Abstract
Previously, using primary hepatocytes residing in early G1 phase, we demonstrated that expression of the cyclin-dependent kinase (CDK) inhibitor protein p21Cip-1/WAF1/mda6 (p21) enhanced the toxicity of deoxycholic acid (DCA) + MEK1/2 inhibitor. This study examined the mechanisms regulating this apoptotic process. Overexpression of p21 or p27Kip-1 (p27) enhanced DCA + MEK1/2 inhibitor toxicity in primary hepatocytes that was dependent on expression of acidic sphingomyelinase and CD95. Overexpression of p21 suppressed MDM2, elevated p53 levels, and enhanced CD95, BAX, NOXA, and PUMA expression; knockdown of BAX/NOXA/PUMA reduced CDK inhibitor-stimulated cell killing. Parallel to cell death processes, overexpression of p21 or p27 profoundly enhanced DCA + MEK1/2 inhibitor-induced expression of ATG5 and GRP78/BiP and phosphorylation of PKR-like endoplasmic reticulum kinase (PERK) and eIF2α, and it increased the numbers of vesicles containing a transfected LC3-GFP construct. Incubation of cells with 3-methyladenine or knockdown of ATG5 suppressed DCA + MEK1/2 inhibitor-induced LC3-GFP vesicularization and enhanced DCA + MEK1/2 inhibitor-induced toxicity. Expression of dominant negative PERK blocked DCA + MEK1/2 inhibitor-induced expression of ATG5, GRP78/BiP, and eIF2α phosphorylation and prevented LC3-GFP vesicularization. Knock-out or knockdown of p53 or CD95 abolished DCA + MEK1/2 inhibitor-induced PERK phosphorylation and prevented LC3-GFP vesicularization. Thus, CDK inhibitors suppress MDM2 levels and enhance p53 expression that facilitates bile acid-induced, ceramide-dependent CD95 activation to induce both apoptosis and autophagy in primary hepatocytes.
Bile acids are detergent molecules, synthesized from cholesterol in the liver, that are released into the gut upon feeding and are essential for digestion (1). In the intestine, bile acids function in the solubilization and absorption of fats, certain vitamins, and cholesterol (2). Bile acids, post-feeding, re-enter the liver via the portal vein together with digested nutrients and are re-circulated back into the gallbladder for use during the next feeding cycle (3). Individually, when retained within the liver because of impaired secretion into the bile canaliculi, bile acids are also known to cause hepatocellular toxicity both in vivo and in vitro (4–8).
Treatment of primary rodent and human hepatocytes as well as hepatoma cells with a physiologic concentration of the bile acid deoxycholic acid (DCA)2 has been shown to cause activation of the ERK1/2 pathway (9–12). Blockade of DCA-induced ERK1/2 and AKT activation, with inhibitors of RAS, phosphatidylinositol 3-kinase, or MEK1/2, increased apoptosis ∼10-fold within 6 h of exposure. Apoptosis was dependent on bile acid-induced, ligand-independent, and ceramide-dependent activation of the CD95 death receptor. Other studies demonstrated that overexpression of the cyclin-dependent kinase inhibitor p21Cip-1/WAF1/mda6 (p21) enhanced DCA toxicity in hepatocytes that was due to enhanced expression of the tumor suppressor p53 (9, 13). Elevated expression of p53 correlated with a p21-dependent reduction in the expression of MDM2, the E3 ligase known to regulate p53 protein levels. MDM2 is also known to be a negative regulator of p21 expression, independently of p53 function (14–22). These findings suggested that under endogenous promoter control p21 and MDM2 may potentially titrate the expression of each other to maintain a steady state amount of p53 within the cell.
This study was designed, initially, to determine the mechanisms by which the CDK inhibitor stimulated expression of p53, via reduction of MDM2 levels, and promoted bile acid toxicity in primary hepatocytes. However, based on our recent discovery using the novel cancer therapeutics sorafenib and vorinostat, activation of CD95 can promote endoplasmic reticulum (ER) stress as well as PERK- and ATG5-dependent autophagy, and reduced expression of an E3 ligase, such as MDM2, will also be predicted to increase the levels of unfolded proteins in cells. Therefore, we subsequently examined whether CDK inhibitors promoted bile acid-induced ER stress and autophagy in primary hepatocytes (23–26).
Autophagy is a ubiquitous process in which cells degrade cytosolic materials such as proteins and organelles, and this process continuously occurs at a basal level in eukaryotic cells. In this process, cytoplasmic constituents are sequestered into forming membrane vesicles referred to as autophagosomes, which then fuse with lysosomes to form an autolysosome. In the autolysosome the contents of the vesicle are degraded and recycled. Autophagy has been primarily researched in yeast as a response to nutrient depletion, and there are at least 25 yeast genes specifically involved in the autophagic process, and the levels of their gene products are directly elevated when autophagy is up-regulated. Recent studies have shown that yeast ATGs have very similar mammalian homologues arguing that autophagy is a conserved mechanism throughout evolution.
