myoblasts as well as myosatellite cells will therefore prove invaluable in determining the underlying mechanisms involved. Myostatin gene expression increases as primary chicken myosatellite cells differentiate into mature myotubes (111), suggesting a positive role for myostatin at some point in the differentiation process. Targeted ablation of myostatin transcripts using RNA interference reduces myostatin protein and mRNA by 55 and 75%, respectively, and concomitantly delays, rather than stimulates, differentiation (112). Silencing myostatin in differentiating cells ultimately produced myotubes that were morphologically distinct from control cells. These data further suggest that myostatin’s growth inhibitory actions are natural to the differentiation process. They also once again illustrate the utility of the primary chicken myosatellite cell culture model.
Skeletal muscle growth in mammals and avians is primarily limited to increases in cell size, rather than to increases in cell number. Although undifferentiated myosatellite cells contribute to hypertrophic growth after muscle injury or exercise, their numbers remain relatively constant after birth (113,114). Most fish species, however, have abundant myosatellite cells because hyperplastic growth accounts for approximately 50% of the changes in growing fish skeletal muscle (115). Primary myosatellite cells are readily isolated from adult fish and are easily cultured in vitro (116,117,118,119,120,121). These cells proliferate and fully differentiate into mature multinucleated myotubes and express skeletal muscle-specific markers that are recognized by mammalian antisera (122,123,124). We are now exploiting this underutilized model system to determine whether myostatin’s actions are conserved in more distant vertebrates. Indeed, recombinant mouse myostatin (MetaMorphix, Inc., Beltsville, MD) inhibits the proliferation of primary myosatellite cells isolated from rainbow trout (Fig. 2). The recombinants were generated in CHO cells and were used at concentrations equivalent to the physiologically relevant concentrations used with mammalian systems (14,46,71). Thus, myostatin’s most fundamental biological action, the growth inhibition of proliferating skeletal myoblasts, appears to be widely conserved among several, if not all, vertebrate classes. Extensive studies are nevertheless needed to determine the degree of conservation and the underlying mechanisms involved as several aspects of myostatin biology in the fishes appear to be quite different from those in mammals. Some of these differences question whether myostatin’s role in fish, and possibly mammals as well, is truly limited to skeletal muscle. They also illustrate the need for both in vitro and in vivo assessment of myostatin action in lower vertebrates.
IV. Evolution of the Myostatin/GDF-11 Gene Subfamily
A. Phylogenetic analysis
Although the most influential and defining functional studies have been performed with medically important model systems, a surprising number of myostatin sequences have been described in different vertebrate species not normally associated with biomedical research. These naturally include commercially important mammalian species, although many nondomesticated species are also represented, most of which are bony fish (Osteichthyan, not Chondrichthyan). In fact, after accounting for the redundancy of sequences for a particular species, the numbers of fish and mammalian species with CoreNucleotide sequences in GenBank are about the same. Interest in fish, however, is not purely academic. Over 20,000 fish species are currently known, and many are potential candidates for culture. Assuming that myostatin’s actions are conserved in these species, the successful manipulation of myostatin expression and/or bioactivity could significantly impact the aquaculture industry, if not revolutionize it. Skeletal muscle in fish constitutes a far greater percentage of total body weight than it does in mammals, and seemingly minor improvements in animal growth can disproportionately enhance product yield. Many technological barriers, however, still exist within the aquaculture industry due in many instances to a poor understanding of fish biology. In fact, many cultivars have not been heavily selected for commercially important traits such as growth, and none have been selected to the degree of most domestic mammalian species. Thus, the potential impact of enhancing muscle growth by disrupting myostatin’s actions is likely far greater in fish.
A zebrafish homolog was the first fish sequence to be described (4), although sequences from commercially important species were first described at the fourth International Symposium on Fish Endocrinology in 2000 (125,126). The following year could accurately be described as the “Year of the Fish” because 16 myostatin homologs were described from 12 different species (127,128,129,130,131,132,133). Thus, some fish species expressed two myostatins from distinctly different loci. These additional genes were presumed by some to be ancestors of GDF-11, a structurally similar and highly conserved TGFβ superfamily sibling. However, the different fish myostatins lacked epitopes characteristic of mammalian GDF-11 peptides. Indeed, GDF-11 homologs have since been described in zebrafish (134) and can be identified by BLAST searches of different fish genomic databases. Thus, the additional myostatins arose independently and after the divergence of myostatin and GDF-11. This was subsequently supported by an extensive phylogenetic analysis of the entire myostatin/GDF-11 subfamily (135).
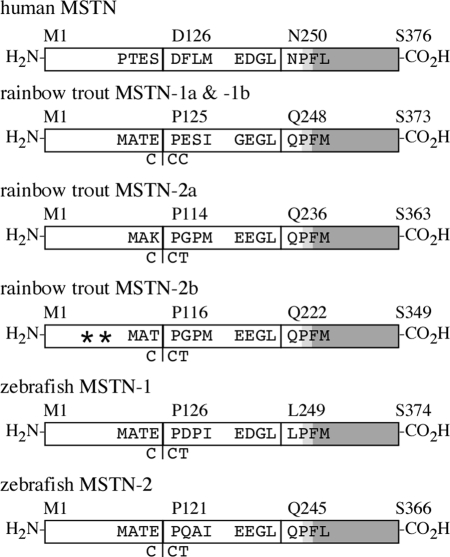
Alignments of myostatin amino acid sequences from multiple vertebrate species indicate that the proteins are fairly well conserved overall, particularly within related taxa (132). Sequences are best conserved within the carboxy-terminal bioactive domains (88–100%) and differ significantly in their respective LAP domains (50–90%). It is unknown, however, whether the intermolecular interactions that occur between mammalian myostatin and LAP peptides also occur in other vertebrates. Subtle, yet notable, differences within the bioactive domains of fish myostatin (MSTN)-1 and -2 orthologs exist and include methionine for lysine and histidine for arginine substitutions in the carboxy-terminal domains of fish MSTN-2 proteins. The former substitution occurs in a region hypothesized to have contributed to enhanced musculature in domesticated and wild bovids (4,132,136). Myostatin’s actions are mediated by binding to activin receptors (31,77), and perturbations in activin signaling increases muscle mass (14,31,48). The carboxy-terminal domain of activin is nearly identical to that of myostatin and is critical to receptor activation (137,138). Thus, differences within this domain could potentially impact bioactivity. Future functional studies are therefore needed to determine whether MSTN-1 and -2 proteins bind Acvr2 or Acvr2b equally or to other receptors.
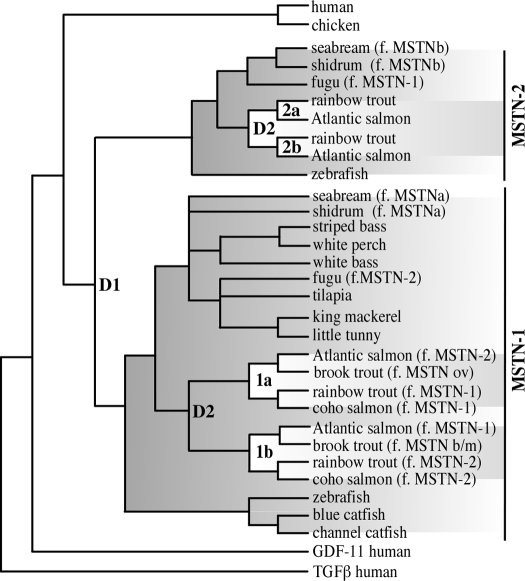
Three of the original cloning studies that reported novel fish myostatins constructed neighbor-joining trees to evaluate myostatin phylogenies (127,128,133). Each of these studies sampled only a limited number of sequences and employed only elementary methods, a Poisson-corrected distance matrix calculated from amino acid sequences, that are known to underestimate distances with increased sequence divergence (139). By contrast, a more rigorous model-based approach was later conducted and included substantially more sequences and outgroups (135). This study also analyzed nucleotide sequences aligned to an inferred amino acid alignment, which allowed for a more detailed exploration of clade membership and timing of gene copy duplication. The analysis identified two myostatin sister clades, MSTN-1 and MSTN-2, within the teleosts that arose from a well-characterized genome duplication event (D1 in Fig. 3) in the fish lineage (140,141). This likely occurred before the divergence of teleosts or recently thereafter because MSTN-2 homologs have been identified in two separate teleost Superorders: Acanthopterygii and Ostariophysi. The recent tetraploidization of salmonids (142) resulted in an additional duplication, and the production of two more paralogs (D2 in Fig. 3). Barring losses within specific taxa, this study indicates that all fish should possess at least two myostatin genes, whereas salmonids should have four, namely MSTN-1a, -1b, -2a, and -2b. Indeed, all four genes were identified in rainbow trout (143,144) and recently in Atlantic salmon (145).
Figure 3.
Phylogenetic relationship of vertebrate myostatin (MSTN) homologs. The tree was constructed from previously published Maximum Likelihood and Bayesian Inference analyses (135,144,145). Clades for the two teleost fish paralogs, MSTN-1 and MSTN-2, are shaded. Within each clade are the additional salmonid paralogs (1a, MSTN-1a; etc.), which are indicated by reverse shading. Genome duplication events within bony fish and salmonid lineages are indicated as D1 and D2, respectively. The nomenclature for the fish homologs has been revised (146). Thus, the former (f.) names are indicated in parentheses (ov, ovarian; b/m, brain/muscle; GDF, growth/differentiating factor; TGF, transforming growth factor).
These analyses revealed that the vast majority of previously described fish myostatin genes were actually MSTN-1 orthologs despite names that suggested otherwise. The former nomenclature was extremely misleading and, for the most part, was based on the order by which each gene was identified rather than true phylogenetic relationships. This resulted in MSTN-1 orthologs inaccurately being named MSTN-2 and vice versa. A small consortium of comparative biologists in the field, including the current authors, therefore proposed a standardized nomenclature for the entire myostatin subfamily that is based solely on the phylogenies described (146). A description of the specific genes whose names have been changed as well as their respective GenBank accession numbers is included in Table 1. The nomenclature revisions also included the adoption of “myostatin” as the official gene name, rather than “GDF-8” (a known alias), and all of the recommendations were ultimately accepted by the Mouse Genomic Nomenclature Committee and the Human Genome Organization (HUGO) Gene Nomenclature Committee, which subsequently revised their databases. This change was also propagated to other public databases including those managed by the National Center for Biotechnology Information (NCBI) and Ensemble.
Table 1.
Revisions to fish myostatin gene nomenclature
| Species | MSTN-1
|
MSTN-2
|
||||
|---|---|---|---|---|---|---|
| Suffix | Alias | GenBank accession no. | Suffix | Alias | GenBank accession no. | |
| Atlantic salmon | a/b | 2/1 | AJ344158/AJ297267 | |||
| Blue catfish | AY540992 | |||||
| Brook trout | a/b | ov/bm | AF313912/AF247650 | |||
| Channel catfish | AF396747 | |||||
| Coho salmon | a/b | 1/2 | AY434465/AF394687 | |||
| Fugu | 2 | AY445322 | 1 | AY445321 | ||
| King mackerel | AF317667 | |||||
| Little tunny | AF317666 | |||||
| Rainbow trout | a/b | 1/2 | AF273035/AF273036 | a/b | DQ417326/DQ417327 | |
| Seabream | a | AF258448 | b | AY046314 | ||
| Shidrum | a | AF316881 | b | AY059386 | ||
| Striped bass | AF290910 | |||||
| Tilapia | AF197193 | |||||
| White bass | AF197194 | |||||
| White perch | AF290911 | |||||
| Zebrafish | AY258034 | AY687474 | ||||
Alias, Former name; ov, ovarian; bm, brain/muscle.
Putative homologs of myostatin have also been described in two invertebrates, the fruit fly (Drosophila melanogaster) (147) and bay scallop (Argopeten irradians) (148), and in two chordates, the urochordate sea squirt (Ciona intestinalis) (148) and the cephalochordate amphioxus (Branchiostoma belcheri tsingtauense, also known as lancelets) (149). The notion that these genes could represent ancestral forms of vertebrate myostatin and GDF-11 genes is intriguing because putative homologs for GDF-11 have not been specifically identified in any of these species. However, their coding sequences are only minimally conserved with other vertebrate myostatin or GDF-11 homologs (∼35–40%), and the genomic organization of each gene also substantially differs (see Section V.A). It is difficult, therefore, to determine whether these genes are true myostatin/GDF-11 ancestors or whether they are simply novel TGFβ superfamily homologs. More in-depth phylogenetic analyses that include myostatin and other TGFβ superfamily genes from basal vertebrate groups and from representative chordates will help determine the true identity of the ancestral gene as well as the timing of gene duplication.
B. Impact of natural and artificial selection
Four studies have investigated the impact of selection pressures on myostatin coding sequence polymorphisms and have discovered evidence for naturally occurring positive selection that may have influenced the evolution of modern bovids (136) and potentially salmonids (145). As with many gene families, multiple duplications have provided genetic materials for such evolutionary processes as differential selection, pseudogenization, subfunctionalization, and neofunctionalization, all of which contribute to functional divergence (150,151,152,153,154,155). Liberles et al. (156) and Tellgren et al. (136) identified several instances of positive (diversifying) selection in bovids, some of which corresponded to similar regions in fish myostatins (132). Analysis of nonsynonynmous/synonymous nucleotide substitution rates indicated that selection occurred during the divergence of modern bovids and, therefore, was not a product of artificial selection among domesticated species. However, Pie and Alvares (157) found insufficient evidence for selection in these lineages, although the differing results are likely due to differences in analytical methods. Where positive selection has been found, early counting methods (158,159) or maximum likelihood methods (160) were employed and have been criticized for not robustly adjusting for codon usage bias (e.g., counting methods) (157,161) or for their susceptibility to producing false-positives (157,162). However, similar methods also identified evidence for positive selection among salmonid myostatins (145), supporting the conclusion that strong evolutionary forces have helped influence the functionalization of myostatin in different vertebrates and quite possibly the functional divergence in salmonids.
