Abstract
Begomoviruses are single-stranded DNA viruses and cause severe diseases in major crop plants worldwide. Based on current genome sequence analyses, we found that synonymous codon usage variations in the protein-coding genes of begomoviruses are mainly influenced by mutation bias. Base composition analysis suggested that the codon usage bias of AV1 and BV1 genes is significant and their expressions are high. Fourteen codons were determined as translational optimal ones according to the comparison of codon usage patterns between highly and lowly expressed genes. Interestingly the codon usages between begomoviruses from the Old and the New Worlds are apparently different, which supports the idea that the bipartite begomoviruses of the New World might originate from bipartite ones of the Old World, whereas the latter evolve from the Old World monopartite begomoviruses.
Keywords: Begomovirus, Synonymous codon usage, Evolution, Bioinformatics
INTRODUCTION
Synonymous codon usage bias has been investigated in many organisms, as the genetic code is degenerate. The synonymous codons are also non-randomly used in viruses infecting living organisms. Several factors such as mutational bias (Jenkins and Holmes, 2003; Gu et al., 2004; Zhou et al., 2005), translational selection (Sau et al., 2005a; 2005b; 2005c), gene function (Wang et al., 2002; Gu et al., 2004; Zhou et al., 2005), gene length (Sau et al., 2005a), and CpG island (Shackelton et al., 2006) were found to influence codon usage in animal viruses and phages, and mutational bias was found as the major determinant factor. Adams and Antoniw (2004) also suggested that mutational bias rather than translational selection was the major determinant of codon usage variation amongst plant viruses.
Geminiviruses (family Geminiviridae) are single-stranded DNA (ssDNA) viruses that cause severe disease in major crop plants worldwide. Most geminiviruses belong to the genus Begomovirus, which are transmitted exclusively by the whitefly Bemisia tabaci (Harrison and Robinson, 1999). Many begomoviruses have bipartite genomes known as DNA A and DNA B. DNA A contains the AV1 (coat protein) and AV2 ORFs (open reading frames) in the virus strand and, on the complementary strand, four ORFs: AC1 (replication initiation protein), AC2 (transcriptional activator protein), AC3 (replication enhancer protein) and AC4. The virus and complementary strands of DNA B contain two ORFs: BV1 (nuclear shuttle protein) and BC1 (movement protein). Some begomoviruses have a monopartite genome and lack DNA B. Phylogenetic analysis shows that begomoviruses can be generally divided into two groups, the Old World begomoviruses (eastern hemisphere, Asia, Africa, Europe, the Mediterranean areas) and the New World begomoviruses (western hemisphere, the Americas). All the New World begomoviruses are evolved to bipartite with lack of AV2 ORF in DNA A, whereas both bipartite and monopartite begomoviruses in the Old World encode AV2 ORF (Harrison and Robinson, 1999).
Because of their destructive effect on cash crops (Moffat, 1999; Moriones and Navas-Castillo, 2000; Mansoor et al., 2006), numerous studies on begomoviruses have been conducted to understand their symptoms, host range, distribution, genome structure, gene function, and so on (Fauquet et al., 2003; Zhou et al., 2003). In this paper, we report the analysis of codon usage bias in begomoviruses and also perform an evolutionary analysis based on their codon usage pattern.
MATERIALS AND METHODS
Dataset
The complete genomic sequences of 147 begomovirus species were downloaded from the GenBank database, from which a total of 932 variants of the 8 known protein-coding genes were extracted. To minimize sampling errors, 915 variants were selected for further analysis by the following sifting standard: (1) the selected genes should be complete coding DNA sequences (CDS) with correct initial and terminal codons; (2) only those CDS including at least 80 codons were selected in the dataset; (3) those CDS with uncertain annotation or annotated as hypothetical protein-coding genes were excluded from this study.
Measures of synonymous codon usage bias
GC content is the frequency of G+C in a coding gene. GC1, GC2 and GC3 contents are the frequencies of G+C at the first, second and third positions of codons, respectively. A3, T3, G3 and C3 contents are the frequencies of A, T, G and C at the synonymous third position of codons, respectively. Effective number of codons (N c), ranging from 20 to 61, is generally used to measure the bias of synonymous codons. When N c value approaches 20, only one codon is used with extreme bias for one amino acid and, if the value is up to 61, the anonymous codons are used equally with no bias (Wright, 1990).
