Summary
The relay domain of myosin is hypothesized to function as a communication pathway between the nucleotide-binding site, actin-binding site and the converter domain. In Drosophila melanogaster, a single myosin heavy chain gene encodes three alternative relay domains. Exon 9a encodes the indirect flight muscle isoform (IFI) relay domain whereas exon 9b encodes one of the embryonic body wall isoform (EMB) relay domains. To gain a better understanding of the function of the relay domain and the differences imparted by the IFI and the EMB versions, we constructed two transgenic Drosophila lines expressing chimeric myosin heavy chains in indirect flight muscles lacking endogenous myosin. One expresses the IFI relay domain in the EMB backbone (EMB-9a) while the second expresses the EMB relay domain in the IFI backbone (IFI-9b). Our studies reveal that the EMB relay domain is functionally equivalent to the IFI relay domain when it is substituted into IFI. Essentially no differences in ATPase activity, actin-sliding velocity, flight ability at room temperature or muscle structure are observed in IFI-9b compared to native IFI. However, when the EMB relay domain is replaced with the IFI relay domain, we find a 50% reduction in actin-activated ATPase activity, a significant increase in actin affinity, abolition of actin sliding, defects in myofibril assembly and rapid degeneration of muscle structure compared to EMB. We hypothesize that altered relay domain conformational changes in EMB-9a impair intramolecular communication with the EMB-specific converter domain. This decreases transition rates involving strongly bound actomyosin states, leading to a reduced ATPase rate and loss of actin motility.
Keywords: Myosin, Drosophila, relay domain, protein isoforms, muscle
Introduction
A single-copy gene, Mhc, encodes Drosophila melanogaster muscle myosin heavy chain (MHC).1,2 Alternative splicing of Mhc transcripts produces myosin isoforms with different tissue specificity and functional properties.3–6 We have shown through P element-mediated transformation that expression of an embryonic myosin isoform (EMB) in indirect flight muscle (IFM) results in decreased myofibril stability and a flightless phenotype.7 The EMB myosin has lower ATPase activity and reduced in vitro actin sliding velocity compared to the native indirect flight muscle isoform (IFI).6 Further, IFM mechanical power is decreased but force output is increased when EMB is present.8 Thus the isoform-specific domains encoded by alternative splicing of myosin transcripts appear to dictate fast (IFI) versus slow (EMB) muscle properties.
The isoform-specific domains of MHC are encoded by five sets of exons that are alternatively spliced and one exon that is either included or excluded.3 Four of the alternative exon sets, exons 3, 7, 9 and 11, encode portions of the S-1 head.9 S-1 contains several domains critical for myosin function, including the actin-binding region, the ATP binding domain and the lever arm, which amplifies the power stroke to generate force and movement. We are using an integrative approach to study transgenic organisms expressing chimeric myosins in order to understand how isoform-specific domains influence functional properties of S-1. In previous studies, we determined that alternative versions of a region near the N-terminus of MHC (encoded by exons 3a or 3b) influence the rate of the cross-bridge cycling as seen by differences in actin-activated ATPase activity and actin sliding velocity, most likely through local conformational changes.10 These myosin kinetic adjustments help achieve optimal locomotory performance.11 We also showed that two of the four alternative versions of exon 7, which encode a region at the lip of the ATP binding pocket, influence ATPase activity but not actin sliding velocity, suggesting that a transition rate between detached states or the rate of myosin attachment to actin is affected.12 A series of locomotory and mechanical assays demonstrated that this domain is important in establishing optimal fiber oscillation speed, hence influencing flight ability.13 In addition, we analyzed two of the five alternative versions of exon 11 that encode a portion of the converter domain. The converter domain is coupled to the lever arm, which rotates during the power stroke.14,15 We found that the EMB and IFI exon 11 domains influence the kinetic properties of myosin both in vivo and in vitro.8,16
Three different copies of exon 9 of Drosophila Mhc encode alternative versions of the relay domain, which is located at the C-terminal end of the 50-kDa proteolytic fragment. The 57 amino acid peptide encoded by each version of exon 9 corresponds to residues 472–528 of chicken skeletal muscle MHC. The relay domain is hypothesized to play a key role in the communication pathway that couples ATP hydrolysis with actin-binding and mechanical movement of the lever arm.17,18 The relay domain can be divided into the relay helix (residues 472–507) and the relay loop (residues 508–518).18 In chicken myosin structures, the relay domain adopts different conformations at the end of the power stroke compared to when the lever arm assumes the pre-power stroke state.19,20,21 During the transition from the post-stroke state to the pre-stroke state, the relay helix becomes partially unwound and kinked.18 A distortion of the central seven-stranded β-sheet within the molecule causes rotation of the SH1 and SH2 helices, which provides part of the mechanism for relieving the strain on the kinked relay helix. The subsequent straightening of the relay helix is theorized to be required for driving the power stroke, as the relay’s interactions with the converter domain are at least partially responsible for lever arm movement.18, 22
Drosophila alternative exons encoding the relay domain are differentially expressed. Exon 9a is used in the IFM and tergal depressor of the trochanter (TDT) muscle; exon 9b is found in most embryonic body wall muscles, cardioblasts and pharyngeal muscles; exon 9c is expressed in transverse embryonic body wall muscles.4,5,23 Despite their tissue-specific expression, the three versions of the relay domain are highly conserved. There are only 5 different amino acid residues encoded by exons 9a and exon 9b, three of which are conserved changes (see Fig. 4). Two of the five different amino acids are located in the relay loop and flank an ATP-sensitive invariant tryptophan (Trp-510). This tryptophan residue changes orientation during the transition from weakly bound to tightly bound states and during MgATP hydrolysis.24 The other three differing residues are in the relay helix.
Figure 4. Location of isoform specific amino acid residues in the myosin molecule.
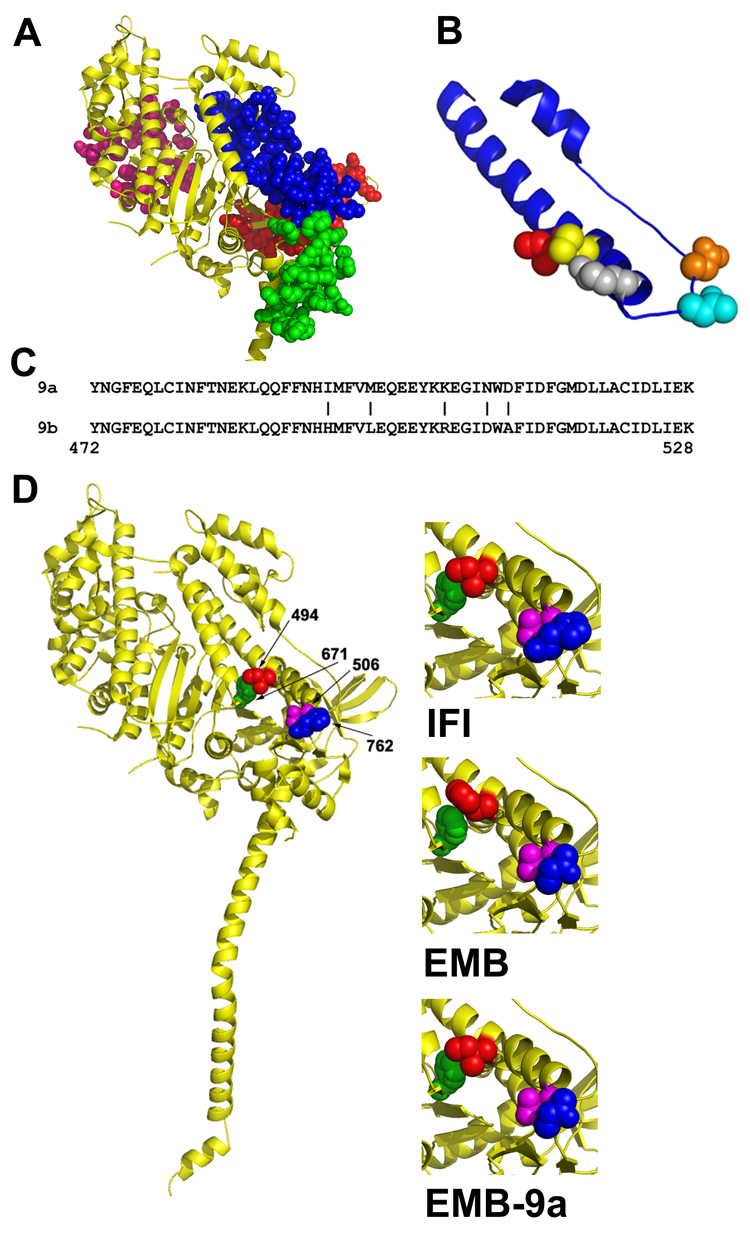
A, Location of the domains encoded by alternative exons in Drosophila MHC. Exon 3 (red), exon 7 (pink), exon 9 (blue) and exon 11 (green) domains are mapped onto the crystal structure of scallop myosin S-1 in the pre-power stroke state.31 The only interactions observable between the relay domain (encoded by exon 9) and other alternative domains are with the converter domain (encoded by exon 11). B, Close-up view of the relay domain (blue) encoded by Drosophila alternative exon 9. The five IFI-specific amino acids encoded by exon 9a are mapped on the molecule as red (Ile-494), yellow (Met-498), grey (Lys-505), cyan (Asn-509) and orange (Asp-511). C, The amino acid sequence of the two alternative relay domains; a vertical line (|) marks each of the five amino acid differences encoded by exon 9a and exon 9b. The chicken pectoralis muscle myosin numbering system is used. D, Mapping of Drosophila amino acids onto the crystal structure of scallop myosin S-1 in the pre-power stroke state.31 Interacting amino acids in IFI (left and upper right) are Ile-494 (red) with Phe-671 (green) and Glu-506 (magenta) with His-762 (blue). The same color scheme is used to map the corresponding amino acids in EMB (middle right; His-494, Phe-671, Glu-506, Asn-762) and in the chimeric EMB-9a myosin (lower right; Ile-494, Phe-671, Glu-506, Asn-762).