As predicted by Kerr et al. (135), four myostatin genes were identified in rainbow trout (rtMSTN-1a, -1b, -2a, and -2b) (143,144). These genes are differentially expressed (see Section V.B), possess clade-specific polymorphisms, and have distinct promoter regions. The Atlantic salmon MSTN-2 orthologs were also recently identified (145), and our lab has cloned orthologs of all four genes from many salmonid groups, including the genus Thymallus (grayling), which belongs to a subfamily separate from the more readily recognized Salmoninae subfamily (trout and salmon), and in every taxa, MSTN-2b is a pseudogene (see Section V.A). Ostbye et al. (145) recently calculated the nucleotide substitution rates of a very small subset of salmonid myostatins and determined that the MSTN-1 genes, most likely MSTN-1a, evolved under strong positive selection, whereas MSTN-2b evolved under relaxed selection. Expanded sampling of salmonid lineages will provide a well-resolved myostatin gene family phylogeny, particularly by incorporating coding and noncoding sequences with rigorous likelihood-based analyses. This will provide a robust framework for the analysis of selection patterns in terms of coding regions as well as branch location. The MSTN-2b paralogs have clearly become nonfunctional, although it is not known whether this event predates lineage diversification or if it occurred more recently. Nevertheless, further analysis of myostatin gene phylogenies will help explain overall patterns of functional divergence, particularly as the salmonid genes are currently diverging, and will provide a highly unique opportunity to investigate the underlying mechanisms of molecular evolution in “pseudo real-time”.
V. Functional Divergence and Comparative Genomics of Myostatin
A. Genomic organization
Myostatin genes have been mapped in several vertebrate species, albeit to different degrees of resolution, and are located on chromosomes 2 in humans (2q32.2), chimps (2B), cattle (2q14-q15), and sheep, 1 in mice (C1.1), 9 in rats (9q22), 15 in pigs, 18 in horse, 37 in dogs, and 7 in chicken (7p11) (2,3,5,10,12,19,163,164,165,166) (see also Entrez Gene at www.ncbi.nlm.nih.gov/sites/entrez?db=gene). The fish MSTN-1 and -2 genes have also been cloned in zebrafish (28,135) and map to chromosomes 9 and 22, respectively (see Entrez). They are also located on separate chromosomes in the green-spotted pufferfish (Tetraodon nigroviridis), chromosomes 2 and 3, which share more duplicated genes than any other chromosome pair, and are putative paralogs themselves (167). These data are consistent with the phylogenetic distribution of the two fish myostatin clades and further suggest that MSTN-1 and -2 arose from an early genome duplication event that occurred approximately 350,000,000 yr ago (140,141). Ostbye et al. (145) recently mapped Atlantic salmon MSTN-1a and -1b to separate linkage groups indicating that these paralogs, and likely the MSTN-2a and -2b paralogs as well, arose from the recent (25,000,000–100,000,000 yr ago) tetraploidization of the salmonid genome (142), which again is consistent with the well-described phylogenies (135,144).
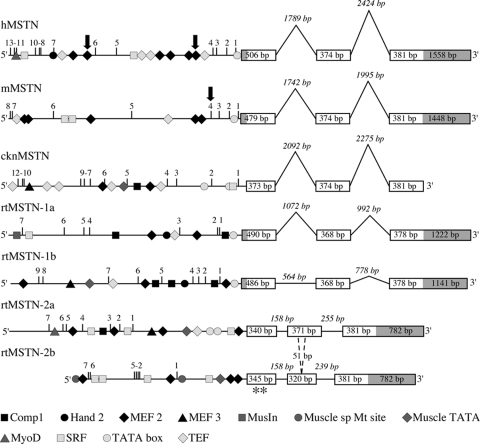
In all vertebrate clones to date, the genomic organization includes three similarly sized exons separated by two intervening introns whose sizes appear to be conserved (∼2 kb) in birds and mammals (Fig. 4). Intron sizes are smaller among salmonid MSTN-2 genes (only rainbow trout shown), which is consistent with an increased susceptibility to relaxed selection because the probable development of null mutations within coding regions is greater among genes with smaller noncoding regions. Both rainbow trout and Atlantic salmon MSTN-2b paralogs contain in-frame stop codons in their first exons and lack a 51-bp cassette from the second exon that is common to all other fish MSTN genes. It is unknown whether similar indels and polymorphisms exist in myostatin genes from other salmonid species. However, these data suggest that only three of the four myostatin genes are functionally active in modern salmonids because rtMSTN-2b appears to be a pseudogene. At first glance, the putative Drosophila myostatin/GDF-11 homolog, myoglianin, appears to possess a genomic organization similar to the fish MSTN-1 genes (147). It also contains three exons and is approximately the same size. However, the ratio of coding to noncoding sequence is significantly different because myoglianin has smaller introns and a disproportionately large first exon that is almost the same size as the entire coding sequence for each fish gene. The putative amphioxus myostatin/GDF-11 gene differs considerably from other known and putative homologs because it is much larger, over twice the size of fish MSTN-1 genes, and contains five exons of variable sizes (149). Amphioxus is the closest extant relative of vertebrata and is an important model for understanding the evolution of all chordates including vertebrates. If the amphioxus gene is indeed a true ancestor, then substantial changes in the genomic organization must have occurred subsequent to the rise of modern vertebrates. Further studies are therefore needed to clarify the phylogenetic relationship between the vertebrate and nonvertebrate homologs.
Figure 4.
Comparative mapping of vertebrate myostatin genes and promoters. The genomic structure and organization of human (h), mouse (m), chicken (ckn) and rainbow trout (rt) myostatin (MSTN) genes are shown. Exons are boxed with open reading frames in white and untranslated regions in gray (if known). Exon sizes are indicated within the boxes, intron sizes in italics. Two in-frame stop codons within rtMSTN-2b are indicated by asterisks, and a 51-bp cassette missing from the second exon is indicated by dashed lines. The locations of putative cis elements within each gene’s promoter region (−2.4 and −1.5 kb for rtMSTN-2a and -2b, respectively; −2 kb for others) are indicated by the shapes in the key; only known myogenic elements are shown. Putative E-boxes are numbered and motifs determined to be functionally active in the human and mouse promoters are indicated by arrows.
Comparative mapping of coding regions of different myostatin genes reveals a similar organization in all species. Codons flanking the first and the second exon are highly conserved among the fishes despite minor differences in rtMSTN-2a and -2b, whereas the codons flanking the second and third exons are highly conserved among all vertebrate species (Fig. 5). A previously identified consensus sequence (143) for the codons flanking the first and second exon, MAT(E/K) - PXXI (X= any amino acid), was slightly different from a recently described consensus (144) that incorporated the rtMSTN-2 proteins, MAT(E/K) vs. MA(T/K). By contrast, the consensus between the second and third exons, (G/E)(E/D)GL - XPFφ (φ + hydrophobic amino acid), is consistently found in all vertebrate myostatin homologs to date (Fig. 5). Furthermore, the mature myostatin peptide, as well as the precluding RXXR (Fig. 2) proteolytic processing site, is encoded by the third exon in all vertebrate homologs. The high level of conservation among vertebrate myostatin genes is therefore reflected not only in coding sequences, but in overall gene organization as well.
Figure 5.
Comparative mapping of myostatin coding domains. For each gene, sequential boxes represent coding domains from each of three exons. The first amino acid coded by each exon and the last of each protein are shown above. The sequences adjacent to each exonic boundary are shown inside the boxes. In all fish genes, the codon of the proline residue located at the first exonic boundary is partially coded by nucleotides in the first and second exons as shown. In-frame stop codons in the rainbow trout myostatin (MSTN)-2b paralog are indicated with asterisks. The location of each RXXR proteolytic processing domain is indicated in light gray, and the highly conserved domain that is eventually processed into the mature myostatin dimer is in dark gray.
Coding and noncoding regions of a genome evolve at different rates (168), and as a result, alignments of noncoding regions are often difficult to interpret and sometimes to even perform. Indeed, homologous promoters with similar functions in different species often do not align. However, subsequence analysis can identify consensus sequences of known motifs within noncoding regions including gene promoters. Such analyses identified a rich distribution of putative muscle-specific transcription factor binding sites in the promoters of different vertebrate myostatin genes (Fig. 4), including some of those previously determined to be functionally active in humans, mice, and cattle (169,170,171,172). Earlier studies indicated that myostatin transactivation is stimulated by the activated glucocorticoid receptor and MEF2 in humans (169) and by MyoD and Myf5 in mice and cattle (170,171). More recent studies indicate that FoxO1 and Smad2, -3, and -4 also directly stimulate myostatin gene expression (172). Although glucocorticoids appear to mediate some stress-induced increases in mammalian myostatin gene expression (173,174), stress and/or glucocorticoids were reported to have the opposite effect in two fish species (175,176) and in chickens (177). These reports were among the first indications that myostatin promoters have significantly diverged in mammals. The value of in silico comparisons, however, rests in the ability to identify such evolutionary changes in promoter organization and to predict possible differences in activity. Functional significance of these differences, however, must be directly assessed. Nevertheless, gene expression patterns can be correlated to changes in promoter structure. For example, myostatin genes that are readily expressed in skeletal muscle, all except for the rtMSTN-2 genes, contain several E-box motifs within the proximal half of the promoter. These motifs are potential binding sites for basic helix-loop-helix (bHLH) transcription factors and are necessary for transactivation in vitro and in vivo (170). Many of these factors, including MyoD and Myf5, heterodimerize with E-proteins to promote myogenesis by stimulating the transactivation of genes necessary for muscle cell determination, differentiation and, maturation (178). The zebrafish MSTN-2 promoter was similarly analyzed and was found to also lack E-box motifs in this region, whereas the zfMSTN-1 promoter contained 6 (135). This is consistent with other fish myostatin genes because the MSTN-1 orthologs are all highly expressed in skeletal muscle, whereas the MSTN-2 orthologs are not (see Section V.B).
The structure and distribution of putative cis elements in the rainbow trout gene promoters revealed both clade- and gene-specific differences (Fig. 4). Although the rtMSTN-1 promoters are structurally similar and share putative Comp1, HAND2, MEF2, MusIn, SRF, and TEF-1 binding sites, they differ significantly from both rtMSTN-2 promoters, which themselves are also quite different. This does not suggest that these specific elements are biologically active, but rather that the promoters of these closely related genes are indeed diverging. Mutations in the rtMSTN-2b promoter would be inconsequential because the altered expression of a pseudogene would not be phenotypically expressed. The expression of fish MSTN-2 genes is far more limited than that of the MSTN-1 genes (see Section V.B) and appears to occur mostly in the brain. Thus, selective pressures have profoundly influenced the evolution of myostatin gene promoters independent of their effects on coding sequences. This resulted in the divergence of MSTN-1 and -2 expression patterns, which occurred after the Actinopterygian-Sarcopterygian split (140,141), as well as after the presalmonid diversifying tetraploidization (142) because the promoters and expression patterns of the salmonid MSTN-1 genes are similar, but not identical.
Sequence differences in noncoding regions can also contribute to functional divergence by mechanisms qualitatively different from those in coding regions and are based on two fundamental hypotheses (179): changes in cis regulatory elements 1) are more likely to have phenotypic consequences, and 2) are more sensitive to selection pressures. Environmental factors and physiological responses greatly and rapidly influence gene transcription, whereas changes in protein structure occur more slowly. Mutations in cis elements can also be codominant (180,181) and are thus more sensitive to natural selection because these changes are often expressed in heterozygotes, whereas similar changes in coding regions are usually recessive (182,183). Thus, polymorphisms in orthologous gene promoters contribute to functional divergence by substantially changing gene expression patterns that ultimately affect fitness. Indeed, differences in promoter activity are inherently indicative of functional divergence because altered expression in different tissues or development stages similarly alters function. Although the precise function for each fish myostatin paralog is not known, they are clearly diverging because the gene expression patterns are very different. Further analysis of myostatin function and molecular evolution will provide a unique opportunity to better understand fundamental mechanisms of evolutionary change. They may also help understand novel functions for myostatin in other vertebrates, including mammals, because recent studies indicate that myostatin expression in mammals is more diverse, and more similar to fish, than originally presumed.
B. Differential gene expression
Surprisingly little is known about myostatin expression in developing mammalian embryos. It is first detected in the myotome compartment of developing mouse somites (1) and presumably continues in developing myogenic cells. By contrast, many studies have both qualitatively and quantitatively assessed myostatin expression in developing embryos of different fish species (27,28,127,130,133,134,135,143,144,175,184,185,186,187) and in chickens (188,189,190). The most comprehensive of these studies used extensive RNA panels and gene-specific “real-time” assays to correlate expression levels of all myostatin genes in rainbow trout and zebrafish to key ontological events (143,144,187). All transcripts were detected in unfertilized and newly fertilized embryos. This is consistent with maternal deposition and with the expression of MSTN-1 and -2 genes in other fish species and even in chicken embryos (188,189,190). It also suggests that myostatin plays a significant role during early development, but not during gastrulation because all studies to date report a rapid decline in myostatin expression during this stage. Expression levels of all MSTN-1 genes in both fish species progressively increased during somitogenesis, which is consistent with myostatin’s myogenic role in mice. However, myostatin is also expressed in many developing chicken tissues and in most adult fish tissues (see below). Further studies are clearly needed to determine whether this dynamic regulation of myostatin expression occurs in mammalian embryos and whether myostatin is also expressed in developing mammalian tissues other than skeletal muscle. If conserved, the different fish model systems, particularly zebrafish, will prove invaluable to elucidating myostatin’s different developmental functions.
