Abstract
The aim of this study was to purify and characterize a keratinase produced by a new isolated Bacillus subtilis KD-N2 strain. The keratinase produced by the isolate was purified using ammonium sulphate precipitation, Sephadex G-75 and DEAE (diethylaminoethyl)-Sepharose chromatographic techniques. The purified enzyme was shown to have a molecular mass of 30.5 kDa, as determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis. The optimum pH at 50 °C was 8.5 and the optimum temperature at pH 8.5 was 55 °C. The keratinase was partially inactivated by some metal ions, organic solvents and serine protease inhibitor phenylmethanesulfonyl fluoride (PMSF). Sodium dodecyl sulfate (SDS) and ethylene diamine tetraacetic acid (EDTA) had positive effect on the keratinase activity. Reducing agents including dithiothreitol (DTT), mercaptoethanol, L-cysteine, sodium sulphite, as well as chemicals of SDS, ammonium sulfamate and dimethylsulfoxide (DMSO) stimulated the enzyme activity upon a feather meal substrate. Besides feather keratin, the enzyme is active upon the soluble proteins ovalbumin, bovine serum albumin (BSA), casein and insoluble ones as sheep wool and human hair. Calf hair, silk and collagen could not be hydrolyzed by the keratinase.
Keywords: Ammonium sulfatate, Bacillus subtilis, Characterization, Feather, Keratin, Keratinase, Purification, Reducing agents
INTRODUCTION
Feathers are largely produced as a waste byproduct at poultry plants (Williams et al., 1991). They are insoluble structural proteins cross-linked by disulfide, hydrogen and hydrophobic bonds but could represent a rich protein resource because they contain over 90% (w/w) keratins. Keratins cannot be degraded by the usual proteolytic enzymes such as pepsin, trypsin and papain. Nevertheless, feathers do not accumulate in nature because keratins could be degraded by keratinases (EC 3.4.21/24/99.11) produced by some microorganisms (Onifade et al., 1998). Many keratinases from species of Bacillus (Williams et al., 1990; Riffel et al., 2003; Lucas et al., 2003), fungi (El-Naghy et al., 1998; Gradišar et al., 2000; Friedrich et al., 2005) and Actinomycetes (Ignatova et al., 1999; Gushterova et al., 2005) had been reported and some of them were purified and characterized (Lin et al., 1992; Böckle et al., 1995; Nam et al., 2002). Different keratinases vary in their characteristics. Until now, only a few keratinases from Bacillus sp. had been purified.
Keratinases from microorganisms have many applications in the feed, fertilizer, detergent, leather and pharmaceutical industries (Gupta and Ramnani, 2006). For example, the feather hydrolysates of Bacillus licheniformis PWD-1 and Vibrio sp. strain kr2 (Williams et al., 1991; Grazziotin et al., 2006) can be used as animal food. Indeed, addition of the crude keratinase from B. licheniformis PWD-1 improved the poultry growth (Odetallah et al., 2003). Also, the keratinase can degrade the infectious form of prion, PrPsc, in the presence of detergents and heat treatment (Langeveld et al., 2003), which could be important for the utilization of animal meal as feed. As for leather industry, the keratinase from Bacillus subtilis S14 exhibits remarkable dehairing capabilities (Macedo et al., 2005) without the degradation of collagen and this ecofriendly dehairing approach shows great utilization potential.
We had screened a new feather-degrading Bacillus subtilis KD-N2 strain which degraded feathers completely within 30 h in submerged cultivation (Cai et al., 2008), and the crude keratinase from the isolate showed capabilities of removing calf hair and sheep wool from the skins. The aim of this study was to purify and characterize the keratinase produced in feathers substrate by this Bacillus spp. isolate.
MATERIALS AND METHODS
Bacteria and growth conditions
Strain KD-N2 was screened from a local poultry plant and kept in our laboratory (Cai et al., 2008). The medium (pH 7.2) used for keratinase production contained the following constituents (g/L): NaCl 0.5, KH2PO4 0.7, K2HPO4 1.4, MgSO4 0.1 and feathers 10. Cultivation was performed using 500 ml Erlenmeyer flasks containing 100 ml medium for 24 h at 28 °C with constant shaking at 200 r/min. As inocula, 5% (v/v) bacteria grew in Luria-Bertani broth [peptone 1% (w/v), yeast extract 0.3% (w/v) and NaCl 0.5% (w/v), pH 7.2] for 20 h. Culture supernatants obtained after centrifugation at 8000×g for 20 min were used for further study.
