Abstract
Proteinase-activated receptor 2 (PAR2), a seven-transmembrane G protein-coupled receptor, is activated at inflammatory sites by proteolytic cleavage of its extracellular N terminus by trypsin-like enzymes, exposing a tethered, receptor-activating ligand. Synthetic agonist peptides (AP) that share the tethered ligand sequence also activate PAR2, often measured by Ca2+ release. PAR2 contributes to inflammation through activation of NF-κB-regulated genes; however, the mechanism by which this occurs is unknown. Overexpression of human PAR2 in HEK293T cells resulted in concentration-dependent, PAR2 AP-inducible NF-κB reporter activation that was protein synthesis-independent, yet blocked by inhibitors that uncouple Gi proteins or sequester intracellular Ca2+. Because previous studies described synergistic PAR2- and TLR4-mediated cytokine production, we hypothesized that PAR2 and TLR4 might interact at the level of signaling. In the absence of TLR4, PAR2-induced NF-κB activity was inhibited by dominant negative (DN)-TRIF or DN-TRAM constructs, but not by DN-MyD88, findings confirmed using cell-permeable, adapter-specific BB loop blocking peptides. Co-expression of TLR4/MD-2/CD14 with PAR2 in HEK293T cells led to a synergistic increase in AP-induced NF-κB signaling that was MyD88-dependent and required a functional TLR4, despite the fact that AP exhibited no TLR4 agonist activity. Co-immunoprecipitation of PAR2 and TLR4 revealed a physical association that was AP-dependent. The response to AP or lipopolysaccharide was significantly diminished in TLR4–/– and PAR –/–2 macrophages, respectively, and SW620 colonic epithelial cells exhibited synergistic responses to co-stimulation with AP and lipopolysaccharide. Our data suggest a unique interaction between two distinct innate immune response receptors and support a novel paradigm of receptor cooperativity in inflammatory responses.
Extracellular proteases (or proteinases), e.g. thrombin and trypsin, play crucial roles as direct regulators of cellular functions, in addition to their long recognized importance in digestion, hormone processing, and hemostasis (reviewed in Refs. 1, 2). Serine proteases released or generated during tissue injury, malignancy, infection, or inflammation can activate cell signaling by triggering proteinase-activated receptors (PARs),3 a novel, four-member family of seven-transmembrane G protein-coupled receptors (GPCR) (reviewed in Refs. 3–10). In general, PAR1, PAR3, and PAR4 are targeted by thrombin; PAR2 responds to trypsin and trypsin-like serine proteases, including mast cell tryptase, tissue kallikreins, and coagulation factors VIIa and Xa. These serine proteases cleave PARs irreversibly at a specific site in the extracellular N terminus to expose a tethered “neo-ligand” that, in turn, binds intramolecularly to PARs to trigger receptor autoactivation. Synthetic peptides that correspond to the sequences of the tethered neo-ligands can also activate uncleaved PARs (except in the case of PAR3) and serve as valuable experimental tools to study the consequence of PAR activation in the absence of exogenous proteases. To date, it has been widely reported that the activation of PARs by serine proteases contributes to normal physiology as well as to various pathophysiological states, including autoimmunity, cancer, tissue injury and repair, infection, and inflammation (reviewed in Refs. 3–10).
PARs are distributed ubiquitously throughout the body, with relatively high expression in the gastrointestinal and respiratory tracts (reviewed in Refs. 11, 12). Importantly, the presence of PARs on epithelia, endothelia, monocytes, and macrophages (reviewed in Ref. 7) suggests a possible role for PARs in the innate immune defense and immune surveillance (reviewed in Refs. 4, 11–13). However, innate immunity has largely been attributed to the capacity of “pattern-recognition receptors” (PRRs) to respond to conserved “pathogen-associated molecular patterns” (PAMPs) (reviewed in Ref. 14), whereas PARs have generally been considered to be “sensors” of the extracellular proteolytic environment (reviewed in Refs. 12, 13). Specifically, PAR2 has been associated with the inflammatory response to infection and microbial proteases and, in this context, may also act as a PRR. In mice, PAR2 activation plays a pivotal role in the pathogenesis of periodontitis caused by Porphyromonas gingivalis (15) and in the development of infectious colitis induced by Citrobacter rodentium (16). Exposure of PAR2 to proteases derived from P. gingivalis (17) or Serratia marcescens (18) induces expression of the antimicrobial peptide β-defensin 2 (17), pro-inflammatory cytokines and chemokines, e.g. interleukin (IL)-6 and IL-8 (18), as well as activation of the transcription factors AP-1, C/EBPβ, and NF-κB (18) in human gingival cells (17) and lung epithelial cells (18). The house dust mite allergens, Der p3 and Der p9 serine proteases, also induce pro-inflammatory responses in human airway epithelial cells via PAR2 activation (19, 20). Thus, PAR2 is involved in the innate immune response at anatomic sites that interact with protease-rich environments, such as the gut lumen and respiratory tract. Consequently, several microbes evade PAR2-mediated immune surveillance by directly disabling the receptor. For example, proteases from Pseudomonas aeruginosa (21) or Treponema denticola (15) have been reported to inactivate PAR2 and thereby alter its function.
The Toll-like receptors (TLRs) are important sentinels of the innate immune response through their ability to respond to PAMPs (e.g. lipopolysaccharide (LPS), double-stranded RNA, flagellin, and unmethylated CpG DNA) (reviewed in Refs. 14, 22). TLRs are germ line-encoded, evolutionarily conserved type I transmembrane glycoproteins and represent a family of closely related PRRs that respond to many different microbial PAMPs. More recently, TLRs have also been implicated in the response to endogenous molecules released by damaged cells or chemical substances generated during tissue injury or highly inflammatory processes (reviewed in Refs. 23–27). In particular, TLR4 responds not only to bacterial LPS but also to an array of other exogenous and endogenous agonists, e.g. respiratory syncytial virus F protein, chlamydial heat-shock protein 60, fibrinogen, surfactant protein A, murine β-defensin 2, and the serine protease elastase (reviewed in Refs. 24–30). To date, 10 functional murine and 9 functional human TLRs have been identified.
Direct or indirect interaction of TLR ectodomains with cognate ligands results in receptor oligomerization leading to conformational changes within intra-cytoplasmic Toll/interleukin-1 receptor resistance (TIR) domains that create docking platforms for the recruitment of TIR-containing adapter proteins and downstream kinases (22). TLR4 is the only known TLR that is capable of recruiting all four adapter proteins (i.e. myeloid differentiation factor 88 (MyD88), TIR domain-containing adapter protein/MyD88-adapter-like (TIRAP/Mal), TIR domain-containing adapter inducing IFN-β (TRIF), and TRIF-related adapter molecule (TRAM)) through TIR-TIR interactions. TLR4 engagement by its prototype agonist, LPS, results in the activation of two major signaling pathways, i.e. “MyD88-dependent” and “MyD88-independent” (reviewed in Refs. 22, 31, 32). Activation of the MyD88-dependent pathway results predominantly in the “early” nuclear translocation of NF-κB that is required for expression of most pro-inflammatory cytokines, e.g. IL-1, IL-6, and TNF-α. In contrast, the MyD88-independent pathway activates a “delayed” NF-κB response, as well as rapid activation of another key transcription factor, IRF-3, that up-regulates transcription of IRF-3-responsive genes, e.g. RANTES and IFN-β.
Inflammation caused by C. rodentium infection has been reported to depend on both TLR4 (33) and PAR2 (16). Given that PAR2 has been recently associated with several infectious and noninfectious models leading to inflammation (reviewed in Refs. 4–7, 9, 10, 18, 34–36), we hypothesized that PAR2 might interact with TLR4, perhaps at the level of cross-talk between signaling pathways. Indeed, several studies have investigated the possible connection between PAR2- and TLR4-mediated signaling pathways. For example, concurrent activation of PAR2 and TLR4 by PAR2 AP and LPS, respectively, amplifies NF-κB activation and IL-6 production in endothelial cells (37). In contrast, the inflammatory response induced by Aspergillus infection involves TLR4-mediated suppression of PAR2 signaling (36). Thus, signaling cross-talk between PAR2 and TLR4 has the potential to augment or mitigate an ongoing inflammatory response when both receptors are accessible.
In this study, we demonstrate PAR2 activation by its AP, as measured by ELAM (NF-κB)- or RANTES (IRF-3)-reporter activation and IL-8 secretion, in transiently transfected HEK293T cells. PAR2-induced NF-κB activation by AP was MyD88-independent but TRIF- and TRAM-dependent. Moreover, a synergistic response to PAR2 AP was observed when PAR2 was co-expressed with the TLR4 receptor complex (TLR4/MD-2/CD14), despite the fact that AP exhibited no TLR4 activity. The observed synergy was fully MyD88-dependent. These results suggest that PAR2 and TLR4 interact functionally at the level of intracellular adapter utilization for the activation of NF-κB. Co-immunoprecipitation of PAR2 and TLR4 in HEK293T transfectants was observed only in the presence of AP. In contrast to wild-type macrophages, macrophages derived from TLR4–/– mice responded poorly to PAR2 AP to elicit IL-1β mRNA, and conversely, PAR –/–2 macrophages exhibited a significant down-regulation of iNOS gene expression induced by LPS. The colonic epithelial cell line, SW620, responded synergistically to AP and LPS, further supporting the conclusion that functional synergy also occurs in nontransfected cells. Collectively, these data support a novel model of heterophilic receptor interaction between TLR4 and PAR2 that centers on shared utilization of TLR adapter proteins, leading to enhanced NF-κB activation upon PAR2 engagement. The findings in this study also imply a previously under-appreciated role of PARs as an additional level of the innate immune defense, apart from classical pattern-recognition receptors.