Our present findings demonstrate that CDK-stimulated expression of p53 promoted bile acid toxicity by causing p53 to translocate from the cytoplasm to the nucleus and to increase the expression of BAX, PUMA, NOXA, and CD95. Overexpression of p21 or p27Kip-1 enhanced bile acid-induced autophagy signaling in an acidic sphingomyelinase- and CD95-dependent fashion, which was a protective event, compared with bile acid-induced acidic sphingomyelinase- and CD95-dependent apoptosis. Collectively, these findings argue that CDK inhibitors can promote a protective autophagy response in response to toxic bile acid treatment.
EXPERIMENTAL PROCEDURES
Materials—All bile acids were obtained from Sigma. Phospho-/total-ERK1/2 were purchased from Cell Signaling Technologies (Worcester, MA). Jo2 hamster anti-mouse CD95 IgG was from Pharmingen. All the other secondary antibodies (anti-rabbit, anti-mouse, and anti-goat horseradish peroxidase) and rhodamine-conjugated goat anti-Armenian hamster IgGs were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). 3-Methyladenine was supplied by Calbiochem as powder, dissolved in sterile PBS, and stored frozen under light-protected conditions at –80 °C. Enhanced chemiluminescence (ECL) kits were purchased from Amersham Biosciences and PerkinElmer Life Sciences. Trypsin-EDTA, Williams Medium E, and penicillin/streptomycin were purchased from Invitrogen. Other reagents were as described previously (9–11, 23–26).
Primary Culture of Rodent Hepatocytes—Hepatocytes were isolated from adult male Sprague-Dawley rats and wild type or CD95–/– or ASMase–/– or p21–/– or p53–/– C57/BL6 mice by the two-step collagenase perfusion technique. The freshly isolated hepatocytes were plated on rat tail collagen (Vitrogen)-coated plate at a density of 2 × 105 cells/well of a 12-well plate, and cultured in Williams E medium supplemented with 0.1 nm dexamethasone, 1 nm thyroxine, and 100 μg/ml of penicillin/streptomycin, at 37 °C in a humidified atmosphere containing 5% CO2. The initial medium change was performed 3 h after cell seeding to minimize the contamination of dead or mechanically damaged cells. In some studies, where noted, hepatocytes were cultured in the presence of 50 nm insulin, 0.1 nm dexamethasone, 1 nm thyroxine, and 100 μg/ml of penicillin/streptomycin.
Recombinant Adenoviral Vectors; Generation and Infection in Vitro—Two adenoviral technologies were used. Replication-defective adenovirus was conjugated to poly-l-lysine and a cDNA plasmid construct, as indicated in each legend and as described previously (9–11). Next we used recombinant adenoviruses. Hepatocytes were transfected/infected with these adenoviruses at an approximate multiplicity of infection of 250 and 30, respectively. Cells were further incubated for 24 h to ensure adequate expression of transduced gene products prior to bile acid/drug treatments.
SDS-PAGE and Western Blot Analysis—At various time points after indicated treatment, hepatocytes were lysed in whole-cell lysis buffer (0.5 m Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 1% β-mercaptoethanol, 0.02% bromphenol blue), and the samples were boiled for 30 min. The boiled samples were loaded onto 14% SDS-PAGE, and electrophoresis was run overnight. Proteins were electrophoretically transferred onto 0.22-μm nitrocellulose and immunoblotted with various primary antibodies against different proteins. All immunoblots were visualized by ECL. For presentation, immunoblots were digitally scanned at 600 dpi using Adobe PhotoShop CS2; the color was removed, and figures were generated in Microsoft PowerPoint.
LC3-GFP Visualization—Cells were plated as described above and transfected 24 h after plating. For mouse embryonic fibroblasts (2–5 μg) or other hepatocytes (0.5 μg), plasmids expressing a specific mRNA (or siRNA) or appropriate vector control plasmid DNA were diluted in 50 μl of serum-free and antibiotic-free medium (1 portion for each sample). Concurrently, 2 μl of Lipofectamine 2000 (Invitrogen) was diluted into 50 μl of serum-free and antibiotic-free medium (1 portion for each sample). Diluted DNA was added to the diluted Lipofectamine 2000 for each sample and incubated at room temperature for 30 min. This mixture was added to each well/dish of cells containing 200 μl of serum-free and antibiotic-free medium for a total volume of 300 μl, and the cells were incubated for 4 h at 37 °C. An equal volume of 2× medium was then added to each well. Cells were incubated for 48 h and then treated with DCA ± PD184352. For analysis of cells transfected with GFP-LC3 constructs, the GFP-LC3-positive vacuolated cells were examined under the ×40 objective of a Zeiss Axiovert fluorescent microscope. In control studies, expression of β-galactosidase using Ad.β-gal did not induce GFP-LC3 vacuoles in hepatocytes in the presence or absence of DCA ± PD184352.3 Forty LC3-GFP-positive cells were analyzed per condition. Each vacuole was counted, and the average number of vacuoles per cell for each, including cells that did not exhibit vacuolization, was calculated.