Assay of keratinase activity
Keratin azure (Sigma-Aldrich, USA) was used as the substrate. It was first frozen at −20 °C and then ground into a fine powder. The 5 mg keratin azure powder was suspended in 1 ml 50 mmol/L Tris-HCl buffer (pH 8.0). The reaction mixture contained 1 ml keratin azure suspension and 1 ml appropriately diluted enzyme. The reactions were carried out at 50 °C in a water bath with constant agitation of 200 r/min for 30 min. After incubation, the reactions were stopped by adding 2 ml 0.4 mol/L trichloroacetic acid (TCA) and followed by centrifuging at 3000×g for 20 min to remove the substrate. The supernatant was spectrophotometrically measured for release of the azo dye at 595 nm. The 1 ml keratin azure suspension in the same buffer (like that of the sample) was agitated for 30 min at 50 °C, then was added 2 ml 0.4 mol/L TCA and 1 ml enzyme solution as a control. One unit (U) keratinase activity was defined as the amount of enzyme causing 0.01 absorbance increase between the sample and control at 595 nm under the conditions given.
Protein determination
Protein concentration was measured by the method of Bradford (1976), using bovine serum albumin (BSA) (Sigma, USA) as standard. The specific activity was expressed as the enzymatic activity (U) per mg of protein.
Purification of keratinase
All operations were performed at room temperature. After centrifugation at 8000×g for 20 min, solid ammonium sulphate was added to the supernatant to achieve 30% saturation, and then centrifuged to remove the pellet. The enzyme was precipitated from the supernatant by addition of solid ammonium sulphate, with gentle stirring until 80% saturation, and then allowed to stand for 12 h followed by centrifugation at 8000×g. The pellet was dissolved in 20 mmol/L Tris-HCl buffer (pH 8.0) and applied to a Sephadex G-75 column (6.0 cm×60.0 cm), which was processed at a flow rate of 10 ml/h with 20 mmol/L Tris-HCl buffer (pH 8.0) and every 3 ml fraction was collected. Active fractions were collected, concentrated by polyethylene glycol 2000 (PEG 2000) and applied to a DEAE (diethylaminoethyl)-Sepharose Fast Flow (FF) column (1.9 cm×20.0 cm). Samples were eluted at a flow rate of 6 ml/min with different concentrations of sodium chloride solution in 20 mmol/L Tris-HCl buffer (pH 8.0) and every 6 ml fraction was collected. Then the active fractions with PEG 2000 were concentrated and applied to a Sephadex G-75 column (1.0 cm×60.0 cm) at a flow rate of 6 ml/h. The 3 ml fractions were collected. The active fractions were then concentrated by PEG 2000 for further analysis.
Molecular mass determination
Polyacrylamide gel electrophoresis (PAGE) (12%, w/w) in the presence of sodium dodecyl sulfate (SDS) was carried out by the method of Laemmli (1970). The electrophoresed protein gels were stained with Coomassie brilliant blue R250 (Sigma, USA).
Characterization of the keratinase
To determine the optimal temperature for keratinolysis, enzyme reactions were carried out at different temperatures for 30 min. In order to investigate thermostability, the enzyme solution was pre-incubated for 20, 40, 60, 80, 100 and 120 min at 55 °C, and then the residual activity was measured. The optimum pH was determined at 55 °C using the following buffers (50 mmol/L): sodium phosphate buffer (pH 6.0~7.5), Tris-HCl buffer (pH 7.5~9.0), and Glycine/NaOH buffer (pH 9.0~10.0).
The effects of metal ions, enzyme inhibitor, detergent and organic solvents on keratinase activity were studied by assaying the enzyme activity as described above after pre-incubation with each chemical for 10 min at room temperature. The concentrations of the chemicals are as follows: 5.0 mmol/L of Ca2+, Mg2+, Cu2+, Mn2+, Zn2+, Al3+, SDS and phenylmethanesulfonyl fluoride (PMSF); 2.5, 5.0 and 10.0 mmol/L ethylene diamine tetraacetic acid (EDTA); 1% (v/v) of methanol, ethanol, dimethyl sulfoxide and isopropyl alcohol.
Substrate specificity of keratinase
To determine the keratinase specificity, the following substrates were used: casein (Sigma, USA), BSA (Sigma, USA), ovalbumin, type I collagen (Worthington, USA), chicken feather, goat hair, calf hair and human hair. The incubation procedures were the same as described for keratinase activity determination except that every incubation contained 20 mg substrate and 50 μl purified keratinase. The extent of hydrolysis was determined spectrophotometrically at 280 nm by measuring TCA-soluble peptides released from the substrates during incubation.
Effects of chemicals on feather meal hydrolysis
To determine the effects of chemicals on feather meal hydrolysis, 1 or 5 mmol/L dithiothreitol (DTT), SDS, sodium sulphite, L-cysteine, ammonium sulfamate and 1% (v/v) or 5% (v/v) mercaptoethanol, dimethylsulfoxide (DMSO) were added to 2 ml of 50 mmol/L Tris-HCl (pH 8.5) buffer containing 20 mg feather meal and 50 μl purified enzyme. After incubation for 1 h, the reaction was stopped by adding 2 ml 0.4 mol/L TCA. An incubation of feather meal and TCA for 1 h before enzyme addition was carried out as a control. The extent of feather meal hydrolysis was determined spectrophotometrically at 280 nm by measuring TCA-soluble peptides released from the substrate during incubation.
