Abstract
All mammalian eggs are surrounded by a relatively thick extracellular coat, the zona pellucida, that plays vital roles during oogenesis, fertilization, and preimplantation development. The mouse zona pellucida consists of three glycoproteins that are synthesized solely by growing oocytes and assemble into long fibrils that constitute a matrix. Zona pellucida glycoproteins are responsible for species-restricted binding of sperm to unfertilized eggs, inducing sperm to undergo acrosomal exocytosis, and preventing sperm from binding to fertilized eggs. Many features of mammalian and non-mammalian egg coat polypeptides have been conserved during several hundred million years of evolution.
The plasma membrane of mammalian and non-mammalian eggs is surrounded by at least one extracellular coat. A zona pellucida (ZP)2 surrounds mammalian oocytes, ovulated eggs, and embryos up to the early blastocyst stage of development. The ZP appears during oocyte growth, and blastocyst stage embryos hatch from the ZP prior to implanting in the uterus.
Important roles have been ascribed to the ZP during oogenesis, fertilization, and preimplantation development (1, 2). For example, during fertilization, the ZP regulates binding of sperm to ovulated eggs and induces bound sperm to undergo cellular exocytosis, the acrosome reaction. The ZP also prevents binding of sperm to fertilized eggs. Removal of the ZP results in direct exposure of the egg plasma membrane to sperm and allows sperm from heterologous species to fuse with eggs. During preimplantation development, the ZP influences alignment of the embryonic-abembryonic axis and first cleavage plane (3). The ZP also ensures the integrity of preimplantation embryos as they traverse the female reproductive tract.
Characteristics of the ZP
All mammalian eggs possess a ZP, although its thickness (∼1–25 μm) and protein content (∼1–30 ng) vary considerably for eggs from different species (4). The mouse egg ZP is ∼6.2 μm thick and contains ∼3.5 ng of protein (Fig. 1). The ZP is an elastic porous coat penetrable by antibodies, enzymes, and small viruses. All ZPs are composed of long interconnected fibrils that can be solubilized by mild acid or base, heat, or reducing agents. ZP glycoproteins (ZPGs) are held together in fibrils by noncovalent interactions.
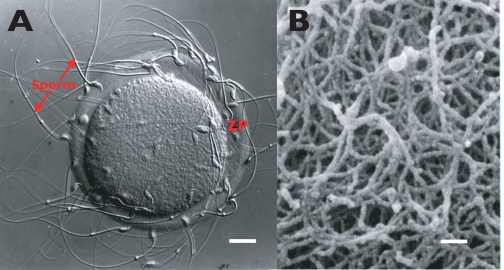
FIGURE 1.
Light and electron micrographs of the mouse ZP. A, light micrograph (Nomarski-differential interference contrast) of sperm bound to the egg ZP. Scale bar ∼ 13 μm. B, scanning electron micrograph of the egg ZP (taken from Ref. 56 with permission). Scale bar ∼ 200 nm.
Characteristics of ZPGs
Three glycoproteins, mZP1 (∼200 kDa), mZP2 (∼120 kDa), and mZP3 (∼83 kDa), compose the mouse egg ZP (4). The human egg ZP consists of four glycoproteins, hZP1 (∼100 kDa), hZP2 (∼75 kDa), hZP3 (∼55 kDa), and hZP4 (∼65 kDa) (5, 6). Three or more glycoproteins related to mZP1–3 are found in the ZP of eggs from a wide variety of mammalian species. In mice, mZP2 and mZP3 account for >80% of the mass of the ZP. ZPGs are heterogeneously glycosylated with asparagine-linked (N-) and serine/threonine-linked (O-) oligosaccharides. These cause ZPGs to migrate as broad bands on SDS-PAGE, and because the oligosaccharides are sialylated and sulfated, ZPGs have low isoelectric points. mZP1–3 possess four, six, and five N-linked oligosaccharides, respectively, and at least two sites of O-linked oligosaccharides are present on mZP3 (7, 8). Under nonreducing conditions mZP2 and mZP3 migrate as monomers on SDS-PAGE; mZP1 migrates as a disulfide-linked homodimer. Polypeptides of mouse and human ZPGs are encoded by single-copy genes located on different chromosomes (mZP1–3, chromosomes 19, 7, and 5; and hZP1–4, chromosomes 11, 16, 7, and 1) (9–11). ZP2- and ZP3-related ZPGs from different mammals are well conserved (∼65–98% identity); ZP1-related ZPGs are conserved to a lesser degree (∼40% identity). ZPGs have regions of polypeptide in common, suggesting that they may be derived from a common ancestral gene. Also, ZPGs are related to vitelline envelope (VE) glycoproteins (VEGs) of eggs from birds, frogs, fish, ascidians, and molluscs.