Immunofluorescence—Hepatocytes were cultured at 10–20% confluence on collagen-coated glass coverslips in 4-well dishes for 24 h. After treatment, the cells were washed with PBS and fixed in cold methanol:acetone (1:1) for 10 min at –20 °C. After washing three times in PBS for 5 min each, the cells were blocked in 0.5% bovine serum albumin in PBS for 30 min at room temperature and incubated with the primary antibody, Jo2, diluted in PBS, 0.5% bovine serum albumin (1:500) overnight at 4 °C. For measurements of total cellular CD95, hepatocytes were fixed in 3.7% formaldehyde in PBS for 10 min at 4 °C and permeabilized with 0.1% Triton X-100 in PBS for 3 min at 4 °C prior to incubation with the primary antibody. Cells were washed again with PBS and then incubated with rhodamine-conjugated goat anti-Armenian hamster IgG at 1:100 dilution for 60 min at room temperature. After washing with PBS, the cells were stained with 5 μm 4′, 6′-diamidino-2-phenylindole dihydrochloride in PBS for 5 min. The cells were washed again with PBS, followed by distilled water, and mounted in Crystal-Mount. Fluorescence staining was viewed with a Zeiss Axiovert fluorescent microscope. Fluorescence was quantified via Image-Pro® Plus analysis software and expressed as integrated optical density.
Measurement of Endogenous Ceramide—Lipids were extracted, and mass amounts of ceramide in cellular extracts were measured by the diacylglycerol kinase enzymatic method. Briefly, hepatocytes were washed with PBS and scraped into 1 ml of cold methanol containing 2.5 μl of concentrated HCl. Lipids were extracted by adding 2 ml of chloroform, 1 m NaCl (1:1, v/v), and phases were separated. An aliquot (100 μl) of the chloroform phase was dried under nitrogen gas. Bovine brain type III ceramide was used as a standard. The enzymatic reaction was started by the addition of 100 μl of 50 mm imidazole, 1 mm diethylenetriaminepentaacetic acid, 12.5 mm MgCl2, 50 mm NaCl, 1 mm EGTA, 10 mm dithiothreitol, 1 mm ATP, 1.5% N-octyl-β-d-glucopyranoside, 1 mm cardiolipin, diacylglycerol kinase (0.01 unit), and [γ-32P]ATP (1 μCi). After incubation at 37 °C for 35 min with 15 min of sonication at room temperature in-between, lipids were extracted by the addition of 500 μl of chloroform:methanol:HCl (100:100:1) and 100 μl of 1 m NaCl. Labeled ceramide 1-phosphate and phosphatidic acid (50-μl organic layer) were resolved by TLC with chloroform:acetone: methanol:acetic acid:water (10:4:3:2:1). Spots corresponding to ceramide were quantified with a Bio-Rad PhosphorImager.
Detection of Cell Death by Giemsa and/or Hoescht 33342 Assay—Cells were harvested by trypsinization with trypsin/EDTA for ∼10 min at 37 °C. As some apoptotic cells detached from the culture substratum into the medium, these cells were also collected by centrifugation of the medium at 1,500 rpm for 5 min. The pooled cell pellets were resuspended and spun onto glass slides (cytospin). Cells were stained using a commercial Giemsa kit (DiffQuick) and visualized under light microscopy (×40 magnification), scoring the number of cells exhibiting the “classic” morphological features of apoptosis and necrosis. For each condition, 10 randomly selected fields per slide were evaluated, encompassing at least 1500 cells. In parallel for confirmation, cells fixed to slides were stained with Hoechst 33342 dye followed by PBS washing to remove excess dye, air-dried, and mounted in Fluoro-Guard Antifade (Bio-Rad). Nuclear morphology was evaluated. Apoptotic cells were identified as those whose nuclei exhibited brightly staining condensed chromatin or nuclear fragmentation.
Data Analysis—Comparison of the effects of various treatments was performed using one-way analysis of variance and a two-tailed t test. Differences with a p value of < 0.05 were considered statistically significant. Experiments shown are the means of multiple points (±S.E.).