RESULTS AND DISCUSSION
Feather degradation and keratinase production
B. subtilis KD-N2 strain produced inducible keratinase in feathers substrate and degraded feathers completely in 30 h (Cai et al., 2008). Feathers were the mostly utilized substrate for keratinase production. Accompanied by feather degradation and keratinase production, the pH value of the culture increased to about 8.5. B. subtilis strains had been widely utilized for enzyme production, including the keratinases (Lal et al., 1999; Suh and Lee, 2001; Kim et al., 2001; Macedo et al., 2005). Other Bacillus species including B. licheniformis, B. pumilis, B. cereus, B. halodurans and B. pseudofirmus (Williams et al., 1990; Takami et al., 1999; Rozs et al., 2001; Kim et al., 2001; Gessesse et al., 2003; El-Refai et al., 2005) had also been reported for their keratinolytic activity. Among these strains, B. subtilis KD-N2 degraded feathers more quickly than any other reported until now, except for B. licheniformis RG1, which degraded feathers completely in 24 h (Ramnani and Gupta, 2004). Among the keratinases reported, only two of them had been purified (Suh and Lee, 2001; Macedo et al., 2005).
Enzyme purification
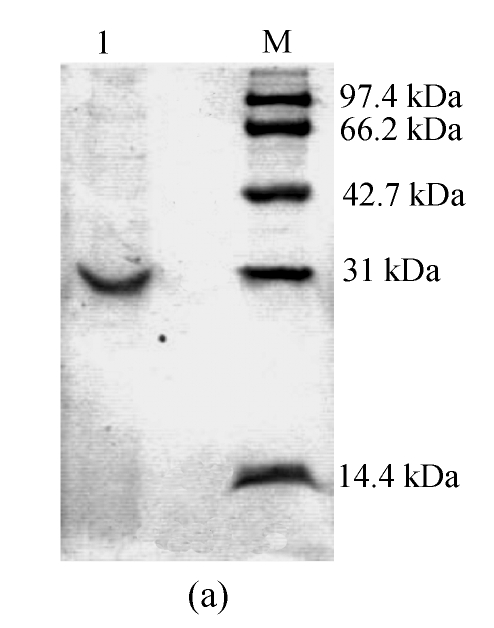
The results of the purification procedures are summarized in Table 1. SDS-PAGE analysis of the sample revealed a single band (Fig.1a), indicating that the keratinase was purified. The overall purification factor was about 12.7-fold, and the final yield was 4.6%. The final product had a specific activity of about 63.3 U/mg.
Table 1.
Purification of keratinase from B. subtilis KD-N2
| Step | Total protein (mg) | Total activity (U) | Specific activity (U/mg) | Purification | Yield (%) |
| Crude enzyme | 206.40 | 1040.0 | 5.0 | 1.0-fold | 100.0 |
| Sephadex G-75 (6.0 cm×60.0 cm) | 10.20 | 132.6 | 13.0 | 2.6-fold | 12.8 |
| DEAE-Sepharose FF (1.9 cm×20.0 cm) | 1.40 | 73.0 | 51.9 | 10.4-fold | 7.0 |
| Sephadex G-75 (1.0 cm×60.0 cm) | 0.76 | 48.0 | 63.3 | 12.7-fold | 4.6 |
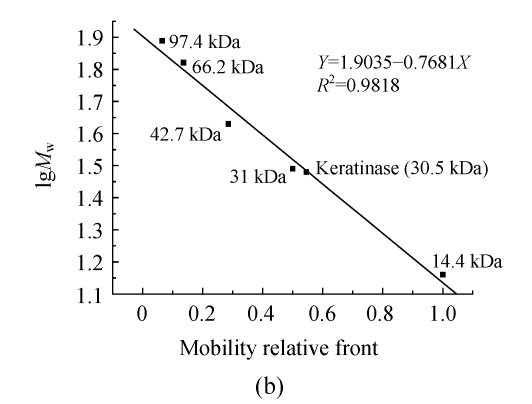
Fig. 1.
(a) SDS-PAGE of the purified keratinase and marker proteins and (b) molecular weight determination of the purified keratinase
Lane 1: Purified keratinase; Lane M: Molecular mass of marker proteins (Promega, USA): rabbit muscle phosphorylase b 97.4 kDa, bovine serum albumin 66.2 kDa, chicken egg ovalbumin 42.7 kDa, bovine erythrocytes carbonic anhydrase 31 kDa and chicken egg white lysozyme 14.4 kDa
Molecular mass of the keratinase
The molecular mass of the keratinase was estimated by comparing the electrophoretic mobility of the enzyme with the electrophoretic mobilities of marker proteins. The apparent molecular mass was 30.5 kDa (Fig.1b). Molecular masses of keratinases range from 18 to 200 kDa, except for a unique enzyme from a pathogenic fungi (Gupta and Ramnani, 2006). For Bacillus species, they are of medium size, such as 33 kDa (B. licheniformis) (Lin et al., 1992), 25.4 kDa (B. subtilis) (Suh and Lee, 2001) and 24 kDa (B. paeudofirmis) (Gessesse et al., 2003).
