Abstract
Huntington’s disease (HD) is caused by progressive loss of striatal medium spiny neurons (MSN). The molecular trigger of HD is a polyglutamine expansion in the Huntingtin protein (Htt). The mutant Htt protein forms insoluble nuclear aggregates which have been proposed to play a key role in causing neuronal cell death in HD. Other lines of investigation suggest that expression of mutant Htt facilitates activity of the NR2B subtype of NMDA receptors and the type 1 inositol 1,4,5-trisphosphate receptors (InsP3R1), and that disturbed calcium (Ca2+) signaling causes apoptosis of MSNs in HD. The YAC128 transgenic HD mouse model expresses the full-length human Htt protein with 120Q CAG repeat expansion and displays age-dependent loss of striatal neurons as seen in human HD brain. In contrast, the shortstop mice express an amino-terminal fragment of the mutant Htt protein (exons 1 and 2) and display no behavioral abnormalities or striatal neurodegeneration despite widespread formation of neuronal inclusions. Here we compared Ca2+ signals in primary MSN neuronal cultures derived from YAC128 and shortstop mice to their wild type non-transgenic littermates. Repetitive application of glutamate results in supranormal Ca2+ responses in YAC128 MSNs, but not in shortstop MSNs. In addition, while currents mediated by the NR2B subtype of NMDA receptors were increased in YAC128 MSNs, currents in SS MSNs were found to be similar to WT. Furthermore, YAC128 MSNs were sensitized to glutamate-induced apoptosis. Consistent with these findings, we found that application of glutamate induced rapid loss of mitochondrial membrane potential in YAC128 MSNs. In contrast, SS MSNs do not show increased cell death post glutamate treatment nor cause loss of mitochondrial membrane potential. Glutamate-induced loss of mitochondrial membrane potential in YAC128 MSNs could be prevented by inhibitors of NR2B NMDA receptors and mGluR1/5 receptors. Our results are consistent with the hypothesis that disturbed neuronal Ca2+ signaling plays a significant role in the degeneration of MSN containing full length mutant Httexp. Furthermore, the results obtained with neurons from shortstop mice provide additional evidence that not all fragments of mutant Httexp are toxic to neurons.
Keywords: calcium, NMDAR, Huntington’s, disease, transgenic mouse, mitochondria, apoptosis, excitotoxicity
Introduction
Huntington’s disease (HD) has onset usually between 35 and 50 years with chorea and psychiatric disturbances and gradual but inexorable intellectual decline to death after 15–20 years (Vonsattel and DiFiglia, 1998). Neuropathological analysis reveals selective and progressive neuronal loss in the striatum (Vonsattel and DiFiglia, 1998), particularly affecting the GABAergic medium spiny striatal neurons (MSN). At the molecular level, the cause of HD is a polyglutamine (polyQ) expansion (exp) in the amino-terminus of huntingtin (Htt), a 350 kDa ubiquitously expressed cytoplasmic protein (The. et al., 1993). The cellular mechanisms that link the Httexp mutation with the disease are under intense investigation (Tobin and Signer, 2000). The mutant Httexp protein forms insoluble nuclear aggregates which have been proposed to play a key role in causing neuronal cell death in HD (Cooper et al., 1998; Ross, 2002). A number of laboratories are focused on screening for compounds which are able to inhibit the formation of Htt aggregates (Heiser et al., 2002; Apostol et al., 2003; Zhang et al., 2005; Hockly et al., 2006). A decrease in nuclear inclusion formation has been interpreted as a positive outcome in preclinical therapeutic trials with HD mouse models (Smith et al., 2001; Ferrante et al., 2002; Tanaka et al., 2004). However, more recent evidence suggested that Htt-formed nuclear inclusions may not be toxic and may even protect MSN neurons from cell death (Kuemmerle et al., 1999; Arrasate et al., 2004).
Several lines of evidence indicate that glutamate-mediated excitotoxicity plays a role in neurodegeneration of HD MSNs (DiFiglia, 1990). Striatal injection of kainic acid induced death of MSNs and yielded one of the first animal models of HD (Coyle and Schwarcz, 1976; McGeer and McGeer, 1976). More direct evidence for an involvement of NMDAR was obtained when HD-like lesions were observed following striatal injection of the NMDAR agonist quinolinic acid (Beal et al., 1986; Hantraye et al., 1990; Beal et al., 1991). It has been reported that Httexp facilitates activity of the NR2B subtype of NMDA receptors (Chen et al., 1999; Sun et al., 2001; Zeron et al., 2002; Zeron et al., 2004; Fan et al., 2007) and the type 1 inositol 1,4,5-trisphosphate receptors (InsP3R1) (Tang et al., 2003). Moreover, elevated Ca2+ signals have been directly linked to the cell death of striatal MSNs cultured from HD mouse models (Zeron et al., 2004; Tang et al., 2005; Shehadeh et al., 2006). These studies suggested that excitotoxicity and abnormal neuronal Ca2+ signaling plays an important role in HD pathogenesis (Bezprozvanny and Hayden, 2004).
A number of transgenic HD mouse models have been generated which reproduce some features of the disease (Menalled and Chesselet, 2002; Rubinsztein, 2002). However, none of the previously generated HD mouse models reproduced selective MSN degeneration, which is a hallmark of HD. Recently, a yeast artificial chromosome (YAC) mouse model of HD has been generated (YAC128) (Slow et al., 2003). In this model, the full-length human Htt protein with 120Q expansion is expressed under the control of the endogenous promoter and regulatory elements. The early-onset motor deficit, and striatal neuronal loss observed in the YAC128 mouse model accurately recapitulate the progression of HD (Slow et al., 2003). Thus, the YAC128 mouse model is ideal for understanding cellular mechanisms that lead to neurodegeneration in HD, as well as for validating potential therapeutic agents. Our laboratory have been extensively using the YAC128 HD mouse model in studies of HD pathogenesis and in the testing of potential HD therapeutic agents (Tang et al., 2005; Wu et al., 2006; Tang et al., 2007).
In the process of generating the YAC128 HD mouse model, a mouse expressing a short fragment (exons 1 and 2 of 67) of human Htt with 120 CAG repeat expansion was serendipitously established (shortstop mouse) (Slow et al., 2005). Analysis of the shortstop mice revealed widespread Htt nuclear inclusions. However, despite widespread inclusion burden, these mice do not manifest a behavioral, HD-related phenotype as assessed by rotarod or decreases in brain weight, striatal volume, and striatal neuronal count at 12 or 18 months of age (Slow et al., 2005). The shortstop mouse therefore illustrated in vivo the inability of Htt inclusions to have a toxic effect over the lifespan of an organism, clearly demonstrating that Htt nuclear inclusions are not toxic.
Here we took an advantage of the YAC128 and shortstop HD mouse models to further evaluate the importance of Ca2+ signaling in HD pathogenesis. The results obtained are consistent with the hypothesis that disturbed neuronal Ca2+ signaling plays a significant role in the degeneration of MSNs containing full length mutant Httexp. Furthermore, the results derived from our analysis of SS MSNs support the premise that not all fragments of mutant Httexp are toxic to neurons.
Materials and Methods
Materials
Propidium iodide (PI), Fura-2 acetoxymethyl ester (Fura-2 AM) and tetramethylrhodamine methyl ester (TMRM+) were obtained from Molecular Probes. Glutamate, NMDA, CNQX,TTX, MPEP, CPCCOEt, (+)-MK801 maleate, and ifenprodil were purchased from Tocris. Cell culture reagents were all from Life Technologies. All other reagents were from Sigma.
Primary neuronal cultures
Generation and characterization of YAC128 and shortstop mice has been previously described (Slow et al., 2003; Slow et al., 2005). In our experiments heterozygous male YAC128 (line 53) or shortstop (SS) mice were crossed with the wild-type (WT) female mice and resulting litters were collected at postnatal days 1–2. The pups were genotyped by PCR with primers specific for exons 44 and 45 of human Htt gene for YAC128 mice (Tang et al., 2005), and with primers specific for intron 1 and exon 2 of human Htt gene for the shortstop mice. The wild type littermates of YAC128 are indicated as WT. The wild type littermates of shortstop mice are indicated as ss-WT. The medium spiny neuronal (MSN) cultures of WT, YAC128, shortstop and ss-WT mice were established from P1-P2 pups and maintained in culture according to published procedures (Mao and Wang, 2001; Tang et al., 2005).