Initial studies suggested that in adult mammals, myostatin expression was limited primarily to skeletal muscle (1), although subsequent studies identified low levels of expression in mammary gland (191) and heart (192). By contrast, many nonmammalian vertebrates do not share this limited expression pattern because myostatin mRNA and/or protein is expressed in most fish tissues (28,125,129,131,133,175,184,185,187,193,194) and in many different developing chicken tissues as well (188,195,196). In fish, the MSTN-1 genes are widely expressed, whereas MSTN-2 expression is more limited and occurs mostly in the brain (127,135,144,145,187,197). Expression of the former is also dynamically regulated during development, whereas MSTN-2 expression changes very little (127,144,187). These data strongly suggest that in fish, myostatin’s actions are not restricted to the negative regulation of skeletal muscle growth and development, but may additionally influence these processes in many other tissues through the differential expression of each paralog. The similarly ubiquitous expression pattern in chickens suggests that myostatin’s more limited expression pattern in mammals evolved more recently. A more thorough analysis of myostatin’s tissue-specific expression pattern in other avians is needed to determine whether limited expression in general is unique to mammals. Nevertheless, the comparative analysis of myostatin expression suggests that myostatin originally functioned as a general differentiation factor, which is consistent with the wide distribution of activin receptors in nonmammalian vertebrates (198,199,200,201), and recently adopted a more specialized role in mammals.
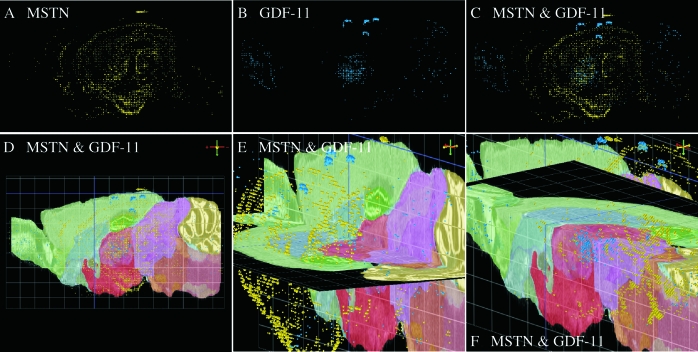
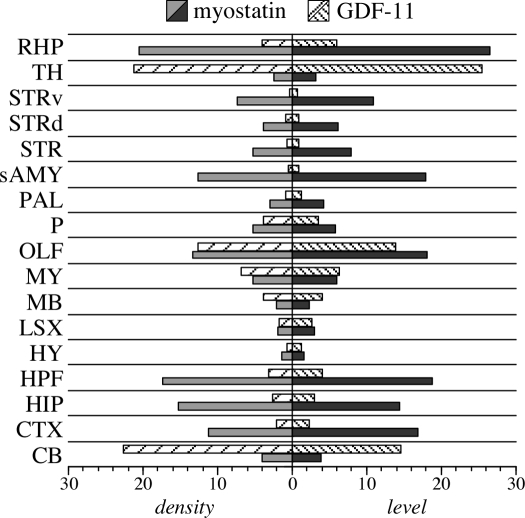
Despite what appears to be a mostly skeletal muscle phenotype in the Mighty Mouse, growing evidence suggests that myostatin may influence other mammalian tissues as well. Indeed, the initial report by McPherron et al. also identified myostatin expression in adipose tissue (1), although this and possibly other phenotypes were likely overshadowed by the extreme nature of the enhanced musculature. The ubiquitous expression pattern of myostatin in fish and chicken together with its expression in mammalian adipose tissue, hearts, and mammary glands suggests that its expression and in turn, its function, may not be as limited in mammals as originally presumed. In fact, Helterline et al. (187) identified myostatin expression in mouse spleens, and a recently published genomewide survey of the mouse brain transcriptome identified several regions where myostatin expression occurs (202,203). A “neuroinformatic pipeline” for assessing in situ expression and localization of neural transcripts was recently developed by the Allen Brain Institute and is available to the general public (www.brain-map.org). Using this three-dimensional tool, myostatin expression was mapped to many brain regions that are clearly distinguishable from GDF-11 (Fig. 6). Myostatin is expressed evenly throughout the hippocampal formation and the cerebral cortex, including the cortical plate. Within the olfactory areas, myostatin expression is confined to the piriform and cortical amygdalar areas. By contrast, GDF-11 is primarily expressed in the main olfactory bulb, the thalamus, and dorsal nuclei of the cerebral cortex above the cortical plate. Analysis of expression levels and density within specific brain regions further demonstrates distinctly different spatial expression patterns for each gene (Fig. 7). It is therefore highly unlikely that either expression pattern was influenced by cross-hybridization of probes because both patterns are unique. The functional significance of myostatin in the brain is currently unknown. However, GDF-11 appears critical to proper neural development (see Section VI.C) (204), and thus myostatin may possess overlapping and compensatory roles. Recent studies also indicate that myostatin influences cardiac muscle growth processes (91,192,205,206,207,208) and adipogenesis (57,209,210) in mammals. Thus, some of myostatin’s more ubiquitous functions may not have been entirely lost in mammals.
Figure 6.
Differential localization of myostatin and GDF-11 expression in mouse brains. Expression of both genes was determined by the Allen Brain Atlas project (202,203) using in situ hybridization. Brains from adult (56-d-old) male C56/B57 mice were hybridized with gene-specific probes as described (www.brain-map.org). Three-dimensional expression patterns were reconstructed using Brain Explorer 1.4 and sagittal section data files, both of which are available at the indicated website. A and B, Individual expression patterns of myostatin (MSTN) and GDF-11 expression, respectively. C, Overlay of A and B. D, Combined expression patterns on top of saggital nissl (left to right) with color-coded anatomical features (see website for key). E and F, Three-dimensional images of combined expression patterns with sagittal and horizontal nissls. Image and nissl orientation is indicated by the color-coded compass in upper right of panels D–F.
Figure 7.
Expression levels of myostatin and GDF-11 in different brain regions. Expression of both genes was determined by the Allen Brain Atlas project (202,203) using in situ hybridization and gene-specific probes. Levels of expression were replotted from data available at www.brain-map.org and are scaled to normalized densities (number of expressing cells/anatomical space) and intensities (total level of expression/anatomical space) of 0 to 100. GDF, Growth/differentiation factor; RHP, retrohippocampal region; TH, thalamus; STRv, ventral striatum; STRd, dorsal striatum; STR, striatum; sAMY, striatum-like amydalar nuclei; PAL, pallidum; P, pons; OLF, olfactory bulb; MY, medulla; MB, midbrain; LSX, lateral septal complex; HY, hypothalamus; HPF, hippocampal formation; HIP, hippocampal region; CTX, cerebral cortex; CB, cerebellum.
C. Alternative processing
Processing of pre-mRNA involves the removal of intronic sequences and modifications to the 5′ and 3′ ends. This is coordinated by small nuclear RNA that directs the spliceosome to target sequences within the primary transcript. Alternative splicing, the generation of multiple transcripts from a single unprocessed pre-mRNA, occurs with poorly conserved splice sites and is influenced by cis and trans elements (sequences within the transcript and their binding proteins, respectively) that either enhance or silence spliceosomal activity. It also explains how over 60,000 transcripts are generated from approximately 30,000 genes in most vertebrates and is the primary source of protein complexity (211,212,213). Alternative splicing of myostatin transcripts has been described in rainbow trout (144) and in developing chicken embryos (189). Although the precise functional significance remains to be determined for both animal models, preliminary evidence suggests that in rainbow trout, it enhances myostatin’s effects in the brain, whereas in the chicken it helps control bioavailability.
Both intact and truncated myostatin transcripts are expressed in chicken embryos (189). The latter lacks the coding sequence for the C-terminal mature peptide. Translation of this transcript would therefore block myostatin bioavailability because only the LAP domain would be produced. In rainbow trout, MSTN-2a and -2b are lowly expressed in most tissues except for brain where rtMSTN-2a is highly expressed (144). Nearly all tissues express mostly unprocessed transcripts that retain both introns, which themselves contain several in-frame stop codons. By contrast, rtMSTN-2a, but not -2b, is fully processed in the brain. Tissue-specific alternative processing is rare and is mediated by cis recognition of tissue-specific proteins that bind to repeating motifs in the pre-mRNA (214). It occurs most often in the brain where it is commonly mediated by Nova proteins that act as splice enhancers or silencers, depending on their proximity to the splice site, and is required with transcripts possessing poorly conserved splice sites (215,216,217,218). Nova proteins recognize YCAY motifs and are known to enhance the alternative splicing of two neurotransmitter receptors, GABAARg2 and GlyRa2, and to silence the splicing of some Nova transcripts themselves, specifically Nova-1 (219,220,221,222). Both rtMSTN-2 genes lack conserved splice sites, although rtMSTN-2a has almost twice the number of expected YCAY motifs throughout and three times the expected motifs in the second intron alone. Whether Nova regulates the alternative splicing of rtMSTN-2a remains to be determined. Nevertheless, a comparative analysis of rtMSTN-2a and -2b processing provides a perfectly controlled system to investigate the basic mechanisms responsible for tissue-specific pre-mRNA processing.
As the mature myostatin peptide is encoded entirely by the third exon, the introduction of alternative transcription start sites in either intron of rtMSTN-2b, or any other salmonid MSTN-2b, could presumably result in the expression of a mature myostatin. Therefore, the inability to remove expressed introns contributes to the pseudogenization of rtMSTN-2b as it prohibits the “accidental” translation of an otherwise silenced gene. This is particularly important as the rtMSTN-2b gene still possess a functionally active promoter. Contributions from noncoding regions to the functional divergence of duplicated genes or to the subfunctionalization of a particular allele are not necessarily limited to gene promoters. Gene expression and pre-mRNA processing are both regulated by protein:DNA interactions that depend upon highly specific binding sites. Thus, alterations in such binding sites over time could not only influence gene function, but ultimately organismal complexity and speciation as well.
VI. Novel Actions
A. Adipose tissue
Myostatin is minimally expressed in adipose tissue, and myostatin-null animals have less total and im body fat than wild-type animals (1,56,223,224,225). Increases in muscle mass have long been known to similarly increase resting energy expenditure (REE), which in turn can reduce fat free mass (226) and is inversely correlated to negative outcomes of patients with type 2 diabetes mellitus (227). Thus, the reduced adiposity in myostatin-null animals could simply be due to the caloric draw from enhanced musculature. Circulating levels of leptin, an adipokine and satiety factor that controls body fat, are reduced rather than elevated in these animals (56,228). This suggests that the increased REE is indeed responsible. However, myostatin has been shown to directly influence the cellular physiology of three different adipocyte culture systems with conflicting results. It inhibits the differentiation of 3T3-L1 preadipocytes (209,229) and primary preadipocytes from cattle (230) or humans (231). It also down-regulated the expression of several adipogenic markers and transcription factors in these cells, including peroxisome proliferator-activated receptor γ, C/EBPα, aP2, and leptin.
By contrast, myostatin promotes adipogenesis in a multipotent mesenchymal cell line (C3H10T1/2) that can differentiate into many cell lineages including muscle-like cells (210,232). Although the latter results are consistent with reduced adiposity in myostatin-null animals, the studies’ physiological relevance is somewhat questionable in light of the consistent results obtained with primary cells and with a very well-established cell line (3T3-L1). Transgenic mice overexpressing myostatin specifically in adipocytes fail to develop obesity when fed a high-fat diet and respond better to glucose tolerance tests than do obese nontransgenic mice (232). Their adipocytes, however, express preadipocyte markers, suggesting that adipogenic determination may be stimulated by myostatin, but not differentiation. Reduced adiposity in myostatin-null animals is, therefore, likely due to the indirect effects of enhanced musculature on REE rather than to direct effects on the adipocytes themselves. In fact, myostatin fails to influence lipolysis in vitro or fat mass in vivo when administered pharmacologically to wild-type or obese mice (229).
McPherron and Lee (56) directly tested myostatin’s ability to influence the development of obesity by crossing the Mighty Mouse with two murine models of obesity and insulin resistance: the agouti lethal yellow (Ay) and leptin-deficient obese (Lepob/ob) mice. Introducing a myostatin-null background into either strain increased muscle mass, suppressed adiposity (as indicated by reduced mass of different fat pads), and vastly improved glucose tolerance. Similar results were also observed in transgenic mice overexpressing LAP and fed a high-fat diet (57,58). Thus, enhancing muscle mass by blocking myostatin bioactivity and/or bioavailability can prevent the development of obesity and insulin resistance. These results do not, however, prove that myostatin’s effects are mediated entirely by increasing REE because the myostatin-null environment could have influenced the development of preadipocytes in utero or even the de novo synthesis of triglycerides in already differentiated adipocytes. Several additional questions also remain unanswered. In particular, will disrupting myostatin production and/or bioavailability be similarly beneficial in animals that are already obese and displaying signs of insulin resistance? Regardless, these results are extremely exciting and potentially introduce a new therapeutic target for treating obesity and type 2 diabetes mellitus.
B. Cardiac muscle
Cardiac expression of myostatin has been documented in sheep (192), chickens (195,196), mice (192), rats (205), and different fish species (129,130,143,144,187). In developing chicken hearts, myostatin expression is detected early and progressively increases until morphogenesis is complete, suggesting a functional role for myostatin in the development of cardiac as well as skeletal muscle (195). High levels of myostatin expression in primary mouse cardiomyoblasts are correlated with a low proliferative index, whereas recombinant myostatin inhibits the growth of these cells, as well as rat H9C2 cardiomyoblast growth, without inducing apoptosis (205,233). Myostatin also suppresses cardiomyocyte hypertrophic growth responses, specifically protein synthesis, induced by either phenylepinephrine (205) or IGF-I (208). Recent studies with Akt transgenic mice (234), in vitro models of cyclic stretch, and IGF-I-stimulated cardiomyocytes (208) all suggest that myostatin not only regulates some cardiac muscle growth process, but that it may function as a cardiac chalone (235) just as it does in skeletal muscle (8). Indeed, myostatin expression is elevated in all of these models and may therefore provide a negative feedback mechanism to limit cardiac muscle growth, and thus hypertrophy.