RESULTS
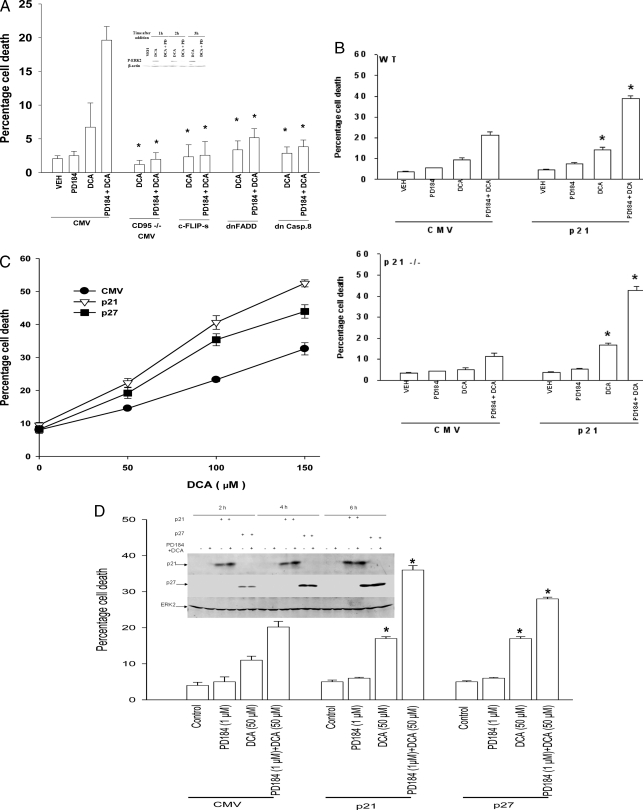
Bile Acid Toxicity Is CD95-dependent and Is Enhanced by Overexpression of p21 or p27—Initial studies characterized the regulation of rodent hepatocyte survival in vitro after exposure to the cell-permeable bile acid DCA and a MEK1/2 inhibitor. Treatment of mouse hepatocytes with DCA and a MEK1/2 inhibitor enhanced cell killing within 6 h that was blocked by expression of dominant negative FADD or dominant negative caspase 8, overexpression of the caspase 8 inhibitor c-FLIP-s, or was blocked in CD95–/– hepatocytes (Fig. 1A). Hence, the primary death signal generated by DCA + MEK1/2 inhibitor treatment emanated from CD95 (11).
FIGURE 1.
Overexpression of p21 or p27 enhances the lethality of DCA in primary hepatocytes. Primary hepatocytes were isolated as described under “Experimental Procedures.” Cells were treated with vehicle (VEH) control (DMSO), DCA (50 μm), MEK1/2 inhibitor (PD184352 (PD184, PD 2 μm)) or both agents combined, as indicated in each panel. A, in triplicate wild type mouse hepatocytes were transfected/infected using the poly-l-lysine adenoviral technique 4 h after plating to express dominant negative FADD and dominant negative caspase 8 (dnCasp.8) or to overexpress c-FLIP-s. In parallel, wild type and CD95–/– mouse hepatocytes were plated, and vector control was transfected. Cells were treated 24 h after plating with vehicle, PD184, DCA, or the agents in combination, and 6 h later cells were isolated and spun onto glass slides for determination of apoptosis as described under “Experimental Procedures” (n = 3 studies, ±S.E.). Inset panel, ERK1/2 phosphorylation at each time point in vector control cells. *, p < 0.05 apoptosis value less than corresponding value in cells infected with empty vector (CMV) plasmid/virus. B, in triplicate wild type and p21–/– mouse hepatocytes were infected 4 h after plating to express nothing (vector, CMV) or p21. Cells were treated 24 h after plating with vehicle, PD184352, DCA, or the agents in combination, and 6 h later cells were isolated and spun onto glass slides for determination of apoptosis as described under “Experimental Procedures” (n = 3 studies, ±S.E.). *, p < 0.05 apoptosis value greater than corresponding value in cells infected with empty vector (CMV) virus. C, rat hepatocytes were infected 4 h after plating to express nothing (vector, CMV) or p21 or p27. Cells were treated 24 h after plating with vehicle or increasing concentrations of DCA, and 6 h later cells were isolated and spun onto glass slides for determination of apoptosis as described under “Experimental Procedures” (n = 3 studies, ±S.E.). D, primary rat hepatocytes plated in triplicate were infected to express nothing (vector, CMV) or p21 or p27, as indicated. Twenty four hours after plating cells were treated with vehicle, DCA, PD184352, or both agents in combination for 6 h after which cells were isolated and spun onto glass slides for determination of apoptosis as described under “Experimental Procedures” (n = 3, ±S.E.). *, p < 0.05 apoptosis value greater than corresponding value in cells infected with empty vector (CMV) virus. Inset panel, expression of p21 or p27 in primary hepatocytes infected with recombinant adenoviruses to express p21 or p27.
Loss of basal p21 expression in p21–/– hepatocytes lowered the toxicity of DCA ± MEK1/2 inhibitor, and overexpression of p21 in hepatocytes enhanced DCA ± MEK1/2 inhibitor lethality (Fig. 1B). We next determined whether expression of a member of the same CDK inhibitor family as p21 would also enhance bile acid lethality, p27Kip-1 (p27). Overexpression of p27 also promoted DCA lethality and DCA + MEK1/2 inhibitor lethality in primary rat hepatocytes (Fig. 1, C and D).