Relative synonymous codon usage (RSCU) is defined as the ratio of the observed frequency of codons to the expected frequency given that all the synonymous codons for the same amino acids are used equally. RSCU values have no relation to the amino acids usage and the abundance ratio of synonymous codons, which can directly reflect the bias of synonymous codon usage (Sharp and Li, 1986). The codon adaptation index (CAI) was used to estimate the extent of bias towards codons that were known to be preferred in highly expressed genes (Sharp and Li, 1987). It is now proved that CAI values mostly approach the theoretical values to reflect the expression level of a gene. Thus it has been widely utilized to measure the gene expression level (Naya et al., 2001; Gupta et al., 2004). A CAI value ranges from 0 to 1.0, and a higher value indicates a stronger codon usage bias and a higher expression level.
Statistical analysis
CodonW version 1.4.2 (John Peden, available at http://sourceforge.net/projects/codonw/), an integrated program, was utilized for calculating GC, GC3 contents and N c values and then carrying out correspondence analysis (CA), while GC, GC1, GC2 and GC3 contents were calculated by practical extraction and report language (PERL) scripts which were written by us. A3, T3, G3 and C3 contents, as well as RSCU and CAI values, were also calculated by using PERL scripts.
Correspondence analysis (CA) on synonymous codon usage
CA is the most commonly used multivariate statistical analysis at present (Greenacre, 1984). This method can successfully present variation trends among genes, and then distribute them along the continuous axis by using RSCU value as variable data. In CA, all genes were plotted in a 59-dimensional hyperspace, according to the usage of the 59 sense codons. Major variation trends can be determined using these RSCU values and genes ordered according to their positions along the major axis, which can also be used to distinguish the major factors influencing the codon usage of a gene.
Determination of optimal codons
The set of reference sequences used to calculate CAI values in this study were the genes coding for coat proteins. According to the calculated CAI values, 5% of the total genes with extremely high and low CAI values were regarded as the high and low dataset, respectively. Then we calculated the average RSCU values of the two gene samples and subtracted them subsequently in each dataset group (ΔRSCU). If the ΔRSCU values are larger than 0.08, then this codon will be defined as the optimal codon (Duret and Mouchiroud, 1999).
RESULTS
Base composition analysis
In order to examine the base composition variation among different genes, the base composition of different protein-coding genes was calculated. Table 1 shows that with the exception of the AC3 gene that has a lower average GC content (0.391), no obvious difference in GC content was found among other tested genes. However, differences in GC content at the different synonymous positions of codons were apparent. For example, GC content at the synonymous first position of codons for the AV2 gene is 0.563, while that for the AC4 gene is only 0.412. GC content at the synonymous third position of codons for the AC4 gene is 0.534, while that for the BV1 gene is only 0.399.