Ca2+ Imaging Experiments
Fura-2 Ca2+ imaging experiments with 13–14 DIV MSN cultures were performed as previously described (Tang et al., 2005; Tang et al., 2007). Briefly, the cells were maintained in artificial cerebrospinal fluid (aCSF) (140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 10 mM Hepes, pH 7.3) at 37°C during measurements (PH1 heater, Warner Instruments, Hamden, CT). Fura-2 340/380 ratio images were collected every 6 sec for the duration of the experiment using a DeltaRAM illuminator, an IC-300 camera, and IMAGEMASTER PRO software (all from PTI, South Brunswick, NJ). Baseline (1–3 min) measurements were obtained before first pulse of glutamate. The 20 µM glutamate solution was dissolved in aCSF and 1-min pulses of 37°C glutamate solution (SH-27B in-line solution heater, Warner Instruments) were applied by using a valve controller (VC-6, Warner Instruments) driven by a square-pulse electrical waveform generator (Model 148A, Wavetek, San Diego).
TUNEL Staining Experiments
The TUNEL staining experiments have been performed as previously described (Tang et al., 2005; Wu et al., 2006; Tang et al., 2007). Briefly, 13–14 DIV MSN were exposed for 8 h to a range of glutamate concentrations (0, 100 µM, 250 µM) added to the culture medium. During exposure to glutamate, the cells were maintained in a cell culture incubator (humidified 5% CO2, 37°C). Immediately after exposure to glutamate, neurons were fixed for 30 min in 4% paraformaldehyde plus 4% sucrose in PBS (pH 7.4), permeabilized for 5 min in 0.25% Triton X-100, treated with RNase for 30 min, then stained by using the DeadEnd Fluorometric TUNEL System (Promega) according to the manufacturer's instructions. Nuclei were counterstained with 5 µM propidium iodide (PI). Coverslips were extensively washed with PBS and mounted in Mowiol 4–88 (Polysciences). FITC- and PI-fluorescent images were collected with an Olympus IX70 microscope with x40 objectives, using a Cascade:650 camera (Roper Scientific) and METAFLUOR software (Universal Imaging, Downingtown, PA). Four to six randomly chosen microscopic fields containing 100–200 MSN were captured, and the numbers of TUNEL-positive neuronal nuclei were calculated as a fraction of PI-positive neuronal nuclei in each microscopic field. Nuclei of glial cells, identified by large size and weak PI staining, were not counted in the analysis.
Electrophysiology for NMDA currents
Whole-cell patch-clamp recordings of currents in cultured DIV9–10 MSN were performed according to published procedures (Zeron et al., 2002; Fan et al., 2007). The MSNs were easily distinguished from the large-sized cells based on morphological identification and membrane capacitance ranging from 2–10 pF. A multibarrel perfusion system (“sewer pipe”) was employed to achieve a rapid exchange of extracellular solutions during recordings. All drugs were prepared according to the specifications of the manufacturers and applied with a gravity-fed "sewer pipe" capillary array. Whole-cell currents were recorded using Axopatch 200B amplifiers (Axon Instruments). Data were filtered at 2 kHz and digitized at 5 Hz using a Digidata 1200 DAC unit (Axon Instruments). The online acquisition was done using pCLAMP software (Version 8, Axon Instruments). For NMDA current recordings the MSN were clamped at VH = −60 mV membrane potential. The extracellular solution contained 140 mM NaCl, 5 mM KCl, 2.0 mM CaCl2, 10 mM HEPES (pH 7.4; 320 to ~330 mOsm). The pipette solution contained 135 mM CsMeSO4, 10 mM HEPES, 5 mM 1,2-bis(2-aminophenoxy)ethane N,N,N′,N′-tetraacetic acid, 3 mM MgATP, 1 mM MgCl2, 0.3 mM GTP-tris. In all experiments, 50 µM glycine was added to both control and NMDA-containing extracellular solutions. 10 µM CNQX and 0.1 µM TTX were added to the extracellular recording solution immediately before each experiment to block AMPA/kainate-type glutamate receptors and voltage-gated sodium currents, respectively.
Mitochondrial membrane potential (∆ψm) measurement
The procedure for mitochondrial membrane potential measurements was adapted from (Oliveira et al., 2006). Briefly, DIV13–14 MSN were loaded with 25 nM TMRM in Tyrode solution (150 mM NaCl, 4 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 10 mM glucose, 10 mM Hepes, pH 7.4) at room temperate for 30 min, and during the whole image process, 25 nM TMRM present in all the solution. Cells were stimulated with 100 µM glutamate at room temperature. For the inhibitor experiments, 20 µM ifenprodil, 20 µM MPEM, 50 µM CPCCOEt and /or 10 µM (+) MK801 were added at the loading buffer and image buffer as indicated in the text. The images were collected by confocal Zeiss LS800 microscope using 543 nm laser line for TMRM+ excitation (emission, > 560 nm). The mitochondria puncta within neuronal cell soma was chosen randomly as a region of interest (ROI). The initial TMRM+ fluorescence intensity in the ROI was defined as F0. The TMRM+ fluorescence intensity in the same ROI was measured each 10 sec as a function of time, yielding F(t). To compare results from different experiments F(t) was normalized to F0 for each puncta (F/F0) and the data from multiple cells were averaged together. For quantitative analysis we measured the time for F(t) to reach 0.8F0, defined as t0.8. For the puncta that did not reach 0.8F0 by the end of the experiment, then total recording time (typically 800 sec) was used as t0.8.
Statistical analysis
Statistical comparison of results obtained in experiments with YAC128 and WT cultures and shortstop and ss-WT cultures was performed by Student’s unpaired t-test. The p values are indicated in the text and figure legends as appropriate. The differences between control and experimental groups were determined to be non-significant in cases where p > 0.05.
Results
Ca2+ signaling is destabilized in YAC128 MSNs but not in shortstop MSNs
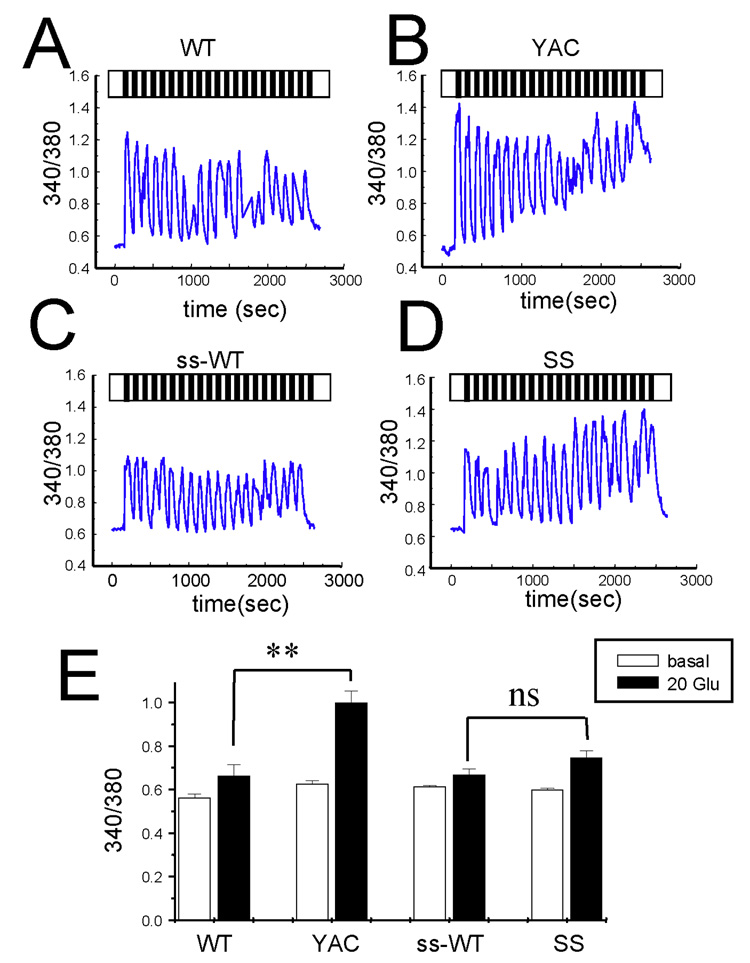
In the first series of experiments we compared Ca2+ responses induced by glutamate application to YAC128 and shortstop (ss) MSNs at 13–14 DIV. The corresponding wild type littermate cultures (WT and ss-WT) were used as an internal control in these experiments. In our studies we adapted an experimental paradigm that we used previously in studies of Ca2+ signals in YAC128 MSNs (Tang et al., 2005; Tang et al., 2007). To mimic physiological conditions more closely, we applied repetitive pulses of 20 µM glutamate 1 min in duration, followed by a 1-min washout. The intracellular Ca2+ concentration in these experiments was continuously monitored by Fura-2 imaging, and the data were presented as average 340/380 ratios for each group of cells (Figs 1A–1D). On average, the basal Ca2+ levels before glutamate application were not significantly different between all four cultures (Fig 1E). Consistent with our previous findings (Tang et al., 2005; Tang et al., 2007), repetitive pulses of glutamate caused large elevation of Ca2+ levels in the YAC128 MSNs (Fig 1B), but much smaller Ca2+ increases in the littermate WT MSNs cultures (Fig 1A). In contrast, glutamate evoked similar Ca2+ responses in shortstop MSNs (Fig 1D) and ss-WT littermate MSNs cultures (Fig 1C). On average, Ca2+ levels after 20 pulses of glutamate were not significantly different between shortstop MSNs and their WT littermates (ss-WT), but were statistically higher in YAC128 MSNs (p < 0.01) when compared with their WT littermates (Fig 1E). This evidence demonstrates that Ca2+ signaling is destabilized in YAC128 MSNs but not in shortstop MSNs.