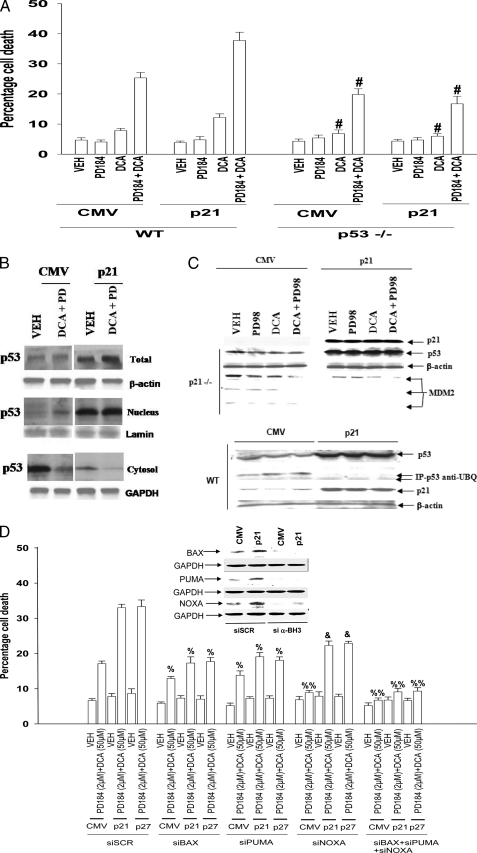
The Promotion of Bile Acid Toxicity by CDK Inhibitors Is p53-dependent and Correlates with Reduced p53 Ubiquitination, with Increased p53 Nuclear Localization, and Increased CD95, BAX, PUMA, and NOXA Levels—The tumor suppressor p53 is widely regarded as a key transcriptional regulator of p21 expression. In p53–/– hepatocytes, loss of p53 expression reduced DCA lethality, whereas overexpression of p21 did not promote DCA ± MEK1/2 inhibitor killing in p53–/– cells (Fig. 2A).
FIGURE 2.
CDK inhibitors enhance p53 expression and promote apoptosis in part via elevated BH3 domain protein expression. Primary hepatocytes were isolated as described under “Experimental Procedures.” Cells were treated with vehicle (VEH) control (DMSO), DCA (50 μm; 150 μm), MEK1/2 inhibitor (PD98059, PD98, 50 μm; PD184352, PD184, 2 μm), or both agents combined, as indicated in each panel. A, in triplicate wild type (WT) and p53–/– mouse hepatocytes were infected 4 h after plating to express nothing (vector, CMV) or p21. Cells were treated 24 h after plating with vehicle, PD184352, DCA, or the agents in combination, and 6 h later cells were isolated and spun onto glass slides for determination of apoptosis as described under “Experimental Procedures.” (n = 3 studies, ±S.E.). #, p < 0.05 apoptosis value less than corresponding value in wild type cells. B, primary rat hepatocytes were infected to express nothing (vector, CMV) or p21 or p27, as indicated. Cells were treated 24 h after plating with vehicle or with PD184352 and DCA and isolated 2 h after treatment. Cells lysed and then were separated into cytosolic and nuclear fractions as noted under “Experimental Procedures.” B, levels of p53 in each fraction were determined in parallel with a protein loading control (n = 2). C, upper section, primary mouse hepatocytes (p21–/–) were infected to express nothing (vector, CMV) or p21, as indicated, and incubated for 24 h. Twenty four hours after infection cells were treated with vehicle (VEH), DCA, PD98059, or both agents combined. Six hours after the addition of DCA/MEK1/2 inhibitor, cells were lysed in SDS-PAGE running buffer, and total cell lysates were subjected to SDS-PAGE and immunoblotting. Data are from a representative experiment (n = 5). Overexpression of p21 for 24 h enhanced total p53 expression in untreated wild type and p21–/– hepatocytes by 3.5 ± 0.4- and 4.6 ± 0.7-fold (S.E.; p < 0.05; n = 5), respectively. Overexpression of p21 reduced total “p90” MDM2 expression in untreated p21–/– hepatocytes by 37 ± 6% (S.E.; p < 0.05; n = 5). Lower section, primary mouse hepatocytes in triplicate were infected to express nothing (vector, CMV) or p21, as indicated and incubated for 24 h. Cells were isolated, lysed, and p53 immunoprecipitated from the lysates. Immunoblotting was performed to determine the amount of ubiquitinated p53 (n = 2). D, primary rat hepatocytes 4 h after plating were transfected with scrambled siRNA (siSCR) or siRNA molecules to knock down PUMA, BAX, or NOXA (or a combination of these siRNA molecules, as indicated). Thirty six hours after transfection, cells were treated with vehicle, PD184352, DCA, or the agents in combination, and 6 h later cells were isolated and spun onto glass slides for determination of apoptosis as described under “Experimental Procedures” (n = 3 studies, ±S.E.). % p < 0.05 apoptosis value less than corresponding value in siSCR cells; %, p < 0.01 apoptosis value less than corresponding value in siSCR cells; &, p < 0.05 apoptosis value greater than corresponding value in siBAX or siPUMA cells. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Overexpression of p21 in rat hepatocytes enhanced the expression of p53 (Fig. 2B). In cells overexpressing p21, the majority of p53 was located in the nuclear fraction, whereas in vector control infected cells, p53 was mostly cytosolic. Treatment of hepatocytes with DCA + MEK1/2 inhibitor promoted modest nuclear translocation of p53, regardless of p21 overexpression. Overexpression of p21 increased the protein levels of several established/bona fide p53 target genes that have been implicated by many investigators in enhanced cell death responses, including the BH3 domain proteins BAX, PUMA, and NOXA (Table 1). In contrast to prior studies from this laboratory, which did not observe any significant changes in CD95 expression during these treatments, more definitive analyses using different antibody reagents that generated immunoblots with less nonspecific background bands demonstrated that overexpression of p21 increased CD95 protein levels (Table 1) (9).