EXPERIMENTAL PROCEDURES
Reagents and Cell Culture—Human embryonic kidney (HEK) 293T cells (ATCC, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium (BioWhittaker, Walkersville, MD) supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. PAR2 AP (SLIGRL-NH2 (rodent); SLIGKV-NH2 (human)) or inactive peptides (LSIGRL-NH2 and VKGILS-NH2) were purchased from Sigma or synthesized by the Biopolymer Laboratory (University of Maryland, Baltimore) or Phoenix Pharmaceuticals, Inc. (Burlingame, CA). PAR1 AP (TFLLR-NH2) and scrambled control peptide were synthesized at the University of Calgary. Cycloheximide (CHX) and 1,2-bis-(2-aminophenoxy) ethane-N,N,N′,N′-tetraacetic acid tetra(acetomethyl) ester (BAPTA-AM) were purchased from Sigma. Polyclonal anti-hemagglutinin (HA) antibody (clone Y-11) was obtained from Santa Cruz Biotechnology (Santa Cruz, CA), and anti-GFP polyclonal antibody was purchased from Invitrogen. Recombinant human TNF-α was kindly provided by Cetus Corp. (Emeryville, CA). Recombinant human IL-1β was obtained from R & D Systems (Minneapolis, MN). Protein-free, phenol/water-extracted Escherichia coli K235 LPS was prepared as described elsewhere (38). S-[2,3-Bis(palmitoyloxy)-(2-RS)-propyl]-N-palmitoyl-(R)-Cys-(S)-Ser-(Lys)4-OH, trihydrochloride was purchased from EMC Microcollections (Tuebingen, Germany), and polyinosinic:polycytidylic acid (poly(I:C)) was from Amersham Biosciences. Purified Bordetella pertussis toxin (PT) was a kind gift of Dr. Nicholas Carbonetti (University of Maryland, Baltimore). RNeasy total RNA extraction kit and SuperFect transfection reagent were from Qiagen (Valencia, CA). RT-PCR kit was purchased from Promega Corp. (Madison, WI). All expression plasmid constructs were prepared using EndoFree plasmid maxi kit (Qiagen). Oligonucleotide synthesis and DNA sequencing analyses were carried out by the Biopolymer and Genomics Core Facility (University of Maryland, Baltimore). Cell-permeable BB loop peptides (BPs) corresponding to the 14 amino acid sequences of the BB loops of human MyD88, TRIF, and TRAM, as well as a scrambled control peptide (CP), were synthesized and used as described elsewhere (39). Primary peritoneal macrophages from C57BL/6J mice, PAR –/–2 mice (The Jackson Laboratory, Bar Harbor, ME), or TLR4–/– mice (kindly provided by Dr. S. Akira, Osaka, Japan), which had been backcrossed ≥8 times onto a C57BL/6 background, were collected for in vitro studies as described elsewhere (40). The human colonic epithelial cell line SW620 (ATCC) was propagated in tissue culture flasks in RPMI media supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm l-glutamine, and 2% fetal calf serum.
Plasmid Constructs—We used a wild-type (WT) human PAR2 expression vector, pcDNA3-PAR2, that encodes a native, untagged PAR2 protein (41). Expression plasmids pCMV1-FLAG-TLR4, which encodes human TLR4 with an N-terminal FLAG epitope tag, pcDNA3-huCD14, pEFBOS-HA-huMD-2, and pELAM-1 luciferase (pELAM-luc; an NF-κB reporter) and pCMV1-β-galactosidase reporter plasmids were kindly provided by Dr. Douglas Golenbock (University of Massachusetts Medical School, Worcester) and have been described previously (42–44). Expression constructs for human dominant negative (DN)-MyD88, DN-TRIF, and DN-TRAM, and the RANTES promoter-luciferase reporter construct, FLAG-TLR2, and FLAG-TLR3 expression constructs were kindly provided by Drs. Douglas Golenbock and Katherine Fitzgerald (University of Massachusetts Medical School, Worcester) (45). pcDNA3-WT YFP-TLR4 was kindly provided by Dr. Golenbock, and the plasmid encoding the P714H YFP-TLR4 mutant was generated by site-directed mutagenesis, using Quick-Change site-directed mutagenesis kit (Stratagene) (44). A C-terminally HA-tagged human PAR2 vector (46) was the kind gift of Dr. Steven Compton (University of Hull, Hull, UK). The coding sequences of all plasmid constructs were confirmed by sequencing.
Reporter Assay—HEK293T cells were seeded into 12-well Costar plates (Corning Inc., Corning, NY) at 2 × 105 cells/well and incubated overnight at 37 °C, 5% CO2 atmosphere. Using SuperFect reagent, cells were transfected for 4 h with PAR2 expression vector (200 ng/well; determined in preliminary studies to result in optimal responsiveness to AP (data not shown)), TLR4 complex (pFLAG-huTLR4, 7.5 ng/well; pEF-BOS-HA-huMD-2, 3 ng/well; and pcDNA3-huCD14, 30 ng/well), or both as indicated under “Results,” together with the ELAM-luc reporter (500 ng/well) or RANTES-luc reporter (200 ng/well) and pCMV1-β-galactosidase (100 ng/well). The final input DNA was adjusted to 1.5 μg/well with the pcDNA3 blank vector (Invitrogen). After overnight recovery, cells were washed with 1× PBS and stimulated with PAR2 AP or LPS at the indicated concentrations for 5 or 16 h, as indicated. Cells were lysed in 1× reporter assay lysis buffer (Promega). β-Galactosidase (Tropix, Galacto-Light System, Bedford, MA) and luciferase (Promega, Luciferase Assay System) activities were analyzed using a Berthold LB 9507 luminometer (Berthhold Technologies, Bad Wildbad, Germany). “Relative luciferase activity,” represented as relative luciferase units, was calculated by normalizing luciferase activity for each sample to constitutive β-galactosidase activity measured within the same sample. Based on staining of β-galactosidase activity in individual cells, the transfection efficiency was consistently >90%.
Immunoprecipitation and Western Analysis—HEK293T cells were plated in 100-mm Petri dishes (3 × 106 cells/dish), grown overnight, and co-transfected for 4 h with YFP-TLR4 WT (5 μg/dish) and pcDNA3-huCD14 (450 ng/dish) and pEF-BOS-HA-huMD-2 (45 ng/dish) expression vectors, without or with HA-PAR2 expression construct (5 μg/dish) using the SuperFect transfection reagent. The cells were incubated for an additional 72 h before treating the cells with AP for the times indicated. To prepare cell extracts for immunoprecipitation, cells were washed twice with cold 1× PBS, resuspended in 1 ml of ice-cold lysis buffer (M-PER buffer (Pierce), with 1 mm phenylmethylsulfonyl fluoride and protease inhibitor mixture), transferred to 1.5-ml Eppendorf tubes, and mixed by rotation at 4 °C for 1 h. Lysates were centrifuged at maximum speed for 10 min at 4 °C; the supernatant was transferred to another Eppendorf tube, and the protein concentration was measured. Cell lysates containing ∼8 mg of total protein were precleared with 40 μl of 1 × PBS-washed EZview Red Protein G affinity gel (Sigma) for 1 h at 4°C. Precleared supernatant was then mixed with 5 μl of anti-HA polyclonal antibody and incubated at 4 °C overnight. Forty μl of protein G beads were added, and the mixture was incubated for an additional 5 h at 4°Cona rocker. For the anti-GFP antibody (5 μl/sample), the lysate and antibody mixture was incubated at 4 °C for 4 h, and then the protein G beads were added, and the incubation was continued overnight. The samples were centrifuged at 3600 rpm at 4 °C for 1 min; the supernatant was removed, and the red affinity beads were washed two times with cold lysis buffer and four times with cold 1× PBS. The final red bead pellet was resuspended in 20 μl of 2 × SDS sample buffer, vortexed, and boiled for 10 min.
For Western analysis, the entire immunoprecipitate was resolved on 4–15% Tris-glycine SDS-polyacrylamide gel (Bio-Rad), transferred onto Immobilon P membrane, and developed by Western analysis using the methods described elsewhere (42). Anti-HA antibody was used at 1:500 dilution, and anti-GFP antibody at 1:1000 dilution. Horseradish peroxidase-conjugated secondary antibodies were used at appropriate dilutions and conditions as suggested by the manufacturer.
RT-PCR—Total RNA from HEK293T transfectants was isolated and quantified spectrophotometrically, and cDNA was prepared by using Moloney murine leukemia virus reverse transcriptase kit (Promega, Madison, WI). Relative mRNA quantities of firefly luciferase (230-bp PCR product) and huGAPDH (internal control, 600-bp PCR product) were determined by RT-PCR using the following primers: luciferase, 5′-TCA AAG AGG CGA ACT GTG TG-3′ (sense), 5′-GGT GTT GGA GCA AGA TGG AT-3′ (antisense); huGAPDH, 5′-CTG ATG CCC CCA TGT TCG TCA-3′ (sense), 5′-CCA CCA CCC TGT TGC TGT AG-3′ (antisense), followed by electrophoresis of amplified products and visualization by ethidium bromide staining. The optimal cycle number for each gene (luciferase, 45 cycles; GAPDH, 35 cycles) was chosen to detect amplified products under nonsaturating conditions. Each cycle consisted of 1 min at 95 °C, 1 min at a gene-specific annealing temperature (luciferase, 52 °C; huGAPDH, 58 °C), and 2 min at 72 °C. mRNA expression in macrophages and SW620 cells was detected by quantitative real time PCR as described elsewhere (40).
Statistical Analyses—Data are presented as means ± S.E. and analyzed using a one-way analysis of variance with repeated measures, followed by post hoc comparisons using Tukey's multiple paired comparison test of the GraphPad PRISM 4.0 (GraphPad Software, Inc., San Diego, CA).
RESULTS
PAR2-stimulated ELAM (NF-κB) Reporter Activation: Specificity, Kinetics, and Dose Dependence—Although PAR2 has been strongly implicated in the induction of inflammatory responses in a variety of species (reviewed in Refs. 4–7, 9, 10, 34, 35), little is known about the molecular mechanisms by which PAR2 activates the inflammatory response. To delineate the intracellular signaling pathways elicited by PAR2, we initially sought to develop a transient transfection system in HEK293T cells. The HEK293T cell line is highly transfectable and, under our conditions of culture, did not express detectable endogenous PAR2 or TLR4 mRNA (data not shown) but, as demonstrated below, responded to PAR2 AP with NF-κB activation only following transfection with a human PAR2 expression construct.