Synthesis of ZPGs
The ZP of mouse and human eggs appears during oocyte growth, such that ZP thickness and oocyte diameter increase concomitantly (Fig. 2). mZP1–3 and hZP1–4 are synthesized coordinately and exclusively by growing and fully grown oocytes (4). Synthesis of mouse and human ZPGs reflects oocyte-specific and therefore female-specific gene expression. mZP1–3 and hZP1–4 are synthesized as precursor polypeptides possessing an N-terminal signal sequence (SS) and C-terminal propeptide (CTP) (Fig. 3). The former is removed during transit of nascent protein from the endoplasmic reticulum to the Golgi, and the latter at the egg plasma membrane. Synthesis of ZPGs ceases during conversion of oocytes to eggs.
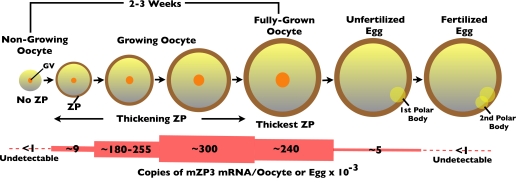
FIGURE 2.
Schematic representation of mouse oocyte growth and levels of mZP3 mRNA in oocytes and eggs. GV, germinal vesicle.
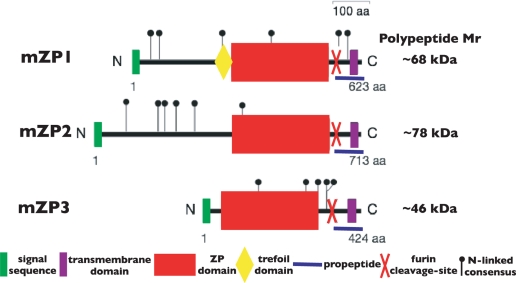
FIGURE 3.
Schematic representation of the overall organization of mZP1–3 precursor polypeptides. The precursor polypeptide of each ZPG is drawn to scale with the N and C termini indicated. Key features of the polypeptides and the number of amino acids in each polypeptide are indicated. aa, amino acids.
ZPG genes exhibit conserved organization, with exon/intron boundaries defining distinct domains in ZPGs. ZPG genes share TATAA boxes ∼30 bp upstream of transcription start sites, as well as E-box sequences (CANNTG) at –200 bp. E-boxes are involved in oocyte-specific expression of ZPG genes that occurs upon binding E12/FIGα heterodimers (12). Female mice that are homozygous nulls for FIGα do not express ZPG genes and are infertile because of massive depletion of oocytes (13). As little as 153 nucleotides of mZP3 5′-flanking sequence target expression of a luciferase reporter to growing oocytes; enhancer elements are present between nucleotides –153 and –470 of the 5′-flanking sequence (14, 15). Two ovary-specific DNA-binding proteins have been identified; ZAP-1 binds to the sequence 5′-CAC(G/C) TG-3′ within 250 bp upstream of the mZP2 and mZP3 promoter TATAA box, and OSP-1 binds to the sequence 5′-GATAA-3′ within the first 100 bp of the mZP3 promoter (16, 17).
During mouse oocyte growth, the absolute rate of protein synthesis increases ∼40-fold, and ZPG synthesis represents ∼5% of the total. The number of copies of mZP3 mRNA per oocyte or egg increases from undetectable levels in non-growing oocytes to ∼300,000 copies in mid-stage growing oocytes, to ∼240,000 copies in fully grown oocytes, and to almost undetectable levels (∼5,000 copies) in unfertilized eggs (18). There is a dramatic fall in mZP3 mRNA levels (∼98%) when oocytes become unfertilized eggs. ZPG mRNA is undetectable in cleavage stage embryos (<1,000 copies/zygote) (Fig. 2).