Figure 1. Glutamate-induced Ca2+ signals in YAC128 and shortstop MSN.
(A–D) Repetitive application of 20 µM glutamate induces Ca2+ signals in MSN from the WT (A), YAC128 (B), ss-WT (C) and shortstop (D)) mice. Cytosolic Ca2+ levels are presented as a 340/380 Fura-2 ratio. The traces shown are representative cell traces. (E) The average 340/380 values (mean ± SE) before and after 20 pulses of glutamate are shown for WT (n = 56), YAC128 (n = 79), ss-WT (n=100) and shortstop (n = 100) MSN as indicated. After 20 pulses of glutamate the Ca2+ levels in YAC128 MSN are significantly (**, p < 0.01) higher than those in WT, but shortstop MSN do not have significant difference with their littermate ss-WT MSN.
YAC128 MSN, but not shortstop MSN are sensitized to glutamate-induced apoptosis
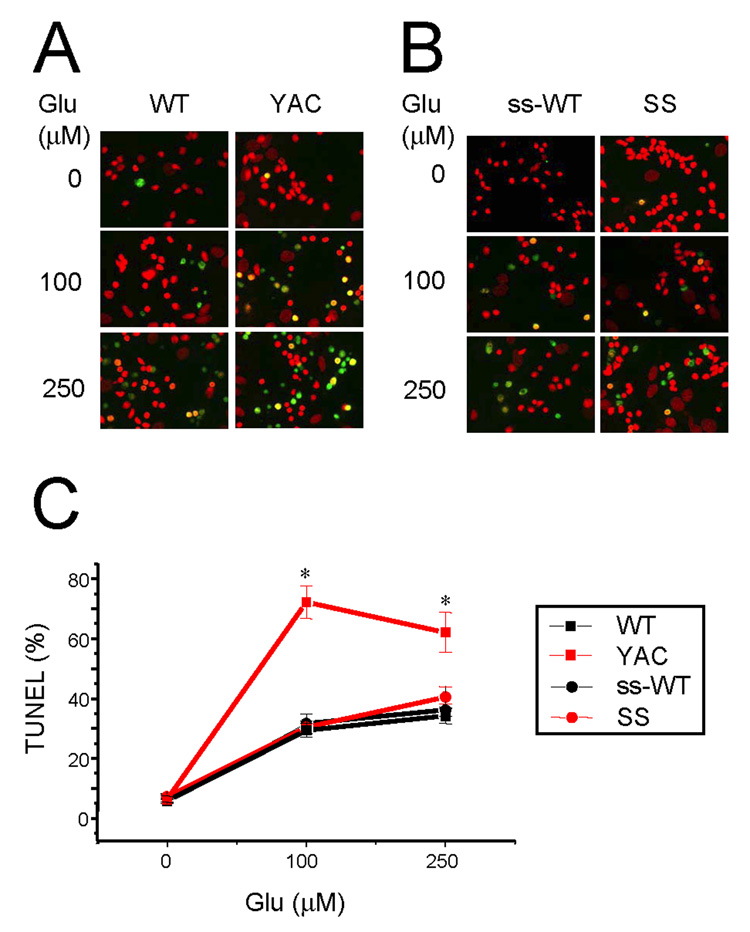
In the next series of experiments we utilized a previously developed “in vitro HD” assay (Tang et al., 2005; Wu et al., 2006; Tang et al., 2007) to compare sensitivity of YAC128 and shortstop MSNs to glutamate-induced apoptosis. As in the Ca2+ imaging experiments described above, wild type littermate cultures (WT and ss-WT) were used as an internal control in these experiments. The 13–14 DIV MSN cultures were challenged by an 8 hour application of glutamate (0, 100 µM and 250 µM) as previously described (Tang et al., 2005; Wu et al., 2006; Tang et al., 2007). After exposure to glutamate, the MSNs were fixed, permeabilized, and scored for apoptotic cell death by TUNEL staining with propidium iodide counterstaining (Fig 2). We determined that in basal conditions (no glutamate added), approximately 10% of MSNs in all four experimental groups were apoptotic (TUNEL-positive) (Fig 2A and 2B, first row). As previously described (Tang et al., 2005; Wu et al., 2006), addition of 100 µM or 250 µM glutamate increased apoptotic cell death to 60–70% for YAC128 MSNs, but only to 30–40% for WT MSNs (Fig 2A, second and third rows). In contrast, addition of 100 µM or 250 µM glutamate to SS MSNs resulted in a similar level of apoptotic cell death as observed for wild type cultures (Fig 2B, second and third rows). On average, the fraction of apoptotic cells following 100 µM or 250 µM glutamate stimulation was not significantly different between shortstop MSNs and their ss-WT littermates. However, it was much higher in YAC128 MSNs (p < 0.05) when compared with their WT littermates (Fig 2C). Thus, we concluded that YAC128 MSNs, but not shortstop MSNs are sensitized to glutamate-induced apoptosis. This conclusion is consistent with the previous in vitro and in vivo analysis of NMDA-induced excitotoxicity in YAC128 and shortstop MSNs (Slow et al., 2005; Graham et al., 2006b). Importantly, both YAC128 and SS MSN express endogenous mouse Huntingtin, and therefore observed differences in apoptotic cell death (Fig 2) must be due to different nature of transgenes expressed in these cells (full-length vs truncated Htt-128Q).
Figure 2. Glutamate-induced apoptosis of YAC128 and shortstop MSN.
(A) DIV13–14 MSN from WT, YAC128, ss-WT and shortstop mice were exposed to a range of glutamate concentrations for 8 h, fixed, permeabilized, and analyzed by TUNEL staining (green) and PI counterstaining (red). (B) The fraction of TUNEL-positive MSN nuclei was determined as shown in panel A and plotted against glutamate concentration for WT (black square), YAC128 (red square), ss-WT (black circles), and shortstop (red circles). At each glutamate concentration, the data are shown as mean ± SE for each genotype (n = 4–6 microscopic fields, 100–200 MSN per field). At 100 and 250 µM glutamate, the fraction of TUNEL-positive MSN is significantly (*, p < 0.05) higher for YAC128 than for WT, ss-WT or shortstop. Similar results were obtained with 3 independent MSN preparations.