Table 1.
The GC content at the different codon positions and the A, T, G and C contents at the third position of begomovirus genes
| Genes | No. | GC | GC1 | GC2 | GC3 | A3 | T3 | G3 | C3 |
| AV1 (V1) | 147 | 0.452 (0.021)* | 0.465 (0.015) | 0.408 (0.013) | 0.485 (0.059) | 0.159 (0.030) | 0.357 (0.044) | 0.266 (0.028) | 0.219 (0.041) |
| AV2 (V2) | 92 | 0.484 (0.018) | 0.563 (0.040) | 0.404 (0.020) | 0.487 (0.042) | 0.220 (0.030) | 0.294 (0.033) | 0.245 (0.030) | 0.241 (0.028) |
| AC1 (C1) | 147 | 0.448 (0.018) | 0.497 (0.018) | 0.395 (0.012) | 0.452 (0.040) | 0.275 (0.027) | 0.273 (0.028) | 0.190 (0.027) | 0.262 (0.023) |
| AC2 (C2) | 143 | 0.471 (0.022) | 0.521 (0.031) | 0.466 (0.026) | 0.427 (0.037) | 0.293 (0.029) | 0.280 (0.028) | 0.185 (0.029) | 0.241 (0.032) |
| AC3 (C3) | 145 | 0.391 (0.021) | 0.421 (0.028) | 0.308 (0.027) | 0.442 (0.040) | 0.283 (0.034) | 0.275 (0.026) | 0.200 (0.030) | 0.242 (0.028) |
| AC4 (C4) | 117 | 0.483 (0.023) | 0.412 (0.026) | 0.502 (0.055) | 0.534 (0.030) | 0.265 (0.031) | 0.201 (0.039) | 0.331 (0.038) | 0.203 (0.025) |
| BV1 | 53 | 0.423 (0.028) | 0.459 (0.026) | 0.411 (0.025) | 0.399 (0.059) | 0.218 (0.030) | 0.383 (0.050) | 0.218 (0.036) | 0.181 (0.043) |
| BC1 | 53 | 0.447 (0.028) | 0.457 (0.017) | 0.412 (0.017) | 0.473 (0.068) | 0.300 (0.046) | 0.227 (0.041) | 0.230 (0.047) | 0.243 (0.053) |
Values in parenthesis indicate standard deviation (SD)
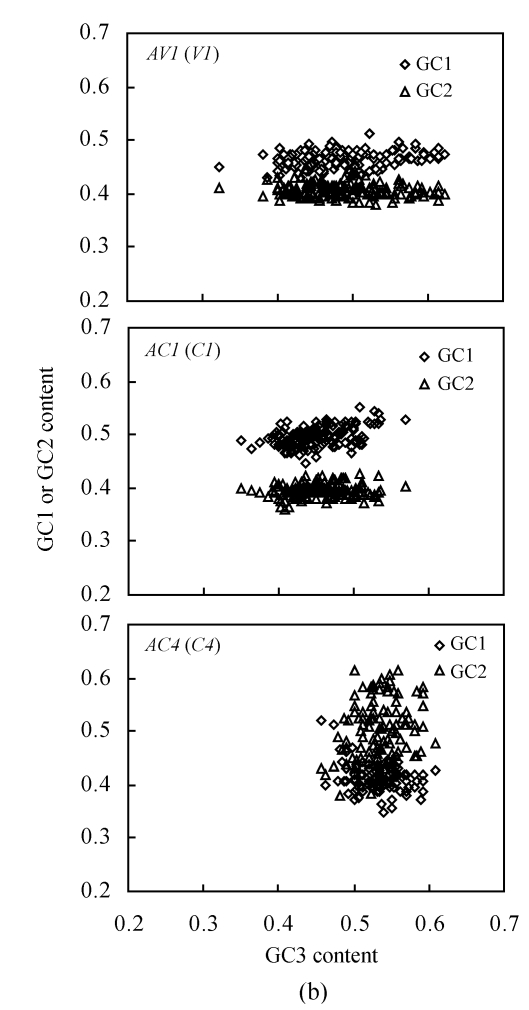
It was also observed that the average percentage of GC content was generally higher at the first than at the second codon position (Table 1), except for the AC4 gene whose GC2 content was larger than GC1 content. This result suggested that the AC4 gene might have a special codon usage pattern.
In addition we found that AV1 and BV1 genes had a tendency to usage bias at the synonymous third codon position. The AV1 gene does not tend to use A(T)-ending or G(C)-ending codons but tends to use T-ending codons relative to A-ending codons. However, BV1 gene apparently uses T-ending codons and seldom used C-ending codons. Therefore, AV1 and BV1 genes should show a stronger codon usage bias among the begomovirus genes.
Analysis of factors shaping codon usage
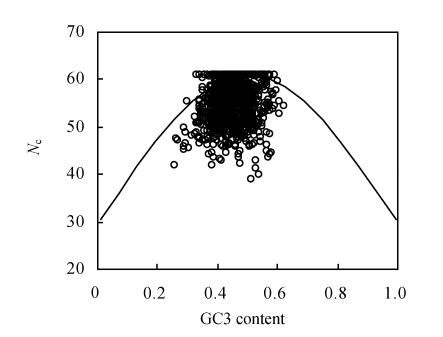
An N c plot (a plot of N c vs GC3 content) was widely used to investigate the determinants of the codon usage variations among genes in different organisms. It was suggested that if GC3 content was the only determinant of the codon usage variation among the genes, then the N c value would fall on the continuous curve between N c value and GC3 content (Wright, 1990). In general, if genes are distributed in N c plots approaching the expected continuous curve with no selection, then codon usage bias of genes is mainly influenced by compositional constraints. Otherwise the codon usage bias of genes is more affected by other factors such as translational selection, etc. The N c plot for genes of the 147 begomoviruses showed that although a small number of genes were located on the expected curve, most points lay far below the expected curve (Fig.1), suggesting that apart from mutation bias, other factors might also play a role in shaping the codon usage bias of begomoviruses (Guo et al., 2007).