TABLE 1.
CDK inhibitors enhance the expression of multiple BH3 domain proteins
Primary rat hepatocytes were infected to express nothing (vector, CMV) or p21 or p27, as indicated. Cells were treated 24 h after plating with vehicle (VEH) or with PD184352 (PD) and DCA and isolated 2 h after treatment. Total cell lysates were subjected to immunoblotting to determine the expression of CD95, FAS-L, PUMA, NOXA, BAX, BCL-2, BCL-XL, and glyceraldehyde-3-phosphate dehydrogenase (n = 3, ±S.E.). Data in the table are normalized to the expression of the protein in vehicle-treated vector control (CMV)-infected cells, and in addition, all values are then normalized to the total protein content of the lysate (glyceraldehyde-3-phosphate dehydrogenase value).
|
Infection/treatment |
||||||
|---|---|---|---|---|---|---|
| CMV/VEH | CMV/PD + DCA | p21/VEH | p21/PD + DCA | p27/VEH | p27/PD + DCA | |
| BAX | 1.00 | 1.05 ± 0.03 | 1.55 ± 0.06a | 3.01 ± 0.42b | 1.40 ± 0.17a | 2.88 ± 0.16b |
| NOXA | 1.00 | 1.52 ± 0.13b | 1.67 ± 0.05a | 2.59 ± 0.08b | 1.91 ± 0.24a | 3.11 ± 0.40b |
| CD95 | 1.00 | 1.51 ± 0.33b | 1.80 ± 0.20a | 3.03 ± 0.36b | 3.09 ± 0.09a | 4.23 ± 0.27b |
| p53 | 1.00 | 1.08 ± 0.17 | 2.09 ± 0.26a | 5.27 ± 1.01b | 3.91 ± 0.69a | 5.42 ± 1.22b |
| PUMA | 1.00 | 7.42 ± 0.07b | 6.24 ± 0.54a | 12.97 ± 0.14b | 7.87 ± 0.07a | 10.91 ± 0.59b |
p < 0.05 expression value greater than corresponding value in vector control (CMV) infected cells.
p < 0.05 expression value is greater than corresponding value in parallel untreated cells.
In prior studies overexpression of p21 in p21–/– hepatocytes decreased the expression of MDM2 and increased the expression of p53 (Fig. 2C, upper panel) (9). We have now noted that overexpression of p21 correlated with decreased ubiquitination of p53 in immunoprecipitates of p53 (Fig. 2C, lower panel). These findings suggest that p21 interferes with the ability of MDM2 to ubiquitinate p53. As p21 overexpression increased the levels of the pro-apoptotic proteins BAX, PUMA, and NOXA, we determined whether the enhanced expression of these proteins played any role in the elevated levels of the cell killing we were observing. Knockdown of BAX, NOXA, or PUMA expression significantly reduced p21-stimulated DCA + MEK1/2 inhibitor toxicity (Fig. 2D). Knockdown of NOXA expression reduced DCA + MEK1/2 inhibitor lethality to a greater extent than knockdown of either BAX or PUMA in vector (CMV) control infected cells. Knockdown of either BAX or PUMA expression reduced DCA + MEK1/2 inhibitor lethality to a greater extent than knockdown of NOXA in cells infected to express a CDK inhibitor. Combined knockdown of BAX and PUMA and NOXA expression abolished DCA + MEK1/2 inhibitor toxicity.
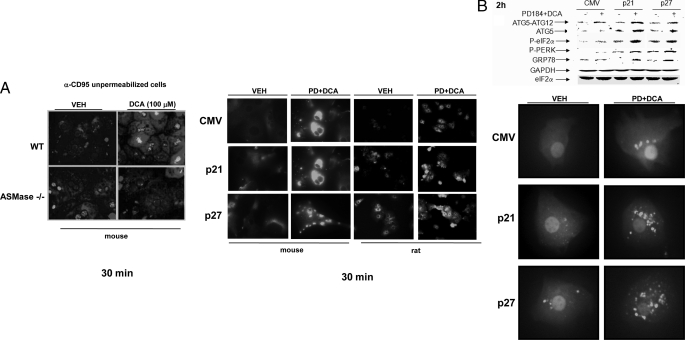
Overexpression of CDK Inhibitors Facilitates a Bile Acid-induced Autophagy Response That Is ASMase- and CD95-dependent—In Fig. 1 we demonstrated that DCA + MEK1/2 inhibitor treatment failed to kill CD95–/– hepatocytes. Recent studies have noted in gastrointestinal and genitourinary tumor cell types that activation of CD95 can be causal in chemotherapy-induced death processes as well as in the induction of ER stress signaling and autophagy (23–26). Based on our findings in Fig. 1, as well as our data showing that CD95 expression levels were being enhanced by p21 overexpression, we next determined whether overexpression of CDK inhibitors altered ER stress signaling and the levels of autophagy in hepatocytes treated with DCA + MEK1/2 inhibitor, and whether these effects were CD95-dependent.