Activity of NR2B NMDA receptors is enhanced in YAC128 MSNs, but not in shortstop MSNs
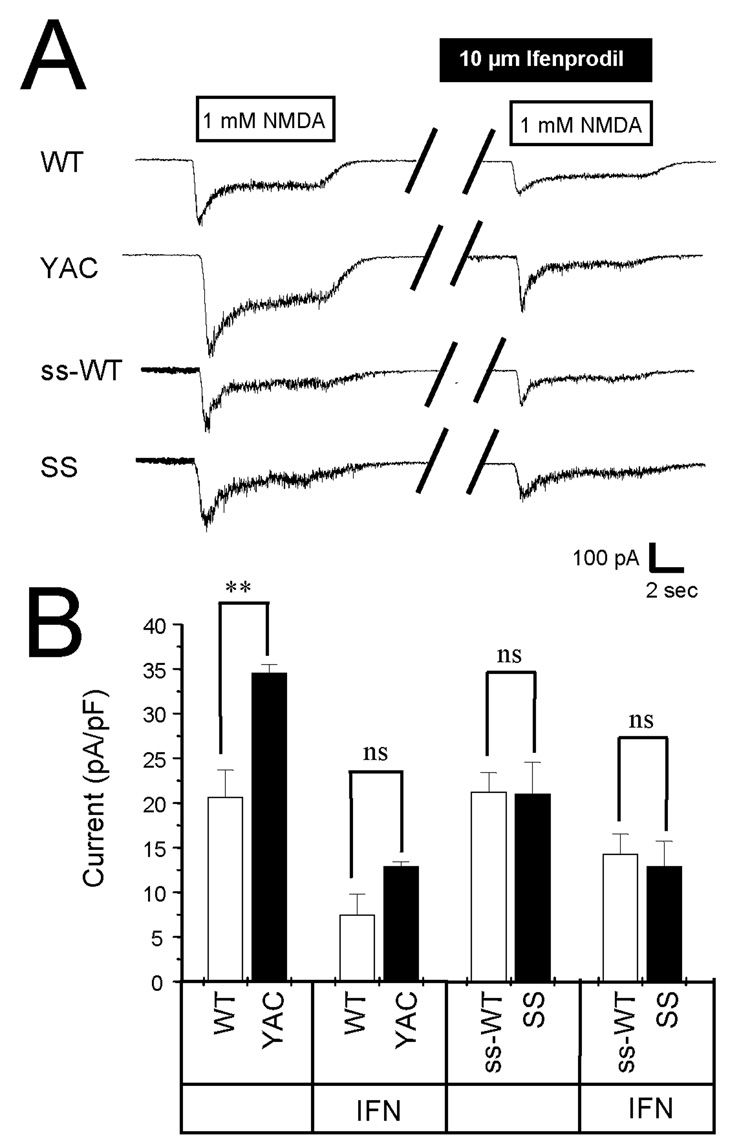
Previous studies indicated that activity of NMDA receptors is enhanced in MSNs from the YAC72 HD mouse model (Cepeda et al., 2001; Zeron et al., 2002; Li et al., 2004; Zeron et al., 2004; Fan et al., 2007). Surprisingly, a recent report indicated that NMDAR activity was not potentiated in MSNs from YAC128 line 55 (Fernandes et al., 2007). The analysis of NMDAR currents in YAC128 line 53 MSNs or shortstop MSNs has not been previously reported. Thus, in the next series of experiments we used whole-cell patch clamp recording technique to measure the size of the NMDA-evoked current in DIV9–10 MSN cultures from YAC128 mice (line 53), shortstop mice and corresponding wild type littermates (WT and ss-WT). The MSNs in these experiments were voltage-clamped at −60 mV and the currents were evoked by local and rapid application of 1 mM NMDA using multibarrel perfusion system (“sewer pipe”). We found that NMDA induced much larger currents (p<0.01) in YAC128 MSNs than in the WT littermate cultures (Fig 3A). In contrast, the size of the NMDA-evoked current was similar in shortstop and ss-WT MSNs (Fig 3A). To compare the results from different experiments, we normalized the peak amplitude of NMDA-evoked currents to capacitance of each cell and averaged data from multiple experiments (min 6 independent cultures established from YAC128, WT, shortstop and ss-WT mice). Cell capacitance for most cells in these experiments was less than 10 pF which was not significantly different between the four groups tested. We found that the normalized size of NMDA-evoked peak currents was significantly (p < 0.01) higher in YAC128 MSNs than in WT MSNs, but that the size of normalized NMDA-evoked peak current was not significantly different between shortstop and ss-WT MSNs (Fig 3B). To investigate which subtype of NMDA receptors is upregulated in YAC128 MSNs we repeated NMDA application in the presence of 10 µM ifenprodil, a NR2B-specific antagonist. We found that ifenprodil inhibited a substantial fraction of the NMDA-evoked currents in all four groups of cells (Fig 3A). To quantitatively compare the results from different experiments, the size of ifenprodil-resistant component of the peak NMDA current (mediated by NR1/NR2A subtypes of NMDAR) was normalized to the MSN cell capacitance, and the data were averaged for each group of cells. We found that the average size of ifenprodil-resistant NMDA currents was not significantly different between YAC128 and WT MSNs and between shortstop and ss-WT MSNs (Fig 3B). This data suggests that potentiation of NMDA-evoked currents in YAC128 MSNs is largely due to an increase in NR2B current density, consistent with the previous findings in MSN from the YAC72 mice (Cepeda et al., 2001; Zeron et al., 2002; Li et al., 2004; Zeron et al., 2004; Fan et al., 2007).
Figure 3. NMDA-evoked currents in YAC128 and shortstop MSN.
(A). Representative traces of currents evoked by application of NMDA to WT, YAC128, ss-WT and shortstop MSN at DIV8–10. Bar represents application of 1 mM NMDA and application of 1 mM NMDA in the presence of 10 µM Ifenprodil to the same cell. (B) The peak of NMDA-evoked current was normalized to cell capacitance and averaged across multiple experiments obtained with 6 different YAC128 and shortstop cultures. The average normalized peak current is shown as mean ± SE (n = 17~25 different cells) for WT, YAC128, ss-WT and shortstop MSN. Peak NMDA-induced current in was significantly larger in YAC128 MSN than in WT MSN (**, p<0.01). No significant difference was observed for the size of the peak NMDA current between ss-WT and shortstop MSN. The size of ifenprodil-resistant peak NMDA current was similar in all 4 cultures.
Glutamate induces rapid loss of mitochondrial membrane potential in YAC128 MSNs, but not in shortstop MSNs
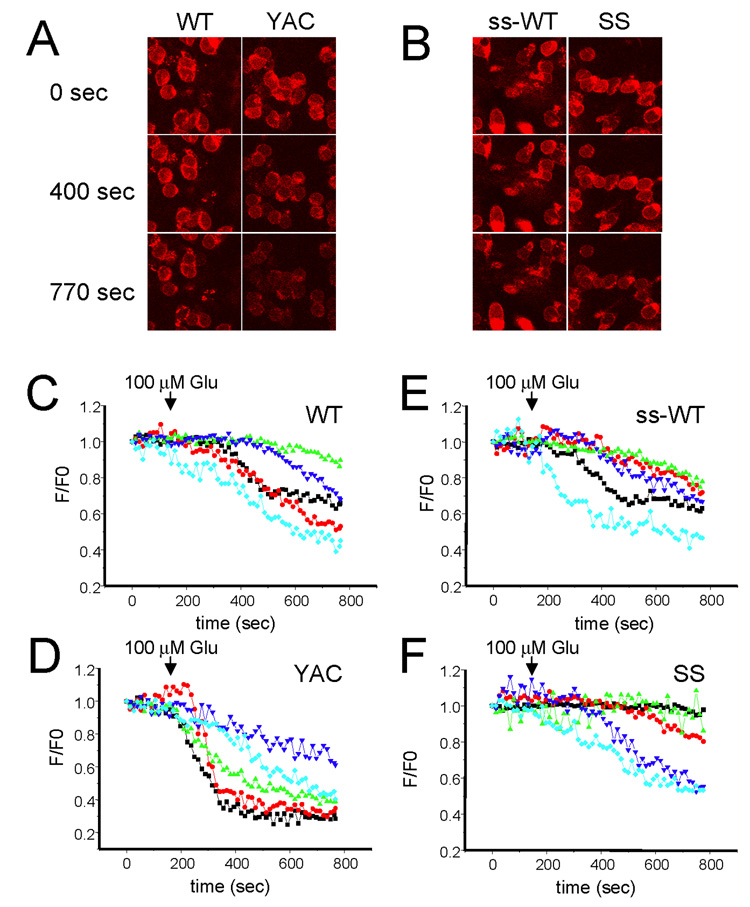
It has been proposed that mitochondrial dysfunction plays an important role in HD pathogenesis (Albin and Greenamyre, 1992; Beal, 1992; Jenkins et al., 1993; Sawa et al., 1999; Panov et al., 2002; Choo et al., 2004; Tang et al., 2005; Oliveira et al., 2006; Shehadeh et al., 2006; Fernandes et al., 2007; Oliveira et al., 2007; Trushina and McMurray, 2007). To determine whether mitochondrial dysfunction plays a role in the enhanced sensitivity of YAC128 to glutamate-induced apoptosis, in the next series of experiments we compared glutamate-induced changes in mitochondrial membrane potential in YAC128, shortstop, WT and ss-WT MSNs at 13–14 DIV. The mitochondrial membrane potential in these experiments was determined by confocal microscopy using a TMRM imaging dye as described in Methods. In control experiments we confirmed that TMRM+ signal co-localized with the signal from mitochondrial marker MitoTracker Green (data not shown). We determined that the TMRM+ fluorescence was concentrated in individual puncta, most likely corresponding to mitochondrial clusters (Fig 4A). In our experiments the TMRM+ fluorescence intensity was determined for individual puncta at the beginning of the experiment (time = 0 sec) and defined as F0. The TMRM+ fluorescence intensity for the same puncta was measured as a function of time F(t) with 10 sec interval. In all experiments the neuronal cultures were stimulated with 100 µM glutamate at time = 100 sec from the start of the imaging session. We found that addition of 100 µM glutamate resulted in much more rapid loss of TMRM+ fluorescence in YAC128 MSNs when compared to WT MSNs (Fig 4A). In contrast, TMRM+ fluorescence changes were similar in ss-WT and shortstop MSNs (Fig 4B).