To determine if the alternative relay domains encoded by exons 9a and 9b are responsible for any of the functional differences between IFI and EMB myosins and to increase our understanding of the role of the relay domain, we genetically engineered two Drosophila transgenes and expressed them by P element-mediated germline transformation. One transgene forces the expression of IFI exon 9a in the EMB backbone (EMB-9a). The other forces the expression of EMB exon 9b in the IFI backbone (IFI-9b). In contrast to the expectation of making EMB myosin more like the faster IFI, our biochemical, biophysical and ultrastructural studies showed that the IFI version of exon 9 drastically decreases functionality of the EMB isoform. Myosin from EMB-9a shows a significant decrease in actin-activated MgATPase activity, with an increase in actin affinity compared to EMB. In vitro motility assays show that the chimera is unable to move actin. In vivo, myofibrils of EMB-9a have assembly defects and rapidly degenerate compared to EMB. We further found that IFI does not acquire EMB-like properties when the IFI relay domain is replaced by the EMB relay. Instead we detect virtually no difference between native (IFI) and chimeric (IFI-9b) myosins. Our studies show that the structural differences in alternative relay domains can differentially affect the functional properties of specific myosin isoforms.
Results
P Element Transformation and Generation of Transgenic Lines
Transgenic lines that express either the IFI-9b or EMB-9a chimeric transgenes were obtained by injecting embryos with DNA constructs described in Materials and Methods. For IFI-9b we injected 892 embryos and obtained 5 independent lines that express the transgene. Two of the five IFI-9b inserts mapped to the X chromosome, two to the second chromosome and one to the fourth chromosome. For EMB-9a we injected 1548 embryos and obtained 10 independent lines that express the transgene. Two of the EMB-9a inserts mapped to the X chromosome, seven to the second chromosome, and one to the third chromosome.
To eliminate endogenous MHC expression from the transgenic stocks, we crossed each into the Mhc10 background, which is null for MHC in IFM and TDT muscle.25 The transgenic lines that mapped to the second chromosome were not useful for this purpose since the endogenous myosin gene is located on this chromosome. We examined three transgenic lines in detail for each construct. For EMB-9a, these were lines 1 (X-linked), 18 (third chromosome), and 34 (X-linked). For IFI-9b, these were lines 1 (X-linked), 2 (fourth chromosome), and 3 (X-linked).
Verification of Transgene RNA Splicing and Protein Expression
The EMB-9a transgenic lines were examined for myosin gene expression and correct exon usage by RT-PCR. RNA was isolated from upper thoraces of 2-day-old adults of wild type (yw), EMB and EMB-9a. cDNAs were produced for each using a forward primer to exon 8 and primers specific to embryonic-specific exon 11c or IFM-specific exon 11e. As expected, the exon 11e primer generated a 1050 bp cDNA from wild-type mRNA, whereas no cDNA was produced from EMB and EMB-9a mRNA. The complementary pattern was observed when the exon 11c reverse primer was used. The presence of exon 9a in both wild-type and EMB-9a transcripts was verified by digestion at an exon-specific Pst I site. These results confirmed that EMB-9a transgenic flies express adult-specific exon 9a in conjunction with the expected embryonic version of exon 11, i.e. exon 11c (data not shown).
RT-PCR and northern blots of RNA isolated from upper thoraces of 2-day-old adults were used to confirm that IFI-9b mRNAs have the same alternative exons as yw, except for containing exon 9b instead of 9a. Since IFI-9b contains a partial cDNA insert for Mhc in which the IFI specific exons 7d and 11e are included, we examined whether the other IFI exons (exons 3b, 15a and 18) were spliced correctly in the IFI-9b transcripts. RT-PCR of fragments containing exon 3 and exon 15 showed no difference between IFI-9b and yw with the expected inclusion of exons 3b and 15a. Northern blot analysis showed the inclusion of exon 18 in both IFI-9b and yw. RT-PCR of the exon 9 region confirmed that IFI-9b expresses exon 9b with no detectable amounts of exon 9a and that yw expresses exon 9a, as expected. Our results show that the only difference in alternative exon usage for IFI-9b compared to yw is the use of exon 9b instead of 9a (data not shown).
We next determined whether transgene expression yielded normal levels of MHC. MHC expression levels in upper thoraces relative to actin accumulation were determined for three transgenic lines expressing each construct by densitometry of polyacrylamide gels. Levels of MHC expression were essentially identical to wild type in all transgenic lines tested (Table 1).
Table 1.
Myosin expression levels
| Fly line | Protein amount ± S.E.M. |
|---|---|
| Mhc10 | 0.08 ± 0.04a |
| EMB-9a-1 | 0.91 ± 0.05 |
| EMB-9a-18 | 0.98 ± 0.04 |
| EMB-9a-34 | 0.94 ± 0.05 |
| IFI-9b-1 | 0.91 ±0.04 |
| IFI-9b-2 | 1.00 ± 0.03 |
| IFI-9b-3 | 0.93 ± 0.03 |
| yw | 1.00 ± 0.03 |
Transgenes were crossed into the Mhc10 (IFM myosin null) background to determine myosin protein accumulation (relative to actin levels) in upper thoraces, as detailed in Materials and Methods. Values are compared to yw (wild type). Protein amounts are means ± standard error of the mean (S.E.M.).
Student’s t test p < 0.001, statistically different from yw.
Basal and Actin-activated ATPase Activities
We determined the CaATPase, basal MgATPase, and actin-activated MgATPase activities of wild-type IFI and EMB myosin and of the IFI-9b and EMB-9a chimeric isoforms (Table 2 and Fig. 1). As shown previously10, CaATPase, basal MgATPase, and Vmax of actin-activated ATPase activities for EMB myosin were ~1/2 that of IFI myosin, while Km values for actin determined in the actin-activated ATPase assay were similar for both IFI and EMB myosins.10 Substitution of the embryonic exon 9b domain into IFI did not alter any of these parameters compared to IFI. On the other hand, expression of adult exon 9a in EMB myosin reduced actin-activated ATPase activities by ~50% compared to EMB. Apparent affinity for actin of EMB-9a increased significantly compared to other isoforms, as suggested by the ~3-fold decrease in Km (Table 2).
Table 2.
ATPase kinetics and actin-sliding velocities of IFI, EMB, IFI-9b, and EMB-9a myosins
| Isoform | Basal MgATPase ± SD head−1 s−1 | Actin-activated Vmax ± SD head−1 s−1 | Km (actin) ± SD µM | Basal Ca-ATPase ± SD head−1 s−1 | Actin sliding velocity ± SD µm s−1 |
|---|---|---|---|---|---|
| IFI | 0.20 ± 0.06 (n=7) | 1.83 ± 0.61 (n=7) | 0.28 ± 0.05 (n=7) | 6.95 ± 1.43 (n=7) | 6.4 ± 0.6 (n=4) |
| IFI-9b | 0.18 ± 0.05 (n=4) | 1.91 ± 0.19 (n=4) | 0.41 ± 0.09 (n=4) | 9.00 ± 3.22 (n=4) | 6.4 ± 0.6 (n=4) |
| EMB | 0.08 ± 0.04 (n=7) | 1.10 ± 0.14 (n=7) | 0.29 ± 0.07 (n=7) | 4.47 ± 2.20 (n=6) | 0.7 ± 0.2 (n=3) |
| EMB-9a | 0.06 ± 0.01 (n=5) | 0.46 ± 0.24 (n=5)a | 0.10 ± 0.14 (n=5)b | 2.27 ± 0.92 (n=4) | 0 (n=5) |
Statistically different from EMB (p = 0.0037, Student’s t test)
Statistically different from EMB (p = 0.039, Student’s t test)
Figure 1. Actin-stimulated Mg-ATPase activity of wild-type and chimeric myosins.