Structural Features of ZPGs
The primary structures of ZPGs from mouse eggs to human eggs have been determined during the past 25 years. These have permitted construction of phylogenetic trees for the ZP gene family, including genes encoding VEGs from fish, frogs, and birds (19). Certain common features have emerged for polypeptides of ZPGs. These include an N-terminal SS, a ZP domain (ZPD), a CTP with a consensus furin cleavage site (CFCS) and a transmembrane domain (TMD), in some cases a trefoil domain, and frequently multiple copies of the N-terminal subdomain (NTS) of the ZPD (Fig. 3) (20–22).
The ZPD is a hallmark of ZPGs and represents ∼80% of mZP3. It is an ∼260-amino acid sequence (containing 8 conserved Cys residues present as intramolecular disulfides) that is found in hundreds of proteins of diverse functions from a wide variety of tissues in all multicellular eukaryotes (20, 21). Other unique domains are present on different ZPD proteins that give each its distinctive character and function. ZPD proteins are often glycosylated modular structures consisting of multiple types of domains. The only invariant residues within the ZPDs of mZP1–3 are 8 Cys, 2 Gly, and 3 aromatic residues; however, several regions exhibit conserved physiochemical character. mZP1–3 are predicted to be relatively rich in β-structure (∼25%) but poor in α-helix content (<2%), whereas their ZPDs probably adopt an all β-fold (20, 21).
The ZPD is a bipartite structure divided by a short protease-sensitive region (23, 24). mZP1–3 have 4 Cys residues in the ZPD NTS (Cys1–Cys3, Cys2–Cys4); mZP3 has 4 Cys residues, and mZP1 and mZP2 have 6 Cys residues in the C-terminal subdomain. Consequently, the C-terminal subdomain adopts two alternative disulfide bond connectivities (mZP3, Cys5– Cys7, Cys6–Cys8 (Type 1); and mZP1 and mZP2, Cys5–Cys6, Cys7–Cysa, Cys8–Cysb (Type 2)) (21). Mutations in ZPDs, especially in the NTS, can result in severe pathologies, such as infertility, deafness, and cancer. In this context, it was proposed that ZPDs might play a role in polymerization of proteins into fibrils or matrices (25). Indeed, it was found that ZPDs polymerize into fibrils on their own (26) and that the NTS is responsible for polymerization (27). For example, the ZPD from uromodulin polymerizes into a double helical structure. Rotary-shadowed mouse ZP fibrils exhibit similar structural features, suggesting that polymers assembled by different ZPDs may share a similar three-dimensional structure (21).
Ultrastructural analyses of solubilized mouse egg ZPs have revealed their fibrillar nature. Fibrils are interconnected by mZP1 and exhibit a structural repeat attributable to mZP2/mZP3 dimers (28). Ultrastructural results obtained with uromodulin ZPDs suggest a model in which mZP2 and mZP3 homopolymers form a double helical structure. In this context, it has been found that ZPGs can assemble into homopolymers under nondenaturing conditions (29).
Secretion and Assembly of ZPGs
Features of the CTP (Fig. 4) regulate ZPG secretion and assembly (30). Nascent ZPGs are transported to oocyte plasma membrane in secretory vesicles and released into the extracellular space following cleavage of the CTP at its CFCS by a furin family member (31–34). Nascent ZPGs are incorporated into the thickening ZP at its innermost surface, implying that the ZP thickens from the inside to the outside (33). Mutation of the CFCS causes an enhanced retention of ZPGs within growing oocytes, whereas replacement of the native TMD with a heterologous TMD or mutation of the conserved Cys residue or the charged patch of the CTP has no effect on secretion of nascent ZPGs (23).
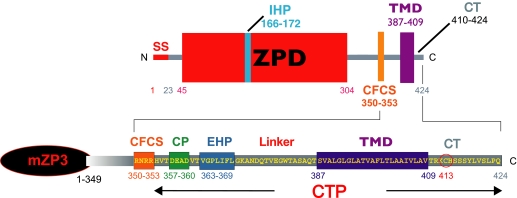
FIGURE 4.