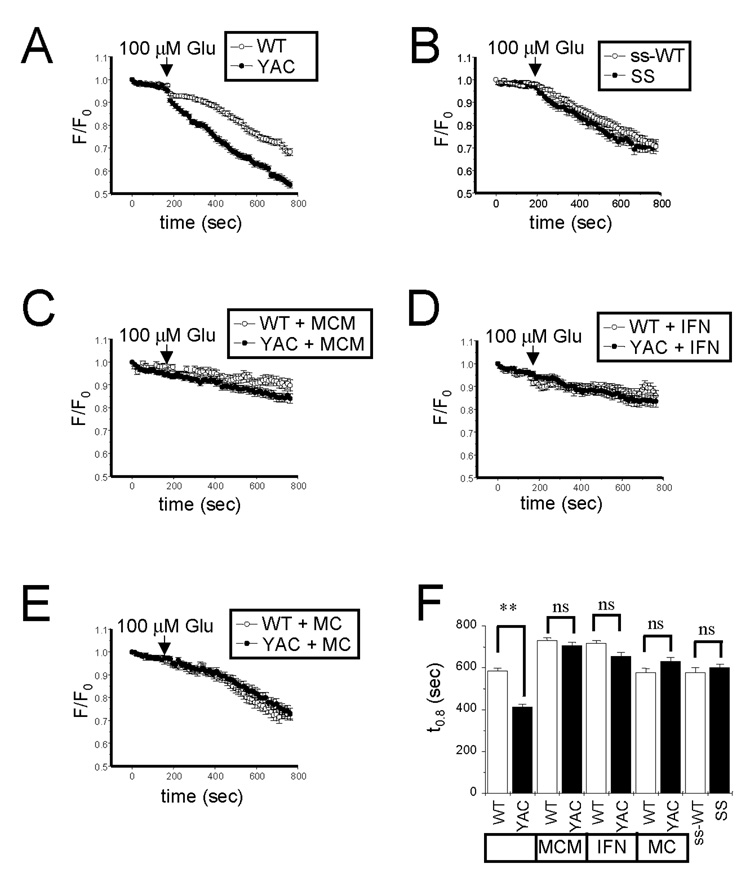
Figure 4. Glutamate-induced mitochondrial depolarization in YAC128 and shortstop MSN.
(A, B). TMRM+ confocal images of WT, YAC128, ss-WT and shortstop MSN at DIV13–14. The images before (0 sec) and after (400 sec and 770 sec) addition of 100 µM glutamate are shown as indicated. (C, D, E, F) Representative traces for individual puncta are shown for WT (C), YAC128 (D), ss-WT (E) and shortstop (F) MSNs. 100 µM Glutamate was added to each MSN culture at t = 100 sec as indicated. The representative data for five individual pucta are shown as normalized F/F0 traces for each MSN culture.
For quantitative analysis individual mitochondria puncta within neuronal cell soma were chosen randomly as a region of interest (ROI). Each of these puncta most likely corresponds to a separate “mitochondrial cluster”. The TMRM+ fluorescence intensity in the same ROI was measured each 10 sec as a function of time, yielding F(t). To compare results obtained with different puncta, the F(t) was normalized to F0 value recorded for the same puncta at the beginning of the imaging session (t = 0). The time course of TMRM+ signal changes was highly heterogeneous when analyzed at the single puncta level in all 4 MSN cultures. The responses of individual puncta could be classified into 3 categories: (i) soma puncta did not show any significant response to addition of glutamate; (ii) soma puncta responded by a slow reduction in TMRM+ fluorescence (corresponding to a slow mitochondrial depolarization); (iii) some puncta displayed very rapid loss of TMRM+ fluorescence (most likely corresponding to rapid loss of mitochondrial membrane potential due to opening of mitochondrial permeability transition pore). Although all 3 kinds of responses could be observed in all 4 MSN cultures (Figs 4C, 4D, 4E, 4F), it was apparent that a fraction of “rapid responders” is higher in the YAC128 MSNs cultures (Fig 4D) than in WT cultures (Fig 4C). Thus, we concluded that addition of glutamate induces opening of the mitochondrial permeability transition pore more easily in YAC128 MSNs than in WT MSNs. In contrast, shortstop and ss-WT cultures had similar fraction of “rapid responders” (Figs 4E and 4F), indicating that opening of mitochondrial permeability transition pore is induced with similar frequency in these two cultures.
To compare results from multiple experiments, we plotted an average F/F0 ratio as a fraction of time for WT, YAC128, ss-WT and shortstop MSNs. These averaging procedure resulted in a smooth time-dependent curves (Figs 5A and 5B) with reflects a weighted average of 3 types of responses described above. These data further confirmed that addition of 100 µM glutamate causes rapid loss of TMRM+ fluorescence in YAC128 MSNs, but not in WT ss-WT or shortstop MSNs (Figs 5A and 5B). For each puncta we determined a time (t0.8) necessary for F(t) to become equal to 0.8 of F0. We determined that average t0.8 values are significantly reduced in experiments with YAC128 MSNs when compared to WT MSNs (Fig 5F). In contrast, we did not observe significant difference in average t0.8 values for ss-WT and shortstop MSNs (Fig 5F). Thus, we concluded that glutamate specifically promotes mitochondrial depolarization in YAC128 MSNs, but not in shortstop MSNs.
Figure 5. Analysis of glutamate-induced mitochondrial depolarization.
(A–E). The average normalized F/F0 traces for (A) WT (n=196) and YAC128 (n=160) MSN; (B) ss-WT (n=73) and shortstop (n=114) MSN; (C) WT (n=54) and YAC128 (n=64) MSN in the presence of 20 µM MPEP, 50 µM CPCCOEt, and 10 µM(+)MK801; (D) WT (n=87) and YAC128 (n=74) MSN in the presence of 20 µM ifenprodil; (E) WT (n=79) and YAC128 (n=87) in the presence of 20 µM MPEP and 50 µM CPCCOEt. (F) The average t0.8 time is shown for each cell group as mean ± SE (same n as above). The YAC128 MSN mitochondrial membrane potential depolarize much more readily than in WT cells (**, p<0.01). The difference is abolished in the presence of NMDAR and mGluR1/5 inhibitors. The mitochondria in shortstop MSN depolarize at the same rate as mitochondria in MSN form ss-WT littermates.
In the next series of experiments we utilized a similar paradigm to determine which signaling pathways induce loss of mitochondrial membrane potential in YAC128 MSNs. By using known MPTP inhibitor bongkrekic acid (BkA) we confirmed that addition of 10 µM BkA caused significant delay in glutamate-induced mitochondrial membrane potential depolarization in YAC128 MSN (Supplementary Fig 1). These results are consistent with our interpretation of t0.8 values as quantitative measure of intrinsic apoptotic pathway activation in cultured MSN. We further determined that mixture of NMDA and mGluR1/5 inhibitors (20 µM MPEP, 50 µM CPCCOEt, and 10 µM(+)MK801) (MCM) completely prevented glutamate-induced mitochondrial membrane depolarization in both WT and YAC128 MSNs (Fig 5C, 5F). The remaining reduction in TMRM+ fluorescence was due to continuous dye photo bleaching. Addition of 20 µM of NR2B-specific antagonist of ifenprodil (IFN) also prevented glutamate-induced loss of mitochondrial membrane potential in WT and YAC128 MSNs (Fig 5D, 5F). Interestingly, addition of mGluR1/5 inhibitors (20 µM MPEP, 50 µM CPCCOEt) (MC) had no significant effect on glutamate-induced mitochondrial depolarization in WT MSNs, but abolished the difference between WT and YAC128 MSNs (Fig 5E, 5F). These data are consistent with our previous findings that inhibitors of NMDA receptors ((+) MK801 or ifenprodil) or inhibitors of mGluR1/5 receptors (MPEP and CPCCOEt) protect YAC128 MSNs from glutamate-induced apoptosis (Tang et al., 2005). These data further indicate that NR2B-mediated Ca2+ influx plays a major role in inducing mitochondrial membrane depolarization in MSNs neurons, consistent with the previous findings (Shehadeh et al., 2006). Furthermore, it appears that activation of mGluR1/5 receptors and resulting InsP3R1-mediated Ca2+ release may play an important role in loss of mitochondrial membrane potential in HD MSNs, in agreement with our earlier hypothesis (Tang et al., 2003; Tang et al., 2005).