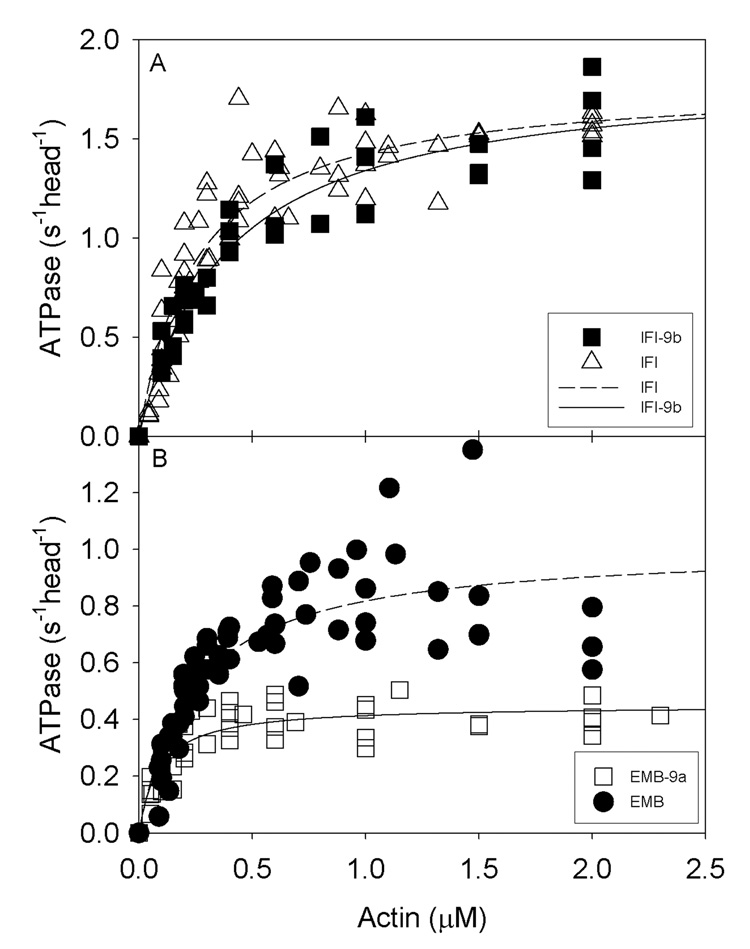
Mg-ATPase activities of myosins isolated from the four transgenic lines were determined as described in Materials and Methods. The concentrations of chicken actin used ranged from 0–2 µM. Basal Mg-ATPase activities obtained in the absence of actin were subtracted from all data points, which were fitted with the Michaelis-Menten equation (rectangular hyperbola, y = ax/(b + x)). A, IFI (n = 9) and IFI-9b (n = 4) actin-activated MgATPase activities show negligible differences. B, EMB (n = 7) and EMB-9a (n = 5) actin-activated Mg-ATPase activities. Vmax and Km for EMB-9a are substantially reduced compared to EMB (see Table 2).
In vitro Motility
IFI-9b myosin translocates fluorescently labeled actin filaments at a velocity of 6.42 µm/sec at 22.5° C, which is statistically similar to IFI (wild-type) induced motility (Table 2). In contrast, EMB myosin drives actin filament movement at a rate of 0.7 µm/sec (Table 2). Velocity for EMB is stimulated six-fold by decoration with smooth muscle tropomyosin.6 EMB-9a myosin was unable to generate movement of actin filaments in vitro. Upon addition of ATP, actin filament ends began to wiggle, however no translocation was detected. Filaments were observed for at least 20 minutes to insure absolutely no movement occurred. This result was reproducible under all of the following conditions: with the addition of smooth muscle tropomyosin, with varying myosin concentrations (from 0.2 to 1 µg/µl), with increased ATP concentrations (from 1.4 to 8 mM), or with altered ionic strength in the assay solution (~20 to ~60 mM).
Flight and Jump Ability
The EMB isoform when expressed in a Mhc10 null background results in flightless Drosophila.7 Substituting the IFI relay domain into the EMB isoform did not rescue flight (Table 3). This was not surprising, since the actin sliding velocity and ATPase rates (Table 2) of EMB-9a were even lower than EMB. In contrast, the flight ability of IFI-9b was normal at room temperature compared to PwMhc2 (Table 3), which expresses the wild-type IFI isoform in a myosin null (Mhc10) background.26, 27
Table 3.
Flight and jump ability of wild-type, mutant and transgenic flies
| Line | No. flight tested | Up (%) | Horizontal (%) | Down (%) | Not at all (%) | Flight Index ± S.E. | Jump distance ± S.E. (cm) |
|---|---|---|---|---|---|---|---|
| EMB | 123 | 0 | 0 | 0.8 | 99.2 | 0 | 2.2 ± 0.1a |
| EMB-9a-1 | 129 | 0 | 0 | 1.5 | 98.5 | 0 | 1.9 ± 0.1 |
| EMB-9a-18 | 119 | 0 | 0 | 1.7 | 98.3 | 0 | 2.1 ± 0.1 |
| EMB-9a-34 | 108 | 0 | 0 | 0.9 | 99.1 | 0 | 1.8 ± 0.2 |
| PwMhc2 | 116 | 57.8 | 14.6 | 15.5 | 12.1 | 4.4 ± 0.1 | 4.9 ± 0.2a |
| IFI-9b-1 | 131 | 50.8 | 28.0 | 15.1 | 6.1 | 4.5 ± 0.2 | 5.0 ± 0.1 |
| IFI-9b-2 | 146 | 44.6 | 34.2 | 17.1 | 4.1 | 4.4 ± 0.1 | 5.1 ± 0.1 |
| IFI-9b-3 | 129 | 58.1 | 32.6 | 7.8 | 1.5 | 4.9 ± 0.3 | 5.0 ± 0.2 |
Flight abilities expressed as percentages. Transgenic flies were assayed for the ability to fly up (U), horizontal (H), down (D) or not at all (N). Flight index equals 6U/T + 4H/T + 2D/T + 0N/T; T is the total number of flies tested.40
Jump distance for PwMhc2 and EMB are from Swank et al.8
Student's t test showed no significant difference between PwMhc2 and IFI-9b flight indexes and no significant difference between EMB and EMB-9a jumping ability (p<0.05 was considered statistically significant).
Drosophila expressing EMB in an Mhc10 background show reduced jump ability compared to wild-type flies.7 Our results show that there was no difference in the jump ability of EMB-9a compared to EMB. Thus substitution of exon 9b in EMB by exon 9a of IFI did not rescue jump ability (Table 3). Adults expressing the embryonic version of exon 9 (IFI-9b) show normal jump ability compared to PwMhc2 flies (Table 3).
Ultrastructure
We examined the ultrastructure of the IFM of each transgenic line at various stages of development: late pupae, two-day-old adults and one-week-old adults. As in IFI-expressing PwMhc2 (Fig. 2A), IFI-9b late pupae show the normal hexagonal arrangement of thick and thin filaments (Fig. 2B). One-week-old PwMhc2 and IFI-9b adults maintain normal myofibril morphology (Fig. 2C, 2D) and have comparable sarcomere structures, with normal Z- and M-lines (Fig. 2E, 2F). This is also the case for two-day-old adults (data not shown). Thus, substituting the EMB relay domain into IFI did not affect myofibril assembly, structure or stability.
Figure 2. Effects of the IFI-9b myosin isoform on myofibril assembly and stability in dorsal longitudinal IFM.
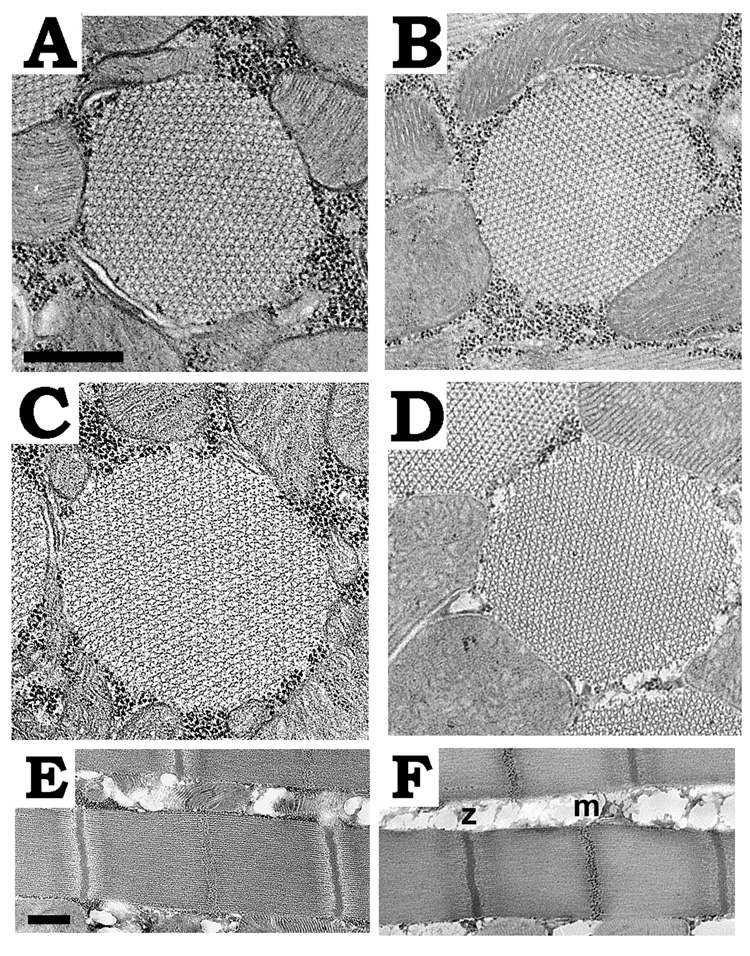
A, Transverse section from PwMhc2 late stage pupa (expressing IFI wild-type isoform). B, Transverse section from IFI-9b late stage pupa. Myofibril structure for IFI-9b pupa is similar to PwMhc2 pupa, with normal hexagonal packing of thick and thin filaments. C, Transverse section from PwMhc2 one-week-old adult. D, Transverse section from IFI-9b one-week-old adult. Myofibril structures are similar in one-week-old adults. E, Longitudinal section from PwMhc2 one-week-old adult. F, Longitudinal section from IFI-9b one-week-old-adult. Sarcomere structures are similar to PwMhc2. z, Z-line; m, M-line. Scale bars, 0.5 µm.