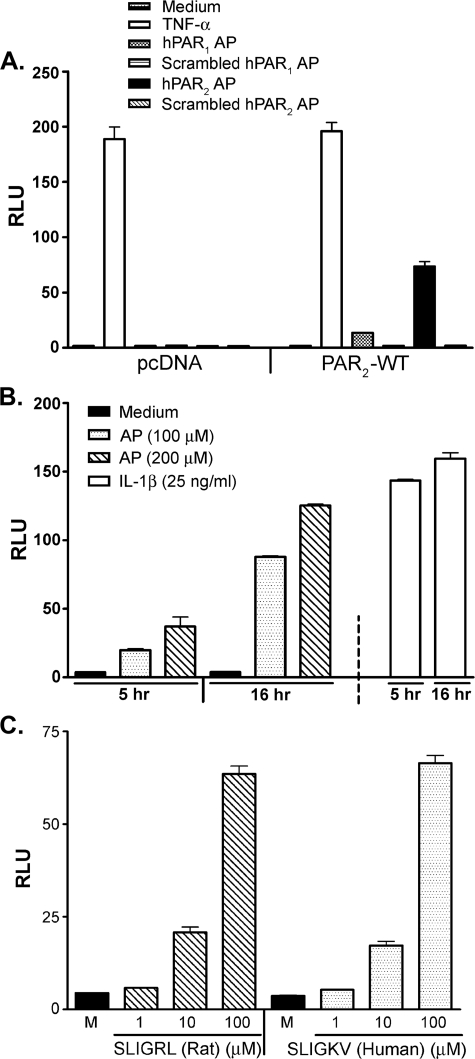
HEK293T cells transiently transfected with the pcDNA3.1 control vector did not elicit NF-κB reporter activation in response to PAR2 AP, but were responsive when stimulated with TNF-α via the endogenous TNF receptor (Fig. 1A, left side of graph). Introduction of an optimized input concentration (i.e. 200 ng/well) of the PAR2 expression vector resulted in ELAM (NF-κB)-driven luciferase activity in cells stimulated with huPAR2 AP (SLIGKV-NH2) but not with scrambled control peptides (Fig. 1A, right side of graph). PAR2 transfectants also exhibited a weak response to huPAR1 AP (TFLLR-NH2) that was not seen in pcDNA-transfected cells, suggesting a slight degree of cross-reactivity because no PAR1-mediated response was observed in pcDNA control transfectants (Fig. 1A). The response to TNF-α was unperturbed in PAR2 transfectants (Fig. 1A). Activation of PAR2 by AP was concentration-dependent within a 10–200 μm range (supplemental Fig. 1A) and plateaued at higher concentrations. Treatment of PAR2 transfectants, but not pcDNA3.1-transfectants, with trypsin (10 nm) induced a level of NF-κB-luciferase activity comparable with that induced by AP (100 μm) (supplemental Fig. 1B).
FIGURE 1.
HEK293T/PAR2 transfectants respond in a specific manner independent of species of origin of AP. A, HEK293T cells were co-transfected with either the control pcDNA or the untagged human PAR2 construct, together with ELAM (NF-κB)-luciferase and pCMV1-β-galactosidase reporter constructs. Transfected cells were treated with TNF-α (50 ng/ml), 200 μm huPAR1 AP (TFLLR-NH2), or its inactive scrambled peptide or 200 μm huPAR2 AP (SLIGKV-NH2) or its inactive scrambled peptide for 16 h. Reporter activities were measured in cell lysates as described under “Experimental Procedures.” A representative experiment is shown (n = 3). B, HEK293T cells, transfected as described in A, were treated with huPAR2 AP at the indicated concentrations or with human rIL-1β for 5 or 16 h. Cell lysates were assayed for ELAM-luc and β-galactosidase reporter activities. A representative experiment is shown (n = 8). C, HEK293T cells, transfected as described in A, were treated with medium only, rat/mouse PAR2 AP (SLIGRL-NH2), or huPAR2 AP for 16 h. ELAM-luc and β-galactosidase reporter activities were measured in cell lysates. A representative experiment is shown (n = 5). RLU, relative luciferase units; M, medium.
We next sought to optimize the treatment time necessary to achieve the maximum stimulation of PAR2 at different AP concentrations. Fig. 1B illustrates that a 5-h treatment of PAR2 transfectants with AP elicited ELAM reporter activity that was significantly lower than observed after 16 h. The luciferase activity induced after 16 h of PAR2 stimulation was of a similar magnitude to that induced by rIL-1β after only 5 or 16 h of stimulation (Fig. 1B, far right bar). A 16-h treatment period was also necessary to achieve optimal trypsin-induced NF-κB (supplemental Fig. 1B) or AP-induced RANTES-luciferase in PAR2 transfectants (data not shown).
The PAR2 APs, SLIGRL-NH2 (mouse/rat) and SLIGKV-NH2 (human), have been shown to activate PAR2 in both human and rat cells (47, 48). We observed an equivalent degree of activation of the ELAM reporter when the PAR2 HEK293T cell transfectants were stimulated with either the rat- or human-derived APs over a broad concentration range (Fig. 1C), indicating that these APs can be used interchangeably in this system. Previous studies have shown that in contrast to SLIGRL-NH2, a peptide with the first two amino acids inverted, i.e. LSIGRL-NH2, is inactive (47). The supplemental Fig. 1C confirms this observation in our transfection system and also illustrates that pretreatment with the inactive LSIGRL-NH2 peptide failed to block the ability of the active PAR2 AP, SLIGRL-NH2, to induce the ELAM (NF-κB) reporter.
AP-induced, PAR2-mediated ELAM (NF-κB)-driven Luciferase Accumulation Does Not Require de Novo Protein Synthesis and Is Sensitive to Pharmacologic Inhibitors—To date, most of the signaling and structure-activity work on PAR2 activation by APs has used intracellular calcium (Ca2+) release as an index of receptor activation (47–49). In contrast to the Ca2+ signaling response, which occurs within seconds of stimulation with AP (47–49), optimal AP-induced NF-κB-luciferase reporter activation observed in HEK293T cells transfected with PAR2 required a substantially longer period of incubation (i.e. ≥5 h versus 0.2 min). This observation prompted us to consider the possibility that AP stimulated the synthesis of an intermediate that, in turn, was required to elicit the delayed NF-κB-driven luciferase accumulation. To determine whether de novo protein synthesis was required for PAR2 AP-stimulated NF-κB luciferase activity, transfectants were pretreated for 1 h with medium only or with 5 μg/ml CHX, a mammalian protein synthesis inhibitor, followed by treatment of cells with 100 μm huPAR2 AP for different time intervals in the absence or presence of CHX. Total RNA was prepared at each time point, and the relative levels of luciferase mRNA were determined. Based on the results of three separate experiments, we failed to observe any significant differences in the relative quantities of steady-state luciferase mRNA at any time point, regardless of whether or not cells were treated with CHX (supplemental Fig. 2A).
To confirm this observation, two sets of HEK293T cells co-transfected with the PAR2 and reporter constructs were prepared. One set of transfectants was treated with either tissue culture medium only or 100 μm of huPAR2 AP for 11 h (supplemental Fig. 2B, left panel). Supernatants from this first set of cells were transferred to a second set of untreated PAR2 transfectants and incubated for an additional 5 h, for a total of 16 h treatment (supplemental Fig. 2B, right panel). NF-κB-driven luciferase activity was measurable after 11 h of treatment, whereas the supernatants derived from these cells failed to transfer the capacity to induce a rapid (5 h) or delayed (16 h; data not shown) NF-κB activation in the second set of transfectants. Taken together, these data strongly suggest that activation of PAR2-mediated NF-κB-driven luciferase accumulation does not require the induction of a secreted protein intermediate.
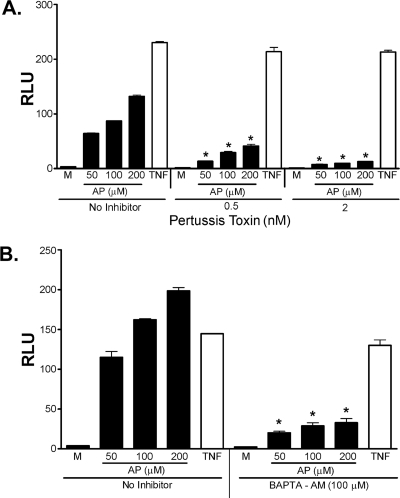
PAR2 is a seven-transmembrane GPCR that has been found to signal not only via Gq but also via Gi (reviewed in Ref. 3), a recognized target for inhibition by PT. To test the hypothesis that PAR2 activated NF-κB through Gi, PAR2 transfectants were pretreated with PT (0.5 or 2.0 nm) for 1 h, and then stimulated with PAR2 AP for an additional 16 h. PT attenuated PAR2 activation induced by AP, whereas the ability of TNF-α to activate NF-κB was not affected (Fig. 2A). This result indicates that PAR2 signal transduction leading to NF-κB-induced luciferase expression was strongly Gi-dependent.
FIGURE 2.
PAR2 activation by AP in HEK293T/huPAR2 transfectants is inhibited by PT and BAPTA-AM. HEK293T cells were co-transfected with huPAR2 construct together with the ELAM-luc and β-galactosidase reporter constructs. Transfectants were pretreated with medium (M) or the indicated concentrations of PT (A) or BAPTA-AM (100 μm)(B) for 1 h. Cells were then further treated with indicated concentrations of huPAR2 AP or TNF-α (TNF, 50 ng/ml) for an additional 16 h. Cell lysates were prepared and reporter activities measured. A representative experiment is shown (n = 5). *, p < 0.001 versus medium-pretreated controls. RLU, relative luciferase units.
PAR2 activation by its agonists has been observed to elevate and release intracellular Ca2+ rapidly and transiently (47–49). To test the involvement of elevated intracellular Ca2+ in NF-κB signaling induced by PAR2 activation, we used BAPTA-AM to sequester intracellular Ca2+ stores and block the release of intracellular Ca2+ induced by stimulation of transfectants with AP. We first confirmed that BAPTA-AM would indeed inhibit AP-induced elevation of intracellular Ca2+ in PAR2 transfectants (data not shown). To determine whether AP-stimulated NF-κB activation in PAR2-expressing HEK293T cells also depended on Ca2+ mobilization, PAR2 transfectants were pretreated with 100 μm of BAPTA-AM for 1 h, and then further treated with PAR2 AP for an additional 16 h. TNF-α treatment was again used as a control for PAR2-independent NF-κB signaling in the cells. As was observed for PT treatment, incubation of cells with BAPTA-AM inhibited NF-κB activation induced by AP, but it failed to inhibit TNF-α-mediated signaling (Fig. 2B).