Discussion
The shortstop mouse, which expresses a short fragment (exons 1 and 2) of human mutant Httexp with 120 CAG repeat expansion was serendipitously established (Slow et al., 2005). The striatal and cortical neurons in the shortstop mice accumulate abundant Htt nuclear aggregates. However, these mice do not manifest a behavioral phenotype or striatal neuronal loss (Slow et al., 2005). These results are in contrast to the full-length YAC128 mouse generated on identical FVB/N background, which displays early-onset motor deficits and MSNs loss (Slow et al., 2003). Moreover, YAC128 MSNs, but not shortstop MSNs, have been shown to be sensitized to NMDA-induced excitotoxicity in vivo and in vitro (Slow et al., 2005). To explain these findings, we performed a series of experiments with primary MSN cultures established form YAC128 and shortstop mice. The MSN cultures form wild type littermates (WT and ss-WT, respectively) were used as an internal control in all experiments. As a result of these experiments we determined: (1) repetitive application of glutamate causes supernormal Ca2+ signals in YAC128 MSNs, but not in shortstop MSNs (Fig 1); (2) addition of glutamate causes apoptosis of YAC128 MSNs, but not shortstop MSNs (Fig 2); (3) NR2B-supported NMDA currents are enhanced in YAC128 MSNs, but not in shortstop MSNs (Fig 3); (4) addition of glutamate causes rapid loss of mitochondrial membrane potential in YAC128 MSNs, but not in shortstop MSNs (Fig 4 and Fig 5). In general, these results indicate that cytosolic and mitochondrial Ca2+ signaling is abnormal in YAC128 MSNs, but not in shortstop MSNs. A possible explanation of these findings is that rapid nuclear translocation and/or aggregation of the Htt-120 amino-terminal fragment in shortstop mice MSNs prevents pathological association of this fragment with the InsP3R1 (Tang et al., 2003), impairs its ability to affect NMDAR currents (Cepeda et al., 2001; Zeron et al., 2002; Zeron et al., 2004; Fan et al., 2007) and mitochondrial Ca2+ handling (Panov et al., 2002; Choo et al., 2004). Further studies are required to determine if SS-MSNs demonstrate accelerated inclusions formation in vitro.
The enhanced NMDAR-mediated currents have been reported in subpopulations of MSNs obtained from other HD fragment models, such as R6/2 (Cepeda et al., 2001; Starling et al., 2005) and HD100 (Laforet et al., 2001). Thus, the Httexp amino-terminal fragment expressed in the shortstop mice does not affect NMDAR currents as it has been found for the Httexp amino-terminal fragments expressed in R6/2 and HD100 mice. However, in vivo assessment of susceptibility to excitotoxic stress has revealed a resistance to QA-mediated toxicity in R6/1, R6/2, N171-82Q and shortstop fragment models (Hansson et al., 1999; Hickey and Morton, 2000; Morton and Leavens, 2000; Hansson et al., 2001; MacGibbon et al., 2002; Jarabek et al., 2004; Slow et al., 2005). No change in QA-induced excitotoxicity was observed in HD100 mouse model (Petersen et al., 2002). In contrast, the YAC128 model demonstrates enhanced susceptibility to excitotoxic stress both in vtiro and in vivo (Slow et al., 2005; Tang et al., 2005; Graham et al., 2006b; Graham et al., 2006a; Shehadeh et al., 2006; Fernandes et al., 2007) (Fig 2, Fig 4, Fig 5). These data suggest that the specific size of the Httexp fragment plays a critical role in the excitotoxic phenotype and reinforces the importance of the context dependent nature of a specific, disease relevant, Httexp fragment in the pathogenesis of HD. Furthermore, comparison of the excitotoxic phenotype in truncated HD models with HD mouse models which express the full length Httexp protein highlight the link between enhanced NMDAR currents and HD pathology.
In our experiments we observed an increased NMDAR-mediated currents in the MSN cultured from YAC128 line 53 full-length model (Fig 3). In contrast, the MSN from YAC128 line 55 homozygous mice show similar NMDAR current densities as WT (Fernandes et al., 2007). Both YAC128 lines 53 and 55 have been generated using the same transgenic construct and maintained on identical FVB/N background (Slow et al., 2003). The line 53 express approximately 2-fold higher levels of transgenic mutant Htt-120 protein that the line 55 (Graham et al., 2006a). The 2-fold difference in Httexp may account for differences observed between our experiments with line 53 (Fig 3) and the previous experiments with line 55 (Fernandes et al., 2007). Indeed, since many protein kinases and phosphatases, as well as proteases such as calpains, modulate NMDAR surface expression and activity, there may be differences in the balance of activities of these enzymes secondary to stress-induced pathways in these two lines or else induced by culturing technique differences. In general, our recording conditions are similar to ones used by Fernandes at al (2007), but the average size of the normalized peak NMDA current induced by application of 1 mM NMDA was 20–30 pA/pF in our experiments (Fig 3B) and 50–60 pF/pA in experiments by Fernandes at al (2007). The differences in the size of the NMDA current may reflect differences in the culture conditions, such as for example variation in cell plating density. In a recent report the MSN NMDA and AMPA currents were systematically evaluated in acute corticostriatal slices obtained from 25–27 days old YAC18, YAC72, YAC128 (line 53) and YAC128 (line 55) mice (Milnerwood and Raymond, 2007). It was discovered that the size of NMDA eEPSC amplitudes was similar across all genotypes, but that the NMDAR:AMPAR current ratios were significantly enhanced in YAC72, YAC128 (line 55) and YAC128 (line 53) cortico-striatal synapses when compared to YAC18 mouse (Milnerwood and Raymond, 2007). Furthermore, the same authors reported that decay time constant for NMDAR-mediated eEPSC was significantly increased in YAC128 (line 53) MSN when compared to YAC18 MSN (Milnerwood and Raymond, 2007). These results potentially indicate that NMDAR subunit composition is altered in YAC128 (line 53) MSN, most likely in favor of NR2B-containing complexes (Milnerwood and Raymond, 2007). The changes in the NMDA eEPSC decay time constant were specific for YAC128 line 53 and were not observed for YAC128 line 55 or for YAC72 MSN (Milnerwood and Raymond, 2007). All these results are consistent with changes of NMDAR function in YAC128 MSN lines, with changes in YAC128 line 53 being more dramatic than changes in YAC128 line 55. Consistent with this conclusion, the neurodegenerative phenotype developed significantly faster in YAC128 line 53 than in YAC128 line 55 (Graham et al., 2006a). Interestingly, cultured MSNs from both lines show similarly enhanced sensitivity to NMDAR-mediated apoptosis when compared with WT MSNs (Tang et al., 2005; Graham et al., 2006a; Shehadeh et al., 2006; Fernandes et al., 2007). Thus, with expression of mutant Httexp with an extreme expansion of 120 polyQ, it appears that differences in regulation of cell death signaling downstream of NMDAR activation also play an important role in augmenting apoptosis, as was previously reported (Fernandes et al., 2007).
In conclusion, our data provide additional support for the importance of deranged Ca2+ signaling in HD pathology (Bezprozvanny and Hayden, 2004). Ca2+ signaling abnormalities observed in YAC128 MSNs neurons in our experiments correlate with the HD-like in vivo pathology described for this mice (Slow et al., 2003). In contrast, shortstop MSNs display normal Ca2+ signaling properties, consistent with the lack of neurodegeneration reported for this mice (Slow et al., 2005). Furthemore, in agreement with our earlier studies (Cepeda et al., 2001; Zeron et al., 2002; Tang et al., 2003; Zeron et al., 2004; Tang et al., 2005; Shehadeh et al., 2006; Wu et al., 2006), these latest results (Fig 5D, 5E, 5F) further indicate that NR2B subtype of NMDA receptors and mGluR1/5 receptors should be considered as potential therapeutic targets for HD treatment.