In EMB, myofibrils assemble nearly normally, but degenerate as flies age.7,27 In contrast, EMB-9a displays a severe defect in myofibril assembly and shows faster degeneration than EMB (Figure 3). Compared to EMB (Fig. 3A), EMB-9a late pupae (Fig. 3B) show frayed and cracked myofibrils with some disruption of the hexagonal arrangement of thick and thin filaments. In contrast to the regular arrangements of myofilaments in EMB late pupae (Fig. 3C), EMB-9a sarcomeres often appear “cracked” or misaligned (Fig. 3D). While EMB two-day-old adults maintain some normal myofilament packing (Fig. 3E), EMB-9a myofibrils are completely disrupted (Fig. 3F). The myofibril disruption seen in EMB-9a two-day-old adults is similar to that of EMB one-week-old adults (Fig. 3G). Thus substitution of the IFI exon 9a relay domain into EMB exacerbates the abnormalities of myofibril stability associated with EMB.
Figure 3. Effects of the EMB-9a myosin isoform on myofibril assembly and stability in dorsal longitudinal IFM.
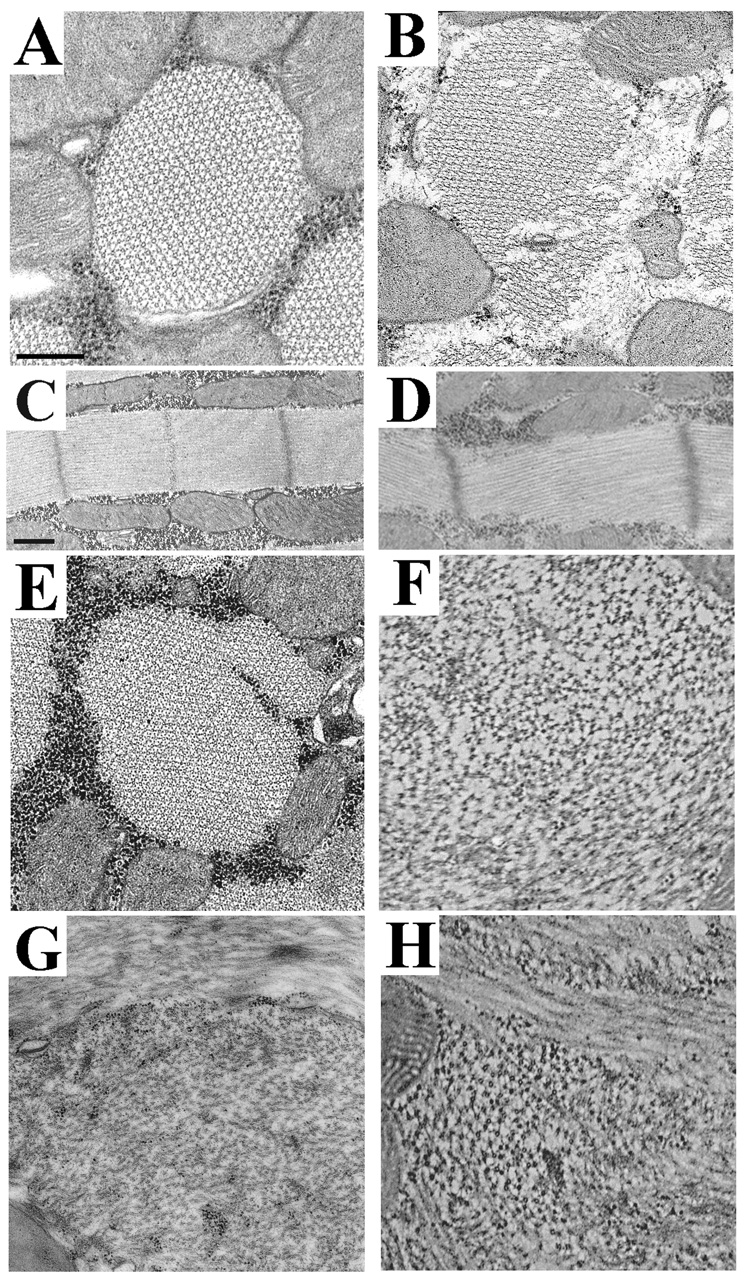
A, Transverse section from EMB late stage pupa. B, Transverse section from EMB-9a late stage pupa. Myofibril structure of EMB-9a shows cracking and disruption of the normal hexagonal packing of thick and thin filaments compared to EMB. The phenotype of EMB-9a pupa resembles that of EMB 2-day-old adult (E). C, Longitudinal section from EMB late stage pupa. D, Longitudinal section from EMB-9a late stage pupa. The sarcomere structure of EMB-9a is abnormal, Z-bands are slightly disrupted and M-lines are greatly disrupted compared to EMB sarcomeres. E, Transverse section from EMB two-day-old adult. Myofibrils show some abnormal hexagonal packing of thick and thin filaments. F, Transverse section from EMB-9a two-day-old adult. There is complete loss of myofibril shape, similar to the degeneration seen in EMB one-week-old adult (G). G, Transverse section from EMB one-week-old adult. H, Transverse section from EMB-9a one-week-old adult. In one-week-old adults, EMB and EMB-9a shown complete loss of myofibril shape and hexagonal packing of thick and thin filaments. Scale bars, 0.5 µm.
Discussion
Drosophila muscle Mhc is a single copy gene1,2 that utilizes alternative RNA splicing to produce different myosin isoforms.3,9 Exon 9 is one of the four sets of alternatively spliced exons encoding portions of the myosin S-1 head region (Fig. 4A). Alternative versions of exon 9 encode the relay domain, which is thought to act as a communication pathway between the converter domain, nucleotide-binding site and actin binding domain.28 Exon 9a encodes the indirect flight muscle isoform (IFI) relay domain, while exon 9b encodes a relay domain within one of the embryonic body wall isoforms (EMB). Analysis of myosin crystal structures in the pre- and post-power stroke states indicates that the converter domain (encoded by exon 11) is the only alternative domain that interacts with the relay domain. This interaction is discussed more fully below.
We generated chimeric myosins (IFI-9b and EMB-9a) by exchanging relay domains between the two native isoforms, IFI and EMB. We have previously shown that there are large functional differences between these myosins including a 9-fold difference in actin motility and a 2-fold difference in actin-activated Mg2+ ATPase.6 Surprisingly, exchanging relay regions did not make functional characteristics of EMB become more like IFI or vice versa, as has been the result of some of our previous alternative exons exchanges.8,10,12,16 Expression of IFI-9b in flies yielded wild-type structure and stability of IFM myofibrils, while expression of EMB-9a resulted in significant disruption of muscle structure and myofibril stability compared to EMB flies. The flight and jump ability of flies expressing IFI-9b were similar to wild type, while the EMB-9a isoform failed to rescue the flightless and impaired jumping phenotypes observed in flies expressing the EMB isoform. Assays with full-length IFI-9b myosin showed that its basal and actin-stimulated ATPase activities, as well as its in vitro actin sliding velocity, are similar to those of IFI. In contrast, the EMB-9a isoform shows a reduction of actin-stimulated ATPase activity compared to EMB and a marked increase in Km, suggesting an increased actin affinity. Most notably, EMB-9a is not capable of translocating actin filaments in vitro.