Features of the CTP of mZP3. Polypeptide boundaries are marked by gray bars. Key features of the CTP and amino acid numbers are indicated. CP, charged patch; CT, cytoplasmic tail.
Analysis of ZPG mutations suggests that elements required for secretion are located between the CFCS and TMD (23, 30). A short conserved motif consisting of an almost invariant Gly-Pro sequence immediately followed by 4–5 hydrophobic amino acids is present C-terminal to the charged patch (Fig. 4) (23, 35). This external hydrophobic patch (EHP) is found in ZPGs, VEGs, and many other ZPD proteins. In the absence of a TMD, removal of the EHP prevents secretion of ZPGs. However, the presence of an EHP is not required for secretion of ZPGs when a TMD is present. On the other hand, deletion of the EHP from full-length ZPGs that have a TMD abolishes their assembly into the ZP. An EHP and TMD are required for incorporation of nascent ZPGs into the ZP.
An internal hydrophobic patch (IHP), located within the ZPD (Fig. 4), is also implicated in the secretion and assembly of ZPGs (23). Mutation of the IHP prevents incorporation of full-length ZPGs into the ZP without affecting secretion. However, secretion is inhibited when the IHP is mutated in ZPGs lacking a TMD, identical to results obtained with the EHP. This suggests that the EHP and IHP are functionally related to each other and, together with the CFCS and TMD, control incorporation of nascent ZPGs into the ZP. A mechanism for activation of polymerization of ZPGs has been proposed in which proteolytic processing of the CTP disrupts interactions between the IHP and EHP and leads to polymerization of mature ZPGs in the extracellular space (21, 23).
Elimination of ZPG Synthesis
mZP1–3 null mutant mice have been produced by homologous recombination in embryonic stem cells. Mutant females heterozygous for mZP3 are fertile, although their egg ZPs are half the thickness of the wild-type ZP (36). However, homozygous mutant females missing either mZP2 or mZP3 fail to lay down a ZP during oocyte growth, possess oocytes and eggs that lack a ZP, and are infertile (37–39). Failure to synthesize either ZPG prevents assembly of the synthesized ZPGs into a ZP. mZP2–/– and mZP3–/– females possess fewer growing oocytes, Graafian follicles, and ovulated eggs compared with wild-type females (40). On the other hand, oocytes and eggs from mZP1–/– females have a ZP, but are not as fertile as wild-type mice (41). Although mZP2 and mZP3 assemble into a ZP in mZP1–/– females, it exhibits large pores because of insufficient cross-linking of ZP fibrils. hZP2 and hZP3 can replace mZP2 and mZP3 and restore a ZP to oocytes in homozygous null female mice, but the mosaic ZP does not permit binding of human sperm (42).
Functions of ZPGs during Fertilization
ZPGs participate at several steps in the fertilization pathway (1, 2, 43, 44). Principal among these are roles for mZP2 and mZP3 as receptors for sperm and for mZP3 as inducer of sperm exocytosis, the acrosome reaction. Evidence in mice suggests that acrosome-intact sperm bind to mZP3, complete the acrosome reaction, bind to mZP2, penetrate the ZP, and fuse with the egg plasma membrane. Fusion of sperm and egg leads to modifications of ZPGs such that free-swimming sperm cannot bind to the ZP and previously bound sperm cannot penetrate the ZP. A region of mZP3 encoded by exon 7 is the sperm combining site (45–47), but whether sperm binding is supported by mZP3 polypeptide, oligosaccharides, or both remains controversial (48). This particular region of mZP3 has undergone many changes during evolution compared with the remainder of the polypeptide and is a site of positive Darwinian selection (49). Many sperm proteins recognize and bind to the ZP and, in some cases, specifically to mZP3. Fertilization may be so important to the organism that binding of sperm to eggs is supported by several redundant mechanisms (50).