Supplementary Material
Acknowledgments
We thank Xiangmei Kong and Huarui Liu for help with maintaining the YAC128 and shortstop mouse colony, Janet Young and Leah Benson for administrative assistance, IB, a Carla Cocke Francis Professor in Alzheimer's Research, is supported by the McKnight Neuroscience of Brain Disorders Award, the Robert A. Welch Foundation, the HighQ foundation, and NINDS R01NS38082 and R01NS056224. MRH, a Killam University Professor, holds a Canada Research Chair in Human Genetics and is supported by grants from the Huntington Disease Society of America, High Q Foundation, Canadian Institutes of Health Research and Michael Smith Foundation for Health Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albin RL, Greenamyre JT. Alternative excitotoxic hypotheses. Neurology. 1992;42:733–738. doi: 10.1212/wnl.42.4.733. [DOI] [PubMed] [Google Scholar]
- Apostol BL, Kazantsev A, Raffioni S, Illes K, Pallos J, Bodai L, Slepko N, Bear JE, Gertler FB, Hersch S, Housman DE, Marsh JL, Thompson LM. A cell-based assay for aggregation inhibitors as therapeutics of polyglutamine-repeat disease and validation in Drosophila. Proc Natl Acad Sci U S A. 2003;100:5950–5955. doi: 10.1073/pnas.2628045100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Beal MF. Does impairment of energy metabolism result in excitotoxic neuronal death in neurodegenerative illnesses? Ann Neurol. 1992;31:119–130. doi: 10.1002/ana.410310202. [DOI] [PubMed] [Google Scholar]
- Beal MF, Ferrante RJ, Swartz KJ, Kowall NW. Chronic quinolinic acid lesions in rats closely resemble Huntington's disease. J Neurosci. 1991;11:1649–1659. doi: 10.1523/JNEUROSCI.11-06-01649.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF, Kowall NW, Ellison DW, Mazurek MF, Swartz KJ, Martin JB. Replication of the neurochemical characteristics of Huntington's disease by quinolinic acid. Nature. 1986;321:168–171. doi: 10.1038/321168a0. [DOI] [PubMed] [Google Scholar]
- Bezprozvanny I, Hayden MR. Deranged neuronal calcium signaling and Huntington disease. Biochem Biophys Res Commun. 2004;322:1310–1317. doi: 10.1016/j.bbrc.2004.08.035. [DOI] [PubMed] [Google Scholar]
- Cepeda C, Ariano MA, Calvert CR, Flores-Hernandez J, Chandler SH, Leavitt BR, Hayden MR, Levine MS. NMDA receptor function in mouse models of Huntington disease. J Neurosci Res. 2001;66:525–539. doi: 10.1002/jnr.1244. [DOI] [PubMed] [Google Scholar]
- Chen N, Luo T, Wellington C, Metzler M, McCutcheon K, Hayden MR, Raymond LA. Subtype-specific enhancement of NMDA receptor currents by mutant huntingtin. J Neurochem. 1999;72:1890–1898. doi: 10.1046/j.1471-4159.1999.0721890.x. [DOI] [PubMed] [Google Scholar]
- Choo YS, Johnson GV, MacDonald M, Detloff PJ, Lesort M. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum Mol Genet. 2004;13:1407–1420. doi: 10.1093/hmg/ddh162. [DOI] [PubMed] [Google Scholar]
- Cooper JK, Schilling G, Peters MF, Herring WJ, Sharp AH, Kaminsky Z, Masone J, Khan FA, Delanoy M, Borchelt DR, Dawson VL, Dawson TM, Ross CA. Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture. Hum Mol Genet. 1998;7:783–790. doi: 10.1093/hmg/7.5.783. [DOI] [PubMed] [Google Scholar]
- Coyle JT, Schwarcz R. Lesion of striatal neurones with kainic acid provides a model for Huntington's chorea. Nature. 1976;263:244–246. doi: 10.1038/263244a0. [DOI] [PubMed] [Google Scholar]
- DiFiglia M. Excitotoxic injury of the neostriatum: a model for Huntington's disease. Trends Neurosci. 1990;13:286–289. doi: 10.1016/0166-2236(90)90111-m. [DOI] [PubMed] [Google Scholar]
- Fan MM, Fernandes HB, Zhang LY, Hayden MR, Raymond LA. Altered NMDA receptor trafficking in a yeast artificial chromosome transgenic mouse model of Huntington's disease. J Neurosci. 2007;27:3768–3779. doi: 10.1523/JNEUROSCI.4356-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes HB, Baimbridge KG, Church J, Hayden MR, Raymond LA. Mitochondrial sensitivity and altered calcium handling underlie enhanced NMDA-induced apoptosis in YAC128 model of Huntington's disease. J Neurosci. 2007;27:13614–13623. doi: 10.1523/JNEUROSCI.3455-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante RJ, Andreassen OA, Dedeoglu A, Ferrante KL, Jenkins BG, Hersch SM, Beal MF. Therapeutic effects of coenzyme Q10 and remacemide in transgenic mouse models of Huntington's disease. J Neurosci. 2002;22:1592–1599. doi: 10.1523/JNEUROSCI.22-05-01592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RK, Slow EJ, Deng Y, Bissada N, Lu G, Pearson J, Shehadeh J, Leavitt BR, Raymond LA, Hayden MR. Levels of mutant huntingtin influence the phenotypic severity of Huntington disease in YAC128 mouse models. Neurobiol Dis. 2006a;21:444–455. doi: 10.1016/j.nbd.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, Warby SC, Doty CN, Roy S, Wellington CL, Leavitt BR, Raymond LA, Nicholson DW, Hayden MR. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006b;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Hansson O, Petersen A, Leist M, Nicotera P, Castilho RF, Brundin P. Transgenic mice expressing a Huntington's disease mutation are resistant to quinolinic acid-induced striatal excitotoxicity. Proc Natl Acad Sci U S A. 1999;96:8727–8732. doi: 10.1073/pnas.96.15.8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson O, Guatteo E, Mercuri NB, Bernardi G, Li XJ, Castilho RF, Brundin P. Resistance to NMDA toxicity correlates with appearance of nuclear inclusions, behavioural deficits and changes in calcium homeostasis in mice transgenic for exon 1 of the huntington gene. Eur J Neurosci. 2001;14:1492–1504. doi: 10.1046/j.0953-816x.2001.01767.x. [DOI] [PubMed] [Google Scholar]
- Hantraye P, Riche D, Maziere M, Isacson O. A primate model of Huntington's disease: behavioral and anatomical studies of unilateral excitotoxic lesions of the caudate-putamen in the baboon. Exp Neurol. 1990;108:91–104. doi: 10.1016/0014-4886(90)90014-j. [DOI] [PubMed] [Google Scholar]
- Heiser V, Engemann S, Brocker W, Dunkel I, Boeddrich A, Waelter S, Nordhoff E, Lurz R, Schugardt N, Rautenberg S, Herhaus C, Barnickel G, Bottcher H, Lehrach H, Wanker EE. Identification of benzothiazoles as potential polyglutamine aggregation inhibitors of Huntington's disease by using an automated filter retardation assay. Proc Natl Acad Sci U S A. 2002;99 Suppl 4:16400–16406. doi: 10.1073/pnas.182426599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey MA, Morton AJ. Mice transgenic for the Huntington's disease mutation are resistant to chronic 3-nitropropionic acid-induced striatal toxicity. J Neurochem. 2000;75:2163–2171. doi: 10.1046/j.1471-4159.2000.0752163.x. [DOI] [PubMed] [Google Scholar]
- Hockly E, Tse J, Barker AL, Moolman DL, Beunard JL, Revington AP, Holt K, Sunshine S, Moffitt H, Sathasivam K, Woodman B, Wanker EE, Lowden PA, Bates GP. Evaluation of the benzothiazole aggregation inhibitors riluzole and PGL-135 as therapeutics for Huntington's disease. Neurobiol Dis. 2006;21:228–236. doi: 10.1016/j.nbd.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Jarabek BR, Yasuda RP, Wolfe BB. Regulation of proteins affecting NMDA receptor-induced excitotoxicity in a Huntington's mouse model. Brain. 2004;127:505–516. doi: 10.1093/brain/awh058. [DOI] [PubMed] [Google Scholar]
- Jenkins BG, Koroshetz WJ, Beal MF, Rosen BR. Evidence for impairment of energy metabolism in vivo in Huntington's disease using localized 1H NMR spectroscopy. Neurology. 1993;43:2689–2695. doi: 10.1212/wnl.43.12.2689. [DOI] [PubMed] [Google Scholar]
- Kuemmerle S, Gutekunst CA, Klein AM, Li XJ, Li SH, Beal MF, Hersch SM, Ferrante RJ. Huntington aggregates may not predict neuronal death in Huntington's disease. Ann Neurol. 1999;46:842–849. [PubMed] [Google Scholar]
- Laforet GA, Sapp E, Chase K, McIntyre C, Boyce FM, Campbell M, Cadigan BA, Warzecki L, Tagle DA, Reddy PH, Cepeda C, Calvert CR, Jokel ES, Klapstein GJ, Ariano MA, Levine MS, DiFiglia M, Aronin N. Changes in cortical and striatal neurons predict behavioral and electrophysiological abnormalities in a transgenic murine model of Huntington's disease. J Neurosci. 2001;21:9112–9123. doi: 10.1523/JNEUROSCI.21-23-09112.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Murphy TH, Hayden MR, Raymond LA. Enhanced striatal NR2B-containing N-methyl-D-aspartate receptor-mediated synaptic currents in a mouse model of Huntington disease. J Neurophysiol. 2004;92:2738–2746. doi: 10.1152/jn.00308.2004. [DOI] [PubMed] [Google Scholar]