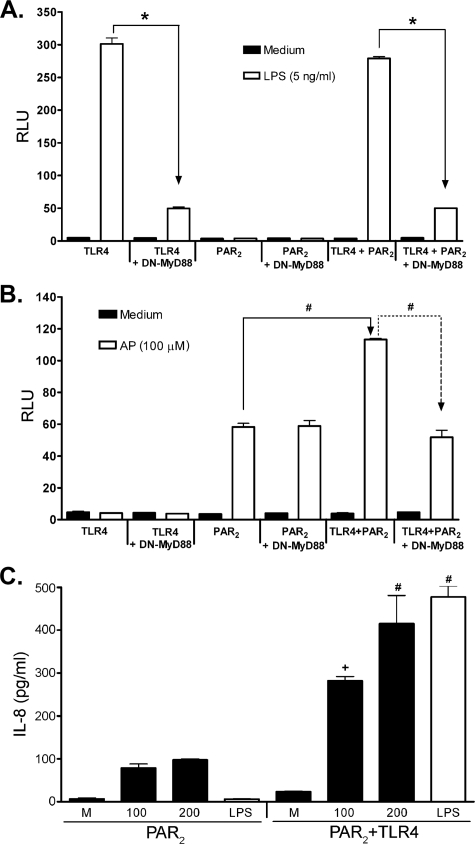
Cooperation between PAR2 and TLR4 Results in Synergistic, MyD88-dependent NF-κB Activation of PAR2 by Its AP—TLR4 activation by LPS has been shown to be partially dependent upon MyD88, a key adapter shared by all known TLRs (except TLR3) and the IL-1/IL-18 receptor signaling pathways (22, 31, 32). Co-transfection of cells with constructs that encode the TLR4 signaling complex (TLR4/MD-2/CD14) and a DN-MyD88 mutant construct (i.e. that lacks the death domain) significantly inhibited LPS-induced NF-κB activation (Fig. 3A, far left panels), as reported previously (50). Neither HEK293T cells transfected with pcDNA, in lieu of the TLR4 complex constructs (data not shown), nor the PAR2 construct alone responded to LPS, and there was no effect of DN-MyD88 co-expression (Fig. 3A, middle panels). Furthermore, no augmentation of LPS-induced signaling was observed in cells expressing both TLR4 and PAR2 (even at suboptimal concentrations of LPS or suboptimal input concentrations of TLR4 vector; data not shown), and this response was as dependent on MyD88 as were cells transfected with TLR4/MD-2/CD14 alone (Fig. 3A, right panels). Thus, the presence of PAR2 does not affect TLR4 response to LPS in PAR2/TLR4 HEK293T co-transfectants.
FIGURE 3.
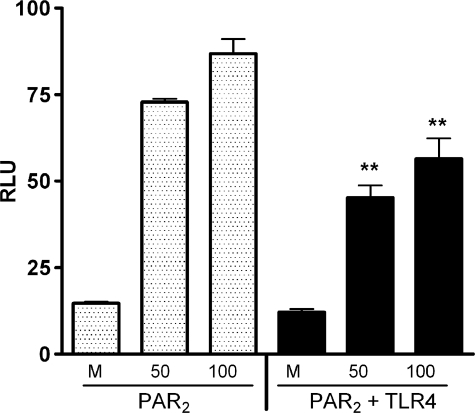
Co-expression of TLR4 and PAR2 results in synergistic, MyD88-dependent NF-κB activation in response to PAR2 AP. A and B, HEK293T cells were co-transfected with TLR4 signaling complex (pFLAG-huTLR4, pEF-BOS-HA-huMD-2, pcDNA3-huCD14) and/or huPAR2 constructs, without or with DN-MyD88 construct (200 ng/well). Cells were also co-transfected with the NF-κB-luciferase and β-galactosidase reporter constructs. A, cells were treated with medium only or LPS, although in B the cells were treated with medium only or AP. A representative experiment is shown for A and B (n = 6). C, HEK293T cells were transfected as described above in the absence of reporter constructs. Levels of IL-8 in the supernatants were determined by enzyme-linked immunosorbent assay. A representative experiment is shown (n = 2). RLU, relative luciferase units. *, P < 0.001, #, P < 0.01, and +, P, < 0.05.
In contrast to the data shown in Fig. 3A, HEK293T cells transfected with TLR4/MD-2/CD14, but not PAR2, were not activated by PAR2 AP (Fig. 3B, left panels), even after 16 h of treatment. This result represents a very important control because it indicates that the PAR2 AP used in these studies is not LPS-contaminated. PAR2-expressing cells responded to AP as expected, and activation of NF-κB luciferase was not affected by the co-expression of DN-MyD88 (Fig. 3B, middle panels). Although no synergy was observed for LPS when both TLR4 and PAR2 were co-expressed (Fig. 3A), co-expression of TLR4 and PAR2 resulted in a statistically significant increase in activation of ELAM (NF-κB) reporter activity in response to PAR2 AP (Fig. 3B), compared with cells expressing only PAR2. Fig. 3B (far right) also shows that the observed synergy between TLR4 and PAR2 was reversed by DN-MyD88 to the level induced by AP in cells transfected with PAR2 only. In parallel with these findings, levels of secreted IL-8 were significantly higher in AP-stimulated HEK293T cells transfected with the PAR2 and TLR4 than PAR2 alone (Fig. 3C).
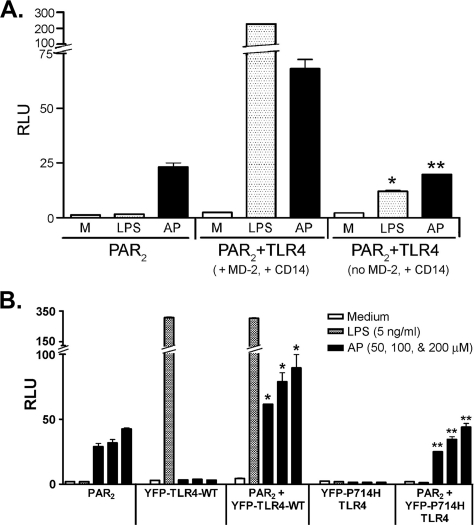
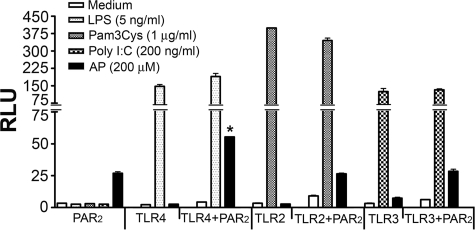
The observed synergy between TLR4 and PAR2 in response to AP was dependent upon the presence of MD-2 (Fig. 4A), a necessary co-receptor for TLR4 signaling (42, 51). The observed synergy between TLR4 and PAR2 was also lost when HEK293T cells were transfected with the signaling-incompetent, mutant TLR4 construct (P714H mutant) and treated with PAR2 AP (Fig. 4B). Taken together, these two findings indicate that TLR4 must be able to signal to enable the synergistic response to AP. In contrast to TLR4, neither TLR2 nor TLR3 facilitated a similar synergistic PAR2 response to AP (Fig. 5).
FIGURE 4.
Synergy between PAR2 and TLR4 induced by PAR2 AP requires MD-2 and a signaling-competent TLR4. A, HEK293T cells were co-transfected with the huPAR2 construct only or huPAR2, pFLAG-huTLR4, and huCD14, without or with the huMD-2 expression construct. All wells were co-transfected with ELAM-lucandβ-galactosidase reporter constructs. Transfected cells were treated with huPAR2 AP or LPS, and reporter activities were measured. A representative experiment is shown (n = 4). B, HEK293T cells were co-transfected with the huPAR2 construct only, or PAR2 co-transfected with YFP-TLR4-WT or YFP-TLR4-P714H mutant (200 ng/well each) constructs either individually or co-transfected with huPAR2 construct. All wells were also co-transfected with the reporter constructs. Transfected cells were treated with TNF-α, LPS, or huPAR2 AP, and reporter activities were measured. A representative experiment is shown (n = 3). *, p < 0.001, between the PAR2 and PAR2 + YFP-TLR4-WT transfectants; **, p < 0.001, between PAR2 + YFP-TLR4-WT and PAR2 + YFP-P714H-TLR4 transfectants treated with huPAR2 AP. RLU, relative luciferase units.
FIGURE 5.
Co-expression of PAR2 results in synergistic NF-κB activation induced by PAR2 AP with TLR4 but not with TLR2 and TLR3. HEK293T cells were co-transfected with either TLR4 signaling complex, FLAG-tagged TLR2, or FLAG-tagged TLR3 and/or huPAR2 construct. All wells were transfected with the NF-κB-luciferase and pCMV1-β-galactosidase reporter constructs. Transfectants were treated with medium only or respective TLR agonist (LPS for TLR4, S-[2,3-bis(palmitoyloxy)-(2-RS)-propyl]-N-palmitoyl-(R)-Cys-(S)-Ser-(Lys)4-OH, trihydrochloride (Pam3Cys) for TLR2, and poly(I:C) for TLR3), or AP. A representative experiment is shown (n = 3). *, p < 0.001, between the PAR2 and PAR2 + TLR4-WT transfectants. RLU, relative luciferase units.
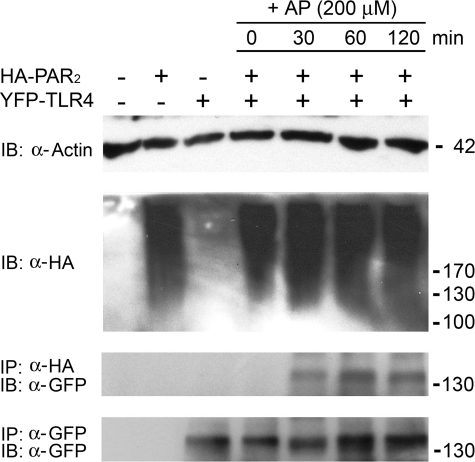
Co-immunoprecipitation of PAR2 with TLR4 in the Presence of AP—To perform co-immunoprecipitation experiments for studying the potential physical interaction of PAR2 with TLR4, HEK293T cells were transfected with HA-PAR2 and/or YFP-TLR4 (plus MD-2 and CD14) constructs. PAR2/TLR4 co-transfectants were treated with AP for the indicated times (Fig. 6). Western blotting for β-actin indicates comparable protein concentration in the original cell lysates (Fig. 6, IB: α-actin). HA-PAR2, shown previously by others to migrate as a very broad band between ∼100 and 200 kDa because of heavy glycosylation (46), was also determined to be equivalent by Western blotting in the same lysates and was detected only in lysates from cells transfected with HA-PAR2 vector (Fig. 6, IB: α-HA). YFP-TLR4 expression was below the level of detection in the lysates (data not shown). When HA-PAR2 was immunoprecipitated with anti-HA antibody, YFP-TLR4 was co-immunoprecipitated in an AP-dependent manner, as detected by Western blotting with anti-GFP antibodies (Fig. 6, IP: α-HA, IBα–GFP). When this blot was stripped and reprobed with anti-HA antibody, we observed equal amounts of PAR2 precipitated from all the respective lysates (data not shown). Equal expression of YFP-TLR4 in cells transfected with this expression vector was confirmed by immunoprecipitation and Western analysis with anti-GFP antibody of YFP-TLR4 in these same lysates (Fig. 6, IP: α-GFP, IB: α-GFP). Thus, PAR2 associates with TLR4, but only in the presence of AP.