Myostatin expression in hearts is also elevated in several pathophysiological conditions. In response to a myocardial infarction, myostatin immunostaining steadily increases in the peri-infarcted region and persists for over 1 month (192). Increased expression also occurs with chronic swim training in rats (236) and with volume-overloaded chronic heart failure (207,237). These changes are similar to those occurring in damaged or regenerating skeletal muscle (174,238,239,240,241) and suggest that myostatin may also influence cardiac muscle repair. Whether or not myostatin’s role in this process is beneficial or detrimental remains to be determined. Two studies report conflicting results on the effect of a myostatin-null environment on heart size (233,242); however, both studies indicate that basic cardiac output parameters (ventricular volume measurements, ejection fraction, etc.) are at least normal. A more in-depth analysis of myostatin’s effects on cardiac function is definitely warranted. These preliminary studies are nevertheless promising and suggest that repairing damaged cardiac muscle by blocking myostatin’s actions could have highly beneficial clinical outcomes.
In mammals, myostatin is clearly expressed in developing and adult cardiac muscle and is capable of manipulating different cardiac muscle growth processes. It is unknown, therefore, why a significant cardiac phenotype has never been described for any myostatin-null animal. Cardiac tissue lacks myoprogenitor cells analogous to the myosatellite cells found in skeletal muscle and may be less sensitive to myostatin’s inhibitory effects. Thus, blocking myostatin’s actions or removing the gene altogether would have very little effect, except under circumstances when myostatin expression is elevated. Several questions remain. Can other cardiac factors compensate for myostatin or the lack of it? Does developing cardiac muscle express factors or mechanisms that block myostatin’s actions (e.g., follistatin, FLRG, etc.)? Do myostatin-null hearts function better or worse when challenged (i.e., stress test)? Most importantly, will blocking myostatin’s actions help regenerating cardiac muscle after an infarction?
C. Brain
Although myostatin is expressed in different brain regions of different vertebrates, its function in the brain is unknown. However, GDF-11, which is closely related to myostatin and has a nearly identical bioactive domain (95% similar), is thought to play a role in neurogenesis. It is more widely expressed in mammalian embryos (52,243) and is secreted by neuroprogenitor cells and fully differentiated neurons of the olfactory epithelium where it inhibits neurogenesis by inducing cell cycle arrest of the neuroprogenitors (204). In the retina, GDF-11 limits the number of ganglion, amacrine, and photoreceptor cells by controlling neuroprogenitor cell competence (244). Thus, GDF-11 functions as a negative autoregulator, or chalone, of neural tissue, which mirrors myostatin action in mammalian skeletal muscle. It is therefore possible that myostatin has similar functions in the brain, albeit in different regions (Figs. 6 and 7), and that compensatory changes in GDF-11 expression and/or availability prohibit the development of neural phenotypes in myostatin-null animals. Fish brains maintain a large number of hyperplastic neuroprogenitor cells that can be easily isolated and even cultured in vitro (245,246). In vivo models for neuroprogenitor cell proliferation and differentiation have also been developed in goldfish and zebrafish (247,248,249,250). These underutilized comparative model systems may therefore prove valuable in distinguishing the neural functions of myostatin and GDF-11.
VII. Final Thoughts: Implications for Biomedical, Agricultural, and Evolutionary Sciences
The most apparent biomedical application for manipulating myostatin action is clearly to enhance muscle growth, although the use of novel “myostatin-blocking” technologies need not be limited to the treatment of skeletal muscle pathologies—injury, sarcopenia, wasting/cachexia, some forms of muscular dystrophy, etc.—because exploiting such technologies may prove beneficial in treating survivors of a myocardial infarction as well. Several of these technologies have already been developed, including immunoneutralizing antisera (251,252,253), a soluble form of Acvr2b’s extracellular domain (48) and a quiver of myostatin binding proteins (see Section III.A), and each successfully stimulates skeletal muscle hypertrophy in wild-type mice and, in some instances, in a murine model for Duchenne muscular dystrophy (44,253,254,255,256). The indirect effect of neutralizing myostatin’s actions on adiposity suggests that these technologies could also be useful in treating obesity and type 2 diabetes mellitus. A promise of “more muscle, less fat” is an attractive marketing campaign and one that these technologies could potentially deliver. Thus, blocking myostatin’s actions for cosmetic purposes is not just probable, but possibly inevitable as the biomedical successes gain visibility among the general public.
Double-muscled cattle like the Belgian Blue and the Piedmontese frequently require cesarean delivery due to large calf size. Piedmontese meat is particularly tender (16,17); nevertheless, the high cost of veterinary care associated with parturition has prevented the widespread acceptance of these and other double-muscled breeds in most cattle production markets. Crossing myostatin-null breeds with normal-muscled breeds or even immunoneutralizing cows before breeding, as has been done with mice (252), could address issues of growth rate and product yield in many animal production industries. It is unknown, however, whether the potential loss of im fat would potentially offset these commercial gains. These issues are not a concern with egg-laying vertebrates. Thus, myostatin technologies may have a greater commercial impact on domestic fish and fowl production, especially because the mass ratio of muscle to nonmuscle tissues in fish is greater than in any other vertebrate class. Such commercial gains are predicated on the assumption that blocking myostatin actions will have no adverse side effects on nonmuscle tissues, many of which express multiple myostatin genes. A more thorough assessment of myostatin’s nonmuscle actions in fish is therefore needed to determine whether blocking its actions is both feasible and commercially beneficial.
Comparative model systems will also help identify novel functions for both myostatin and GDF-11 in ways pertinent to agriculture and medicine as their more basic and conserved actions are defined. The subset of salmonid myostatin genes is a particularly useful model for investigating the functional divergence of duplicated alleles because differences in coding and noncoding sequences have contributed to their evolution in a manner that influences gene expression, transcript processing, protein structure, and pseudogenization. Determining the underlying mechanisms involved presents a unique opportunity to investigate competing evolutionary models—double-recessive vs. duplication-degeneration-complementation (150,257)—that potentially explain the molecular basis of fundamental evolutionary processes. Lack of such mechanistic understanding is an important problem because, without it, we cannot understand how perceivably minor changes in gene structure and function can significantly impact phenotypic differences between species and/or speciation itself. Thus, a growing knowledge of myostatin molecular genetics and bioactivity, in different tissues and in different organisms, has the potential to impact science as well as society.
Footnotes
This work was supported in part by National Institutes of Health Grant 1R03AR051917 and United States Department of Agriculture Grant 2004-34468-15199.
Disclosure Statement: The authors have nothing to disclose.
First Published Online June 30, 2008
Abbreviations: CHO, Chinese hamster ovary; FLRG, follistatin-like related gene; GDF, growth/differentiating factor; IGFBP, IGF binding protein; LAP, latency-associated peptide; LTBP, latent TGFβ binding proteins; MSTN, myostatin; PEMC, porcine embryonic myogenic cells; REE, resting energy expenditure; rt, rainbow trout; T-cap, titin-cap.
References
- McPherron AC, Lawler AM, Lee SJ 1997 Regulation of skeletal muscle mass in mice by a new TGF-β superfamily member. Nature 387:83–90 [DOI] [PubMed] [Google Scholar]
- Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, Riquet J, Schoeberlein A, Dunner S, Menissier F, Massabanda J, Fries R, Hanset R, Georges M 1997 A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle [see comments]. Nat Genet 17:71–74 [DOI] [PubMed] [Google Scholar]
- Kambadur R, Sharma M, Smith TP, Bass JJ 1997 Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res 7:910–916 [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lee SJ 1997 Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA 94:12457–12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TP, Lopez-Corrales NL, Kappes SM, Sonstegard TS 1997 Myostatin maps to the interval containing the bovine mh locus. Mamm Genome 8:742–744 [DOI] [PubMed] [Google Scholar]
- Ramazanov Z, Jimenez del Rio M, Ziegenfuss T 2003 Sulfated polysaccharides of brown seaweed Cystoseira canariensis bind to serum myostatin protein. Acta Physiol Pharmacol Bulg 27:101–106 [PubMed] [Google Scholar]
- Willoughby DS 2004 Effects of an alleged myostatin-binding supplement and heavy resistance training on serum myostatin, muscle strength and mass, and body composition. Int J Sport Nutr Exerc Metab 14:461–472 [DOI] [PubMed] [Google Scholar]
- Lee SJ 2004 Regulation of muscle mass by myostatin. Annu Rev Cell Dev Biol 20:61–86 [DOI] [PubMed] [Google Scholar]
- Varga L, Szabo G, Darvasi A, Muller G, Sass M, Soller M 1997 Inheritance and mapping of Compact (Cmpt), a new mutation causing hypermuscularity in mice. Genetics 147:755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo G, Dallmann G, Muller G, Patthy L, Soller M, Varga L 1998 A deletion in the myostatin gene causes the compact (Cmpt) hypermuscular mutation in mice. Mamm Genome 9:671–672 [DOI] [PubMed] [Google Scholar]
- Varga L, Pinke O, Muller G, Kovacs B, Korom E, Szabo G, Soller M 2005 Mapping a syntenic modifier on mouse chromosome 1 influencing the expressivity of the compact phenotype in the myostatin mutant (MstnCmpt-dl1Abc) compact mouse. Genetics 169:489–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga L, Muller G, Szabo G, Pinke O, Korom E, Kovacs B, Patthy L, Soller M 2003 Mapping modifiers affecting muscularity of the myostatin mutant (Mstn(Cmpt-dl1Abc)) compact mouse. Genetics 165:257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisz-Porszasz S, Bhasin S, Artaza JN, Shen R, Sinha-Hikim I, Hogue A, Fielder TJ, Gonzalez-Cadavid NF 2003 Lower skeletal muscle mass in male transgenic mice with muscle-specific overexpression of myostatin. Am J Physiol Endocrinol Metab 285:E876—E888 [DOI] [PubMed] [Google Scholar]
- Zimmers TA, Davies MV, Koniaris LG, Haynes P, Esquela AF, Tomkinson KN, McPherron AC, Wolfman NM, Lee SJ 2002 Induction of cachexia in mice by systemically administered myostatin. Science 296:1486–1488 [DOI] [PubMed] [Google Scholar]
- Page BT, Casas E, Heaton MP, Cullen NG, Hyndman DL, Morris CA, Crawford AM, Wheeler TL, Koohmaraie M, Keele JW, Smith TP 2002 Evaluation of single-nucleotide polymorphisms in CAPN1 for association with meat tenderness in cattle. J Anim Sci 80:3077–3085 [DOI] [PubMed] [Google Scholar]
- Wheeler TL, Cundiff LV, Shackelford SD, Koohmaraie M 2001 Characterization of biological types of cattle (cycle V): carcass traits and longissimus palatability. J Anim Sci 79:1209–1222 [DOI] [PubMed] [Google Scholar]
- Wheeler TL, Shackelford SD, Casas E, Cundiff LV, Koohmaraie M 2001 The effects of Piedmontese inheritance and myostatin genotype on the palatability of longissimus thoracis, gluteus medius, semimembranosus, and biceps femoris. J Anim Sci 79:3069–3074 [DOI] [PubMed] [Google Scholar]
- Johnson PL, McEwan JC, Dodds KG, Purchas RW, Blair HT 2005 Meat quality traits were unaffected by a quantitative trait locus affecting leg composition traits in Texel sheep. J Anim Sci 83:2729–2735 [DOI] [PubMed] [Google Scholar]
- Walling GA, Visscher PM, Wilson AD, McTeir BL, Simm G, Bishop SC 2004 Mapping of quantitative trait loci for growth and carcass traits in commercial sheep populations. J Anim Sci 82:2234–2245 [DOI] [PubMed] [Google Scholar]
- Marchitelli C, Savarese MC, Crisa A, Nardone A, Marsan PA, Valentini A 2003 Double muscling in Marchigiana beef breed is caused by a stop codon in the third exon of myostatin gene. Mamm Genome 14:392–395 [DOI] [PubMed] [Google Scholar]