Previous studies from our laboratories have demonstrated that bile acid-induced activation of CD95 in hepatocytes is dependent on ASMase and the generation of ceramide (27, 28). Treatment of primary mouse hepatocytes with DCA + MEK1/2 inhibitor enhanced ceramide levels 1.66 ± 0.2-fold (±S.E. n = 3) within 30 min after exposure, an effect that was abolished in ASMase–/– cells. In agreement with our former studies, bile acid-induced activation of CD95 in primary mouse hepatocytes was dependent upon expression of ASMase (Fig. 3A, left panels). Treatment of rat or mouse hepatocytes with DCA + MEK1/2 inhibitor promoted cell surface/plasma membrane localization of CD95, indicative of CD95 activation (Fig. 3A, right panels). Overexpression of p21 or p27 modestly increased basal levels of plasma membrane CD95 immunostaining. However, the fold activation of CD95 by DCA + MEK1/2 inhibitor treatment was not profoundly enhanced by overexpression of either p21 or p27.
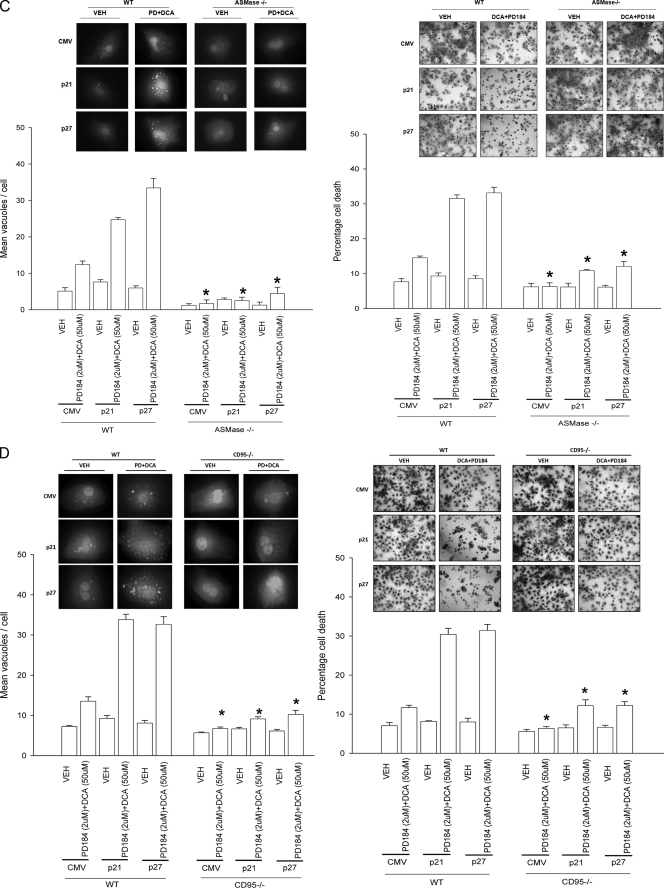
FIGURE 3.
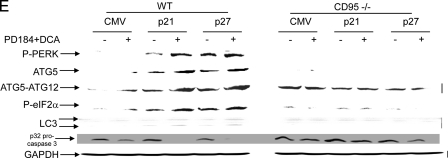
Knock-out of CD95 expression abolishes CDK inhibitor-enhanced autophagy and suppresses DCA + MEK1/2 inhibitor-induced apoptosis. Primary hepatocytes were isolated as described under “Experimental Procedures.” Cells were treated with vehicle (VEH) control (DMSO), DCA (50 μm, or as indicated), MEK1/2 inhibitor (PD184352, PD184, 2 μm) or both agents combined, as indicated in each panel. A, panels to the left, primary hepatocytes from wild type (WT) and ASM knock-out (ASM–/–) mice were treated with 100 μm DCA or vehicle for 30 min. After treatment, the cells were fixed but not permeabilized, and cell surface CD95 was localized by fluorescence microscopy (n = 3 studies). Panels to the right, wild type primary mouse hepatocytes plated in triplicate on glass slides were infected to express nothing (vector, CMV) or p21 or p27, as indicated. Twenty four hours after plating cells were treated with vehicle or with DCA + PD184352. Cells were fixed in situ but not permeabilized 30 min after DCA + PD184352 exposure, and the surface immunofluorescence of CD95 was determined ± S.E. (n = 2 studies). B, upper section, primary rat hepatocytes were infected to express nothing (vector, CMV) or p21 or p27, as indicated. After plating (24 h) cells were treated with vehicle, DCA, PD184352, or both agents in combination for 2 h after which cells were isolated and subjected to SDS-PAGE, and immunoblotting was performed to determine the expression of ATG5, ATG5-ATG12 conjugate, P-eIF2α, P-PERK, and GRP78 (n = 3, a representative study is shown). Lower section, primary rat hepatocytes were infected to express nothing (vector, CMV) or p21 or p27, as indicated. In parallel, cells were transfected with a plasmid to express LC3-GFP. After plating (24 h) cells were treated with vehicle or with DCA and PD184352. Data are from a representative experiment (n = 3). In parallel control studies, expression of β-galactosidase using Ad.β-gal did not induce GFP-LC3 vacuoles in hepatocytes in the presence or absence of DCA ± PD184352.3 C, left graph, primary mouse hepatocytes (wild type and ASMase–/–) plated in triplicate were infected to express nothing (vector, CMV) or p21 or p27, as indicated. In parallel, cells were transfected with a plasmid to express LC3-GFP. After plating (24 h) cells were treated with vehicle or with DCA and PD184352. The mean number of autophagic vesicles per cell (n = 40 cells examined in three fields) was determined 2 h after DCA+PD184352 exposure ± S.E. Data are the means from two independent experiments. *, p < 0.05 apoptosis value less than corresponding value in wild type cells. The upper section shows a representative LC3-GFP microscopic image. Right graph, primary mouse hepatocytes (wild type