Effects of temperature and pH on the keratinase activity
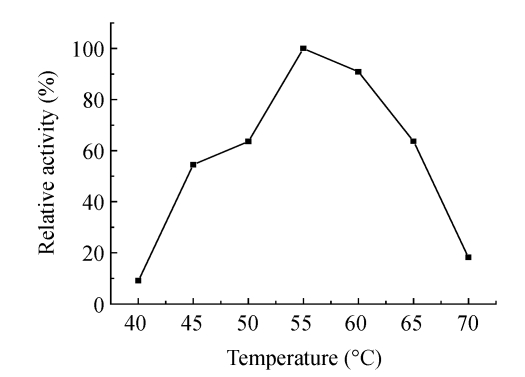
Fig.2 shows that the keratinase was active over a temperature range of 40~70 °C, with an optimum at 55 °C. Fig.3 shows that the keratinase was active at neutral and alkaline conditions, with optimum at pH 8.5, and the most suitable buffer seemed to be Tris-HCl. Most keratinases possess an activity optimum in the range of 30~80 °C, for example, keratinase from B. pseudofirmus AL-89 is of 60~70 °C (Gessesse et al., 2003), Nocardiopsis sp. TOA-1 is of 60 °C (Mitsuiki et al., 2004), and a few have exceptionally high temperature optimum of 100 °C (Nam et al., 2002). Most keratinases are active in neutral to alkali conditions, from pH 7.0 to pH 9.5. For example, the activity optimum of keratinase from Microbacterium kr10 is pH 7.0 (Thys et al., 2004), B. pumilus FH9 of pH 8.0 (El-Refai et al., 2005), Fervidobacterium islandicum AW-1 of pH 9.0 (Nam et al., 2002), and a few of extreme alkalophilic optima at pH 12~13 (Takami et al., 1999) and pH 12.5 (Mitsuiki et al., 2004).
Fig. 2.
Effect of temperature on the activity of keratinase from Bacillus subtilis KD-N2
Assays were performed at different temperatures in 50 mmol/L Tris-HCl buffer (pH 8.0) for 30 min
Fig. 3.
Effect of pH on the activity of keratinase from Bacillus subtilis KD-N2
Assays were performed at 55 °C using the following buffers (50 mmol/L): sodium phosphate buffer (pH 6.0~7.5), Tris-HCl buffer (pH 7.5~9.0), and glycine/NaOH buffer (pH 9.0~10.0)
Effects of metal ions, enzyme inhibitor, detergent and organic solvents on the keratinase activity
The keratinase from B. subtilis KD-N2 was totally inhibited by Cu2+, Mn2+ and partially by Ca2+, Mg2+, Zn2+, Al3+, methanol, ethanol, dimethyl sulfoxide, isopropyl alcohol, and was stimulated by SDS and EDTA (Table 2). Generally, heavy metal ions such as Cu2+ (Nam et al., 2002; Riffel et al., 2003; Thys et al., 2004), Hg2+ (Riffel et al., 2003; Thys et al., 2004) and Zn2+ (Thys et al., 2004) have inhibitory effects on keratinolytic activity. Contrarily, Ca2+, Mg2+ and Mn2+ stimulate some keratinases (Nam et al., 2002; Riffel et al., 2003). But for the keratinase from B. subtilis KD-N2 strain, all metal ions have negative effects on its activity. Serine-proteinase inhibitor PMSF partially inhibited the keratinase activity, which is identical to most of the keratinases studied except for a few from Paecilomyces marquandii, Doratomyces microsporus and Xanthomonas maltophilia, which are fully inhibited (de Toni et al., 2002; Gradišar et al., 2005). EDTA has negative effects on activities of mostly reported keratinases, but we observed in this study that the keratinase from B. subtilis KD-N2 strain was stimulated by 5 mmol/L EDTA, similar to the keratinase from Fervidobacterium islandicum AW-1 (Nam et al., 2002). Further results indicate that 2.5 and 10 mmol/L EDTA could also stimulate the keratinase activity (Table 2). Organic solvents inhibited the keratinase activity to some degree, which is different from keratinases from Chryseobacterium sp. (Riffel et al., 2003) and Nocardiopsis sp. TOA-1 (Mitsuiki et al., 2004). The detergent SDS has positive effects on keratinase activity, which is similar to the keratinase from Streptomyces albidoflavus (Bressollier et al., 1999), but differs from the keratinases from Streptomyces pactum DSM 40530 (Böckle et al., 1995), Chryseobacterium sp. (Riffel et al., 2003) and Nocardiopsis sp. TOA-1 (Mitsuiki et al., 2004). Proteases are widely used in the detergent industry, and it is important for the keratinase to be used in the presence of detergents such as SDS. Compared with previously reported keratinolytic enzymes, the keratinase from B. subtilis KD-N2 showed some novel characterizations and utilization potential.