- MacGibbon GA, Hamilton LC, Crocker SF, Costain WJ, Murphy KM, Robertson HA, Denovan-Wright EM. Immediate-early gene response to methamphetamine, haloperidol, and quinolinic acid is not impaired in Huntington's disease transgenic mice. J Neurosci Res. 2002;67:372–378. doi: 10.1002/jnr.10100. [DOI] [PubMed] [Google Scholar]
- Mao L, Wang JQ. Upregulation of preprodynorphin and preproenkephalin mRNA expression by selective activation of group I metabotropic glutamate receptors in characterized primary cultures of rat striatal neurons. Brain Res Mol Brain Res. 2001;86:125–137. doi: 10.1016/s0169-328x(00)00276-x. [DOI] [PubMed] [Google Scholar]
- McGeer EG, McGeer PL. Duplication of biochemical changes of Huntington's chorea by intrastriatal injections of glutamic and kainic acids. Nature. 1976;263:517–519. doi: 10.1038/263517a0. [DOI] [PubMed] [Google Scholar]
- Menalled LB, Chesselet MF. Mouse models of Huntington's disease. Trends Pharmacol Sci. 2002;23:32–39. doi: 10.1016/s0165-6147(00)01884-8. [DOI] [PubMed] [Google Scholar]
- Milnerwood AJ, Raymond LA. Corticostriatal synaptic function in mouse models of Huntington's disease: early effects of huntingtin repeat length and protein load. J Physiol. 2007;585:817–831. doi: 10.1113/jphysiol.2007.142448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton AJ, Leavens W. Mice transgenic for the human Huntington's disease mutation have reduced sensitivity to kainic acid toxicity. Brain Res Bull. 2000;52:51–59. doi: 10.1016/s0361-9230(00)00238-0. [DOI] [PubMed] [Google Scholar]
- Oliveira JM, Jekabsons MB, Chen S, Lin A, Rego AC, Goncalves J, Ellerby LM, Nicholls DG. Mitochondrial dysfunction in Huntington's disease: the bioenergetics of isolated and in situ mitochondria from transgenic mice. J Neurochem. 2007;101:241–249. doi: 10.1111/j.1471-4159.2006.04361.x. [DOI] [PubMed] [Google Scholar]
- Oliveira JM, Chen S, Almeida S, Riley R, Goncalves J, Oliveira CR, Hayden MR, Nicholls DG, Ellerby LM, Rego AC. Mitochondrial-dependent Ca2+ handling in Huntington's disease striatal cells: effect of histone deacetylase inhibitors. J Neurosci. 2006;26:11174–11186. doi: 10.1523/JNEUROSCI.3004-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, Greenamyre JT. Early mitochondrial calcium defects in Huntington's disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- Petersen A, Chase K, Puschban Z, DiFiglia M, Brundin P, Aronin N. Maintenance of susceptibility to neurodegeneration following intrastriatal injections of quinolinic acid in a new transgenic mouse model of Huntington's disease. Exp Neurol. 2002;175:297–300. doi: 10.1006/exnr.2002.7885. [DOI] [PubMed] [Google Scholar]
- Ross CA. Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington's disease and related disorders. Neuron. 2002;35:819–822. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC. Lessons from animal models of Huntington's disease. Trends Genet. 2002;18:202–209. doi: 10.1016/s0168-9525(01)02625-7. [DOI] [PubMed] [Google Scholar]
- Sawa A, Wiegand GW, Cooper J, Margolis RL, Sharp AH, Lawler JF, Jr., Greenamyre JT, Snyder SH, Ross CA. Increased apoptosis of Huntington disease lymphoblasts associated with repeat length-dependent mitochondrial depolarization. Nat Med. 1999;5:1194–1198. doi: 10.1038/13518. [DOI] [PubMed] [Google Scholar]
- Shehadeh J, Fernandes HB, Zeron Mullins MM, Graham RK, Leavitt BR, Hayden MR, Raymond LA. Striatal neuronal apoptosis is preferentially enhanced by NMDA receptor activation in YAC transgenic mouse model of Huntington disease. Neurobiol Dis. 2006;21:392–403. doi: 10.1016/j.nbd.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Slow EJ, Graham RK, Osmand AP, Devon RS, Lu G, Deng Y, Pearson J, Vaid K, Bissada N, Wetzel R, Leavitt BR, Hayden MR. Absence of behavioral abnormalities and neurodegeneration in vivo despite widespread neuronal huntingtin inclusions. Proc Natl Acad Sci U S A. 2005;102:11402–11407. doi: 10.1073/pnas.0503634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slow EJ, van Raamsdonk J, Rogers D, Coleman SH, Graham RK, Deng Y, Oh R, Bissada N, Hossain SM, Yang YZ, Li XJ, Simpson EM, Gutekunst CA, Leavitt BR, Hayden MR. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet. 2003;12:1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- Smith DL, Portier R, Woodman B, Hockly E, Mahal A, Klunk WE, Li XJ, Wanker E, Murray KD, Bates GP. Inhibition of polyglutamine aggregation in R6/2 HD brain slices-complex dose-response profiles. Neurobiol Dis. 2001;8:1017–1026. doi: 10.1006/nbdi.2001.0438. [DOI] [PubMed] [Google Scholar]
- Starling AJ, Andre VM, Cepeda C, de Lima M, Chandler SH, Levine MS. Alterations in N-methyl-D-aspartate receptor sensitivity and magnesium blockade occur early in development in the R6/2 mouse model of Huntington's disease. J Neurosci Res. 2005;82:377–386. doi: 10.1002/jnr.20651. [DOI] [PubMed] [Google Scholar]
- Sun Y, Savanenin A, Reddy PH, Liu YF. Polyglutamine-expanded huntingtin promotes sensitization of N-methyl-D- aspartate receptors via post-synaptic density 95. J Biol Chem. 2001;276:24713–24718. doi: 10.1074/jbc.M103501200. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Machida Y, Niu S, Ikeda T, Jana NR, Doi H, Kurosawa M, Nekooki M, Nukina N. Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat Med. 2004;10:148–154. doi: 10.1038/nm985. [DOI] [PubMed] [Google Scholar]
- Tang TS, Chen X, Liu J, Bezprozvanny I. Dopaminergic signaling and striatal neurodegeneration in Huntington's disease. J Neurosci. 2007;27:7899–7910. doi: 10.1523/JNEUROSCI.1396-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T-S, Tu H, Chan EY, Maximov A, Wang Z, Wellington CL, Hayden MR, Bezprozvanny I. Huntingtin and huntingtin-associated protein 1 influence neuronal calcium signaling mediated by inositol-(1,4,5) triphosphate receptor type 1. Neuron. 2003;39:227–239. doi: 10.1016/s0896-6273(03)00366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T-S, Slow EJ, Lupu V, Stavrovskaya IG, Sugimori M, Llinas R, Kristal BS, Hayden MR, Bezprozvanny I. Disturbed Ca2+ signaling and apoptosis of medium spiny neurons in Huntington's disease. Proc Natl Acad Sci U S A. 2005;102:2602–2607. doi: 10.1073/pnas.0409402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The., Huntington's., Disease., Collaborative., Research., Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Tobin AJ, Signer ER. Huntington's disease: the challenge for cell biologists. Trends Cell Biol. 2000;10:531–536. doi: 10.1016/s0962-8924(00)01853-5. [DOI] [PubMed] [Google Scholar]
- Trushina E, McMurray CT. Oxidative stress and mitochondrial dysfunction in neurodegenerative diseases. Neuroscience. 2007;145:1233–1248. doi: 10.1016/j.neuroscience.2006.10.056. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- Wu J, Tang T-S, Bezprozvanny I. Evaluation of clinically-relevant glutamate pathway inhibitors in in vitro model of Huntington's disease. Neurosci Lett. 2006;407:219–223. doi: 10.1016/j.neulet.2006.08.036. [DOI] [PubMed] [Google Scholar]
- Zeron MM, Hansson O, Chen N, Wellington CL, Leavitt BR, Brundin P, Hayden MR, Raymond LA. Increased sensitivity to N-methyl-D-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington's disease. Neuron. 2002;33:849–860. doi: 10.1016/s0896-6273(02)00615-3. [DOI] [PubMed] [Google Scholar]
- Zeron MM, Fernandes HB, Krebs C, Shehadeh J, Wellington CL, Leavitt BR, Baimbridge KG, Hayden MR, Raymond LA. Potentiation of NMDA receptor-mediated excitotoxicity linked with intrinsic apoptotic pathway in YAC transgenic mouse model of Huntington's disease. Mol Cell Neurosci. 2004;25:469–479. doi: 10.1016/j.mcn.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Zhang X, Smith DL, Meriin AB, Engemann S, Russel DE, Roark M, Washington SL, Maxwell MM, Marsh JL, Thompson LM, Wanker EE, Young AB, Housman DE, Bates GP, Sherman MY, Kazantsev AG. A potent small molecule inhibits polyglutamine aggregation in Huntington's disease neurons and suppresses neurodegeneration in vivo. Proc Natl Acad Sci U S A. 2005;102:892–897. doi: 10.1073/pnas.0408936102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.