The cross-bridge cycle of EMB-9a myosin is slowed during actin interaction
The decreased rate of transition between cross-bridge states for EMB-9a compared to EMB must involve interaction with actin. This conclusion is based upon our observation that saturating levels of actin increased the MgATPase rate of EMB-9a only 8-fold compared to 14-fold for EMB, even though the basal MgATPase rates of the two isoforms were not significantly different. The basal myosin MgATPase rate is limited by Pi release. The rate of Pi release is accelerated by myosin binding to actin, to the point that the ATP hydrolysis step becomes limiting.29 It is possible that actin binding does not accelerate EMB-9a’s Pi release rate as much as EMB’s, or that one of the subsequent transitions involving actin is not as fast in EMB-9a myosin. The decrease in actin-activated MgATPase Km suggests that initial myosin binding to actin is not impaired and that a conformational change/step following actin binding is slowed. An abnormally high activation energy barrier at one of the strongly bound steps of the cross-bridge cycle could drastically slow a transition between actomyosin states and result in the apparent “stalling” of EMB-9a myosin in the motility assay. Strongly bound heads in the post-power stroke state impose drag on actin, which limits its velocity. The predominant strongly bound transitions involve ADP release and the binding of ATP. Thus the simplest explanation is that EMB-9a spends more time than EMB in one or more of these states, long enough so that the drag by the strongly bound myosins completely inhibits attempts to move actin by other myosins going through their power stroke.30
Upon addition of ATP, we only saw a few actin filament ends wiggling. This is typical of actin held tightly to the surface by strongly bound heads and contrasts with the Brownian-motion-influenced whole actin filament wiggling (moving linearly, forward and back) observed when the myosin density is below the movement threshold in the assay. Stalling in a weakly bound state would have a similar effect as reducing surface myosin concentration as it would decrease the number of heads at any given time that are available to productively interact with actin. If EMB-9a was stalled in a weakly bound state, we should have been able to rescue motility by increasing myosin density, which did not occur.
The more severely compromised myofibril ultrastructure of EMB-9a IFM compared to EMB IFM also suggests that EMB-9a is spending a longer time in strongly bound actin states than EMB. IFMs of flies expressing EMB-based isoforms degenerate7,8,10,12, and we suggested that this is due to the expression of a slower MHC isoform within a very fast fiber type that undergoes stretch-activation.10 EMB myosin’s extended binding to actin would generate high force levels during the IFM’s attempts at contraction, damaging IFM ultrastructure that is designed for low force and high speed rather than high force and slow speed. EMB-9a would produce myofibril degradation sooner and to a much more severe degree than EMB if it binds to actin longer in a strongly bound state. A longer time spent in weakly bound states would not be expected to have such an early, destructive impact on myofibril ultrastructure.
Molecular differences between the relay domains
The substantial difference in biochemical and mechanical properties between EMB-9a and EMB myosin isoforms must be attributed to one or more of the five amino acid residues that differ between the alternative relay domains encoded by exons 9a and 9b (Figs. 4B and 4C). Using the scallop muscle myosin II crystal structure in the pre-power stroke state31 as a model, we highlighted the 57 amino acid long relay domain region encoded by Drosophila exon 9 in blue (Figs. 4A and 4B). The locations of the five residues that differ between the IFI and EMB isoforms are shown as colored space-filling groups of the IFI isoform on the model (Fig. 4B). Three amino acid differences are in the relay helix and two flank Trp-510 in the relay loop.
To examine which amino acid residues might play critical roles in defining the functional differences between EMB-9a and EMB myosins, we substituted the Drosophila residues into the scallop muscle myosin II crystal structure31 in the pre-power stroke state (Figure 4) and in the near rigor state32 (not shown). We then compared interactions of the relay isoform-specific residues with other amino acids by visual inspection using PyMOL. Only amino acid 494 showed obvious interaction with residues outside the relay domain in the crystal structures examined. In IFI (Fig. 4D, top right), Ile-494 (red), a non-conserved residue in the relay helix, likely forms a hydrophobic interaction with Phe-671 (green), a conserved residue in strand three of beta-sheet B in the main body of the molecule. However, in EMB (Fig. 4D, middle right), residue 494 is a positively charged histidine that is expected to have less hydrophobic interaction with Phe-671 than isoleucine. Thus when the IFI relay domain replaces the EMB relay in EMB-9a (Fig. 4D, bottom right), Ile-494 would form a stronger hydrophobic interaction with Phe-671 than that normally present in EMB. Fischer et al.18 predicted that Phe-671 acts as a pivot point for the relay helix during its unwinding and bending. They suggested that two Phe residues located in the relay helix (Phe-490 and Phe-491) have a hydrophobic interaction with Phe-671 that aids in forming this fulcrum. It is thus possible that fulcrum function and hence relay helix conformational changes could be differentially affected by the relay isoform-specific differences.
While four of the isoform-specific relay domain residues do not make obvious inter-domain contacts in the crystal structures examined, the amino acid substitutions could still affect the rigidity of the relay helix or loop and thereby influence the kinetics of the EMB-9a isoform. In particular, the two amino acid changes flanking Trp-510 are worthy of note, since the fluorescence level of this tryptophan residue is altered by the presence of nucleotide, suggesting it reflects conformational changes of the relay loop during the mechanochemical cycle.24
Altering the relay domain likely influences the interactions with the converter region.18 While most of the regions that the relay domain interacts with are identical between IFI and EMB (they are encoded by constitutive exons), the interacting converter regions (encoded by alternative exons 11e and 11c) are very different.8 Exchange of the IFI relay area into the EMB backbone may result in a different interface between the relay area and the converter domain. This could affect the rotation of the converter and ultimately the swing of the lever arm. Indeed, cross-linking experiments have documented the importance of a constant interaction between the relay and converter domains throughout the catalytic cycle.33 There are several amino acid differences between the EMB and IFI converters that could affect the interaction with amino acids of the relay domain. A likely candidate, based on our visual inspection, is a charge difference at residue 762 (blue) of the converter domain (His in IFI vs. Asn in EMB) that likely interacts with Glu-506 (magenta) that is found in both the EMB and IFI relay domains (Fig. 4D).
Our current results regarding EMB-9a and a previous study where the IFI relay was paired with the EMB converter (IFI-EC), show that this combination of relay and converter abolishes or drastically slows in vitro motility. IFI-EC showed in vitro actin motility of 2.7 µm/s compared to 6.4 µm/s for IFI.8 To our knowledge this grouping of relay and converter is not naturally used by Drosophila.5 Perhaps the IFI-EMB relay-converter pairing combined with other mechanism(s) that contribute to the normally slow EMB velocity (0.7 µm/s) severely reduce the probability of a cross-bridge transition, so that no movement of actin is possible by EMB-9a.
It is interesting that exchanging the IFI relay into EMB has such a drastic effect (EMB-9a), but that the opposite exchange, the EMB relay exchanged into IFI (IFI-9b), has no effect on any of the measured molecular parameters. The lack of an effect is consistent with our previous observation that the EMB relay paired with the IFI converter (EMB-IC) nearly retains the IFI’s very fast motility velocity, 5.4 µm/s compared to 6.4 µm/s.8
Several previous studies have documented the importance of relay domain conformational changes in coordinating communication among the converter domain, lever arm, ATP-binding site and actin-binding site. Tsiavaliaris et al.34 showed that mutating a hydrophobic cluster (Phe-496 and Phe-515) resulted in disruption of myosin motor function and increased affinity for actin. They suggested that these hydrophobic residues stabilize the relay helix and help coordinate movements resulting from conformational changes. Ito et al.35 showed that mutating Phe-491 resulted in a myosin that had severely reduced MgATPase activity and failed to move actin. Sasaki et al.17 showed that mutation of Ile-508 results in absence of motor function with an increase of basal MgATPase and that the converter domain was incapable of moving during ATP hydrolysis.
To further examine the possible incompatibility of the IFI relay with the EMB converter and to test possible mechanisms, one could express these two domains together in other backbones besides EMB and IFI. Further, the roles of specific amino acids could be tested. For example one could switch residue 762 in EMB-9a from the EMB-specific converter residue (Asn) to the IFI-specific converter residue (His). This mutation might rescue actin motility and/or improve ATP hydrolysis rate compared to EMB-9a. One could also test an EMB construct in which the naturally occurring His-494 of the relay helix is replaced with the IFI Ile residue with or without replacing the Asn-762 residue of the converter domain with His-762. As these are the residues that appear to form the isoform-specific interactions, they are the most likely candidates for causing functional differences. Another possibility would be to test the importance of each of the amino acids flanking Trp-510, as their proximity to the converter suggests possible transient interactions with it during the mechanochemical cycle. In addition to making point mutations that might define critical residues responsible for the observed incompatibility, it would be useful to employ transient kinetic analyses to support or disprove our hypothesis that decreased ATP affinity or increased ADP affinity are key elements in stalling EMB-9a myosin in a strongly bound state.
The influence of the relay domain compared to other alternative regions
We have now examined all four alternative exon regions in the S-1 head of Drosophila myosin and determined their influence on ATPase rates, in vitro motility and myofibril ultrastructure.8,10,12,16 Paradoxically, the relay region had the most and least effect on myosin molecular functional properties compared to the three other S-1 alternative exon regions we exchanged between the IFI and EMB isoforms. The EMB-9a exchange was the only one of the 8 chimeras to completely abolish actin movement in the motility assay. Two of the other 3 IFI alternative exons exchanged into EMB increased velocity. IFI converter 11e (5.4 µm/s) and N-terminal domain 3b (4.1 µm/s) stimulated higher actin sliding velocity than EMB (0.7 µm/s). IFI domain 7d, near the site of nucleotide exchange did not alter motility. The EMB converter domain was the only one of the EMB alternative regions to affect actin sliding velocity when exchanged into IFI, decreasing it to 2.7 µm/s compared to 6.4 µm/s for IFI myosin. The remaining exon switches, involving EMB exons 7a, 3a, and 9b, did not change IFI velocity.