Fig. 1.
Relationship between effective number of codons (N c) and GC3 content of the begomovirus genes. The continuous curve represents the expected curve between GC3 content and N c under random codon usage
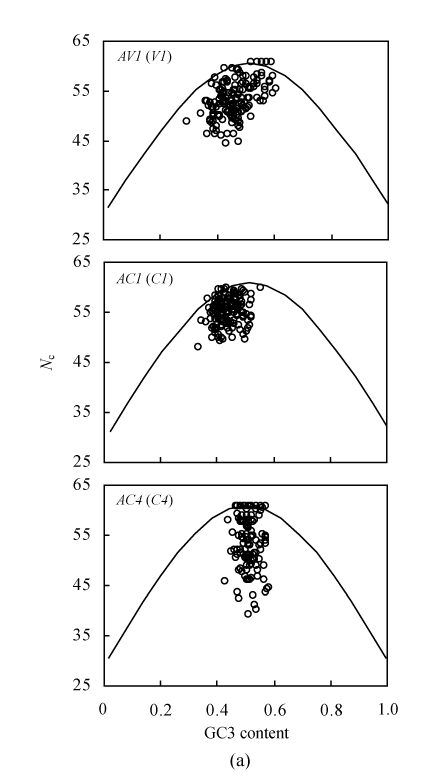
Base composition analysis suggested that the codon usage of the AC4 gene, which is always embedded in the AC1 gene, might differentiate from other genes. In addition, strong codon bias was observed in AV1 gene. Because of the special characteristics of the abovementioned genes, we selected them for examining the influence factors in shaping the codon usage (Fig.2a). The N c plots of AV1, AC1 and AC4 genes suggested that apart from compositional constraints, other factors might play important roles in shaping their codon usage, although high translational selection seems to lay stress on the AC4 gene because of the wide range of N c values for the same GC3 content.
Fig. 2.
(a) Relationship between the effective number of codons (N c) and GC content of the third codon position (GC3) of three different genes (≥80 codons long) in 147 begomoviruses. The continuous curves represent the expected curve between GC3 content and N c under random codon usage; (b) Relationship between the GC content at the first (GC1)/the second (GC2) codon position and that at the third codon position (GC3) of AV1, AC1 and AC4 genes (≥80 codons long) in 147 begomoviruses
To examine the reason for N c variation under the same GC3 content, the relationship between the GC1, GC2 and GC3 contents for AV1, AC1 and AC4 genes was further examined (Fig.2b). It was observed that the GC1 content was always higher than GC2 content for the AV1 and AC1 genes, whereas the GC1 content of the AC4 gene was generally lower than the GC2 content. This result was coincident with the base composition analysis (Table 1). Thus the variations of the synonymous first and second codon positions for the AC4 gene might be caused by the translational selection utilizing G or C at the synonymous second position.
As the AC4 gene is always embedded in the AC1 gene, the codon usage pattern of the AC4 gene might be influenced by the AC1 gene. The sequences of AC1 and AC4 genes were aligned. It was found that the synonymous third codon position of the AC1 gene corresponded to the second codon position of the AC4 gene for all the begomoviruses. Thus the base compositional environment of the AC1 gene might also influence the codon usage pattern of the AC4 gene. It could be postulated that the second codon position of the AC4 gene that tended to use G or C should be influenced by compositional constraints other than translational selection.
Determination of optimal codons
According to the variation analysis of base composition for the genes of begomoviruses, the AV1 gene was selected as a reference dataset to calculate the CAI values of all genes. The correlation analysis between CAI value and the positions of the genes along the first two major axes (generated by correspondence analysis), as well as other indices, was then calculated. It was observed that the CAI values were negatively correlated with the GC3 and GC contents (r=−0.234 and r=−0.535 respectively, P<0.01), while significantly positively correlated with axis 1 (r=0.587, P<0.01). Moreover the CAI value was also significantly negatively correlated with the N c value, indicating a tendency to a higher CAI value or a lower N c value and a higher expression level for begomovirus genes. Thus it is feasible to use the AV1 gene as a reference dataset for our estimation of the CAI value of begomovirus genes.