ZPGs and VEGs
Whereas all ZPGs are synthesized by the ovary, VEGs of non-mammalian eggs are synthesized by the liver, ovary, or both (51). Regardless of their site(s) of synthesis, VEGs resemble ZPGs. For example, trout VEs are composed of VEα, VEβ, and VEγ, which possess an N-terminal SS, a Pro/Gln-rich repeat, a trefoil domain (VEα and VEβ), and a ZPD containing an IHP and a CTP containing an EHP, but lack a TMD (some fish VEGs synthesized by the ovary have a TMD) (52). VEα and VEβ share ∼65% sequence identity and have a Type 2 ZPD; VEγ has a Type 1 ZPD. VEα and VEβ are related to ZP1/ZP4; VEγ is related to ZP3. It is likely that trout VE is assembled from VEα/VEγ and VEβ/VEγ heterodimers, although like ZPGs, VEGs can form homopolymers (53). Despite ∼4 × 108 years separating the appearance of trout and mice and despite the change from external to internal fertilization and development, ZPGs and VEGs have many structural features in common (51).
Concluding Remarks
This Minireview delineates some of the progress that has been made in our understanding of the biochemistry of ZPGs during the past 25 years or so. However, many important issues related to the participation of ZPGs in the fertilization process remain unresolved or controversial. In view of the facts that mutation of ZPG genes remains a potential cause of infertility in women (54) and that ZPGs remain attractive targets for immunocontraception (55), the more we learn about ZPGs, the more effective we will be in addressing these and other aspects of human reproduction. From a personal point of view, ZPGs have proven to be a very challenging and rewarding research endeavor for me and my colleagues for more than three decades.
Supplementary Material
Acknowledgments
I am grateful to Eveline Litscher for reading and improving the manuscript and to Luca Jovine, Costel Darie, Huayu Qi, Zev Williams, and Eveline Litscher for taking our research on ZPGs in new directions.
This work was supported, in whole or in part, by National Institutes of Health Grant HD-35105. This minireview will be reprinted in the 2008 Minireview Compendium, which will be available in January, 2009.
Footnotes
The abbreviations used are: ZP, zona pellucida; ZPG, ZP glycoprotein; mZP, mouse ZP; hZP, human ZP; VE, vitelline envelope; VEG, VE glycoprotein; SS, signal sequence; CTP, C-terminal propeptide; ZPD, ZP domain; CFCS, consensus furin cleavage site; TMD, transmembrane domain; NTS, N-terminal subdomain; EHP, external hydrophobic patch; IHP, internal hydrophobic patch.
References
- 1.Yanagimachi, R. (1994) in Physiology of Reproduction (Knobil, E., and Neill, J. D., eds) Vol. 1, pp. 189–318, Raven Press, Ltd., New York [Google Scholar]
- 2.Florman, H. M., and Ducibella, T. (2006) in Physiology of Reproduction (Neill, J. D., ed.) Vol. 1, pp. 55–112, Elsevier, San Diego [Google Scholar]
- 3.Kurotaki, Y., Hatta, K., Nakao, K., Nabeshima, Y., and Fujimori, T. (2007) Science 316 719–723 [DOI] [PubMed] [Google Scholar]
- 4.Wassarman, P. M. (1988) Annu. Rev. Biochem. 57 415–442 [DOI] [PubMed] [Google Scholar]
- 5.Bauskin, A. R., Franken, D. R., Eberspaecher, U., and Donner, P. (1999) Mol. Human Reprod. 5 534–540 [DOI] [PubMed] [Google Scholar]
- 6.Lefievre, L., Conner, S. J., Salpekar, A., Olufowobi, O., Ashton, P., Pavlovic, B., Lenton, W., Afnan, M., Brewis, I. A., Monk, M., Hughes, D. C., and Barratt, C. L. R. (2004) Human Reprod. 19 1580–1586 [DOI] [PubMed] [Google Scholar]