With respect to actin-activated MgATPase, Vmax of EMB-9a was the lowest of all 8 chimeras. IFI exons 7d and 11e increased EMB’s actin-activated MgATPase, while exon 3b had no effect. For the opposite exchanges, 9b was the only EMB alternative region to have no effect on IFI actin-activated MgATPase. EMB domains encoded by exons 7a and 11c increased IFI ATPase rate, while the exon 3a domain decreased it. As we now have a molecular phenotype for all the exchanges, it will be interesting and challenging to propose and test structural mechanisms that are behind these functional differences.
Materials and Methods
DNA Constructs
We constructed two P element-containing transgenes, IFI-9b and EMB-9a. IFI-9b forces the expression of EMB exon 9b in all MHC isoforms. EMB-9a forces expression of the adult IFM exon 9a isoform in the EMB backbone.
To construct IFI-9b, we removed exons 4 through 12 along with their flanking introns from the wild-type genomic construct pWMHC227 and replaced the deleted region with cDNA encoding constitutive and IFI Mhc exons, except for EMB exon 9b. To accomplish this we digested cDNA clone cD3013 with Pml I and Apa I and then gel isolated a 1.8-kb fragment containing exons 4, 5, 6, 7d, 8, 9b, 10, 11b and 12. pWMHC2 was digested with Xba I and Pml I. A 4.3-kb fragment from this digest (containing the 5’ end of Mhc through exon 4) was gel isolated and ligated with the Pml I-Apa I cD301 fragment and inserted into the pBluescriptKS vector (Stratagene, La Jolla, CA) that had been digested with Xba I and Apa I. The resulting subclone, IFI-9b5’11b, contains a non-IFI version of exon 11. We then switched this version of exon 11 (11b) with the IFI specific exon 11e by using cDNA clone EMB-IC8 that was digested with Nru I and Apa I. The gel-isolated Nru I-Apa I 1.2-kb fragment which contains exons 8, 9b, 10, 11e and 12 was ligated to the vector containing the 5’ portion of IFI-9b5’11b (which had been gel isolated after digestion with Nru I and Apa I). The resulting subclone, IFI-9b5’, was then digested with Apa I and Kpn I and the fragment containing the plasmid plus the 5’ portion of Mhc was gel isolated. We then digested pWMHC2 with Apa I and Kpn I and gel isolated the Apa I-Kpn I 12.4-kb fragment (which contains exon 12 through exon 19 and their flanking introns). This was ligated to the Apa I-Kpn I fragment of IFI-9b5’. The resulting subclone, pIFI-9b, was digested with Xba I and Kpn I and ligated into pCaSpeR 436 which had been digested with the same enzymes, to yield IFI-9b.
To construct EMB-9a, exon 9b from EMB7 was replaced with the adult version of exon 9 (9a). Exon 9a was derived from reverse transcription (RT)-PCR generated cDNA of mRNA from upper thoraces. Specifically we digested EMB with Xba I and Apa I, which produces a 4.7-kb fragment and contains exon 1 through exon 12. This fragment was cloned into pBluescriptKS to produce pEMB5’. Next, RNA was extracted from 50 upper thoraces using a LiCl2 extraction method.37 An aliquot of total RNA (0.5 µg) and 3 µmol of reverse primer specific to exon 12 (5’-GCTTGACCTTCTGCCACAGTT-3’) was used for cDNA synthesis using Roche’s One-Step RT-PCR system. PCR was performed with 1 µl of cDNA and 3 µmol of forward primer specific to exon 8 (5’-CGATACCGCCGAGCTGTACAG-3’) and reverse primer specific to exon 12 (5’-GCTTGACCTTCTGCCACAGTT -3’) using the following PCR conditions: 30 seconds at 94°C, 30 seconds at 55°C and 2 minutes at 72°C, repeated for a total of 30 cycles. The resulting 1.2-kb PCR fragment, which contains exons 8, 9a, 10, 11e and 12 was then digested with Bgl II and Nco I, yielding a 183-bp fragment containing exons 8, 9a and 10. This was ligated into pEMB5’ that had been digested with Bgl II and Nco I to produce the subclone pEMB-9a5’. A 4.3-kb Apa I-Kpn I cDNA fragment containing exons 12 through 19 from EMB was then ligated to pEMB-9a5’ which had been digested with Apa I and Kpn I to generate the subclone, pEMB-9a. The 9-kb cDNA insert from pEMB-9a was then subcloned into pCaSpeR 4 using Xba I and Kpn I to yield EMB-9a.
All cloning sites and PCR generated fragments were sequenced to ensure no changes had taken place in the amplification or cloning process.
P Element Transformation of Chimeric Mhc Genes
P element transformation was carried out as previously described.26 EMB-9a plasmid at 0.4 µg/µl along with the helper plasmid Δ2–3 at 0.08 µg/µl were injected into yw strain embryos. The IFI-9b plasmid was injected in the same manner, but at 0.6 µg/µl. To map the transgene inserts, the transgenic lines were crossed with a balancer line, w1118;CyO/Bl;TM2/TM6B, and standard segregation analysis was carried out. To analyze the expression of myosin in the transgenic lines, each was crossed into the Mhc10 genetic background, which lacks myosin in the IFM and TDT muscle.25,38
RT-PCR
Exon-specific primers were used to determine if the correct exon was being expressed from the EMB-9a transgene. RNA was extracted from 50 upper thoraces. RT-PCR was then performed on RNA (0.5 µg) with 3 µmol of each primer pair as described in the methods for DNA constructs. For EMB-9a and EMB (control), the primer pairs were exon 8 forward primer (5’- CGATACCGCCGAGCTGTACAG-3’) with exon 11c reverse primer (5’- ACCCAAACGGTACTGATCAT-3’) (exon 11c is expressed only by EMB-based genes). For a control, a second set of RT-PCR reactions was performed using exon 8 forward primer (5’-CGATACCGCCGAGCTGTACAG-3’) with exon 11e reverse primer (5’-CGACGGCTTCCAAGCACTTTC-3’) (exon 11e is expressed only in IFI). To confirm that exon 9a was being expressed only from the EMB-9a transgene, the RT-PCR generated fragments were gel isolated and digested with Pst I (a Pst I restriction enzyme site is found only in exon 9a).
Exon-specific primers were used in RT-PCR to determine the expression pattern of the IFI-9b transgene, using RNA extracted from upper thoraces as described above. For IFI-9b and yw (control) the primer pairs were exon 2 forward primer (5’-CCAGTCGCAAATCAGGAG-3’) with exon 4 reverse primer (5’-CAGAGATGGCGAAAATATGG-3’), exon 8 forward (5’-CGATACCGCCGAGCTGTACAGA-3’) with exon 10 reverse (5’-TCGAACGCAGAGTGGTCAT-3’), and exon 14 forward (5’-CTCAAGCTCACCCAGGAGGCT-3’) with exon 16 reverse (5’-GGGTGACAGACGCTGCTTGGTT-3’). The RT-PCR generated fragments were gel isolated and digested at exon specific restriction sites. Exon 3-containing RT-PCR was digested with Bgl II (specific to exon 3b). Exon 15-containing RT-PCR product was digested with Pvu I (Pvu I is specific to exon 15a). The exon 9-containing RT-PCR product was subcloned and sequenced to verify the presence of exon 9b.
Northern Blot Analysis
To determine if the correct exons were being expressed from the IFI-9b transgene, exon-specific probes for each alternative exon in Drosophila Mhc5 were individually used to probe northern blots containing RNA from yw (control) and IFI-9b larvae and pupae. RNA isolation and northern blotting were performed as previously described.37
Myosin Expression Levels
SDS PAGE was performed to determine the expression levels for each transgenic line in the Mhc10 background, as previously described.38 Briefly, upper thoraces were dissected from five 2-day-old female flies and homogenized in 50 µl SDS gel buffer. Ten µl of sample were loaded on a 9% polyacrylamide gel. At least 5 different SDS-PAGE gels were run for each genotype, each containing a newly prepared sample. Stained gels were digitally scanned using an Epson Expression 636 flatbed scanner. Protein accumulation was assessed using NIH image software (http://rsb.info.nih.gov/nih-image/). The myosin to actin ratio was determined for each lane and this was normalized to the ratio for wild-type flies, which was defined as a value of 1.0.
Flight Assay
To determine flight ability, greater than 100 flies for each transgenic line (in a Mhc10 background) were subjected to the flight assay as previously described.7,39 The method determines if flies are capable of flying upward (U), horizontally (H), downward (D) or not at all (N). All flight assays were performed with 2-day-old female flies at 22°C. Flight index was calculated as 6U/T + 4H/T + 2D/T + 0N/T, where T is the total number of flies tested.40
Jump Assay
To determine jump ability, 50 female flies for each transgenic line were assayed as previously described.7 Testing was performed at 22°C after wing removal. Jump ability is defined as the distance a fly jumps horizontally off of a 10-cm platform. Each fly was given 10 jump tests and an average value was determined.