Based on the calculated CAI values, 5% of the total genes with extremely high and low CAI values were regarded as the high and the low datasets, respectively. Then we compared the codon usage of the high dataset to the low dataset. Table 2 shows that 14 codons that code 13 amino acids were apparently used at a high level, and can be determined as translational optimal codons. Out of the 14 optimal codons, 5 were ended with G, 1 with C and 8 with T.
Table 2.
Comparison of codon usage and relative synonymous codon usage (RSCU) between the high and low expressed genes in begomoviruses
| Amino acid | Codon | High | Low | ΔRSCU |
| Ala | GCA | 1.107 | 0.885 | 0.222** |
| GCC | 0.734 | 1.401 | −0.667 | |
| GCG | 0.438 | 0.977 | −0.539 | |
| GCT | 1.722 | 0.737 | 0.985*** | |
| Cys | TGC | 0.060 | 1.556 | −1.495 |
| TGT | 1.940 | 0.444 | 1.495*** | |
| Asp | GAC | 0.387 | 1.043 | −0.657 |
| GAT | 1.613 | 0.957 | 0.657*** | |
| Glu | GAA | 0.859 | 1.087 | −0.228 |
| GAG | 1.141 | 0.913 | 0.228** | |
| Phe | TTC | 0.748 | 1.106 | −0.358 |
| TTT | 1.252 | 0.894 | 0.358*** | |
| Gly | GGA | 0.647 | 1.485 | −0.838 |
| GGC | 0.849 | 0.782 | 0.067 | |
| GGG | 0.696 | 1.042 | −0.347 | |
| GGT | 1.809 | 0.691 | 1.118*** | |
| His | CAC | 0.376 | 1.075 | −0.698 |
| CAT | 1.624 | 0.925 | 0.698*** | |
| Ile | ATA | 0.907 | 0.692 | 0.215** |
| ATC | 0.804 | 1.286 | −0.481 | |
| ATT | 1.288 | 1.022 | 0.266 | |
| Lys | AAA | 0.451 | 1.034 | −0.583 |
| AAG | 1.549 | 0.966 | 0.583*** | |
| Leu | CTA | 0.186 | 1.330 | −1.145 |
| CTC | 0.594 | 0.923 | −0.329 | |
| CTG | 0.755 | 1.045 | −0.291 | |
| CTT | 0.829 | 1.412 | −0.583 | |
| TTA | 1.113 | 0.937 | 0.177* | |
| TTG | 2.524 | 0.353 | 2.171*** | |
| Asn | AAC | 0.866 | 0.835 | 0.031 |
| AAT | 1.134 | 1.165 | −0.031 | |
| Pro | CCA | 1.096 | 1.383 | −0.287 |
| CCC | 1.773 | 0.697 | 1.077*** | |
| CCG | 0.162 | 0.886 | −0.723 | |
| CCT | 0.968 | 1.035 | −0.067 | |
| Gln | CAA | 0.780 | 1.277 | −0.497 |
| CAG | 1.220 | 0.723 | 0.497*** | |
| Arg | AGA | 1.199 | 1.192 | 0.007 |
| AGG | 1.438 | 1.611 | −0.174 | |
| CGA | 0.570 | 1.336 | −0.767 | |
| CGC | 0.526 | 0.537 | −0.011 | |
| CGG | 0.461 | 0.511 | −0.050 | |
| CGT | 1.807 | 0.812 | 0.994*** | |
| Ser | AGC | 1.142 | 0.748 | 0.394*** |
| AGT | 1.042 | 0.830 | 0.212** | |
| TCA | 0.592 | 1.477 | −0.886 | |
| TCC | 0.917 | 0.985 | −0.068 | |
| TCG | 0.867 | 1.094 | −0.228 | |
| TCT | 1.442 | 0.866 | 0.575*** | |
| Thr | ACA | 0.661 | 1.237 | −0.576 |
| ACC | 0.602 | 0.728 | −0.127 | |
| ACG | 0.558 | 1.546 | −0.988 | |
| ACT | 2.179 | 0.489 | 1.690*** | |
| Val | GTA | 0.448 | 1.208 | −0.760 |
| GTC | 0.998 | 0.849 | 0.149* | |
| GTG | 1.034 | 0.784 | 0.250** | |
| GTT | 1.520 | 1.159 | 0.361 | |
| Tyr | TAC | 0.540 | 0.938 | −0.399 |
| TAT | 1.460 | 1.062 | 0.399 |
Major codons, defined by a higher RSCU value in highly expressed genes, are indicated as (ΔRSCU≥0.30)
Major codons, defined by a higher RSCU value in highly expressed genes, are indicated as (ΔRSCU≥0.20)
Major codons, defined by a higher RSCU value in highly expressed genes, are indicated as (ΔRSCU≥0.08)
Codon usage reflecting the evolution process
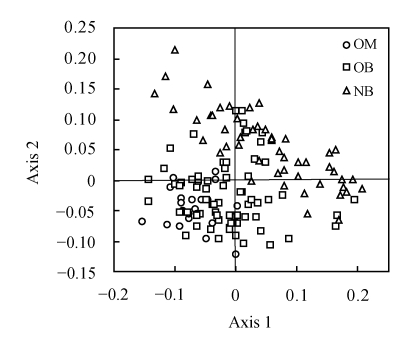
CA was performed on the RSCU value based on the concatenated genes for each begomovirus genome. Fig.3 shows the positions of all tested begomoviruses along the first two major axes. The first major axis accounted for 15.2% of the total variations, while the second major axis accounted for 9.9% of the total variations.