- Berry C, Thomas M, Langley B, Sharma M, Kambadur R 2002 Single cysteine to tyrosine transition inactivates the growth inhibitory function of Piedmontese myostatin. Am J Physiol Cell Physiol 283:C135—C141 [DOI] [PubMed] [Google Scholar]
- Mosher DS, Quignon P, Bustamante CD, Sutter NB, Mellersh CS, Parker HG, Ostrander EA 2007 A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet 3:e79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton GD, Engvall E 2007 Gross muscle hypertrophy in whippet dogs is caused by a mutation in the myostatin gene. Neuromuscul Disord 17:721–722 [DOI] [PubMed] [Google Scholar]
- Johnson PL, McEwan JC, Dodds KG, Purchas RW, Blair HT 2005 A directed search in the region of GDF8 for quantitative trait loci affecting carcass traits in Texel sheep. J Anim Sci 83:1988–2000 [DOI] [PubMed] [Google Scholar]
- Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, Larzul C, Laville E, Meish F, Milenkovic D, Tobin J, Charlier C, Georges M 2006 A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet 38:813–818 [DOI] [PubMed] [Google Scholar]
- Schuelke M, Wagner KR, Stolz LE, Hubner C, Riebel T, Komen W, Braun T, Tobin JF, Lee SJ 2004 Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med 350:2682–2688 [DOI] [PubMed] [Google Scholar]
- Amali AA, Lin CJ, Chen YH, Wang WL, Gong HY, Lee CY, Ko YL, Lu JK, Her GM, Chen TT, Wu JL 2004 Up-regulation of muscle-specific transcription factors during embryonic somitogenesis of zebrafish (Danio rerio) by knock-down of myostatin-1. Dev Dyn 229:847–856 [DOI] [PubMed] [Google Scholar]
- Xu C, Wu G, Zohar Y, Du SJ 2003 Analysis of myostatin gene structure, expression and function in zebrafish. J Exp Biol 206:4067–4079 [DOI] [PubMed] [Google Scholar]
- Acosta J, Carpio Y, Borroto I, Gonzalez O, Estrada MP 2005 Myostatin gene silenced by RNAi show a zebrafish giant phenotype. J Biotechnol 119:324–331 [DOI] [PubMed] [Google Scholar]
- Johnston IA 1999 Muscle development and growth: potential implications for flesh quality in fish. Aquaculture 177:99–115 [Google Scholar]
- Lee SJ, McPherron AC 2001 Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci USA 98:9306–9311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JJ, Davies MV, Pearson AA, Wang JH, Hewick RM, Wolfman NM, Qiu Y 2002 The myostatin propeptide and follistatin-related gene are inhibitory binding proteins of myostatin in normal serum. J Biol Chem 277:40735–40741 [DOI] [PubMed] [Google Scholar]
- Hill JJ, Qiu Y, Hewick RM, Wolfman NM 2003 Regulation of myostatin in vivo by growth and differentiation factor-associated serum protein-1: a novel protein with protease inhibitor and follistatin domains. Mol Endocrinol 17:1144–1154 [DOI] [PubMed] [Google Scholar]
- Nicholas G, Thomas M, Langley B, Somers W, Patel K, Kemp CF, Sharma M, Kambadur R 2002 Titin-cap associates with, and regulates secretion of, myostatin. J Cell Physiol 193:120–131 [DOI] [PubMed] [Google Scholar]
- Koli K, Saharinen J, Hyytiainen M, Penttinen C, Keski-Oja J 2001 Latency, activation, and binding proteins of TGF-β. Microsc Res Tech 52:354–362 [DOI] [PubMed] [Google Scholar]
- Anderson SB, Goldberg AL, Whitman M 2008 Identification of a novel pool of extracellular pro-myostatin in skeletal muscle. J Biol Chem 283:7027–7035 [DOI] [PubMed] [Google Scholar]
- Miura T, Kishioka Y, Wakamatsu J, Hattori A, Hennebry A, Berry CJ, Sharma M, Kambadur R, Nishimura T 2006 Decorin binds myostatin and modulates its activity to muscle cells. Biochem Biophys Res Commun 340:675–680 [DOI] [PubMed] [Google Scholar]
- Nishimura T, Oyama K, Kishioka Y, Wakamatsu J, Hattori A 2007 Spatiotemporal expression of decorin and myostatin during rat skeletal muscle development. Biochem Biophys Res Commun 361:896–902 [DOI] [PubMed] [Google Scholar]
- Velleman SG 1999 The role of the extracellular matrix in skeletal muscle development. Poult Sci 78:778–784 [DOI] [PubMed] [Google Scholar]
- Li Y, Foster W, Deasy BM, Chan Y, Prisk V, Tang Y, Cummins J, Huard J 2004 Transforming growth factor-β1 induces the differentiation of myogenic cells into fibrotic cells in injured skeletal muscle: a key event in muscle fibrogenesis. Am J Pathol 164:1007–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Li Y, Foster W, Fukushima K, Badlani N, Adachi N, Usas A, Fu FH, Huard J 2003 Improvement of muscle healing through enhancement of muscle regeneration and prevention of fibrosis. Muscle Nerve 28:365–372 [DOI] [PubMed] [Google Scholar]
- Zhu J, Li Y, Shen W, Qiao C, Ambrosio F, Lavasani M, Nozaki M, Branca MF, Huard J 2007 Relationships between transforming growth factor-β1, myostatin, and decorin: implications for skeletal muscle fibrosis. J Biol Chem 282:25852–25863 [DOI] [PubMed] [Google Scholar]
- Li Y, Li J, Zhu J, Sun B, Branca M, Tang Y, Foster W, Xiao X, Huard J 2007 Decorin gene transfer promotes muscle cell differentiation and muscle regeneration. Mol Ther 15:1616–1622 [DOI] [PubMed] [Google Scholar]
- Wagner KR, McPherron AC, Winik N, Lee SJ 2002 Loss of myostatin attenuates severity of muscular dystrophy in mdx mice. Ann Neurol 52:832–836 [DOI] [PubMed] [Google Scholar]
- McCroskery S, Thomas M, Platt L, Hennebry A, Nishimura T, McLeay L, Sharma M, Kambadur R 2005 Improved muscle healing through enhanced regeneration and reduced fibrosis in myostatin-null mice. J Cell Sci 118:3531–3541 [DOI] [PubMed] [Google Scholar]
- Thies RS, Chen T, Davies MV, Tomkinson KN, Pearson AA, Shakey QA, Wolfman NM 2001 GDF-8 propeptide binds to GDF-8 and antagonizes biological activity by inhibiting GDF-8 receptor binding. Growth Factors 18:251–259 [DOI] [PubMed] [Google Scholar]
- Yang J, Ratovitski T, Brady JP, Solomon MB, Wells KD, Wall RJ 2001 Expression of myostatin pro domain results in muscular transgenic mice. Mol Reprod Dev 60:351–361 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Reed LA, Davies MV, Girgenrath S, Goad ME, Tomkinson KN, Wright JF, Barker C, Ehrmantraut G, Holmstrom J, Trowell B, Gertz B, Jiang MS, Sebald SM, Matzuk M, Li E, Liang LF, Quattlebaum E, Stotish RL, Wolfman NM 2005 Regulation of muscle growth by multiple ligands signaling through activin type II receptors. Proc Natl Acad Sci USA 102:18117–18122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Hadhazy M, Wehling M, Tidball JG, McNally EM 2000 Dominant negative myostatin produces hypertrophy without hyperplasia in muscle. FEBS Lett 474:71–75 [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Lu N, Vogel H, Sellheyer K, Roop DR, Bradley A 1995 Multiple defects and perinatal death in mice deficient in follistatin. Nature 374:360–363 [DOI] [PubMed] [Google Scholar]
- Winters SJ, Moore JP 2007 Paracrine control of gonadotrophs. Semin Reprod Med 25:379–387 [DOI] [PubMed] [Google Scholar]
- Gamer LW, Wolfman NM, Celeste AJ, Hattersley G, Hewick R, Rosen V 1999 A novel BMP expressed in developing mouse limb, spinal cord, and tail bud is a potent mesoderm inducer in Xenopus embryos. Dev Biol 208:222–232 [DOI] [PubMed] [Google Scholar]
- Lee SJ 2007 Quadrupling muscle mass in mice by targeting TGF-β signaling pathways. PLoS ONE 2:e789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki H, Minegishi T, Nakamura K, Tano M, Miyamoto K, Ibuki Y 1995 Type II and type IIB activin receptors in human placenta. Life Sci 56:1699–1706 [DOI] [PubMed] [Google Scholar]
- Casagrandi D, Bearfield C, Geary J, Redman CW, Muttukrishna S 2003 Inhibin, activin, follistatin, activin receptors and beta-glycan gene expression in the placental tissue of patients with pre-eclampsia. Mol Hum Reprod 9:199–203 [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lee SJ 2002 Suppression of body fat accumulation in myostatin-deficient mice. J Clin Invest 109:595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhao B 2006 Postnatal expression of myostatin propeptide cDNA maintained high muscle growth and normal adipose tissue mass in transgenic mice fed a high-fat diet. Mol Reprod Dev 73:462–469 [DOI] [PubMed] [Google Scholar]
- Zhao B, Wall RJ, Yang J 2005 Transgenic expression of myostatin propeptide prevents diet-induced obesity and insulin resistance. Biochem Biophys Res Commun 337:248–255 [DOI] [PubMed] [Google Scholar]
- Thomas M, Langley B, Berry C, Sharma M, Kirk S, Bass J, Kambadur R 2000 Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. J Biol Chem 275:40235–40243 [DOI] [PubMed] [Google Scholar]
- Rios R, Carneiro I, Arce VM, Devesa J 2001 Myostatin regulates cell survival during C2C12 myogenesis. Biochem Biophys Res Commun 280:561–566 [DOI] [PubMed] [Google Scholar]
- Taylor WE, Bhasin S, Artaza J, Byhower F, Azam M, Willard Jr DH, Kull Jr FC, Gonzalez-Cadavid N 2001 Myostatin inhibits cell proliferation and protein synthesis in C(2)C(12) muscle cells. Am J Physiol Endocrinol Metab 280:E221–E228 [DOI] [PubMed] [Google Scholar]
- Rios R, Carneiro I, Arce VM, Devesa J 2002 Myostatin is an inhibitor of myogenic differentiation. Am J Physiol Cell Physiol 282:C993—C999 [DOI] [PubMed] [Google Scholar]
- Langley B, Thomas M, Bishop A, Sharma M, Gilmour S, Kambadur R 2002 Myostatin inhibits myoblast differentiation by down regulating MyoD expression. J Biol Chem 277:49831–49840 [DOI] [PubMed] [Google Scholar]
- Joulia D, Bernardi H, Garandel V, Rabenoelina F, Vernus B, Cabello G 2003 Mechanisms involved in the inhibition of myoblast proliferation and differentiation by myostatin. Exp Cell Res 286:263–275 [DOI] [PubMed] [Google Scholar]
- McCroskery S, Thomas M, Maxwell L, Sharma M, Kambadur R 2003 Myostatin negatively regulates satellite cell activation and self-renewal. J Cell Biol 162:1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KR, Liu X, Chang X, Allen RE 2005 Muscle regeneration in the prolonged absence of myostatin. Proc Natl Acad Sci USA 102:2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amthor H, Otto A, Macharia R, McKinnell I, Patel K 2006 Myostatin imposes reversible quiescence on embryonic muscle precursors. Dev Dyn 235:672–680 [DOI] [PubMed] [Google Scholar]
- Figeac N, Daczewska M, Marcelle C, Jagla K 2007 Muscle stem cells and model systems for their investigation. Dev Dyn 236:3332–3342 [DOI] [PubMed] [Google Scholar]
- Langley B, Thomas M, McFarlane C, Gilmour S, Sharma M, Kambadur R 2004 Myostatin inhibits rhabdomyosarcoma cell proliferation through an Rb-independent pathway. Oncogene 23:524–534 [DOI] [PubMed] [Google Scholar]
- Forbes D, Jackman M, Bishop A, Thomas M, Kambadur R, Sharma M 2006 Myostatin auto-regulates its expression by feedback loop through Smad7 dependent mechanism. J Cell Physiol 206:264–272 [DOI] [PubMed] [Google Scholar]
- Jiang MS, Liang LF, Wang S, Ratovitski T, Holmstrom J, Barker C, Stotish R 2004 Characterization and identification of the inhibitory domain of GDF-8 propeptide. Biochem Biophys Res Commun 315:525–531 [DOI] [PubMed] [Google Scholar]
- Mittl PR, Priestle JP, Cox DA, McMaster G, Cerletti N, Grutter MG 1996 The crystal structure of TGF-β 3 and comparison to TGF-β 2: implications for receptor binding. Protein Sci 5:1261–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daopin S, Piez KA, Ogawa Y, Davies DR 1992 Crystal structure of transforming growth factor-β 2: an unusual fold for the superfamily [see comments]. Science 257:369–373 [DOI] [PubMed] [Google Scholar]
- Griffith DL, Keck PC, Sampath TK, Rueger DC, Carlson WD 1996 Three-dimensional structure of recombinant human osteogenic protein 1: structural paradigm for the transforming growth factor β superfamily. Proc Natl Acad Sci USA 93:878–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlunegger MP, Grutter MG 1992 An unusual feature revealed by the crystal structure at 2.2 A resolution of human transforming growth factor-β 2. Nature 358:430–434 [DOI] [PubMed] [Google Scholar]
- Attisano L, Wrana JL 2002 Signal transduction by the TGF-β superfamily. Science 296:1646–1647 [DOI] [PubMed] [Google Scholar]
- Rebbapragada A, Benchabane H, Wrana JL, Celeste AJ, Attisano L 2003 Myostatin signals through a transforming growth factor β-like signaling pathway to block adipogenesis. Mol Cell Biol 23:7230–7242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Topouzis S, Liang LF, Stotish RL 2004 Myostatin signaling through Smad2, Smad3 and Smad4 is regulated by the inhibitory Smad7 by a negative feedback mechanism. Cytokine 26:262–272 [DOI] [PubMed] [Google Scholar]
- Xu W, Angelis K, Danielpour D, Haddad MM, Bischof O, Campisi J, Stavnezer E, Medrano EE 2000 Ski acts as a co-repressor with Smad2 and Smad3 to regulate the response to type β transforming growth factor. Proc Natl Acad Sci USA 97:5924–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Yagi K, Kondo M, Kato M, Miyazono K, Miyazawa K 2004 c-Ski inhibits the TGF-β signaling pathway through stabilization of inactive Smad complexes on Smad-binding elements. Oncogene 23:5068–5076 [DOI] [PubMed] [Google Scholar]