Table 2.
Effect of chemicals on the keratinolytic activity of Bacillus subtilis KD-N2
| Chemicals | Concentration | Relative activity* (%) |
| None | 100 | |
| Ca2+ | 5 mmol/L | 44.8±3.4 |
| Mg2+ | 5 mmol/L | 41.4±3.2 |
| Cu2+ | 5 mmol/L | 0 |
| Mn2+ | 5 mmol/L | 0 |
| Zn2+ | 5 mmol/L | 34.5±3.1 |
| Al3+ | 5 mmol/L | 75.9±6.9 |
| SDS | 5 mmol/L | 158.6±13.8 |
| EDTA | 2.5 mmol/L | 131.8±5.6 |
| EDTA | 5 mmol/L | 136.4±6.2 |
| EDTA | 10 mmol/L | 144.8±6.6 |
| Methanol | 1%, v/v | 79.3±7.9 |
| Ethanol | 1%, v/v | 62.1 |
| Dimethyl sulfoxide | 1%, v/v | 51.7±3.5 |
| Isopropyl alcohol | 1%, v/v | 75.9±6.9 |
| PMSF | 5 mmol/L | 79.3±3.4 |
Values are mean of three independent determinations
Substrate specificity of the keratinase
The keratinase from KD-N2 shows activity on the soluble proteins casein, BSA, ovalbumin and the insoluble substrates feather meal, feather keratin, human hair and sheep wool, but no activity was observed upon collagen, calf hair and silk (Table 3). Many keratinases are capable of hydrolyzing a broad range of soluble and insoluble proteins, as the enzymes from Streptomyces pactum DSM40530 (Böckle et al., 1995), B. subtilis KS-1 (Suh and Lee, 2001) and Fervidobacterium islandicum AW-1 (Nam et al., 2002). The keratinase from the isolate had the ability to remove hair from calf and sheep skins (data not shown), but could not hydrolyze collagen, which is the main constituent of the dermis. Indeed, similar result had been reported for keratinase from B. subtilis S14 (Macedo et al., 2005). Type I collagen could be hydrolyzed by keratinases from Streptomyces albidoflavus (Bressollier et al., 1999), B. subtilis KS-1 (Suh and Lee, 2001) and Fervidobacterium islandicum AW-1 (Nam et al., 2002). And keratinases from Paecilomyces marquandii and Doratomyces microsporus were able to hydrolyze different keratin-containing substrates such as stratum corneum keratin, human nail, porcine nail and bovine keratin (Gradišar et al., 2005). Comparing with the substrates listed in Table 3, it seems that insoluble substrates such as human hair and feather meal, which have more disulfide bonds, were more easily hydrolyzed by the purified keratinase than sheep wool, which has less disulfide bonds. Previous studies showed that ball milling could destroy the keratin structure, which makes the substrates easily to be degraded by proteases (Stahl et al., 1949; Noval and Nickerson, 1959). The keratinase from B. subtilis KD-N2 has a broad range of substrates specificity and could degrade substrates containing both α and β keratins.
Table 3.