FIGURE 6.
TLR4 co-immunoprecipitates with PAR2 in a PAR2 AP-dependent manner. HEK293T cells, transfected with either HA-PAR2 vector or YFP-TLR4-WT vector along with MD-2 and CD14 expression vectors, or co-transfected together with HA-PAR2 and YFP-TLR4 complex, were incubated for a further 72 h before washing and lysing as described under “Experimental Procedures.” Immunoprecipitation (IP) and Western blotting (IB) were carried out to analyze the physical interaction between PAR2 and TLR4 proteins. A representative experiment is shown (n = 3). Molecular weight of proteins's represented in kDa (on right).
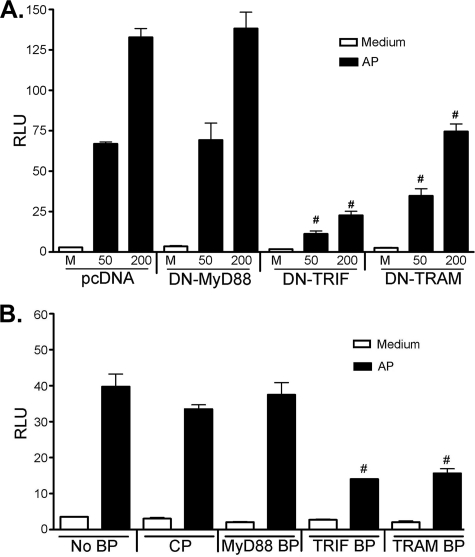
PAR2 Activation by AP Is MyD88-independent but TRIF/TRAM-dependent—Fig. 7A illustrates that the ability of PAR2 to trigger ELAM reporter activity was not affected by co-expression of the DN-MyD88 construct (also shown in Fig. 3B), in contrast to the marked inhibition exerted on LPS-induced signaling (Fig. 3A, left panels). Because the MyD88-independent pathway has been shown to result in delayed NF-κB translocation (52, 53), based on the delayed PAR2-stimulated ELAM (NF-κB)-mediated luciferase accumulation observed in the HEK293T transfectants (Fig. 1B), together with the observed AP-induced synergy and interaction between PAR2 and TLR4, we hypothesized that PAR2 activation of NF-κB might be dependent upon TRIF and/or TRAM, two adapters that have been associated with MyD88-independent signaling through TLR4. To test this hypothesis, HEK293T cells were transfected with PAR2 alone (in the presence of pcDNA3) or were co-transfected with PAR2 along with DN-MyD88 (used as a control), DN-TRIF, or DN-TRAM constructs. After transfection, cells were treated with medium or PAR2 AP. Interestingly, whereas co-expression of PAR2 and DN-MyD88 did not alter PAR2-mediated NF-κB activation, reporter activation was significantly down-regulated in the presence of DN-TRIF and, to a lesser extent, by DN-TRAM (Fig. 7A), using DN input vector concentrations that did not inhibit IL-1β- or TLR2-mediated signaling (data not shown). Recently, we reported that cell-permeable decoy peptides derived from the BB loop sequences of MyD88, TRIF, and TRAM adapter molecules inhibit TLR4-mediated signaling in macrophages, presumably by disrupting TIR-TIR interactions (39). To confirm our findings using the DN-adapter constructs, PAR2 transfectants were pretreated for 1 h with a control peptide (CP) or the BB loop peptides (BPs), corresponding to MyD88, TRIF, or TRAM, and then stimulated with huPAR2 AP (200 μm) for 16 h. Although the CP and MyD88 peptides failed to inhibit AP-induced NF-κB-driven luciferase activity, TRIF and TRAM BPs resulted in significant inhibition (Fig. 7B), mirroring the effects of the DN-TRIF and DN-TRAM vectors (Fig. 7A). TNF-α-induced NF-κB was not inhibited by these cell-permeable inhibitory peptides in this same experiment (data not shown).
FIGURE 7.
PAR2-mediated activation of NF-κB is decreased by inhibitors of TRIF and TRAM. HEK293T cells were co-transfected with huPAR2 construct and DN-MyD88, DN-TRIF, or DN-TRAM (50 ng/well) expression vectors (A). Transfected cells were treated for 16 h with the indicated concentrations of huPAR2 AP and ELAM-luc, and β-galactosidase reporter activities were measured. B, HEK293T cells were transfected with huPAR2 construct, ELAM-luc, and β-galactosidase reporter constructs. Transfected cells were pretreated with medium or 40 μm CP or the indicated cell-permeable adapter blocking peptide (BP), and then treated with AP for an additional 16 h, at which time reporter activities were measured. A representative experiment is shown for each experiment (n = 3). #, p < 0.001. RLU, relative luciferase units.
The RANTES Promoter-Luciferase Construct Is Not Synergistically Activated by AP in HEK293T Cells Expressing PAR2 and TLR4—The results presented thus far indicate that PAR2 and TLR4 synergize to increase the ELAM (NF-κB)-driven luciferase or IL-8 production in response to AP (Fig. 3, B and C) and that the observed synergy is MyD88-dependent (Fig. 3B). It is also clear that the NF-κB promoter activation of PAR2 by AP is sensitive to TRIF or TRAM inhibition but is MyD88-independent (Fig. 7A). Although MyD88-dependent signaling has been largely associated with an early and robust NF-κB response, MyD88-independent signaling gives rise to potent IRF-3 activation with a delayed NF-κB response (52, 53). Therefore, we next sought to determine whether the IRF-3-sensitive RAN-TES-luciferase reporter would also respond to AP stimulation in PAR2 transfectants and synergistically in HEK293T cells co-transfected with TLR4 and PAR2. Activation of the RANTES-luciferase reporter upon AP stimulation of PAR2 transfectants was observed, but it was slightly down-regulated, rather than up-regulated, when TLR4 was co-expressed with PAR2 (Fig. 8). These data indicate that MyD88-dependent NF-κB signaling pathway is favored when PAR2 and TLR4 are concurrently expressed.
FIGURE 8.
Co-transfection of TLR4 receptor complex with PAR2 inhibits RANTES promoter activity. HEK293T cells were co-transfected with either huPAR2 construct or huPAR2 plus TLR4 signaling complex (pFLAG-huTLR4; pEFBOS-HA-huMD-2; pcDNA3-huCD14). The RANTES-luciferase reporter (500 ng/well) and β-galactosidase vector were also co-transfected. Transfectants were treated with the indicated concentrations of huPAR2 AP for 16 h, and luciferase and β-galactosidase activities were measured. A representative experiment is shown (n = 5). **, p < 0.05. RLU, relative luciferase units.
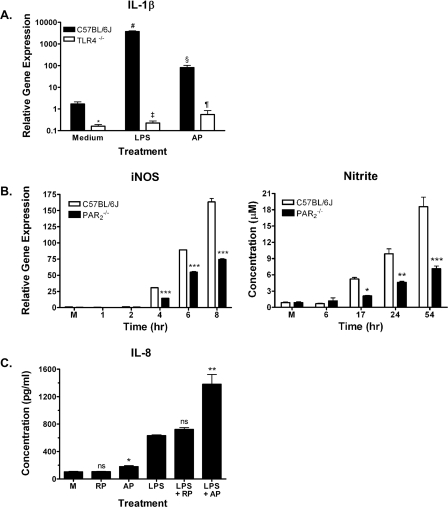
Dysregulated AP and LPS Responses in Macrophages Deficient in PAR2 or TLR4—PAR2 has been found on epithelial and endothelial cells, as well as on monocytes and macrophages (7). To determine whether PAR2 and TLR4 also interacted functionally in primary cells, macrophages from WT and TLR4–/– mice were compared for their sensitivity to PAR2 AP. In WT C57BL/6J macrophages, LPS potently triggered IL-1β gene expression, as measured by real time PCR (Fig. 9A), although in TLR4–/– macrophages, LPS failed to induce IL-1β steady-state mRNA. Compared with the substantial increase in IL-1β mRNA seen in WT macrophages induced by LPS, activation of WT macrophages by AP was relatively modest; nonetheless, in TLR4–/– macrophages, PAR2 AP-induced IL-1β gene expression was consistently lower than in the WT cells. Similarly, exposure of bone marrow-derived macrophages from WT C57BL/6J mice to PAR2 AP induced modest mRNA expression of the chemokines MIP-2 and KC, an effect that was significantly reduced in PAR2 AP-treated TLR4–/– bone marrow-derived macrophages (data not shown). Collectively, these findings support the hypothesis that PAR2 and TLR4 interact functionally in primary murine macrophages to enable optimal signaling by AP.
FIGURE 9.