- Akiyoshi S, Inoue H, Hanai J, Kusanagi K, Nemoto N, Miyazono K, Kawabata M 1999 c-Ski acts as a transcriptional co-repressor in transforming growth factor-β signaling through interaction with smads. J Biol Chem 274:35269–35277 [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Goto K, Horiguchi K, Nagata M, Kawata M, Miyazawa K, Saitoh M, Miyazono K 2007 c-Ski activates MyoD in the nucleus of myoblastic cells through suppression of histone deacetylases. Genes Cells 12:375–385 [DOI] [PubMed] [Google Scholar]
- Zapf J, Froesch ER 1999 Insulin-like growth factor-I actions on somatic growth. In: Kostyo JL, Goodman HM, eds. Handbook of physiology, the endocrine system: hormonal control of growth. New York: Oxford University Press; 663–700 [Google Scholar]
- Foulstone EJ, Savage PB, Crown AL, Holly JM, Stewart CE 2003 Role of insulin-like growth factor binding protein-3 (IGFBP-3) in the differentiation of primary human adult skeletal myoblasts. J Cell Physiol 195:70–79 [DOI] [PubMed] [Google Scholar]
- Pampusch MS, Kamanga-Sollo E, White ME, Hathaway MR, Dayton WR 2003 Effect of recombinant porcine IGF-binding protein-3 on proliferation of embryonic porcine myogenic cell cultures in the presence and absence of IGF-I. J Endocrinol 176:227–235 [DOI] [PubMed] [Google Scholar]
- Johnson BJ, White ME, Hathaway MR, Dayton WR 2003 Effect of differentiation on levels of insulin-like growth factor binding protein mRNAs in cultured porcine embryonic myogenic cells. Domest Anim Endocrinol 24:81–93 [DOI] [PubMed] [Google Scholar]
- Spangenburg EE, Abraha T, Childs TE, Pattison JS, Booth FW 2003 Skeletal muscle IGF-binding protein-3 and -5 expressions are age, muscle, and load dependent. Am J Physiol Endocrinol Metab 284:E340—E350 [DOI] [PubMed] [Google Scholar]
- Lawlor MA, Feng X, Everding DR, Sieger K, Stewart CE, Rotwein P 2000 Dual control of muscle cell survival by distinct growth factor-regulated signaling pathways. Mol Cell Biol 20:3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor MA, Rotwein P 2000 Coordinate control of muscle cell survival by distinct insulin-like growth factor activated signaling pathways. J Cell Biol 151:1131–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor MA, Rotwein P 2000 Insulin-like growth factor-mediated muscle cell survival: central roles for Akt and cyclin-dependent kinase inhibitor p21. Mol Cell Biol 20:8983–8995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morissette MR, Cook SA, Foo S, McKoy G, Ashida N, Novikov M, Scherrer-Crosbie M, Li L, Matsui T, Brooks G, Rosenzweig A 2006 Myostatin regulates cardiomyocyte growth through modulation of Akt signaling. Circ Res 99:15–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Zhang Y, Li Y, Wu Z, Zhu D 2007 Myostatin induces cyclin D1 degradation to cause cell cycle arrest through a phosphatidylinositol 3-kinase/AKT/GSK-3 β pathway and is antagonized by insulin-like growth factor 1. J Biol Chem 282:3799–3808 [DOI] [PubMed] [Google Scholar]
- Yang W, Chen Y, Zhang Y, Wang X, Yang N, Zhu D 2006 Extracellular signal-regulated kinase 1/2 mitogen-activated protein kinase pathway is involved in myostatin-regulated differentiation repression. Cancer Res 66:1320–1326 [DOI] [PubMed] [Google Scholar]
- Huang Z, Chen D, Zhang K, Yu B, Chen X, Meng J 2007 Regulation of myostatin signaling by c-Jun N-terminal kinase in C2C12 cells. Cell Signal 19:2286–2295 [DOI] [PubMed] [Google Scholar]
- Yaffe D, Saxel O 1977 Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 270:725–727 [DOI] [PubMed] [Google Scholar]
- Katagiri T, Yamaguchi A, Komaki M, Abe E, Takahashi N, Ikeda T, Rosen V, Wozney JM, Fujisawa-Sehara A, Suda T 1994 Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol 127:1755–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi G, Hathaway MR, Dayton WR, White ME 2007 Growth factor messenger ribonucleic acid expression during differentiation of porcine embryonic myogenic cells. J Anim Sci 85:143–150 [DOI] [PubMed] [Google Scholar]
- Kamanga-Sollo E, Pampusch MS, White ME, Dayton WR 2003 Role of insulin-like growth factor binding protein (IGFBP)-3 in TGF-β- and GDF-8 (myostatin)-induced suppression of proliferation in porcine embryonic myogenic cell cultures. J Cell Physiol 197:225–231 [DOI] [PubMed] [Google Scholar]
- Firth SM, Baxter RC 2002 Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev 23:824–854 [DOI] [PubMed] [Google Scholar]
- Xi G, Kamanga-Sollo E, Hathaway MR, Dayton WR, White ME 2006 Effect of constitutive expression of porcine IGFBP-3 on proliferation and differentiation of L6 myogenic cells. Domest Anim Endocrinol 31:35–51 [DOI] [PubMed] [Google Scholar]
- Pampusch MS, Xi G, Kamanga-Sollo E, Loseth KJ, Hathaway MR, Dayton WR, White ME 2005 Production of recombinant porcine IGF-binding protein-5 and its effect on proliferation of porcine embryonic myoblast cultures in the presence and absence of IGF-I and Long-R3-IGF-I. J Endocrinol 185:197–206 [DOI] [PubMed] [Google Scholar]
- Xi G, Kamanga-Sollo E, Pampusch MS, White ME, Hathaway MR, Dayton WR 2004 Effect of recombinant porcine IGFBP-3 on IGF-I and long-R3-IGF-I-stimulated proliferation and differentiation of L6 myogenic cells. J Cell Physiol 200:387–394 [DOI] [PubMed] [Google Scholar]
- Oufattole M, Lin SW, Liu B, Mascarenhas D, Cohen P, Rodgers BD 2006 Ribonucleic acid polymerase II binding subunit 3 (Rpb3), a potential nuclear target of insulin-like growth factor binding protein-3. Endocrinology 147:2138–2146 [DOI] [PubMed] [Google Scholar]
- Xi G, Hathaway MR, White ME, Dayton WR 2007 Localization of insulin-like growth factor (IGFBP)-3 in cultured porcine embryonic myogenic cells before and after TGF-β1 treatment. Domest Anim Endocrinol 33:422–429 [DOI] [PubMed] [Google Scholar]
- Gu Z, Zhang Y, Shi P, Zhang YP, Zhu D, Li H 2004 Comparison of avian myostatin genes. Anim Genet 35:470–472 [DOI] [PubMed] [Google Scholar]
- Ye X, Brown SR, Nones K, Coutinho LL, Dekkers JC, Lamont SJ 2007 Associations of myostatin gene polymorphisms with performance and mortality traits in broiler chickens. Genet Sel Evol 39:73–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Bobbili NK, Paek KS, Jin HJ 2006 Production of a monoclonal anti-myostatin antibody and the effects of in ovo administration of the antibody on posthatch broiler growth and muscle mass. Poult Sci 85:1062–1071 [DOI] [PubMed] [Google Scholar]
- Kim YS, Bobbili NK, Lee YK, Jin HJ, Dunn MA 2007 Production of a polyclonal anti-myostatin antibody and the effects of in ovo administration of the antibody on posthatch broiler growth and muscle mass. Poult Sci 86:1196–1205 [DOI] [PubMed] [Google Scholar]
- Yang W, Wang K, Chen Y, Zhang Y, Huang B, Zhu DH 2003 [Functional characterization of recombinant myostatin and its inhibitory role to chicken muscle development]. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 35:1016–1022 [PubMed] [Google Scholar]
- McFarland DC, Velleman SG, Pesall JE, Liu C 2007 The role of myostatin in chicken (Gallus domesticus) myogenic satellite cell proliferation and differentiation. Gen Comp Endocrinol 151:351–357 [DOI] [PubMed] [Google Scholar]
- Kocamis H, McFarland DC, Killefer J 2001 Temporal expression of growth factor genes during myogenesis of satellite cells derived from the biceps femoris and pectoralis major muscles of the chicken. J Cell Physiol 186:146–152 [DOI] [PubMed] [Google Scholar]
- Sato F, Kurokawa M, Yamauchi N, Hattori MA 2006 Gene silencing of myostatin in differentiation of chicken embryonic myoblasts by small interfering RNA. Am J Physiol Cell Physiol 291:C538—C545 [DOI] [PubMed] [Google Scholar]
- Arnold HH, Braun T 1996 Targeted inactivation of myogenic factor genes reveals their role during mouse myogenesis: a review. Int J Dev Biol 40:345–353 [PubMed] [Google Scholar]
- Dias P, Dilling M, Houghton P 1994 The molecular basis of skeletal muscle differentiation. Semin Diagn Pathol 11:3–14 [PubMed] [Google Scholar]
- Mommsen TP 2001 Paradigms of growth in fish. Comp Biochem Physiol B Biochem Mol Biol 129:207–219 [DOI] [PubMed] [Google Scholar]
- Castillo J, Ammendrup-Johnsen I, Codina M, Navarro I, Gutierrez J 2006 IGF-I and insulin receptor signal transduction in trout muscle cells. Am J Physiol Regul Integr Comp Physiol 290:R1683—R1690 [DOI] [PubMed] [Google Scholar]
- Castillo J, Codina M, Martinez ML, Navarro I, Gutierrez J 2004 Metabolic and mitogenic effects of IGF-I and insulin on muscle cells of rainbow trout. Am J Physiol Regul Integr Comp Physiol 286:R935—R941 [DOI] [PubMed] [Google Scholar]
- Castillo J, Le Bail PY, Paboeuf G, Navarro I, Weil C, Fauconneau B, Gutierrez J 2002 IGF-I binding in primary culture of muscle cells of rainbow trout: changes during in vitro development. Am J Physiol Regul Integr Comp Physiol 283:R647—R652 [DOI] [PubMed] [Google Scholar]
- Koumans JTM, Akster HA, Dulos GJ, Osse JWM 1990 Myosatellite cells of Cyprinus carpio (Teleostei) in vitro—isolation, recognition and differentiation. Cell Tissue Res 261:173–181 [Google Scholar]
- Fauconneau B, Paboeuf G 2000 Effect of fasting and refeeding on in vitro muscle cell proliferation in rainbow trout (Oncorhynchus mykiss). Cell Tissue Res 301:459–463 [DOI] [PubMed] [Google Scholar]
- Fauconneau B, Paboeuf G 2001 Sensitivity of muscle satellite cells to pollutants: an in vitro and in vivo comparative approach. Aquat Toxicol 53:247–263 [DOI] [PubMed] [Google Scholar]
- Arenas MI, Fraile B, De Miguel M, Paniagua R 1995 Intermediate filaments in the testis of the teleost mosquito fish Gambusia affinis holbrooki: a light and electron microscope immunocytochemical study and western blotting analysis. Histochem J 27:329–337 [DOI] [PubMed] [Google Scholar]
- Koumans JTM, Akster HA, Booms GHR, Osse JWM 1993 Growth of carp (Cyprinus carpio) white axial muscle—hyperplasia and hypertrophy in relation to the myonucleus sarcoplasm ratio and the occurrence of different subclasses of myogenic cells. J Fish Biol 43:69–80 [Google Scholar]
- Greenlee AR, Dodson MV, Yablonkareuveni Z, Kersten CA, Cloud JG 1995 In vitro differentiation of myoblasts from skeletal muscle of rainbow trout. J Fish Biol 46:731–747 [Google Scholar]
- Rodgers BD, Weber GM, Isolation and characterization of myostatin complementary deoxyribonucleic acid clones from two commercially important fish: Oreochromis mossambicus and Morone chrysops. Proc 4th Interantional Symposium on Fish Endocrinology, Seattle, WA, 2000 [DOI] [PubMed] [Google Scholar]
- Roberts SB, Goetz FW, Isolation through cloning of a brook trout myostatin homologue. Proc 4th Interantional Symposium on Fish Endocrinology, Seattle, WA, 2000 [Google Scholar]
- Maccatrozzo L, Bargelloni L, Cardazzo B, Rizzo G, Patarnello T 2001 A novel second myostatin gene is present in teleost fish. FEBS Lett 509:36–40 [DOI] [PubMed] [Google Scholar]
- Maccatrozzo L, Bargelloni L, Radaelli G, Mascarello F, Patarnello T 2001 Characterization of the myostatin gene in the gilthead seabream (Sparus aurata): sequence, genomic structure, and expression pattern. Mar Biotechnol (NY) 3:224–230 [DOI] [PubMed] [Google Scholar]
- Ostbye TK, Galloway TF, Nielsen C, Gabestad I, Bardal T, Andersen O 2001 The two myostatin genes of Atlantic salmon (Salmo salar) are expressed in a variety of tissues. Eur J Biochem 268:5249–5257 [DOI] [PubMed] [Google Scholar]
- Rescan PY, Jutel I, Ralliere C 2001 Two myostatin genes are differentially expressed in myotomal muscles of the trout (Oncorhynchus mykiss). J Exp Biol 204:3523–3529 [DOI] [PubMed] [Google Scholar]
- Roberts SB, Goetz FW 2001 Differential skeletal muscle expression of myostatin across teleost species, and the isolation of multiple myostatin isoforms. FEBS Lett 491:212–216 [DOI] [PubMed] [Google Scholar]
- Rodgers BD, Weber GM 2001 Sequence conservation among fish myostatin orthologues and the characterization of two additional cDNA clones from Morone saxatilis and Morone americana. Comp Biochem Physiol B Biochem Mol Biol 129:597–603 [DOI] [PubMed] [Google Scholar]
- Rodgers BD, Weber GM, Sullivan CV, Levine MA 2001 Isolation and characterization of myostatin complementary deoxyribonucleic acid clones from two commercially important fish: Oreochromis mossambicus and Morone chrysops. Endocrinology 142:1412–1418 [DOI] [PubMed] [Google Scholar]
- Biga PR, Roberts SB, Iliev DB, McCauley LA, Moon JS, Collodi P, Goetz FW 2005 The isolation, characterization, and expression of a novel GDF11 gene and a second myostatin form in zebrafish, Danio rerio. Comp Biochem Physiol B Biochem Mol Biol 141:218–230 [DOI] [PubMed] [Google Scholar]
- Kerr T, Roalson EH, Rodgers BD 2005 Phylogenetic analysis of the myostatin gene sub-family and the differential expression of a novel member in zebrafish. Evol Dev 7:390–400 [DOI] [PubMed] [Google Scholar]