Hydrolysis of various proteins by keratinase from Bacillus subtilis KD-N2
| Substrates | Relative activity (%) |
| Ovalbumin | 100 |
| BSA | 56±5.2 |
| Casein | 68±6.7 |
| Feather keratin | 20±2.4 |
| Feather meal | 80±5.3 |
| Collagen | 0 |
| Human hair | 62±4.5 |
| Wool | 12±0.83 |
| Calf hair | 0 |
| Silk | 0 |
The enzyme was incubated with 20 mg of substrate in 2 ml of 50 mmol/L Tris-HCl buffer (pH 8.5) for 30 min at 55 °C. Feather keratin was prepared by the method of Wawrzkiewicz et al.(1987) with little modification. After solubilization of native feather keratin in dimethyl sulfoxide and precipitation by cold acetone, the keratin was washed twice with distilled water and dried at 40 °C in a vacuum drier to constant weight. Other insoluble substrates were cut into short fragments
Effects of chemicals on feather meal hydrolysis
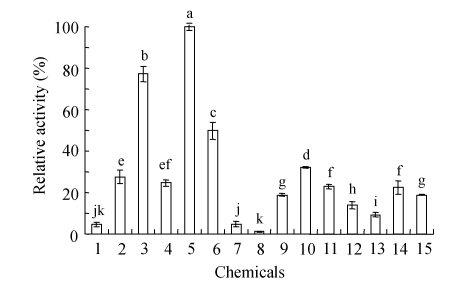
Fig.4 shows that sulphur-containing chemicals, especially reducing agents, stimulated the keratinase activity towards feather meal. All tested chemicals had positive effects on the feather meal hydrolysis except 1 mmol/L sodium sulfite. Five mmol/L DTT, 5 mmol/L ammonium sulfamate and 5% (v/v) β-mercaptoethanol enhanced keratinolytic activity maximally. Five mmol/L of DTT, β-mercaptoethanol, ammonium sulfamate and sodium sulfite had more positive effects than 1 mmol/L, but for L-cysteine, SDS and DMSO, lower concentrations had more effects. Reducing agents such as DTT, β-mercaptoethanol, cysteine and sodium sulfite had been largely discussed on their positive effects on keratinases (Böckle et al., 1995; Bressollier et al., 1999; Ignatova et al., 1999; Nam et al., 2002; Gradišar et al., 2005) and some of the enzymes are thiol-activated (Gupta and Ramnani, 2006). To test the effects of some sulphur-containing chemicals on the keratinase activity, ammonium sulfamate was firstly utilized in the study, and it had positive effects in both low and high concentrations. DMSO is able to solubilize feather keratin (Wawrzkiewicz et al., 1987), and has positive effects on feather meal hydrolysis in this study. Generally, purified keratinases cannot degrade keratin in vitro without the presence of reducing agents (Böckle et al., 1995; Bressollier et al., 1999; Suh and Lee, 2001; Riffel et al., 2003). And until now, detailed mechanisms of keratin hydrolysis has not been elucidated. One of the possible mechanisms is the reduction of disulfide bonds or sulfitolysis of the disulfide bonds by secreted sulfite. In this study, the reducing agents, DTT, β-mercaptoethanol, cysteine, sodium sulfite and sulphur-containing chemicals as SDS, ammonium sulfatate and DMSO stimulated the feather meal hydrolysis, which was probably due to the direct breakdown of the disulfide bonds by the reducing agents or due to reactions caused by the sulphur-containing chemicals.
Fig. 4.
Effects of chemicals on the purified keratinase activity on feather meal
Chemicals were added to the incubation mixtures to reach a final concentration as follows: 1: Control; 2: 1 mmol/L DTT; 3: 5 mmol/L DTT; 4: 1% (v/v) β-mercaptoethanol; 5: 5% (v/v) β-mercaptoethanol; 6: 1 mmol/L SDS; 7: 5 mmol/L SDS; 8: 1 mmol/L sodium sulphite; 9: 5 mmol/L sodium sulphite; 10: 1 mmol/L L-cysteine; 11: 5 mmol/L L-cysteine; 12: 1 mmol/L ammonium sulfamate; 13: 5 mmol/L ammonium sulfamate; 14: 1% (v/v) DMSO; 15: 5% (v/v) DMSO. Bar represents standard error (SE) of the mean. Means with the different letters are significantly different according to Duncan’s multiple range test at P=0.05 using SAS (SAS Institute, version 6.12, Cary, NC)
CONCLUSION
The B. subtilis KD-N2 degraded feathers quickly, and the enzyme produced by this isolate seems to be a new keratinase. The enzyme had a molecular mass of 30.5 kDa and was partially inactivated by PMSF. The keratinase is different from another purified B. subtilis keratinase (Suh and Lee, 2001) in molecular mass and sensitivity to inhibitors, which has a molecular mass of 25.4 kDa and was completely inactivated by PMSF. The keratinase from B. subtilis KD-N2 was inhibited by metal ions and organic solvents to some degree and stimulated by SDS and EDTA. The enzyme showed activity upon feather meal, human hair as well as sheep wool. SDS and some reducing agents had positive effects on the keratinase activity. The keratinase was able to remove hair from the calf and sheep skins. The hydrolysis of collagen and degradation of the dermis should be further characterized, although the enzyme was not able to hydrolyze pure collagen. All the results present potential utilization of the keratinase industrially in keratin hydrolysis, feed nutrient improvement, and hair removing.