Macrophages deficient in TLR4 or PAR2 show dysregulated responses to AP or LPS, respectively, and the SW620 intestinal epithelial cell line exhibits a synergistic response to co-stimulation with AP and LPS. A, TLR4–/– macrophages exhibit diminished IL-1β gene induction in response to huPAR2 AP. Macrophages from C57BL/6J or TLR4–/– mice were stimulated with huPAR2 AP (200 μm) or LPS (10 ng/ml) for 2 h, and steady-state IL-1β mRNA was measured. Results represent mean ± S.E. of six separate experiments. *, p < 0.001; Medium (C57BL/6J versus TLR4–/–); #, p < 0.001 Medium versus LPS (C57BL/6J); ‡, p < 0.001 LPS (C57BL/6J versus TLR4–/–); §, p < 0.001 Medium versus AP (C57BL/6J); and ¶, p < 0.001; AP (C57BL/6J versus TLR4–/–). B, PAR –/–2 macrophages exhibit diminished iNOS gene induction and NO• activity in response to LPS. Macrophages from C57BL/6J or PAR –/–2 mice were stimulated for the indicated times with 10 ng/ml LPS for iNOS mRNA or with 100 ng/ml LPS for supernatant analysis for NO. Data are representative of three separate experiments. M, medium control; *, p < 0.05; **, p < 0.01; ***, p < 0.001 (C). SW620 cells respond synergistically to huPAR2 AP (200 μm) and LPS (100 ng/ml) co-stimulation to augment IL-8 secretion (t = 24 h). Data represent the mean ± S.E. of three separate experiments. M, medium control; RP, reverse peptide; ns, not significant; *, p < 0.05 (Medium versus AP); **, p < 0.01 (LPS versus LPS + AP).
Although the ELAM (NF-κB) response to LPS was not altered in HEK293T cells co-expressing PAR2 and TLR4 versus TLR4 only (Fig. 3A), we tested the responsiveness of PAR –/–2 macrophages to LPS to determine the extent of signaling cross-talk. LPS-induced iNOS mRNA expression was significantly diminished in PAR –/–2 macrophages, as was the release of nitric oxide (NO•), measured by assaying nitrite, a stable metabolite of NO• (Fig. 9B).
PAR2 and TLR4 Signaling Pathways Converge Synergistically in Intestinal Epithelial Cells—The gastrointestinal tract and, in particular, colonic epithelial cells represent a physiological situation in which both PAR2 and TLR4 are expressed in an environment where agonists of each receptor can be found concurrently (reviewed in Refs. 54, 55). The human colonic epithelial cell line, SW620, responded synergistically to combined treatment with AP, but not the reverse peptide, and LPS for secretion of the potent neutrophil chemokine IL-8 (Fig. 9C).
DISCUSSION
The innate immune system has evolved to express PRRs that enable host recognition of conserved structural motifs, or PAMPs, that are shared among classes of microbes (reviewed in Refs. 14, 56). For example, the TLR4/MD-2/CD14 receptor complex is a robust cellular sensor of Gram-negative bacteria through its ability to recognize the lipid A moiety of LPS (reviewed in Refs. 56, 57). This recognition results in the recruitment of adapter molecules and subsequent intracellular signal transduction that evokes a potent pro-inflammatory response. By analogy, the PARs, although structurally unrelated to TLRs, also function in the context of the innate immune response and, in a sense, can be categorized as PRRs; they respond with intracellular signaling to serine proteases through recognition of a molecular motif referred to as the “catalytic triad,” i.e. Ser, His, and Asp, that, in its three-dimensional spatial arrangement, constitutes the active site of this family of proteolytic enzymes (reviewed in Refs. 7, 12, 13). Indeed, in addition to proteases generated at the inflammatory site that are known to activate PAR2 (e.g. trypsin, tryptase, tissue kallikreins, and matriptase), proteases derived from bacteria, dust mites, and fungi have also been reported to induce pro-inflammatory immune responses in host cells (reviewed in Refs. 35, 58), with several having been identified as direct activators of PAR2.
Based on previous studies implicating PAR2 in the inflammatory response (reviewed in Refs. 4–7, 9, 10, 34, 35), it was not surprising that in PAR2-overexpressing HEK293T cells, AP-induced activation of the classical inflammatory transcription factor, NF-κB, compared favorably to the level achieved by other potent stimuli such as IL-1β, albeit with delayed kinetics of luciferase accumulation. The delayed kinetics of PAR2-mediated NF-κB activation was protein synthesis-independent and not the result of a secondary signal from secreted products acting in an autocrine or paracrine fashion. To test the possibility that activation of PAR2 resulted in release of an endogenous TLR4 agonist, the experiment in supplemental Fig. 2B was repeated in HEK293T cells transfected with TLR4/MD-2/CD14. No increase in luciferase activity was observed after treatment of TLR4/MD-2/CD14 transfectants with supernatants from AP-stimulated PAR2 transfectants (data not shown). More importantly, the data contained herein demonstrate for the first time that PAR2, a seven-transmembrane GPCR, is capable of exploiting the evolutionarily ancient and conserved TLR4 signaling apparatus, to induce and enhance the inflammatory response to its AP. In the absence of TLR4, stimulation of HEK293T-PAR2 transfectants with PAR2 AP resulted in activation of both NF-κB and IRF-3 signaling pathways as evidenced by activation of ELAM-luciferase and RANTES-luciferase reporters, respectively, as well as induction of IL-8 secretion. Consistent with its role as a GPCR, PAR2-mediated activation of NF-κB was sensitive to PT and BAPTA-AM treatment. The inhibition of AP-induced ELAM (NF-κB)-luciferase activity by inhibitors of TRIF and TRAM, but not by MyD88 inhibitors, coupled with the delayed accumulation kinetics of NF-κB-driven luciferase support the possibility that PAR2 coopts the MyD88-independent pathway utilized by TLR4 to activate both transcription factors. However, in the presence of TLR4, there is an MyD88-dependent augmentation of the PAR2-mediated NF-κB-driven pathway induced by AP under conditions where even very high concentrations of PAR2 AP fail to activate TLR4 directly, whereas activation of RANTES-luciferase (IRF-3 and NF-κB-dependent) reporter is repressed. Thus, concurrent expression of TLR4 and PAR2 in HEK293T cells alters the capacity of PAR2 to engage specific signaling pathways differentially. Interestingly, no augmentation of PAR2-induced NF-κB was observed when PAR2 was co-expressed either with TLR2 or TLR3 expression constructs. In the case of TLR3, this result was not surprising because TLR3 signaling is MyD88-independent. Previous studies, in which the interactions between MyD88 and TLR4 and TLR2 were modeled, suggested that these TLRs interact with different “faces” of the MyD88 molecule (59). Thus, it is tempting to speculate that the failure of TLR2 to synergize with PAR2 may be due to its inability to form a signaling platform with both PAR2 and MyD88 concurrently, in contrast to TLR4.
The observed increase in PAR2-induced NF-κB in the presence of TLR4 requires a signal-competent TLR4, because MD-2 is required and the TLR4-P714H mutation fails to enable enhanced AP-induced signaling. We have also shown in this study that PAR2 and TLR4 not only interact at the signaling level (Fig. 3) but also interact physically in an AP-dependent manner (Fig. 6). Finally, the data generated in the transfection system are strongly supported by the fact that primary macrophages derived from TLR4–/– mice not only fail to respond to LPS, but also exhibit a significant decrease in the response to PAR2 AP. In support of our observations, Moretti et al. (36) recently reported that PAR2 AP-mediated activation of murine neutrophils was also significantly diminished in the absence of TLR4. Furthermore, the interdependent relationship between PAR2 and TLR4 signaling pathways is strengthened by our observation that in PAR –/–2 macrophages, expression of a subset of LPS-induced genes is dysregulated (e.g. iNOS, see Fig. 9B).4 Finally, a colonic epithelial cell line, which also expresses both PAR2 and TLR4, shows synergistic IL-8 secretion when stimulated by both agonists. Although this differs from the situation observed in the HEK293T transfectants co-expressing both PAR2 and TLR4 in which synergy was observed only in the presence of AP, the data may be explained by differences in the stoichiometry of TLR4 and PAR2 signaling components in non-transfectants versus transfected cells.
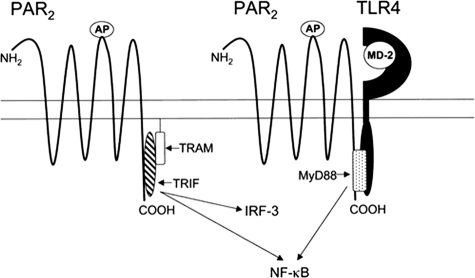
Collectively, these findings establish a novel paradigm of receptor cooperativity in which combinatorial interaction of two distinct pattern-recognition receptors, e.g. PAR2 and TLR4, results in an enhanced NF-κB-mediated inflammatory response. Fig. 10 depicts a hypothetical model of PAR2-TLR4 receptor interaction that accommodates our data. Based on secondary structure analysis of the PAR2 C terminus, we have identified a putative TIR-like interacting domain in PAR2 that, when mutagenized, fails to respond to AP.4 The model suggests that when TLR4 is not present, PAR2 activation leads to delayed NF-κB activity, perhaps through recruitment of TRIF/TRAM to its C terminus. In the presence of a functional TLR4 complex, however, the model predicts enhanced NF-κB signaling and diminished IRF-3-dependent reporter activity in response to PAR2 AP, possibly because of the displacement of TRIF/TRAM by MyD88 (Fig. 3B versus Fig. 8). Whether the putative TIR domain of PAR2 interacts directly with TLR4 or indirectly, through an intermediate adapter, such as MyD88 (as depicted in the Fig. 10), is not known; nonetheless, our data suggest that even in the absence of LPS, TLR4 participates in signaling by facilitating the PAR2 response to AP. Although MyD88 has been shown to associate with other non-TLR molecules, including the IL-1R (60–62), the IL-18R (62), and more recently, the IFN-γ receptor (63), ours is the first study to suggest that TLR4 facilitates augmented GPCR-mediated signaling through an alternative receptor via an MyD88-dependent mechanism. Although the pattern recognition receptor, dectin-1, has been shown to synergize with TLR2 for induction of optimal signaling (64, 65), each receptor signals independently after ligand engagement, i.e. there is no evidence to suggest that these two PRRs share downstream signaling molecules.
FIGURE 10.
Hypothetical model of PAR2 signaling and interaction with TLR4. See “Discussion” for description of model.
Activation of PARs, including PAR2, results in receptor internalization to the early endosomes in a process dependent on the GTPase, dynamin (Ref. 66 and reviewed in Refs. 3, 67). Recently, it was reported that TLR4 activation results in sequential utilization of MyD88- and TRIF-dependent signaling (68). LPS engagement induces TLR4-TIRAP-MyD88 signaling that occurs at the cell surface, but it also results in dynamin-dependent endocytosis of TLR4 that then couples the receptor to the TRAM-TRIF pathway. PAR2, when recruited to the endosomes, may couple to the TRAM-TRIF signaling pathway in a similar manner. However, when TLR4 is present at the cell surface, it may prolong surface retention of PAR2, thereby enhancing the ELAM (NF-κB) activity, while suppressing the RANTES (IRF-3) activity. Further studies will be required to elucidate the trafficking mechanisms involved in PAR2-TLR4 receptor interactions.