- Tellgren A, Berglund AC, Savolainen P, Janis CM, Liberles DA 2004 Myostatin rapid sequence evolution in ruminants predates domestication. Mol Phylogenet Evol 33:782–790 [DOI] [PubMed] [Google Scholar]
- Harrison CA, Gray PC, Fischer WH, Donaldson C, Choe S, Vale W 2004 An activin mutant with disrupted ALK4 binding blocks signaling via type II receptors. J Biol Chem 279:28036–28044 [DOI] [PubMed] [Google Scholar]
- Harrison CA, Gray PC, Koerber SC, Fischer W, Vale W 2003 Identification of a functional binding site for activin on the type I receptor ALK4. J Biol Chem 278:21129–21135 [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S 2000 Molecular evolution and phylogenetics. Oxford, UK: Oxford University Press 17–29. [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH 1998 Zebrafish hox clusters and vertebrate genome evolution. Science 282:1711–1714 [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, Goutel C, Fritz A, Kelsh R, Knapik E, Liao E, Paw B, Ransom D, Singer A, Thomson M, Abduljabbar TS, Yelick P, Beier D, Joly JS, Larhammar D, Rosa F, Westerfield M, Zon LI, Johnson SL, Talbot WS 1998 Vertebrate genome evolution and the zebrafish gene map. Nat Genet 18:345–349 [DOI] [PubMed] [Google Scholar]
- Phillips R, Rab P 2001 Chromosome evolution in the Salmonidae (Pisces): an update. Biol Rev Camb Philos Soc 76:1–25 [DOI] [PubMed] [Google Scholar]
- Garikipati DK, Gahr SA, Rodgers BD 2006 Identification, characterization, and quantitative expression analysis of rainbow trout myostatin-1a and myostatin-1b genes. J Endocrinol 190:879–888 [DOI] [PubMed] [Google Scholar]
- Garikipati DK, Gahr SA, Roalson EH, Rodgers BD 2007 Characterization of rainbow trout myostatin-2 genes (rtMSTN-2a and -2b): genomic organization, differential expression, and pseudogenization. Endocrinology 148:2106–2115 [DOI] [PubMed] [Google Scholar]
- Ostbye TK, Wetten OF, Tooming-Klunderud A, Jakobsen KS, Yafe A, Etzioni S, Moen T, Andersen O 2007 Myostatin (MSTN) gene duplications in Atlantic salmon (Salmo salar): evidence for different selective pressure on teleost MSTN-1 and -2. Gene 403:159–169 [DOI] [PubMed] [Google Scholar]
- Rodgers BD, Roalson EH, Weber GM, Roberts SB, Goetz FW 2007 A proposed nomenclature consensus for the myostatin gene family. Am J Physiol Endocrinol Metab 292:E371—E372 [DOI] [PubMed] [Google Scholar]
- Lo PC, Frasch M 1999 Sequence and expression of myoglianin, a novel Drosophila gene of the TGF-β superfamily. Mech Dev 86:171–175 [DOI] [PubMed] [Google Scholar]
- Kim HW, Mykles DL, Goetz FW, Roberts SB 2004 Characterization of a myostatin-like gene from the bay scallop, Argopecten irradians. Biochim Biophys Acta 1679:174–179 [DOI] [PubMed] [Google Scholar]
- Xing F, Tan X, Zhang PJ, Ma J, Zhang Y, Xu P, Xu Y 2007 Characterization of amphioxus GDF8/11 gene, an archetype of vertebrate MSTN and GDF11. Dev Genes Evol 217:549–554 [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J 1999 Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsune S, Yoshida N, Chen A, Garrett L, Sugiyama F, Takahashi S, Yagami K, Wynshaw-Boris A, Yoshiki A 2003 An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature 423:91–96 [DOI] [PubMed] [Google Scholar]
- Hughes AL 1994 The evolution of functionally novel proteins after gene duplication. Proc Biol Sci 256:119–124 [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS 2000 The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155 [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A 2000 The probability of duplicate gene preservation by subfunctionalization. Genetics 154:459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh JB 1995 How often do duplicated genes evolve new functions? Genetics 139:421–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles DA, Schreiber DR, Govindarajan S, Chamberlin SG, Benner SA 2001 The adaptive evolution database (TAED). Genome Biol 2:RESEARCH0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pie MR, Alvares LE 2006 Evolution of myostatin in vertebrates: is there evidence for positive selection? Mol Phylogenet Evol 41:730–734 [DOI] [PubMed] [Google Scholar]
- Pamilo P, Bianchi NO 1993 Evolution of the Zfx and Zfy genes: rates and interdependence between the genes. Mol Biol Evol 10:271–281 [DOI] [PubMed] [Google Scholar]
- Li WH 1993 Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol 36:96–99 [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R 2002 Codon substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol 19:908–917 [DOI] [PubMed] [Google Scholar]
- Bielawski JP, Dunn KA, Yang Z 2000 Rates of nucleotide substitution and mammalian nuclear gene evolution. Approximate and maximum-likelihood methods lead to different conclusions. Genetics 156:1299–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J 2004 Frequent false detection of positive selection by the likelihood method with branch-site models. Mol Biol Evol 21:1332–1339 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Cadavid NF, Taylor WE, Yarasheski K, Sinha-Hikim I, Ma K, Ezzat S, Shen R, Lalani R, Asa S, Mamita M, Nair G, Arver S, Bhasin S 1998 Organization of the human myostatin gene and expression in healthy men and HIV-infected men with muscle wasting. Proc Natl Acad Sci USA 95:14938–14943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonstegard TS, Rohrer GA, Smith TP 1998 Myostatin maps to porcine chromosome 15 by linkage and physical analyses. Anim Genet 29:19–22 [DOI] [PubMed] [Google Scholar]
- Stratil A, Kopecny M 1999 Genomic organization, sequence and polymorphism of the porcine myostatin (GDF8; MSTN) gene. Anim Genet 30:468–470 [DOI] [PubMed] [Google Scholar]
- Sazanov A, Ewald D, Buitkamp J, Fries R 1999 A molecular marker for the chicken myostatin gene (GDF8) maps to 7p11. Anim Genet 30:388–389 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, Nicaud S, Jaffe D, Fisher S, Lutfalla G, Dossat C, Segurens B, Dasilva C, Salanoubat M, Levy M, Boudet N, Castellano S, Anthouard V, Jubin C, Castelli V, Katinka M, Vacherie B, Biemont C, Skalli Z, Cattolico L, Poulain J, De Berardinis V, Cruaud C, Duprat S, Brottier P, Coutanceau JP, Gouzy J, Parra G, Lardier G, Chapple C, McKernan KJ, McEwan P, Bosak S, Kellis M, Volff JN, Guigo R, Zody MC, Mesirov J, Lindblad-Toh K, Birren B, Nusbaum C, Kahn D, Robinson-Rechavi M, Laudet V, Schachter V, Quetier F, Saurin W, Scarpelli C, Wincker P, Lander ES, Weissenbach J, Roest Crollius H 2004 Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431:946–957 [DOI] [PubMed] [Google Scholar]
- Ellegren H, Smith NG, Webster MT 2003 Mutation rate variation in the mammalian genome. Curr Opin Genet Dev 13:562–568 [DOI] [PubMed] [Google Scholar]
- Ma K, Mallidis C, Artaza J, Taylor W, Gonzalez-Cadavid N, Bhasin S 2001 Characterization of 5′-regulatory region of human myostatin gene: regulation by dexamethasone in vitro. Am J Physiol Endocrinol Metab 281:E1128—E1136 [DOI] [PubMed] [Google Scholar]
- Salerno MS, Thomas M, Forbes D, Watson T, Kambadur R, Sharma M 2004 Molecular analysis of fiber type-specific expression of murine myostatin promoter. Am J Physiol Cell Physiol 287:C1031—C1040 [DOI] [PubMed] [Google Scholar]
- Spiller MP, Kambadur R, Jeanplong F, Thomas M, Martyn JK, Bass JJ, Sharma M 2002 The myostatin gene is a downstream target gene of basic helix-loop-helix transcription factor MyoD. Mol Cell Biol 22:7066–7082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen DL, Unterman TG 2007 Regulation of myostatin expression and myoblast differentiation by FoxO and SMAD transcription factors. Am J Physiol Cell Physiol 292:C188—C199 [DOI] [PubMed] [Google Scholar]
- Lang CH, Silvis C, Nystrom G, Frost RA 2001 Regulation of myostatin by glucocorticoids after thermal injury. FASEB J 15:1807–1809 [DOI] [PubMed] [Google Scholar]
- Ma K, Mallidis C, Bhasin S, Mahabadi V, Artaza J, Gonzalez-Cadavid N, Arias J, Salehian B 2003 Glucocorticoid-induced skeletal muscle atrophy is associated with upregulation of myostatin gene expression. Am J Physiol Endocrinol Metab 285:E363—E371 [DOI] [PubMed] [Google Scholar]
- Vianello S, Brazzoduro L, Dalla Valle L, Belvedere P, Colombo L 2003 Myostatin expression during development and chronic stress in zebrafish (Danio rerio). J Endocrinol 176:47–59 [DOI] [PubMed] [Google Scholar]
- Rodgers BD, Weber GM, Kelley KM, Levine MA 2003 Prolonged fasting and cortisol reduce myostatin mRNA levels in tilapia larvae; short-term fasting elevates. Am J Physiol Regul Integr Comp Physiol 284:R1277—R1286 [DOI] [PubMed] [Google Scholar]
- Guernec A, Chevalier B, Duclos MJ 2004 Nutrient supply enhances both IGF-I and MSTN mRNA levels in chicken skeletal muscle. Domest Anim Endocrinol 26:143–154 [DOI] [PubMed] [Google Scholar]
- Berkes CA, Tapscott SJ 2005 MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol 16:585–595 [DOI] [PubMed] [Google Scholar]
- Wray GA 2007 The evolutionary significance of cis-regulatory mutations. Nat Rev Genet 8:206–216 [DOI] [PubMed] [Google Scholar]
- Pastinen T, Sladek R, Gurd S, Sammak A, Ge B, Lepage P, Lavergne K, Villeneuve A, Gaudin T, Brandstrom H, Beck A, Verner A, Kingsley J, Harmsen E, Labuda D, Morgan K, Vohl MC, Naumova AK, Sinnett D, Hudson TJ 2004 A survey of genetic and epigenetic variation affecting human gene expression. Physiol Genomics 16:184–193 [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG 2004 Evolutionary changes in cis and trans gene regulation. Nature 430:85–88 [DOI] [PubMed] [Google Scholar]
- Thornton K, Long M 2002 Rapid divergence of gene duplicates on the Drosophila melanogaster X chromosome. Mol Biol Evol 19:918–925 [DOI] [PubMed] [Google Scholar]
- Furney SJ, Alba MM, Lopez-Bigas N 2006 Differences in the evolutionary history of disease genes affected by dominant or recessive mutations. BMC Genomics 7:165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SB, Goetz FW 2003 Myostatin protein and RNA transcript levels in adult and developing brook trout. Mol Cell Endocrinol 210:9–20 [DOI] [PubMed] [Google Scholar]
- Roberts SB, McCauley LA, Devlin RH, Goetz FW 2004 Transgenic salmon overexpressing growth hormone exhibit decreased myostatin transcript and protein expression. J Exp Biol 207:3741–3748 [DOI] [PubMed] [Google Scholar]
- Johansen KA, Overturf K 2005 Quantitative expression analysis of genes affecting muscle growth during development of rainbow trout (Oncorhynchus mykiss). Mar Biotechnol (NY) 7:576–587 [DOI] [PubMed] [Google Scholar]
- Helterline DL, Garikipati D, Stenkamp DL, Rodgers BD 2007 Embryonic and tissue-specific regulation of myostatin-1 and -2 gene expression in zebrafish. Gen Comp Endocrinol 151:90–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena VK, Sundaresan NR, Malik F, Ahmed KA, Saxena M, Kumar S, Nandedkar PV, Singh RV 2007 Temporal expression of transforming growth factor-β2 and myostatin mRNA during embryonic myogenesis in Indian broilers. Res Vet Sci 82:50–53 [DOI] [PubMed] [Google Scholar]
- Castelhano-Barbosa EC, Gabriel JE, Alvares LE, Monteiro-Vitorello CB, Coutinho LL 2005 Temporal and spatial expression of the myostatin gene during chicken embryo development. Growth Dev Aging 69:3–12 [PubMed] [Google Scholar]
- Kocamis H, Kirkpatrick-Keller DC, Richter J, Killefer J 1999 The ontogeny of myostatin, follistatin and activin-B mRNA expression during chicken embryonic development. Growth Dev Aging 63:143–150 [PubMed] [Google Scholar]
- Ji S, Losinski RL, Cornelius SG, Frank GR, Willis GM, Gerrard DE, Depreux FF, Spurlock ME 1998 Myostatin expression in porcine tissues: tissue specificity and developmental and postnatal regulation. Am J Physiol 275:R1265—R1273 [DOI] [PubMed] [Google Scholar]
- Sharma M, Kambadur R, Matthews KG, Somers WG, Devlin GP, Conaglen JV, Fowke PJ, Bass JJ 1999 Myostatin, a transforming growth factor-β superfamily member, is expressed in heart muscle and is upregulated in cardiomyocytes after infarct. J Cell Physiol 180:1–9 [DOI] [PubMed] [Google Scholar]
- Kocabas AM, Kucuktas H, Dunham RA, Liu Z 2002 Molecular characterization and differential expression of the myostatin gene in channel catfish (Ictalurus punctatus). Biochim Biophys Acta 1575:99–107 [DOI] [PubMed] [Google Scholar]
- Radaelli G, Rowlerson A, Mascarello F, Patruno M, Funkenstein B 2003 Myostatin precursor is present in several tissues in teleost fish: a comparative immunolocalization study. Cell Tissue Res 311:239–250 [DOI] [PubMed] [Google Scholar]
- Sundaresan NR, Saxena VK, Singh R, Jain P, Singh KP, Anish D, Singh N, Saxena M, Ahmed KA 2008 Expression profile of myostatin mRNA during the embryonic organogenesis of domestic chicken (Gallus gallus domesticus). Res Vet Sci 85:86–91 [DOI] [PubMed] [Google Scholar]