Analysis of Indirect Flight Muscle Ultrastructure
The effects of transgene expression on myofibril assembly and stability were determined using transmission electron microscopy, as previously described.41 At least three different female samples were examined for each transgenic line. Cross-sections and longitudinal sections for each transgenic line were analyzed at various stages of development (pupa, two-day-old adult and one-week-old adult).
Myosin Isolation
Dorsolongitudinal IFM from 120–200 wild-type or transgenic flies were removed via micro dissection, and purified myosin was obtained using methods described previously6 with modifications. In brief, isolated fibers were combined into a 1.75 ml microcentrifuge tube containing 1 ml of York Modified Glycerol (YMG: 20 mM potassium phosphate, pH 7.0, 2 mM MgCl2, 1 mM EGTA, 10 mM DTT, and a protease inhibitor mixture) immediately following dissection, and were subsequently centrifuged at 8,500 × g using a Sorvall Microspin 24S centrifuge for 5 min at 4°C. Muscle fiber pellets were resuspended in 1 ml of YMG solution containing 2% reduced Triton X-100 and allowed to incubate for 30 min on ice. After a 5 min centrifugation (8,500 × g), the permeablized fibers were washed free of detergent and glycerol by briefly suspending them in 1 ml of YM (YMG without glycerol) solution followed by centrifugation at 8,500 × g. The fiber pellets were resuspended in 82 µl of high-salt myosin extraction buffer (MEB: 1 M KCl, 50 mM potassium phosphate, pH 6.8, 5 mM MgCl2, 0.5 mM EGTA, 10 mM sodium pyrophosphate, 10 mM DTT, and a protease inhibitor mixture) and incubated on ice for 15 min to extract myosin. The insoluble material was removed with a 5 min centrifugation (8,500 × g). The high-salt extract was then diluted 25 fold with nano H2O to an ionic strength of 40 mM, and myosin was allowed to precipitate overnight. The following day, the precipitant was centrifuged using a Beckman TLA 100.3 rotor for 30 min at 100,000 × g. The protein pellet was immediately resuspended in 13.5 µl of wash buffer (2.4 M KCl, 100 mM histidine, pH 6.8, 0.5 mM EGTA, 10 mM DTT) and incubated on ice for at least 30 min. After assuring that the protein pellet was completely dissolved, the solution was slowly diluted 8 fold with nano H2O to a KCl concentration of 0.3 M for precipitation of any residual acto-myosin that did not dissociate. After centrifugation at 60,000 × g for 30 min, the resulting supernatant was then diluted 10 fold with nano H2O, and incubated on ice for 1 hour to precipitate the purified myosin. After a 30 min centrifugation at 100,000 × g, the purified myosin pellet was dissolved in myosin storage buffer (0.5 M KCl, 20 mM MOPS, pH 7.0, 2 mM MgCl2, and 10 mM DTT). Myosin concentration was determined by its absorption at 280 nm.42 ATPase and in vitro motility assays were performed immediately following myosin preparation.
Determination of ATPase Activity and in vitro Motility
Myosin ATPase activities were determined using [γ-32P]ATP. CaATPase was measured as previously described.6 Actin-activated ATPase was determined using chicken skeletal muscle actin as previously described.10 G-actin was isolated from acetone powder of chicken skeletal muscle.43 After 1 cycle of polymerization-depolymerization, soluble G-actin obtained after dialysis against 2 mM Tris HCl, pH 8, 0.2 mM ATP, 2 mM CaCl2 and 1 mM DTT was quantified spectophotometrically using an extinction coefficient of 0.62 cm−1 (A310nm-A290nm) for 1 mg ml−1 of G-actin. F-actin was prepared by adding 1 volume of 10X polymerization buffer (50 mM TrisCl, pH 8, 0.5 M KCl, 20 mM MgCl2 and 10 mM ATP) to 9 volumes of G-actin. The working F-actin solution was at a concentration of ~30 µM, so that the amount of non-radioactive ATP added with actin to the ATPase reaction mixture was minimized.
In vitro actin sliding velocity assays were implemented as previously described6, with some alterations. First, the amount of DTT was doubled from 10 mM to 20 mM in all solutions to help prevent rapid photobleaching of fluorescently labeled actin filaments, and for increased protection against oxidation of isolated myosin. Second, 0B/MC/GOC (0 salt motility assay buffer/0.4% methyl cellulose/glucose oxidase and catalase) and 0B/MC/GOC + ATP (same composition as 0B/MC/GOC with 2 mM ATP added) solutions were diluted with nano H20 to 70% of the concentrations used previously. The decreased ionic strength increased acto-myosin interactions, elevating levels of continuous movement by the majority of actin filaments. At least three sets of assays were performed for each isoform studied. Analysis of captured video sequences was performed as previously described44, with the modifications detailed by Swank et al.6
Protein Structure Analysis
The scallop muscle myosin II crystal structures in the pre-power stroke state (pdb #1qvi)31 and the near rigor state (pdb #1SR6)32 were used as templates for protein structure analysis. The entire myosin S1 amino acid sequence for each of the isoforms, IFI, IFI-9b, EMB or EMB-9a was inserted into the scallop myosin S1 backbone. Amino acid differences in relevant areas of the IFI, EMB, and EMB-9a protein models were examined to detect protein interactions unique to each isoform. Structures were viewed using PyMOL (http://pymol.sourceforge.net/, DeLano Scientific, Palo Alto, CA, USA).
Acknowledgements
We appreciate comments on the manuscript from Dr. Michael Geeves and Dr. Marieke Bloemink (University of Kent, Canterbury). We are grateful to Dr. David Shin (the Scripps Research Institute) for help with the PyMOL program. The authors thank Allen Church and Jennifer Suggs for excellent technical assistance. Funds to support this research were provided by NIH grants R01 GM32443 to S.I.B. and R01 AR055611 to D.M.S. and American Heart Association scientist development grant 0635058N to D.M.S. An NSF equipment grant (0308029) to Dr. Steven Barlow of the SDSU Electron Microscope Facility supported the purchase of the electron microscope.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bernstein SI, Mogami K, Donady JJ, Emerson CP., Jr Drosophila muscle myosin heavy chain encoded by a single gene in a cluster of muscle mutations. Nature. 1983;302:393–397. doi: 10.1038/302393a0. [DOI] [PubMed] [Google Scholar]
- 2.Rozek CE, Davidson N. Drosophila has one myosin heavy-chain gene with three developmentally regulated transcripts. Cell. 1983;32:23–34. doi: 10.1016/0092-8674(83)90493-2. [DOI] [PubMed] [Google Scholar]
- 3.George EL, Ober MB, Emerson CP., Jr Functional domains of the Drosophila melanogaster muscle myosin heavy-chain gene are encoded by alternatively spliced exons. Mol Cell Biol. 1989;9:2957–2974. doi: 10.1128/mcb.9.7.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hastings GA, Emerson CP., Jr Myosin functional domains encoded by alternative exons are expressed in specific thoracic muscles of Drosophila. J Cell Biol. 1991;114:263–276. doi: 10.1083/jcb.114.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang S, Bernstein SI. Spatially and temporally regulated expression of myosin heavy chain alternative exons during Drosophila embryogenesis. Mech Dev. 2001;101:35–45. doi: 10.1016/s0925-4773(00)00549-9. [DOI] [PubMed] [Google Scholar]
- 6.Swank DM, Bartoo ML, Knowles AF, Iliffe C, Bernstein SI, Molloy JE, Sparrow JC. Alternative exon-encoded regions of Drosophila myosin heavy chain modulate ATPase rates and actin sliding velocity. J Biol Chem. 2001;276:15117–15124. doi: 10.1074/jbc.M008379200. [DOI] [PubMed] [Google Scholar]
- 7.Wells L, Edwards KA, Bernstein SI. Myosin heavy chain isoforms regulate muscle function but not myofibril assembly. EMBO J. 1996;15:4454–4459. [PMC free article] [PubMed] [Google Scholar]
- 8.Swank DM, Knowles AF, Suggs JA, Sarsoza F, Lee A, Maughan DW, Bernstein SI. The myosin converter domain modulates muscle performance. Nat Cell Biol. 2002;4:312–316. doi: 10.1038/ncb776. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein SI, Milligan RA. Fine tuning a molecular motor: the location of alternative domains in the Drosophila myosin head. J Mol Biol. 1997;271:1–6. doi: 10.1006/jmbi.1997.1160. [DOI] [PubMed] [Google Scholar]
- 10.Swank DM, Knowles AF, Kronert WA, Suggs JA, Morrill GE, Nikkhoy M, Manipon GG, Bernstein SI. Variable N-terminal regions of muscle myosin heavy chain modulate ATPase rate and actin sliding velocity. J Biol Chem. 2003;278:17475–17482. doi: 10.1074/jbc.M212727200. [DOI] [PubMed] [Google Scholar]
