Abstract
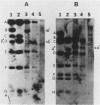
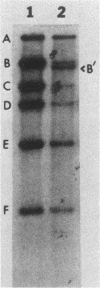
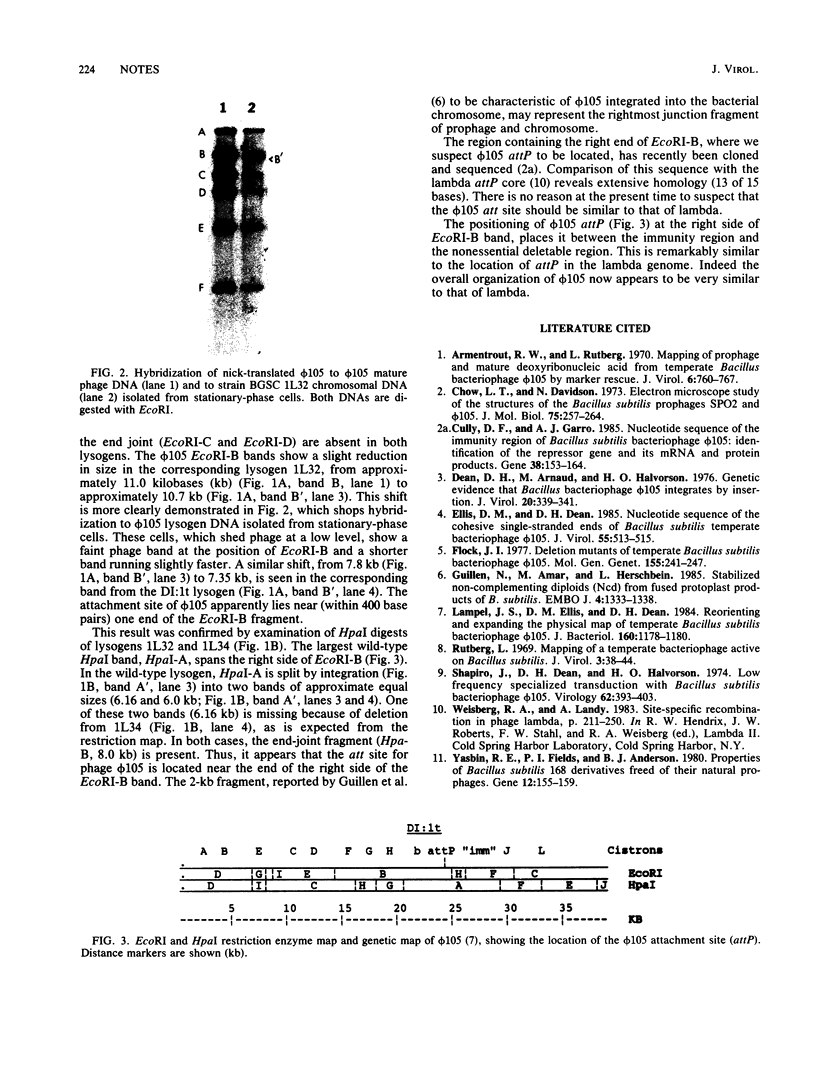
Chromosomal DNAs of lysogens of phi 105 and phi 105 DI:1t were digested with restriction enzymes EcoRI and HpaI and were probed with nick-translated mature phi 105 DNA. Altered bacteriophage-specific bands in the lysogens were detected, indicating that the phage integrates into the host chromosome at a single site, probably via a Campbell-type circular intermediate. The phage attachment site is centrally located in the phage genome and lies between the phage immunity region and the nonessential deletable region of phi 105.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armentrout R. W., Rutberg L. Mapping of prophage and mature deoxyribonucleic acid from temperate Bacillus bacteriophage phi 105 by marker rescue. J Virol. 1970 Dec;6(6):760–767. doi: 10.1128/jvi.6.6.760-767.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L. T., Davidson N. Electron microscope study of the structures of the Bacillus subtilis prophages, SPO2 and phi105. J Mol Biol. 1973 Apr 5;75(2):257–264. doi: 10.1016/0022-2836(73)90019-3. [DOI] [PubMed] [Google Scholar]
- Cully D. F., Garro A. J. Nucleotide sequence of the immunity region of Bacillus subtilis bacteriophage phi 105: identification of the repressor gene and its mRNA and protein products. Gene. 1985;38(1-3):153–164. doi: 10.1016/0378-1119(85)90214-8. [DOI] [PubMed] [Google Scholar]
- Dean D. H., Arnaud M., Halvorson H. O. Genetic evidence that Bacillus bacteriophage phi 105 integrates by insertion. J Virol. 1976 Oct;20(1):339–341. doi: 10.1128/jvi.20.1.339-341.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis D. M., Dean D. H. Nucleotide sequence of the cohesive single-stranded ends of Bacillus subtilis temperate bacteriophage phi 105. J Virol. 1985 Aug;55(2):513–515. doi: 10.1128/jvi.55.2.513-515.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flock J. I. Deletion mutants of temperate Bacillus subtilis bacteriophage phi105. Mol Gen Genet. 1977 Oct 24;155(3):241–247. doi: 10.1007/BF00272803. [DOI] [PubMed] [Google Scholar]
- Guillén N., Amar M., Hirschbein L. Stabilized non-complementing diploids (Ncd) from fused protoplast products of B. subtilis. EMBO J. 1985 May;4(5):1333–1338. doi: 10.1002/j.1460-2075.1985.tb03781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampel J. S., Ellis D. M., Dean D. H. Reorienting and expanding the physical map of temperate Bacillus subtilis bacteriophage phi 105. J Bacteriol. 1984 Dec;160(3):1178–1180. doi: 10.1128/jb.160.3.1178-1180.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutberg L. Mapping of a temperate bacteriophage active on Bacillus subtilis. J Virol. 1969 Jan;3(1):38–44. doi: 10.1128/jvi.3.1.38-44.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A., Dean D. H., Halvorson H. O. Low-frequency specialized transduction with Bacillus subtilis bacteriophage phi 105. Virology. 1974 Dec;62(2):393–403. doi: 10.1016/0042-6822(74)90401-2. [DOI] [PubMed] [Google Scholar]
- Yasbin R. E., Fields P. I., Andersen B. J. Properties of Bacillus subtilis 168 derivatives freed of their natural prophages. Gene. 1980 Dec;12(1-2):155–159. doi: 10.1016/0378-1119(80)90026-8. [DOI] [PubMed] [Google Scholar]