- Kubota K, Sato F, Aramaki S, Soh T, Yamauchi N, Hattori MA 2007 Ubiquitous expression of myostatin in chicken embryonic tissues: its high expression in testis and ovary. Comp Biochem Physiol A Mol Integr Physiol 148:550–555 [DOI] [PubMed] [Google Scholar]
- Helterline DLI, Garikipati D, Stenkamp DL, Rodgers BD 2007 Embryonic and tissue-specific regulation of myostatin-1 and -2 gene expression in zebrafish. Gen Comp Endocrinol 151:90–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg RR, Bally-Cuif L, Lee SE, Gong Z, Ni X, Hew CL, Peng C 1999 Cloning of zebrafish activin type IIB receptor (ActRIIB) cDNA and mRNA expression of ActRIIB in embryos and adult tissues. Mol Cell Endocrinol 153:169–181 [DOI] [PubMed] [Google Scholar]
- Nohno T, Noji S, Koyama E, Myokai F, Ohuchi H, Nishikawa K, Sumitomo S, Taniguchi S, Saito T 1993 Expression patterns of the activin receptor IIA and IIB genes during chick limb development. Prog Clin Biol Res 383B:705–714 [PubMed] [Google Scholar]
- Kos K, Coulombe JN 1997 Activin receptor mRNA expression by neurons of the avian ciliary ganglion. J Neurobiol 32:33–44 [DOI] [PubMed] [Google Scholar]
- Kondo M, Semba K, Shiokawa K, Yamamoto T 1996 Molecular cloning of Xenopus activin type I receptor and the analysis of its expression during embryogenesis. Biochem Biophys Res Commun 218:549–555 [DOI] [PubMed] [Google Scholar]
- Ng L, Pathak SD, Kuan C, Lau C, Dong H, Sodt A, Dang C, Avants B, Yushkevich P, Gee JC, Haynor D, Lein E, Jones A, Hawrylycz M 2007 Neuroinformatics for genome-wide 3D gene expression mapping in the mouse brain. IEEE/ACM Trans Comput Biol Bioinform 4:382–393 [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR 2007 Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176 [DOI] [PubMed] [Google Scholar]
- Wu HH, Ivkovic S, Murray RC, Jaramillo S, Lyons KM, Johnson JE, Calof AL 2003 Autoregulation of neurogenesis by GDF11. Neuron 37:197–207 [DOI] [PubMed] [Google Scholar]
- McKoy G, Bicknell KA, Patel K, Brooks G 2007 Developmental expression of myostatin in cardiomyocytes and its effect on foetal and neonatal rat cardiomyocyte proliferation. Cardiovasc Res 74:304–312 [DOI] [PubMed] [Google Scholar]
- Callis TE, Chen JF, Wang DZ 2007 MicroRNAs in skeletal and cardiac muscle development. DNA Cell Biol 26:219–225 [DOI] [PubMed] [Google Scholar]
- Shyu KG, Lu MJ, Wang BW, Sun HY, Chang H 2006 Myostatin expression in ventricular myocardium in a rat model of volume-overload heart failure. Eur J Clin Invest 36:713–719 [DOI] [PubMed] [Google Scholar]
- Shyu KG, Ko WH, Yang WS, Wang BW, Kuan P 2005 Insulin-like growth factor-1 mediates stretch-induced upregulation of myostatin expression in neonatal rat cardiomyocytes. Cardiovasc Res 68:405–414 [DOI] [PubMed] [Google Scholar]
- Kim HS, Liang L, Dean RG, Hausman DB, Hartzell DL, Baile CA 2001 Inhibition of preadipocyte differentiation by myostatin treatment in 3T3–L1 cultures. Biochem Biophys Res Commun 281:902–906 [DOI] [PubMed] [Google Scholar]
- Artaza JN, Bhasin S, Magee TR, Reisz-Porszasz S, Shen R, Groome NP, Fareez MM, Gonzalez-Cadavid NF 2005 Myostatin inhibits myogenesis and promotes adipogenesis in C3H 10T(1/2) mesenchymal multipotent cells. Endocrinology 146:3547–3557 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M,et al. 2001 Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA,et al. 2001 The sequence of the human genome. Science 291:1304–1351 [DOI] [PubMed] [Google Scholar]
- Shabalina SA, Spiridonov NA 2004 The mammalian transcriptome and the function of non-coding DNA sequences. Genome Biol 5:105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui J, Bindereif A 2005 Alternative pre-mRNA splicing in the human system: unexpected role of repetitive sequences as regulatory elements. Biol Chem 386:1265–1271 [DOI] [PubMed] [Google Scholar]
- Hui J, Reither G, Bindereif A 2003 Novel functional role of CA repeats and hnRNP L in RNA stability. RNA 9:931–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckanovich RJ, Darnell RB 1997 The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol Cell Biol 17:3194–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckanovich RJ, Yang YY, Darnell RB 1996 The onconeural antigen Nova-1 is a neuron-specific RNA-binding protein, the activity of which is inhibited by paraneoplastic antibodies. J Neurosci 16:1114–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckanovich RJ, Posner JB, Darnell RB 1993 Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron 11:657–672 [DOI] [PubMed] [Google Scholar]
- Dredge BK, Darnell RB 2003 Nova regulates GABA(A) receptor γ2 alternative splicing via a distal downstream UCAU-rich intronic splicing enhancer. Mol Cell Biol 23:4687–4700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB 2003 CLIP identifies Nova-regulated RNA networks in the brain. Science 302:1212–1215 [DOI] [PubMed] [Google Scholar]
- Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, Darnell RB 2000 Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 25:359–371 [DOI] [PubMed] [Google Scholar]
- Dredge BK, Stefani G, Engelhard CC, Darnell RB 2005 Nova autoregulation reveals dual functions in neuronal splicing. EMBO J 24:1608–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas E, Stone RT, Keele JW, Shackelford SD, Kappes SM, Koohmaraie M 2001 A comprehensive search for quantitative trait loci affecting growth and carcass composition of cattle segregating alternative forms of the myostatin gene. J Anim Sci 79:854–860 [DOI] [PubMed] [Google Scholar]
- Casas E, Bennett GL, Smith TP, Cundiff LV 2004 Association of myostatin on early calf mortality, growth, and carcass composition traits in crossbred cattle. J Anim Sci 82:2913–2918 [DOI] [PubMed] [Google Scholar]
- Shahin KA, Berg RT 1985 Fat growth and partitioning among the depots in double muscled and normal cattle. Can J Anim Sci 65:295–306 [Google Scholar]
- Wang Z, Heshka S, Gallagher D, Boozer CN, Kotler DP, Heymsfield SB 2000 Resting energy expenditure-fat-free mass relationship: new insights provided by body composition modeling. Am J Physiol Endocrinol Metab 279:E539—E545 [DOI] [PubMed] [Google Scholar]
- Hamilton MT, Hamilton DG, Zderic TW 2007 Role of low energy expenditure and sitting in obesity, metabolic syndrome, type 2 diabetes, and cardiovascular disease. Diabetes 56:2655–2667 [DOI] [PubMed] [Google Scholar]
- Lin J, Arnold HB, Della-Fera MA, Azain MJ, Hartzell DL, Baile CA 2002 Myostatin knockout in mice increases myogenesis and decreases adipogenesis. Biochem Biophys Res Commun 291:701–706 [DOI] [PubMed] [Google Scholar]
- Stolz LE, Li D, Qadri A, Jalenak M, Klaman LD, Tobin JF 2008 Administration of myostatin does not alter fat mass in adult mice. Diabetes Obes Metab 10:135–142 [DOI] [PubMed] [Google Scholar]
- Hirai S, Matsumoto H, Hino N, Kawachi H, Matsui T, Yano H 2007 Myostatin inhibits differentiation of bovine preadipocyte. Domest Anim Endocrinol 32:1–14 [DOI] [PubMed] [Google Scholar]
- Guo W, Flanagan J, Jasuja R, Kirkland J, Jiang L, Bhasin S 2008 The effects of myostatin on adipogenic differentiation of human bone marrow-derived mesenchymal stem cells are mediated through cross-communication between Smad3 and Wnt/β-catenin signaling pathways. J Biol Chem 283:9136–9145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman BJ, Streeper RS, Farese Jr RV, Yamamoto KR 2006 Myostatin modulates adipogenesis to generate adipocytes with favorable metabolic effects. Proc Natl Acad Sci USA 103:15675–15680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artaza JN, Reisz-Porszasz S, Dow JS, Kloner RA, Tsao J, Bhasin S, Gonzalez-Cadavid NF 2007 Alterations in myostatin expression are associated with changes in cardiac left ventricular mass but not ejection fraction in the mouse. J Endocrinol 194:63–76 [DOI] [PubMed] [Google Scholar]
- Cook SA, Matsui T, Li L, Rosenzweig A 2002 Transcriptional effects of chronic Akt activation in the heart. J Biol Chem 277:22528–22533 [DOI] [PubMed] [Google Scholar]
- Gaussin V, Depre C 2005 Myostatin, the cardiac chalone of insulin-like growth factor-1. Cardiovasc Res 68:347–349 [DOI] [PubMed] [Google Scholar]
- Matsakas A, Bozzo C, Cacciani N, Caliaro F, Reggiani C, Mascarello F, Patruno M 2006 Effect of swimming on myostatin expression in white and red gastrocnemius muscle and in cardiac muscle of rats. Exp Physiol 91:983–994 [DOI] [PubMed] [Google Scholar]
- Modesti PA, Vanni S, Bertolozzi I, Cecioni I, Lumachi C, Perna AM, Boddi M, Gensini GF 2004 Different growth factor activation in the right and left ventricles in experimental volume overload. Hypertension 43:101–108 [DOI] [PubMed] [Google Scholar]
- Carlson CJ, Booth FW, Gordon SE 1999 Skeletal muscle myostatin mRNA expression is fiber-type specific and increases during hindlimb unloading. Am J Physiol 277:R601—R606 [DOI] [PubMed] [Google Scholar]
- Lalani R, Bhasin S, Byhower F, Tarnuzzer R, Grant M, Shen R, Asa S, Ezzat S, Gonzalez-Cadavid NF 2000 Myostatin and insulin-like growth factor-I and -II expression in the muscle of rats exposed to the microgravity environment of the NeuroLab space shuttle flight. J Endocrinol 167:417–428 [DOI] [PubMed] [Google Scholar]
- Wehling M, Cai B, Tidball JG 2000 Modulation of myostatin expression during modified muscle use. FASEB J 14:103–110 [DOI] [PubMed] [Google Scholar]
- Zhang P, Chen X, Fan M 2007 Signaling mechanisms involved in disuse muscle atrophy. Med Hypotheses 69:310–321 [DOI] [PubMed] [Google Scholar]
- Cohn RD, Liang HY, Shetty R, Abraham T, Wagner KR 2007 Myostatin does not regulate cardiac hypertrophy or fibrosis. Neuromuscul Disord 17:290–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima M, Toyono T, Akamine A, Joyner A 1999 Expression of growth/differentiation factor 11, a new member of the BMP/TGFβ superfamily during mouse embryogenesis. Mech Dev 80:185–189 [DOI] [PubMed] [Google Scholar]
- Kim J, Wu HH, Lander AD, Lyons KM, Matzuk MM, Calof AL 2005 GDF11 controls the timing of progenitor cell competence in developing retina. Science 308:1927–1930 [DOI] [PubMed] [Google Scholar]
- Evans JJ, Shoemaker CA, Klesius PH 2000 In vivo and in vitro effects of benzothiazole on sheepshead minnow (Cyprinodon variegatus). Mar Environ Res 50:257–261 [DOI] [PubMed] [Google Scholar]
- Sivron T, Eitan S, Schreyer DJ, Schwartz M 1993 Astrocytes play a major role in the control of neuronal proliferation in vitro. Brain Res 629:199–208 [DOI] [PubMed] [Google Scholar]
- Otteson DC, Cirenza PF, Hitchcock PF 2002 Persistent neurogenesis in the teleost retina: evidence for regulation by the growth-hormone/insulin-like growth factor-I axis. Mech Dev 117:137–149 [DOI] [PubMed] [Google Scholar]
- Danks AM, Kim P, Wang Z, Meyer RL 1994 Imaging of individual normal and regenerating optic fibers in the brain of living adult goldfish. J Comp Neurol 345:253–266 [DOI] [PubMed] [Google Scholar]
- Easter Jr SS, Rusoff AC, Kish PE 1981 The growth and organization of the optic nerve and tract in juvenile and adult goldfish. J Neurosci 1:793–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RL, Kageyama GH 1999 Large-scale synaptic errors during map formation by regeneration optic axons in the goldfish. J Comp Neurol 409:299–312 [PubMed] [Google Scholar]
- Whittemore LA, Song K, Li X, Aghajanian J, Davies M, Girgenrath S, Hill JJ, Jalenak M, Kelley P, Knight A, Maylor R, O'Hara D, Pearson A, Quazi A, Ryerson S, Tan XY, Tomkinson KN, Veldman GM, Widom A, Wright JF, Wudyka S, Zhao L, Wolfman NM 2003 Inhibition of myostatin in adult mice increases skeletal muscle mass and strength. Biochem Biophys Res Commun 300:965–971 [DOI] [PubMed] [Google Scholar]
- Liang YC, Yeh JY, Ou BR 2007 Effect of maternal myostatin antibody on offspring growth performance and body composition in mice. J Exp Biol 210:477–483 [DOI] [PubMed] [Google Scholar]
- Bogdanovich S, Krag TO, Barton ER, Morris LD, Whittemore LA, Ahima RS, Khurana TS 2002 Functional improvement of dystrophic muscle by myostatin blockade. Nature 420:418–421 [DOI] [PubMed] [Google Scholar]
- Bogdanovich S, Perkins KJ, Krag TO, Whittemore LA, Khurana TS 2005 Myostatin propeptide-mediated amelioration of dystrophic pathophysiology. FASEB J 19:543–549 [DOI] [PubMed] [Google Scholar]
- Benabdallah BF, Bouchentouf M, Tremblay JP 2005 Improved success of myoblast transplantation in mdx mice by blocking the myostatin signal. Transplantation 79:1696–1702 [DOI] [PubMed] [Google Scholar]
- Nakatani M, Takehara Y, Sugino H, Matsumoto M, Hashimoto O, Hasegawa Y, Murakami T, Uezumi A, Takeda S, Noji S, Sunada Y, Tsuchida K 2008 Transgenic expression of a myostatin inhibitor derived from follistatin increases skeletal muscle mass and ameliorates dystrophic pathology in mdx mice. FASEB J 22:477–487 [DOI] [PubMed] [Google Scholar]
- Maccarthy T, Bergman A 2007 The limits of subfunctionalization. BMC Evol Biol 7:213 [DOI] [PMC free article] [PubMed] [Google Scholar]