- 7.Boja, E. S., Hoodbhoy, T., Fales, H. M., and Dean, J. (2003) J. Biol. Chem. 278 34189–34202 [DOI] [PubMed] [Google Scholar]
- 8.Chalabi, S., Panico, M., Sutton-Smith, M., Haslam, S. M., Patankar, M. S., Lattanzio, F. A., Morris, H. R., Clark, G. F., and Dell, A. (2006) Biochemistry 45 637–647 [DOI] [PubMed] [Google Scholar]
- 9.Epifano, O., Liang, L., and Dean, J. (1995) J. Biol. Chem. 270 27254–27258 [DOI] [PubMed] [Google Scholar]
- 10.Liang, L., and Dean, J. (1993) Dev. Biol. 156 399–408 [DOI] [PubMed] [Google Scholar]
- 11.Hughes, D. C., and Barratt, C. L. (1999) Biochim. Biophys. Acta 1447 303–306 [DOI] [PubMed] [Google Scholar]
- 12.Liang, L., Soyal, S. M., and Dean, J. (1997) Development (Camb.) 124 4939–4947 [DOI] [PubMed] [Google Scholar]
- 13.Soyal, S. M., Amieh, A., and Dean, J. (2000) Development (Camb.) 127 4645–4654 [DOI] [PubMed] [Google Scholar]
- 14.Lira, S. A., Kinloch, R. M., Mortillo, S., and Wassarman, P. M. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 7215–7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lira, S. A., Schickler, M., and Wassarman, P. M. (1993) Mol. Reprod. Dev. 36 49–499 [DOI] [PubMed] [Google Scholar]
- 16.Millar, S. E., Lader, E., and Dean, J. (1993) Dev. Biol. 158 410–413 [DOI] [PubMed] [Google Scholar]
- 17.Schickler, M., Lira, S. A., Kinloch, R. M., and Wassarman, P. M. (1992) Mol. Cell. Biol. 12 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roller, R. J., Kinloch, R. A., Hiraoka, B. Y., Li, S. S., and Wassarman, P. M. (1989) Development (Camb.) 106 251–261 [DOI] [PubMed] [Google Scholar]
- 19.Spargo, S. C., and Hope, R. M. (2003) Biol. Reprod. 68 358–362 [DOI] [PubMed] [Google Scholar]
- 20.Jovine, L., Litscher, E. S., and Wassarman, P. M. (2002) Adv. Dev. Biol. Biochem. 12 31–54 [Google Scholar]
- 21.Jovine, L., Darie, C. C., Litscher, E. S., and Wassarman, P. M. (2005) Annu. Rev. Biochem. 74 83–114 [DOI] [PubMed] [Google Scholar]
- 22.Callebaut, I., Mornon, J. P., and Monget, P. (2007) Bioinformatics 23 1871–1874 [DOI] [PubMed] [Google Scholar]
- 23.Jovine, L., Qi, H., Williams, Z., Litscher, E. S., and Wassarman, P. M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 5922–5927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Llorca, O., Trujillo, A., Blanco, F. J., and Bernabeu, C. (2007) J. Mol. Biol. 365 694–705 [DOI] [PubMed] [Google Scholar]
- 25.Legan, P., Rau, K., Keen, J. N., and Richardson, G. P. (1997) J. Biol. Chem. 272 8791–8801 [DOI] [PubMed] [Google Scholar]
- 26.Jovine, L., Qi, H., Williams, Z., Litscher, E., and Wassarman, P. M. (2002) Nat. Cell Biol. 4 457–461 [DOI] [PubMed] [Google Scholar]
- 27.Jovine, L., Janssen, W. G., Litscher, E. S., and Wassarman, P. M. (2006) BMC Biochem. 7 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wassarman, P. M., and Mortillo, S. (1991) Int. Rev. Cytol. 130 85–110 [DOI] [PubMed] [Google Scholar]
- 29.Litscher, E. S., Janssen, W. G., Darie, C. C., and Wassarman, P. M. (2008) J. Cell. Physiol. 214 153–157 [DOI] [PubMed] [Google Scholar]
- 30.Jovine, L., Qi, H., Williams, Z., Litscher, E. S., and Wassarman, P. M. (2007) in Gamete Biology (Gupta, S. K., Koyama, K., and Murray, J. F., eds) pp. 187–201, Nottingham University Press, Nottingham, UK
- 31.Litscher, E. S., Qi, H., and Wassarman, P. M. (1999) Biochemistry 38 12280–12287 [DOI] [PubMed] [Google Scholar]
- 32.Williams, Z., and Wassarman, P. M. (2001) Biochemistry 40 929–937 [DOI] [PubMed] [Google Scholar]