Fig. 3.
Positions of the 147 begomoviruses in the plot of the first two major axes by correspondence analysis of relative synonymous codon usage (RSCU) values. The first and second major axes can account for 15.2% and 9.9% of the total variations, respectively. The begomoviruses are divided into three groups, the Old World begomoviruses with monopartite genomes (OM), the Old World viruses with bipartite genomes (OB) and the New World viruses with bipartite genomes (NB)
In order to detect the codon usage variation of different genomes, the begomoviruses were divided into three groups including the Old World begomoviruses with monopartite genomes (OM), the Old World begomoviruses with bipartite genomes (OB), and the New World begomoviruses with bipartite genomes (NB). The distribution of the three groups along the first two major axes showed that the Old World monopartite begomoviruses and the New World bipartite ones were located in two independent fields, indicating that the two groups of begomoviruses exhibit a different codon usage pattern. Because the species with a close genetic relationship always present a similar codon usage pattern (Sharp et al., 1988), the genetic relationship of the two groups of begomoviruses should be far removed from each other. As to the Old World begomoviruses with bipartite genomes, we found that the majority of them exhibited a similar codon usage pattern with the Old World monopartite begomoviruses, and a few of them showed a similar codon usage to the New World bipartite begomoviruses. An explication of this result might be that a number of Old World bipartite begomoviruses evolved to adopt the codon usage pattern of some Old World monopartite begomoviruses. Moreover, the New World bipartite begomoviruses were closely related to a small number of the Old World bipartite ones and far removed from the Old World monopartite ones.
DISCUSSION
The results of the N c-plot and base composition analysis indicated that the codon usage pattern of begomoviruses was influenced by mutation bias as well as other factors such as translational selection. Comparative analysis of AC1 and AC4 genes showed that the compositional environment of the former genes might play a role in dictating the codon usage of the latter gene. Thus, although it seems that strong translational selection might have an influence on shaping the codon usage of AC4 genes, the compositional constraints derived from AC1 genes might be the major determinant in determining codon usage. Consequently it can be speculated that mutation bias might play a major role in shaping the codon usage pattern of begomoviruses.
As to the gene itself, the selection pressures from the external environment always act as effective factors in promoting the gene to adapt to the change in the external environment. On the other hand, direct changes to a gene will interfere with or be harmful to the gene. Therefore, the base mutation at the synonymous third codon position may not affect the protein expression for AC1 gene because of the degeneration of genetic codons. But for the embedded AC4 gene, the corresponding base mutation occurs at the second codon position, which probably results in the loss of function of the translated protein. Thus we suggested that AC4 gene might degenerate step by step during the long period of evolution, which might be an important reason for explaining the loss of function for the AC4 gene in the bipartite begomoviruses with DNA B.
The variation analysis of the base composition for begomovirus genes showed that AV1 and BV1 genes exhibit a stronger codon usage bias and a higher gene expression level. Thus CAI values of different gene samples were calculated using the AV1 gene as a reference set. The results of correlation analysis indicated the reliability of choosing AV1 gene as a high expression gene sample for begomoviruses. Then 14 codons were determined as the major optimal codons for begomoviruses. That will be very important during the design of degenerate primers, the introduction of point mutation, the modification of the virus genes and the investigation of the evolution mechanism of species at the molecule level.