- 11.Swank DM, Kronert WA, Bernstein SI, Maughan DW. Alternative N-terminal regions of Drosophila myosin heavy chain tune muscle kinetics for optimal power output. Biophys J. 2004;87:1805–1814. doi: 10.1529/biophysj.103.032078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller BM, Zhang S, Suggs JA, Swank DM, Littlefield KP, Knowles AF, Bernstein SI. An alternative domain near the nucleotide-binding site of Drosophila muscle myosin affects ATPase kinetics. J Mol Biol. 2005;353:14–25. doi: 10.1016/j.jmb.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 13.Swank DM, Braddock J, Brown W, Lesage H, Bernstein SI, Maughan DW. An alternative domain near the ATP binding pocket of Drosophila myosin affects muscle fiber kinetics. Biophys J. 2006;90:2427–2435. doi: 10.1529/biophysj.105.075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell. 1998;94:559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 15.Holmes KC. The swinging lever-arm hypothesis of muscle contraction. Curr Biol. 1997;7:R112–R118. doi: 10.1016/s0960-9822(06)00051-0. [DOI] [PubMed] [Google Scholar]
- 16.Littlefield KP, Swank DM, Sanchez BM, Knowles AF, Warshaw DM, Bernstein SI. The converter domain modulates kinetic properties of Drosophila myosin. Am J Physiol Cell Physiol. 2003;284:C1031–C1038. doi: 10.1152/ajpcell.00474.2002. [DOI] [PubMed] [Google Scholar]
- 17.Sasaki N, Ohkura R, Sutoh K. Dictyostelium myosin II mutations that uncouple the converter swing and ATP hydrolysis cycle. Biochemistry. 2003;42:90–95. doi: 10.1021/bi026051l. [DOI] [PubMed] [Google Scholar]
- 18.Fischer S, Windshugel B, Horak D, Holmes KC, Smith JC. Structural mechanism of the recovery stroke in the myosin molecular motor. Proc Natl Acad Sci U S A. 2005;102:6873–6878. doi: 10.1073/pnas.0408784102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher AJ, Smith CA, Thoden JB, Smith R, Sutoh K, Holden HM, Rayment I. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4. Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 20.Smith CA, Rayment I. X-ray structure of the magnesium(II).ADP.vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 A resolution. Biochemistry. 1996;35:5404–5417. doi: 10.1021/bi952633+. [DOI] [PubMed] [Google Scholar]
- 21.Mesentean S, Koppole S, Smith JC, Fischer S. The principal motions involved in the coupling mechanism of the recovery stroke of the myosin motor. J Mol Biol. 2007;367:591–602. doi: 10.1016/j.jmb.2006.12.058. [DOI] [PubMed] [Google Scholar]
- 22.Geeves MA, Fedorov R, Manstein DJ. Molecular mechanism of actomyosin-based motility. Cell Mol Life Sci. 2005;62:1462–1477. doi: 10.1007/s00018-005-5015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kronert WA, Edwards KA, Roche ES, Wells L, Bernstein SI. Muscle-specific accumulation of Drosophila myosin heavy chains: a splicing mutation in an alternative exon results in an isoform substitution. EMBO J. 1991;10:2479–2488. doi: 10.1002/j.1460-2075.1991.tb07787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yengo CM, Chrin LR, Rovner AS, Berger CL. Tryptophan 512 is sensitive to conformational changes in the rigid relay loop of smooth muscle myosin during the MgATPase cycle. J Biol Chem. 2000;275:25481–25487. doi: 10.1074/jbc.M002910200. [DOI] [PubMed] [Google Scholar]
- 25.Collier VL, Kronert WA, O'Donnell PT, Edwards KA, Bernstein SI. Alternative myosin hinge regions are utilized in a tissue-specific fashion that correlates with muscle contraction speed. Genes Dev. 1990;4:885–895. doi: 10.1101/gad.4.6.885. [DOI] [PubMed] [Google Scholar]
- 26.Cripps RM, Becker KD, Mardahl M, Kronert WA, Hodges D, Bernstein SI. Transformation of Drosophila melanogaster with the wild-type myosin heavy-chain gene: rescue of mutant phenotypes and analysis of defects caused by overexpression. J Cell Biol. 1994;126:689–699. doi: 10.1083/jcb.126.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swank DM, Wells L, Kronert WA, Morrill GE, Bernstein SI. Determining structure/function relationships for sarcomeric myosin heavy chain by genetic and transgenic manipulation of Drosophila. Microsc Res Tech. 2000;50:430–442. doi: 10.1002/1097-0029(20000915)50:6<430::AID-JEMT2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 28.Houdusse A, Kalabokis VN, Himmel D, Szent-Gyorgyi AG, Cohen C. Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. Cell. 1999;97:459–470. doi: 10.1016/s0092-8674(00)80756-4. [DOI] [PubMed] [Google Scholar]
- 29.White HD, Belknap B, Webb MR. Kinetics of nucleoside triphosphate cleavage and phosphate release steps by associated rabbit skeletal actomyosin, measured using a novel fluorescent probe for phosphate. Biochemistry. 1997;36:11828–11836. doi: 10.1021/bi970540h. [DOI] [PubMed] [Google Scholar]
- 30.Tyska MJ, Warshaw DM. The myosin power stroke. Cell Motil Cytoskeleton. 2002;51:1–15. doi: 10.1002/cm.10014. [DOI] [PubMed] [Google Scholar]
- 31.Gourinath S, Himmel DM, Brown JH, Reshetnikova L, Szent-Gyorgyi AG, Cohen C. Crystal structure of scallop myosin S1 in the pre-power stroke state to 2.6 a resolution: flexibility and function in the head. Structure (Camb) 2003;11:1621–1627. doi: 10.1016/j.str.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 32.Himmel DM, Gourinath S, Reshetnikova L, Shen Y, Szent-Gyorgyi AG, Cohen C. Crystallographic findings on the internally uncoupled and near-rigor states of myosin: further insights into the mechanics of the motor. Proc Natl Acad Sci U S A. 2002;99:12645–12650. doi: 10.1073/pnas.202476799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shih WM, Spudich JA. The myosin relay helix to converter interface remains intact throughout the actomyosin ATPase cycle. J Biol Chem. 2001;276:19491–19494. doi: 10.1074/jbc.M010887200. [DOI] [PubMed] [Google Scholar]
- 34.Tsiavaliaris G, Fujita-Becker S, Batra R, Levitsky DI, Kull FJ, Geeves MA, Manstein DJ. Mutations in the relay loop region result in dominant-negative inhibition of myosin II function in Dictyostelium. EMBO Rep. 2002;3:1099–1105. doi: 10.1093/embo-reports/kvf214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito K, Uyeda TQ, Suzuki Y, Sutoh K, Yamamoto K. Requirement of domain-domain interaction for conformational change and functional ATP hydrolysis in myosin. J Biol Chem. 2003;278:31049–31057. doi: 10.1074/jbc.M304138200. [DOI] [PubMed] [Google Scholar]
- 36.Pirrotta V. Vectors for P-mediated transformation in Drosophila. Biotechnology. 1988;10:437–456. doi: 10.1016/b978-0-409-90042-2.50028-3. [DOI] [PubMed] [Google Scholar]
- 37.Becker KD, O'Donnell PT, Heitz JM, Vito M, Bernstein SI. Analysis of Drosophila paramyosin: identification of a novel isoform which is restricted to a subset of adult muscles. J Cell Biol. 1992;116:669–681. doi: 10.1083/jcb.116.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Donnell PT, Collier VL, Mogami K, Bernstein SI. Ultrastructural and molecular analyses of homozygous-viable Drosophila melanogaster muscle mutants indicate there is a complex pattern of myosin heavy-chain isoform distribution. Genes Dev. 1989;3:1233–1246. doi: 10.1101/gad.3.8.1233. [DOI] [PubMed] [Google Scholar]
- 39.Drummond DR, Hennessey ES, Sparrow JC. Characterisation of missense mutations in the Act88F gene of Drosophila melanogaster. Mol Gen Genet. 1991;226:70–80. doi: 10.1007/BF00273589. [DOI] [PubMed] [Google Scholar]
- 40.Tohtong R, Yamashita H, Graham M, Haeberle J, Simcox A, Maughan D. Impairment of muscle function caused by mutations of phosphorylation sites in myosin regulatory light chain. Nature. 1995;374:650–653. doi: 10.1038/374650a0. [DOI] [PubMed] [Google Scholar]
- 41.O'Donnell PT, Bernstein SI. Molecular and ultrastructural defects in a Drosophila myosin heavy chain mutant: differential effects on muscle function produced by similar thick filament abnormalities. J Cell Biol. 1988;107:2601–2612. doi: 10.1083/jcb.107.6.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Margossian SS, Lowey S. Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol. 1982;85(Pt B):55–71. doi: 10.1016/0076-6879(82)85009-x. [DOI] [PubMed] [Google Scholar]
- 43.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85(Pt B):164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 44.Root DD, Wang K. Calmodulin-sensitive interaction of human nebulin fragments with actin and myosin. Biochemistry. 1994;33:12581–12591. doi: 10.1021/bi00208a008. [DOI] [PubMed] [Google Scholar]