Although a role for PAR2 in inflammatory processes has been well documented, the precise mechanisms by which it contributes to the inflammatory response are not fully understood. In the murine blood vessel endothelium, PAR2 activation reduces leukocyte rolling velocity, increases leukocyte rolling flux, and increases leukocyte adhesion (69, 70). These inflammatory responses are delayed in PAR –/–2 mice (70), thereby supporting a crucial role for PAR2 in the inflammatory response in vivo. Furthermore, murine macrophages and human peripheral blood mononuclear cells respond to PAR2 AP with the synthesis of pro-inflammatory cytokines, e.g. IL-1β, IL-6, and TNF-α (71, 72). It has been also reported that PAR2 plays a key role in modulating several major diseases such as experimental autoimmune encephalomyelitis/multiple sclerosis (72) and arthritis (73). Notably, potent inflammatory stimuli such as LPS and pro-inflammatory cytokines such as TNF-α and IL-1α can up-regulate PAR2 expression in human vascular endothelial cells (74–76); thus, a self-sustaining and amplified inflammatory response can be envisioned whereby inflammation-induced up-regulation of PAR2 and coincident generation of PAR2-activating proteases and cytokines contribute to a protracted inflammatory cycle. The independent observations that both PAR2 (16) and TLR4 (33) contribute to colitis induced by C. rodentium further support the notion that these two receptors may cooperate in the generation of a strongly “Th1-type” inflammatory response. Although the focus of recent studies of PAR2 signaling has been on the pro-inflammatory, or Th1-type, response induced by PAR2 activation, Devlin et al. (77) recently provided evidence that PAR2 is also necessary for generating “Th2-skewed” immunity against the helminth Nippostrongylus brasiliensis. The authors concluded that PAR2 serves as a recognition receptor for nematode-derived proteases to invoke the appropriate anti-helminthic Th2 response. Consistent with this observation, PAR2 deficiency in mice attenuates allergic dermatitis (78) and decreases eosinophil infiltration and hyper-reactivity in allergic inflammation of the airway (79). Taken together, the findings suggest that, depending on the environment in which it is found, PAR2 plays an important role in shaping the inflammatory response as either a Th1 or Th2 response (reviewed in Ref. 54).
Differential expression of PAR2 and TLR4, or the availability of the adapter molecules, on different cell types, as well as the availability of proteases capable of activating PAR2, would be expected to contribute to differential capacity for interaction of these two receptor types. Another complexity in cellular responses to the proteolytic environment and LPS exposure arises with the recent observation that pre-exposure of intestinal epithelial cells to trypsin diminishes TLR4 signaling competency as a result of MD-2 cleavage (80). Thus, the interactions between PAR2 and TLR4 demonstrated in this report are likely to occur to different extents depending on the expression of each receptor and the local concentration of proteases and TLR4 agonists.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R37 AI-18797 (to S. N. V.), R01 AI-059524 (to A. E. M.), R01 DK048373 (to A. F.), and T32 AI-07540 (to Q. M. N.), and a Canadian Institutes of Health Research operating grant (to M. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: PAR, proteinase-activated receptor; WT, wild type; GPCR, G protein-coupled receptor; DN, dominant negative, hu, human; HEK, human embryonic kidney; TLR4, Toll-like receptor 4; PRR, pattern recognition receptor; PAMP, pathogen-associated molecular pattern; MyD88, Myeloid differentiation factor 88; TRIF, Toll/IL-1 receptor/resistance (TIR) domain-containing adapter-inducing IFN-β; TRAM, TRIF-related adapter molecule; BP, BB loop peptide; CP, control peptide; AP, agonist peptide; CHX, cycloheximide; IL, interleukin; IFN, interferon; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; TNF, tumor necrosis factor; LPS, lipopolysaccharide; RT, reverse transcription; PBS, phosphate-buffered saline; BAPTA-AM, N,N,N′,N′-tetraacetic acid tetra(acetomethyl) ester; HA, hemagglutinin; RANTES, regulated on activation normal T cell expressed and secreted; PT, pertussis toxin; GFP, green fluorescent protein; iNOS, inducible nitric-oxide synthase; ELAM, endothelial cell-leukocyte adhesion molecule.
Q. Nhu, manuscript in preparation; ELAM, endothelial cell-leukocyte adhesion molecule.
References
- 1.Neurath, H. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 10962–10963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turk, B. (2006) Nat. Rev. Drug Discov. 5 785–799 [DOI] [PubMed] [Google Scholar]
- 3.Macfarlane, S. R., Seatter, M. J., Kanke, T., Hunter, G. D., and Plevin, R. (2001) Pharmacol. Rev. 53 245–282 [PubMed] [Google Scholar]
- 4.Ramachandran, R., and Hollenberg, M. D. (2008) Br. J. Pharmacol. 153 Suppl. 1, S263–S282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollenberg, M. D., and Compton, S. J. (2002) Pharmacol. Rev. 54 203–217 [DOI] [PubMed] [Google Scholar]
- 6.Ossovskaya, V. S., and Bunnett, N. W. (2004) Physiol. Rev. 84 579–621 [DOI] [PubMed] [Google Scholar]
- 7.Steinhoff, M., Buddenkotte, J., Shpacovitch, V., Rattenholl, A., Moormann, C., Vergnolle, N., Luger, T. A., and Hollenberg, M. D. (2005) Endocr. Rev. 26 1–43 [DOI] [PubMed] [Google Scholar]
- 8.Soreide, K., Janssen, E. A., Korner, H., and Baak, J. P. (2006) J. Pathol. 209 147–156 [DOI] [PubMed] [Google Scholar]
- 9.Bushell, T. (2007) J. Physiol. (Lond.) 581 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McIntosh, K. A., Plevin, R., Ferrell, W. R., and Lockhart, J. C. (2007) Curr. Opin. Pharmacol. 7 334–338 [DOI] [PubMed] [Google Scholar]
- 11.Vergnolle, N. (2000) Aliment. Pharmacol. Ther. 14 257–266 [DOI] [PubMed] [Google Scholar]
- 12.Cocks, T. M., and Moffatt, J. D. (2001) Pulm. Pharmacol. Ther. 14 183–191 [DOI] [PubMed] [Google Scholar]
- 13.Cocks, T. M., and Moffatt, J. D. (2000) Trends Pharmacol. Sci. 21 103–108 [DOI] [PubMed] [Google Scholar]
- 14.Janeway, C. A., Jr., and Medzhitov, R. (2002) Annu. Rev. Immunol. 20 197–216 [DOI] [PubMed] [Google Scholar]
- 15.Holzhausen, M., Spolidorio, L. C., Ellen, R. P., Jobin, M. C., Steinhoff, M., Andrade-Gordon, P., and Vergnolle, N. (2006) Am. J. Pathol. 168 1189–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen, K. K., Sherman, P. M., Cellars, L., Andrade-Gordon, P., Pan, Z., Baruch, A., Wallace, J. L., Hollenberg, M. D., and Vergnolle, N. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 8363–8368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung, W. O., Hansen, S. R., Rao, D., and Dale, B. A. (2004) J. Immunol. 173 5165–5170 [DOI] [PubMed] [Google Scholar]
- 18.Kida, Y., Inoue, H., Shimizu, T., and Kuwano, K. (2007) Infect. Immun. 75 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun, G., Stacey, M. A., Schmidt, M., Mori, L., and Mattoli, S. (2001) J. Immunol. 167 1014–1021 [DOI] [PubMed] [Google Scholar]
- 20.Adam, E., Hansen, K. K., Astudillo Fernandez, O., Coulon, L., Bex, F., Duhant, X., Jaumotte, E., Hollenberg, M. D., and Jacquet, A. (2006) J. Biol. Chem. 281 6910–6923 [DOI] [PubMed] [Google Scholar]
- 21.Dulon, S., Leduc, D., Cottrell, G. S., D'Alayer, J., Hansen, K. K., Bunnett, N. W., Hollenberg, M. D., Pidard, D., and Chignard, M. (2005) Am. J. Respir. Cell Mol. Biol. 32 411–419 [DOI] [PubMed] [Google Scholar]
- 22.Akira, S., and Takeda, K. (2004) Nat. Rev. Immunol. 4 499–511 [DOI] [PubMed] [Google Scholar]
- 23.Rifkin, I. R., Leadbetter, E. A., Busconi, L., Viglianti, G., and Marshak-Rothstein, A. (2005) Immunol. Rev. 204 27–42 [DOI] [PubMed] [Google Scholar]
- 24.Mollen, K. P., Anand, R. J., Tsung, A., Prince, J. M., Levy, R. M., and Billiar, T. R. (2006) Shock 26 430–437 [DOI] [PubMed] [Google Scholar]
- 25.Sansonetti, P. J. (2006) Nat. Immunol. 7 1237–1242 [DOI] [PubMed] [Google Scholar]
- 26.Zhang, Z., and Schluesener, H. J. (2006) Cell. Mol. Life Sci. 63 2901–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bianchi, M. E. (2007) J. Leukocyte Biol. 81 1–5 [DOI] [PubMed] [Google Scholar]
- 28.Johnson, G. B., Brunn, G. J., and Platt, J. L. (2004) J. Immunol. 172 20–24 [DOI] [PubMed] [Google Scholar]