References
- 1.Böckle B, Galunsky B, Muller R. Characterization of a keratinolytic serine proteinase from Streptomyces pactum DSM 40530. Appl Environ Microbiol. 1995;61(10):3705–3710. doi: 10.1128/aem.61.10.3705-3710.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1-2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Bressollier P, Letourneau F, Urdaci M, Verneuil B. Purification and characterization of a keratinolytic serine proteinase from Streptomyces albidoflavus . Appl Environ Microbiol. 1999;65(6):2570–2576. doi: 10.1128/aem.65.6.2570-2576.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai CG, Lou BG, Zheng XD. Keratinase production and keratin degradation by a mutant strain of Bacillus subtilis . J Zhejiang Univ Sci B. 2008;9(1):60–67. doi: 10.1631/jzus.B061620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Toni CH, Richter MF, Chagas JR, Henriques JAP, Termignoni C. Purification and characterization of an alkaline serine endopeptidase from a feather-degrading Xanthomonas maltophilia strain. Can J Microbiol. 2002;48(4):342–348. doi: 10.1139/w02-027. [DOI] [PubMed] [Google Scholar]
- 6.El-Naghy MA, El-Ktatny MS, Fadl-Allah EM, Nazeer WW. Degradation of chicken feathers by Chrysosporium georgiae . Mycopathologia. 1998;143(2):77–84. doi: 10.1023/A:1006953910743. [DOI] [PubMed] [Google Scholar]
- 7.El-Refai HA, AbdelNaby MA, Gaballa A, El-Araby MH, Abdel Fattah AF. Improvement of the newly isolated Bacillus pumilus FH9 keratinolytic activity. Process Biochem. 2005;40(7):2325–2332. doi: 10.1016/j.procbio.2004.09.006. [DOI] [Google Scholar]
- 8.Friedrich J, Gradišar H, Vrecl M, Pogacnik A. In vitro degradation of porcine skin epidermis by a fungal keratinase of Doratomyces microsporus . Enzyme Microb Technol. 2005;36(4):455–460. doi: 10.1016/j.enzmictec.2004.09.015. [DOI] [Google Scholar]
- 9.Gessesse A, Rajni HK, Gashe BA. Novel alkaline proteases from alkaliphilic bacteria grown on chicken feather. Enzyme Microb Technol. 2003;32(5):519–524. doi: 10.1016/S0141-0229(02)00324-1. [DOI] [Google Scholar]
- 10.Gradišar H, Kern S, Friedrich J. Keratinase of Doratomyces microsporus . Appl Microbiol Biotechnol. 2000;53(2):196–200. doi: 10.1007/s002530050008. [DOI] [PubMed] [Google Scholar]
- 11.Gradišar H, Friedrich J, Križaj I, Jerala R. Similarities and specificities of fungal keratinolytic proteases: comparison of keratinase of Paecilomyces marquandii and Doratomyces microsporus to some known proteases. Appl Environ Microb. 2005;71(7):3420–3426. doi: 10.1128/AEM.71.7.3420-3426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grazziotin A, Pimentel FA, de Jong EV, Brandelli A. Nutritional improvement of feather protein by treatment with microbial keratinase. Anim Feed Sci Technol. 2006;126(1-2):135–144. doi: 10.1016/j.anifeedsci.2005.06.002. [DOI] [Google Scholar]
- 13.Gupta R, Ramnani P. Microbial keratinase and their prospective application: an overview. Appl Microbiol Biotechnol. 2006;70(1):21–33. doi: 10.1007/s00253-005-0239-8. [DOI] [PubMed] [Google Scholar]
- 14.Gushterova A, Vasileva-Tonkova E, Dimova E, Nedkov P, Haertle T. Keratinase production by newly isolated Antarctic actinomycete strains. World J Microbiol Biotechnol. 2005;21(6-7):831–834. doi: 10.1007/s11274-004-2241-1. [DOI] [Google Scholar]
- 15.Ignatova Z, Gousterova A, Spassov G, Nedkov P. Isolation and partial characterization of extracellular keratinase from a wool degrading Thermophilic actinomycete strain Thermoactinomyces candidus . Can J Microbiol. 1999;45(3):217–222. doi: 10.1139/cjm-45-3-217. [DOI] [PubMed] [Google Scholar]
- 16.Kim JM, Lim WJ, Suh HJ. Feather-degrading Bacillus species from poultry waste. Process Biochem. 2001;37(3):287–291. doi: 10.1016/S0032-9592(01)00206-0. [DOI] [Google Scholar]
- 17.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4 . Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Lal S, Rajak RC, Hasija SK. In vitro degradation of keratin by two species of Bacillus . J Gen Appl Microbiol. 1999;45(6):283–287. doi: 10.2323/jgam.45.283. [DOI] [PubMed] [Google Scholar]
- 19.Langeveld JPM, Wang JJ, van de Wiel DFM, Shih GC, Garssen GJ, Bossers A, Shih JCH. Enzymatic degradation of prion protein in brain stem from infected cattle and sheep. J Infect Dis. 2003;188(11):1782–1789. doi: 10.1086/379664. [DOI] [PubMed] [Google Scholar]