It was speculated that monopartite begomoviruses emerged approximately 130 million years ago (Rybicki, 1994; Mansoor et al., 2006), suggesting that the begomoviruses should have evolved from the original monopartite viruses. Rybicki (1994) suggested that the significant expansion of the New World begomoviruses might have occurred after the transmission of the Old World begomoviruses to those of the New World by whiteflies. Based on codon usage pattern analysis, it could be inferred that there was no direct relationship between the Old World monopartite begomoviruses and the New World bipartite ones. Interestingly, a small number of the Old World bipartite begomoviruses exhibited a similar codon usage pattern to the New World bipartite ones, which suggested that the ancestor of begomoviruses could have evolved from monopartite to bipartite ones before they were transferred to the New World areas. Subsequently it was not the Old World monopartite begomoviruses but the Old World bipartite ones that transmitted to the New World in a certain way and finally evolved into the New World bipartite begomoviruses. In other words, the New World bipartite begomoviruses probably resulted directly from the Old World bipartite ones, while the latter evolved from the Old World monopartite begomoviruses.
It still remains unclear whether the New World begomoviruses directly evolved from the Old World bipartite begomoviruses because of the sharp environmental change after their transfer to the New World, or whether the Old World bipartite begomoviruses had evolved into the New World bipartite ones before their transmission to the New World. Rybicki (1994) suggested that the absence of the AV2 gene in all the New World begomoviruses could be attributed to its earlier loss after the New World begomoviruses arriving in the New World. These results suggest that the current New World begomoviruses evolved from ancient viruses after transferring to the New World. Ha et al.(2006) speculated that the new identified begomovirus termed Corchorus yellow vein virus (CoYVV) might belong to a New World begomovirus group that previously existed in the Old World, which suggested that the common ancestor of the New World begomovirus might originate from the Old World begomovirus. However, both the New World and Old World begomoviruses had began to evolve and coexisted in this area for a long time before the separation of the continents. In other words, the present New World begomoviruses might have evolved in the Old World, and then moved to the New World by some unknown means (Ha et al., 2006).
Footnotes
Project supported by the National Basic Research and Development Program (973) of China (No. 2006CB101903) and the National Excellent Doctoral Dissertation of China (No. 200768)
References
- 1.Adams MJ, Antoniw JF. Codon usage bias amongst plant viruses. Archives of Virology. 2004;149(1):113–135. doi: 10.1007/s00705-003-0186-6. [DOI] [PubMed] [Google Scholar]
- 2.Duret L, Mouchiroud D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis . Proceedings of the National Academy of Sciences of the USA. 1999;96(8):4482–4487. doi: 10.1073/pnas.96.8.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fauquet CM, Bisaro DM, Briddon RW, Brown JK, Harrison BD, Rybicki EP, Stenger DC, Stanley J. Revision of taxonomic criteria for species demarcation in the family Geminiviridae, and an updated list of begomovirus species. Archives of Virology. 2003;148(2):405–421. doi: 10.1007/s00705-002-0957-5. [DOI] [PubMed] [Google Scholar]
- 4.Greenacre MJ. Theory and Applications of Correspondence Analysis. London: Academic Press; 1984. pp. 1–364. [Google Scholar]
- 5.Gu W, Zhou T, Ma J, Sun X, Lu Z. Analysis of synonymous codon usage in SARS Coronavirus and other viruses in the Nidovirales . Virus Research. 2004;101(2):155–161. doi: 10.1016/j.virusres.2004.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo XY, Bao JD, Fan LJ. Evidence of selectively driven codon usage in rice: implications for GC content evolution of Gramineae genes. FEBS Letters. 2007;581(5):1015–1021. doi: 10.1016/j.febslet.2007.01.088. [DOI] [PubMed] [Google Scholar]
- 7.Gupta SK, Bhattacharyya TK, Ghosh TC. Synonymous codon usage in Lactococcus lactis: mutational bias versus translational selection. Journal of Biomolecular Structure and Dynamics. 2004;21(4):527–536. doi: 10.1080/07391102.2004.10506946. [DOI] [PubMed] [Google Scholar]