- 29.Hietaranta, A., Mustonen, H., Puolakkainen, P., Haapiainen, R., and Kemppainen, E. (2004) Biochem. Biophys. Res. Commun. 323 192–196 [DOI] [PubMed] [Google Scholar]
- 30.Devaney, J. M., Greene, C. M., Taggart, C. C., Carroll, T. P., O'Neill, S. J., and McElvaney, N. G. (2003) FEBS Lett. 544 129–132 [DOI] [PubMed] [Google Scholar]
- 31.Doyle, S. L., and O'Neill, L. A. (2006) Biochem. Pharmacol. 72 1102–1113 [DOI] [PubMed] [Google Scholar]
- 32.O'Neill, L. A., and Bowie, A. G. (2007) Nat. Rev. Immunol. 7 353–364 [DOI] [PubMed] [Google Scholar]
- 33.Khan, M. A., Ma, C., Knodler, L. A., Valdez, Y., Rosenberger, C. M., Deng, W., Finlay, B. B., and Vallance, B. A. (2006) Infect. Immun. 74 2522–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawabata, A., and Kawao, N. (2005) J. Pharmacol. Sci. 97 20–24 [DOI] [PubMed] [Google Scholar]
- 35.Chignard, M., and Pidard, D. (2006) Am. J. Respir. Cell Mol. Biol. 34 394–398 [DOI] [PubMed] [Google Scholar]
- 36.Moretti, S., Bellocchio, S., Bonifazi, P., Bozza, S., Zelante, T., Bistoni, F., and Romani, L. (2008) Mucosal Immunol. 1 156–168 [DOI] [PubMed] [Google Scholar]
- 37.Chi, L., Li, Y., Stehno-Bittel, L., Gao, J., Morrison, D. C., Stechschulte, D. J., and Dileepan, K. N. (2001) J. Interferon Cytokine Res. 21 231–240 [DOI] [PubMed] [Google Scholar]
- 38.McIntire, F. C., Sievert, H. W., Barlow, G. H., Finley, R. A., and Lee, A. Y. (1967) Biochemistry 6 2363–2372 [DOI] [PubMed] [Google Scholar]
- 39.Toshchakov, V. U., Basu, S., Fenton, M. J., and Vogel, S. N. (2005) J. Immunol. 175 494–500 [DOI] [PubMed] [Google Scholar]
- 40.Nhu, Q. M., Cuesta, N., and Vogel, S. N. (2006) J. Endotoxin Res. 12 285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Compton, S. J., Cairns, J. A., Palmer, K. J., Al-Ani, B., Hollenberg, M. D., and Walls, A. F. (2000) J. Biol. Chem. 275 39207–39212 [DOI] [PubMed] [Google Scholar]
- 42.Rallabhandi, P., Bell, J., Boukhvalova, M. S., Medvedev, A., Lorenz, E., Arditi, M., Hemming, V. G., Blanco, J. C., Segal, D. M., and Vogel, S. N. (2006) J. Immunol. 177 322–332 [DOI] [PubMed] [Google Scholar]
- 43.Chow, J. C., Young, D. W., Golenbock, D. T., Christ, W. J., and Gusovsky, F. (1999) J. Biol. Chem. 274 10689–10692 [DOI] [PubMed] [Google Scholar]
- 44.Medvedev, A. E., Piao, W., Shoenfelt, J., Rhee, S. H., Chen, H., Basu, S., Wahl, L. M., Fenton, M. J., and Vogel, S. N. (2007) J. Biol. Chem. 282 16042–16053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fitzgerald, K. A., Rowe, D. C., Barnes, B. J., Caffrey, D. R., Visintin, A., Latz, E., Monks, B., Pitha, P. M., and Golenbock, D. T. (2003) J. Exp. Med. 198 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Compton, S. J., Sandhu, S., Wijesuriya, S. J., and Hollenberg, M. D. (2002) Biochem. J. 368 495–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hollenberg, M. D., Saifeddine, M., Al-Ani, B., and Kawabata, A. (1997) Can. J. Physiol. Pharmacol. 75 832–841 [PubMed] [Google Scholar]
- 48.Al-Ani, B., Saifeddine, M., Kawabata, A., Renaux, B., Mokashi, S., and Hollenberg, M. D. (1999) J. Pharmacol. Exp. Ther. 290 753–760 [PubMed] [Google Scholar]
- 49.Kawabata, A., Saifeddine, M., Al-Ani, B., Leblond, L., and Hollenberg, M. D. (1999) J. Pharmacol. Exp. Ther. 288 358–370 [PubMed] [Google Scholar]
- 50.Means, T. K., Jones, B. W., Schromm, A. B., Shurtleff, B. A., Smith, J. A., Keane, J., Golenbock, D. T., Vogel, S. N., and Fenton, M. J. (2001) J. Immunol. 166 4074–4082 [DOI] [PubMed] [Google Scholar]
- 51.Visintin, A., Iliev, D. B., Monks, B. G., Halmen, K. A., and Golenbock, D. T. (2006) Immunobiology 211 437–447 [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto, M., Sato, S., Hemmi, H., Hoshino, K., Kaisho, T., Sanjo, H., Takeuchi, O., Sugiyama, M., Okabe, M., Takeda, K., and Akira, S. (2003) Science 301 640–643 [DOI] [PubMed] [Google Scholar]
- 53.Sato, S., Sugiyama, M., Yamamoto, M., Watanabe, Y., Kawai, T., Takeda, K., and Akira, S. (2003) J. Immunol. 171 4304–4310 [DOI] [PubMed] [Google Scholar]
- 54.Antalis, T. M., Shea-Donohue, T., Vogel, S. N., Sears, C., and Fasano, A. (2007) Nat. Clin. Pract. Gastroenterol. Hepatol. 4 393–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fukata, M., and Abreu, M. T. (2007) Biochem. Soc. Trans. 35 1473–1478 [DOI] [PubMed] [Google Scholar]
- 56.Medzhitov, R., and Janeway, C. A., Jr. (1997) Cell 91 295–298 [DOI] [PubMed] [Google Scholar]
- 57.Munford, R. S., and Varley, A. W. (2006) PLoS Pathog. 2 e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reed, C. E., and Kita, H. (2004) J. Allergy Clin. Immunol. 114 997–1009 [DOI] [PubMed] [Google Scholar]
- 59.Dunne, A., Ejdeback, M., Ludidi, P. L., O'Neill, L. A., and Gay, N. J. (2003) J. Biol. Chem. 278 41443–41451 [DOI] [PubMed] [Google Scholar]
- 60.Mitcham, J. L., Parnet, P., Bonnert, T. P., Garka, K. E., Gerhart, M. J., Slack, J. L., Gayle, M. A., Dower, S. K., and Sims, J. E. (1996) J. Biol. Chem. 271 5777–5783 [DOI] [PubMed] [Google Scholar]
- 61.Wesche, H., Henzel, W. J., Shillinglaw, W., Li, S., and Cao, Z. (1997) Immunity 7 837–847 [DOI] [PubMed] [Google Scholar]
- 62.Adachi, O., Kawai, T., Takeda, K., Matsumoto, M., Tsutsui, H., Sakagami, M., Nakanishi, K., and Akira, S. (1998) Immunity 9 143–150 [DOI] [PubMed] [Google Scholar]
- 63.Sun, D., and Ding, A. (2006) Nat. Immunol. 7 375–381 [DOI] [PubMed] [Google Scholar]
- 64.Gantner, B. N., Simmons, R. M., Canavera, S. J., Akira, S., and Underhill, D. M. (2003) J. Exp. Med. 197 1107–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rogers, N. C., Slack, E. C., Edwards, A. D., Nolte, M. A., Schulz, O., Schweighoffer, E., Williams, D. L., Gordon, S., Tybulewicz, V. L., Brown, G. D., and Reis e Sousa, C. (2005) Immunity 22 507–517 [DOI] [PubMed] [Google Scholar]
- 66.Roosterman, D., Schmidlin, F., and Bunnett, N. W. (2003) Am. J. Physiol. 284 C1319–C1329 [DOI] [PubMed] [Google Scholar]
- 67.Arora, P., Ricks, T. K., and Trejo, J. (2007) J. Cell Sci. 120 921–928 [DOI] [PubMed] [Google Scholar]
- 68.Kagan, J. C., Su, T., Horng, T., Chow, A., Akira, S., and Medzhitov, R. (2008) Nat. Immunol. 9 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vergnolle, N. (1999) J. Immunol. 163 5064–5069 [PubMed] [Google Scholar]
- 70.Lindner, J. R., Kahn, M. L., Coughlin, S. R., Sambrano, G. R., Schauble, E., Bernstein, D., Foy, D., Hafezi-Moghadam, A., and Ley, K. (2000) J. Immunol. 165 6504–6510 [DOI] [PubMed] [Google Scholar]
- 71.Johansson, U., Lawson, C., Dabare, M., Syndercombe-Court, D., Newland, A. C., Howells, G. L., and Macey, M. G. (2005) J. Leukocyte Biol. 78 967–975 [DOI] [PubMed] [Google Scholar]
- 72.Noorbakhsh, F., Tsutsui, S., Vergnolle, N., Boven, L. A., Shariat, N., Vodjgani, M., Warren, K. G., Andrade-Gordon, P., Hollenberg, M. D., and Power, C. (2006) J. Exp. Med. 203 425–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kelso, E. B., Lockhart, J. C., Hembrough, T., Dunning, L., Plevin, R., Hollenberg, M. D., Sommerhoff, C. P., McLean, J. S., and Ferrell, W. R. (2006) J. Pharmacol. Exp. Ther. 316 1017–1024 [DOI] [PubMed] [Google Scholar]
- 74.Nystedt, S., Ramakrishnan, V., and Sundelin, J. (1996) J. Biol. Chem. 271 14910–14915 [DOI] [PubMed] [Google Scholar]
- 75.Cicala, C., Pinto, A., Bucci, M., Sorrentino, R., Walker, B., Harriot, P., Cruchley, A., Kapas, S., Howells, G. L., and Cirino, G. (1999) Circulation 99 2590–2597 [DOI] [PubMed] [Google Scholar]
- 76.Hamilton, J. R., Frauman, A. G., and Cocks, T. M. (2001) Circ. Res. 89 92–98 [DOI] [PubMed] [Google Scholar]
- 77.Devlin, M. G., Gasser, R. B., and Cocks, T. M. (2007) Parasitol. Res. 101 105–109 [DOI] [PubMed] [Google Scholar]
- 78.Kawagoe, J., Takizawa, T., Matsumoto, J., Tamiya, M., Meek, S. E., Smith, A. J., Hunter, G. D., Plevin, R., Saito, N., Kanke, T., Fujii, M., and Wada, Y. (2002) Jpn. J. Pharmacol. 88 77–84 [DOI] [PubMed] [Google Scholar]
- 79.Schmidlin, F., Amadesi, S., Dabbagh, K., Lewis, D. E., Knott, P., Bunnett, N. W., Gater, P. R., Geppetti, P., Bertrand, C., and Stevens, M. E. (2002) J. Immunol. 169 5315–5321 [DOI] [PubMed] [Google Scholar]
- 80.Cario, E., Golenbock, D. T., Visintin, A., Runzi, M., Gerken, G., and Podolsky, D. K. (2006) J. Immunol. 176 4258–4266 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.