- 33.Qi, H., Williams, Z., and Wassarman, P. M. (2002) Mol. Biol. Cell 13 530–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao, M., Gold, L., Ginsberg, A. M., Liang, L. F., and Dean, J. (2002) Mol. Cell. Biol. 22 3111–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao, M., Gold, L., Dorward, H., Liang, L., Hoodbhoy, T., Boja, E. S., Fales, H. M., and Dean, J. (2003) Mol. Cell. Biol. 23 8982–8991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wassarman, P. M., Qi, H., and Litscher, E. S. (1997) Proc. R. Soc. Biol. Sci. 264 323–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, C., Litscher, E. S., Mortillo, S., Sakai, Y., Kinloch, R. A., Stewart, C. L., and Wassarman, P. M. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 5431–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rankin, T., Familiari, M., Lee, E., Ginsburg, A., Dwyer, N., Blanchette-Mackie, J., Drago, J., Westphal, H., and Dean, J. (1996) Development (Camb.) 122 2903–2910 [DOI] [PubMed] [Google Scholar]
- 39.Rankin, T., O'Brien, M., Lee, E., Wigglesworth, K., Eppig, J., and Dean, J. (2001) Development (Camb.) 128 1119–1126 [DOI] [PubMed] [Google Scholar]
- 40.Wassarman, P. M., Liu, C., Chen, J., Qi, H., and Litscher, E. S. (1998) Histol. Histopathol. 13 293–300 [DOI] [PubMed] [Google Scholar]
- 41.Rankin, T., Talbot, P., Lee, E., and Dean, J. (1999) Development (Camb.) 126 3847–3855 [DOI] [PubMed] [Google Scholar]
- 42.Dean, J. (2007) in Gamete Biology (Gupta, S. K., Koyama, K., and Murray, J. F., eds) pp. 359–365, Nottingham University Press, Nottingham, UK
- 43.Wassarman, P. M. (1999) Cell 96 175–183 [DOI] [PubMed] [Google Scholar]
- 44.Wassarman, P. M., Jovine, L., and Litscher, E. S. (2001) Nat. Cell Biol. 3 E59–E64 [DOI] [PubMed] [Google Scholar]
- 45.Kinloch, R. A., Sakai, Y., and Wassarman, P. M. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 263–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams, Z., Litscher, E. S., Jovine, L., and Wassarman, P. M. (2006) J. Cell. Physiol. 207 30–39 [DOI] [PubMed] [Google Scholar]
- 47.Li, D., Cao, S., and Xu, C. (2007) Mol. Reprod. Dev. 74 1327–1336 [DOI] [PubMed] [Google Scholar]
- 48.Clark, G. F., and Dell, A. (2006) J. Biol. Chem. 281 13853–13856 [DOI] [PubMed] [Google Scholar]
- 49.Swanson, W. J., Yang, Z., Wolfner, M. F., and Aquadro, C. F. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shur, B. D., Rodeheffer, C., and Ensslin, M. A. (2004) Curr. Biol. 14 R691–R692 [DOI] [PubMed] [Google Scholar]
- 51.Litscher, E. S., and Wassarman, P. M. (2007) Histol. Histopathol. 22 337–347 [DOI] [PubMed] [Google Scholar]
- 52.Darie, C. C., Biniossek, M. L., Jovine, L., Litscher, E. S., and Wassarman, P. M. (2004) Biochemistry 43 7459–7478 [DOI] [PubMed] [Google Scholar]
- 53.Darie, C. C., Janssen, W. J., Litscher, E. S., and Wassarman, P. M. (2008) Biochim. Biophys. Acta 1784 385–392 [DOI] [PubMed] [Google Scholar]
- 54.Mannikko, M., Tormala, R. M., Tuuri, T., Haltia, A., Martikainen, H., Ala-Kokko, L., Tapanainen, J. S., and Lakkakorpi, J. T. (2005) Human Reprod. 20 1578–1585 [DOI] [PubMed] [Google Scholar]
- 55.Paterson, M., Jennings, Z. A., Wilson, M. R., and Aitken, R. J. (2002) J. Reprod. Immunol. 53 99–107 [DOI] [PubMed] [Google Scholar]
- 56.Familiari, G., Relucenti, M., Heyn, R., Micara, G., and Correr, S. (2006) Microscopy Res. Tech. 69 415–426 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.