- 20.Lin X, Lee CG, Casale ES, Shih JCH. Purification and characterization of a keratinase from a feather-degrading Bacillus licheniformis strain. Appl Environ Microbiol. 1992;58(10):3271–3275. doi: 10.1128/aem.58.10.3271-3275.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucas FS, Broennimann O, Febbraro I, Heeb P. High diversity among feather-degrading bacteria from a dry meadow soil. Microb Ecol. 2003;45(3):282–290. doi: 10.1007/s00248-002-2032-x. [DOI] [PubMed] [Google Scholar]
- 22.Macedo AJ, da Silva WOB, Gava R, Driemerier D, Henriques JAP, Termignoni C. Novel keratinase from Bacillus subtilis S14 exhibiting remarkable dehairing capabilites. Appl Environ Microbiol. 2005;71(1):594–596. doi: 10.1128/AEM.71.1.594-596.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitsuiki S, Ichikawa M, Oka T, Sakai M, Moriyama Y, Sameshima Y, Goto M, Furukawa K. Molecular characterization of a keratinolytic enzyme from an alkaliphilic Nocardiopsis sp. TOA-1. Enzyme Microb Technol. 2004;34(5):482–489. doi: 10.1016/j.enzmictec.2003.12.011. [DOI] [Google Scholar]
- 24.Nam GW, Lee DW, Lee HS, Lee NJ, Kim BC, Choe EA, Hwang JK, Suhartono MT, Pyun YR. Native-feather degradation by Fervidobacterium islandicum AW-1, a newly isolated keratinase-producing thermophilic anaerobe. Arch Microbiol. 2002;178(6):538–547. doi: 10.1007/s00203-002-0489-0. [DOI] [PubMed] [Google Scholar]
- 25.Noval JJ, Nickerson WJ. Decomposition of native keratin by Streptomyces fradiae . J Bacteriol. 1959;77(3):251–259. doi: 10.1128/jb.77.3.251-263.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Odetallah NH, Wang JJ, Garlich JD, Shih JCH. Keratinase in starter diets improves growth of broiler chicks. Poultry Sci. 2003;82(4):664–670. doi: 10.1093/ps/82.4.664. [DOI] [PubMed] [Google Scholar]
- 27.Onifade AA, A1-Sane NA, Al-Musallam AA, Al-Zarban S. Potentials for biotechnological applications of keratin-degrading microorganisms and their enzymes for nutritional improvement of feathers and other keratins as livestock feed resources. Biores Technol. 1998;66(1):1–11. doi: 10.1016/S0960-8524(98)00033-9. [DOI] [Google Scholar]
- 28.Ramnani P, Gupta R. Optimization of medium composition for keratinase production on feather by Bacillus licheniformis RG1 using statistical methods involving response surface methodology. Biotechnol Appl Biochem. 2004;40(11):491–496. doi: 10.1042/BA20030228. [DOI] [PubMed] [Google Scholar]
- 29.Riffel A, Lucas FS, Heeb P, Brandelli A. Characterization of a new keratinolytic bacterium that completely degrades native feather keratin. Arch Microbiol. 2003;179(4):258–265. doi: 10.1007/s00203-003-0525-8. [DOI] [PubMed] [Google Scholar]
- 30.Rozs M, Manczinger L, Vagvolgyi C, Kevei F. Secretion of a trypsin-like thiol protease by a new keratinolytic strain of Bacillus licheniformis . FEMS Microb Lett. 2001;205(2):221–224. doi: 10.1111/j.1574-6968.2001.tb10951.x. [DOI] [PubMed] [Google Scholar]
- 31.Stahl WH, Mcque B, Siu RGH. Decomposition of cystine and wool by treatment in the ball mill and autoclave. J Biol Chem. 1949;177(11):69–73. [PubMed] [Google Scholar]
- 32.Suh HJ, Lee HK. Characterization of a keratinolytic serine protease from Bacillus subtilis KS-1. J Protein Chem. 2001;20(2):165–169. doi: 10.1023/A:1011075707553. [DOI] [PubMed] [Google Scholar]
- 33.Takami H, Nogi Y, Horikoshi K. Reidentification of the keratinase-producing facultativel alkaliphilic Bacillus sp. AH-101 as Bacillus halodurans . Extremophiles. 1999;3(4):293–296. doi: 10.1007/s007920050130. [DOI] [PubMed] [Google Scholar]
- 34.Thys RCS, Lucas FS, Riffel A, Heeb P, Brandelli A. Characterization of a protease of a feather-degrading Microbacterium species. Lett Appl Microbiol. 2004;39(2):181–186. doi: 10.1111/j.1472-765X.2004.01558.x. [DOI] [PubMed] [Google Scholar]
- 35.Wawrzkiewicz K, Lobarzewski J, Wolski T. Intracellular keratinase of Trichophyton gallinae . J Med Veternary Mycol. 1987;25(4):261–268. doi: 10.1080/02681218780000601. [DOI] [PubMed] [Google Scholar]
- 36.Williams CM, Richter CS, Mackenzie JM, Shih JCH. Isolation, identification, and characterization of a feather-degrading bacterium. Appl Environl Microbiol. 1990;56(6):509–1515. doi: 10.1128/aem.56.6.1509-1515.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams CM, Lee CG, Garlich JD, Shih JCH. Evaluation of a bacterial feather fermentation product, feather-lysate, as a feed protein. Poultry Sci. 1991;70(1):85–94. [Google Scholar]