- 8.Ha C, Coombs S, Revill P, Harding R, Vu M, Dale J. Corchorus yellow vein virus, a New World geminivirus from the Old World. Journal of General Virology. 2006;87(4):997–1003. doi: 10.1099/vir.0.81631-0. [DOI] [PubMed] [Google Scholar]
- 9.Harrison BD, Robinson DJ. Natural genomic and antigenic variation in whitefly-transmitted geminiviruses (Begomoviruses) Annual Review of Phytopathology. 1999;37(1):369–398. doi: 10.1146/annurev.phyto.37.1.369. [DOI] [PubMed] [Google Scholar]
- 10.Jenkins GM, Holmes EC. The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Research. 2003;92(1):1–7. doi: 10.1016/S0168-1702(02)00309-X. [DOI] [PubMed] [Google Scholar]
- 11.Mansoor S, Zafar Y, Briddon RW. Geminivirus disease complexes: the threat is spreading. Trends in Plant Science. 2006;11(5):209–212. doi: 10.1016/j.tplants.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Moffat AS. Plant pathology: geminiviruses emerges as serious crop threat. Science. 1999;286(5446):1835. doi: 10.1126/science.286.5446.1835. [DOI] [Google Scholar]
- 13.Moriones E, Navas-Castillo J. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Research. 2000;71(1-2):123–134. doi: 10.1016/S0168-1702(00)00193-3. [DOI] [PubMed] [Google Scholar]
- 14.Naya H, Romero H, Carels N, Zavala A, Musto H. Translational selection shapes codon usage in the GC-rich genomes of Chlamydomonas reinhardtii . FEBS Letters. 2001;501(2-3):127–130. doi: 10.1016/S0014-5793(01)02644-8. [DOI] [PubMed] [Google Scholar]
- 15.Rybicki EP. A phylogenetic and evolutionary justification for three genera of Geminiviridae . Archives of Virology. 1994;139(1-2):49–77. doi: 10.1007/BF01309454. [DOI] [PubMed] [Google Scholar]
- 16.Sau K, Gupta SK, Sau S, Ghosh TC. Synonymous codon usage bias in 16 Staphylococcus aureus phages: implication in phage therapy. Virus Research. 2005;113(2):123–131. doi: 10.1016/j.virusres.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Sau K, Gupta SK, Sau S, Ghosh TC. Comparative analysis of the base composition and codon usages in fourteen mycobacteriophage genomes. Journal of Biomolecular Structure and Dynamics. 2005;23(1):63–71. doi: 10.1080/07391102.2005.10507047. [DOI] [PubMed] [Google Scholar]
- 18.Sau K, Sau S, Mandal CS, Ghosh TC. Factors influencing the synonymous codon and amino acid usage bias in AT-rich Pseudomonas aeruginosa phage PhiKZ . Acta Biochimica et Biophysica Sinica. 2005;37(9):625–633. doi: 10.1111/j.1745-7270.2005.00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shackelton LA, Parrish CR, Holmes EC. Evolutionary basis of codon usage and nucleotide composition bias in vertebrate DNA viruses. Journal of Molecular Evolution. 2006;62(5):551–563. doi: 10.1007/s00239-005-0221-1. [DOI] [PubMed] [Google Scholar]
- 20.Sharp PM, Li WH. An evolutionary perspective on synonymous codon usage in unicellular organisms. Journal of Molecular Evolution. 1986;24(1-2):28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- 21.Sharp PM, Li WH. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Research. 1987;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharp PM, Cowe E, Higgins DG, Shields DC, Wolfe KH, Wright F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens: a review of the considerable within-species diversity. Nucleic Acids Research. 1988;16(17):8207–8211. doi: 10.1093/nar/16.17.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang SF, Xu HL, Lu RJ, Meng X, Wang WL, Wang ZY, Guo W, Ruan L. Analysis of codon usage of vaccinia virus genome. Chinese Journal of Virology. 2002;18(3):227–234. (in Chinese) [Google Scholar]
- 24.Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87(1):23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 25.Zhou T, Gu WJ, Ma JM, Sun X, Lu ZH. Analysis of synonymous codon usage in H5N1 virus and other influenza A viruses. Biosystems. 2005;81(1):77–86. doi: 10.1016/j.biosystems.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Zhou XP, Xie Y, Tao XR, Zhang ZK, Li ZH, Fauquet CM. Characterization of DNAβ associated with begomoviruses in China and evidence for co-evolution with their cognate viral DNA-A. Journal of General Virology. 2003;84(1):237–247. doi: 10.1099/vir.0.18608-0. [DOI] [PubMed] [Google Scholar]