and ASMase–/–) plated in triplicate were infected to express nothing (vector, CMV) or p21 or p27, as indicated. After plating (24 h) cells were treated with vehicle or with DCA and PD184352. Twenty four hours after plating cells were treated with vehicle or with DCA and PD184352. Cells were isolated 6 h after DCA and PD184352 exposure after which cells were isolated and spun onto glass slides for determination of apoptosis as described under “Experimental Procedures” (n = 2, ±S.E.). *, p < 0.05 apoptosis value less than corresponding value in wild type cells. The upper section shows a representative hematoxylin and eosin microscopic image. D, left graph, primary mouse hepatocytes (wild type and CD95–/–) plated in triplicate were infected to express nothing (vector, CMV) or p21 or p27, as indicated. In parallel, cells were transfected with a plasmid to express LC3-GFP. After plating (24 h) cells were treated with vehicle or with DCA and PD184352. The mean number of autophagic vesicles per cell (n = 40 cells examined in three fields) was determined 2 h after DCA+PD184352 exposure ± S.E. Data are the means from three independent experiments. *, p < 0.05 apoptosis value less than corresponding value in wild type cells. The upper section shows a representative LC3-GFP microscopic image. Right graph, primary mouse hepatocytes (wild type and CD95–/–) plated in triplicate were infected to express nothing (vector, CMV) or p21 or p27, as indicated. After plating (24 h) cells were treated with vehicle or with DCA and PD184352. After plating (24 h) cells were treated with vehicle or with DCA and PD184352. Cells were isolated 6 h after DCA and PD184352 exposure after which cells were isolated and spun onto glass slides for determination of apoptosis as described under “Experimental Procedures.” (n = 3, ±S.E.). *, p < 0.05 apoptosis value less than corresponding value in wild type cells. The upper section shows a representative hematoxylin and eosin microscopic image. E, primary mouse hepatocytes (wild type and CD95–/–) were infected to express nothing (vector, CMV) or p21 or p27, as indicated. After plating (24 h) cells were treated with vehicle or DCA and PD184352 in combination for 2 h after which cells were isolated and subjected to SDS-PAGE, and immunoblotting was performed to determine the expression of ATG5, ATG5-ATG12 conjugate, P-eIF2α, P-PERK, LC3 processing, and pro-caspase 3 integrity/cleavage status (n = 2, a representative study is shown). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Expression of p21 or p27 enhanced DCA + MEK1/2 inhibitor lethality, and in parallel we noted that it enhanced the expression of ATG5, ATG5-ATG12 conjugate, GRP78/BiP, and the phosphorylation of PKR-like endoplasmic reticulum kinase (PERK) and eIF2α (Fig. 3B, immunoblot). DCA + MEK1/2 inhibitor treatment rapidly promoted the vesicularization of a transfected LC3-GFP construct in hepatocytes, which was enhanced by p21 or p27 expression but not by expression of β-galactosidase (Fig. 3B, lower immunoblot; data not shown). DCA + MEK1/2 inhibitor treatment did not promote vesicularization of the LC3-GFP construct in either ASMase–/– or in CD95–/– hepatocytes (Fig. 3, C and D, left panels). Overexpression of p21 or p27 did not promote cell killing in either ASMase–/– or CD95–/– hepatocytes (Fig. 3, C and D, right panels). Loss of CD95 expression abolished the ability of DCA + MEK1/2 inhibitor treatment to promote enhanced expression of ATG5, ATG5-ATG12 conjugate, and GRP78/BiP; the processing of LC3 and pro-caspase 3 cleavage; and the phosphorylation of PERK and eIF2α (Fig. 3D). Thus, DCA + MEK1/2 inhibitor treatment simultaneously promotes both an ASMase- and CD95-dependent apoptosis response and an ASMase- and CD95-dependent autophagy response.
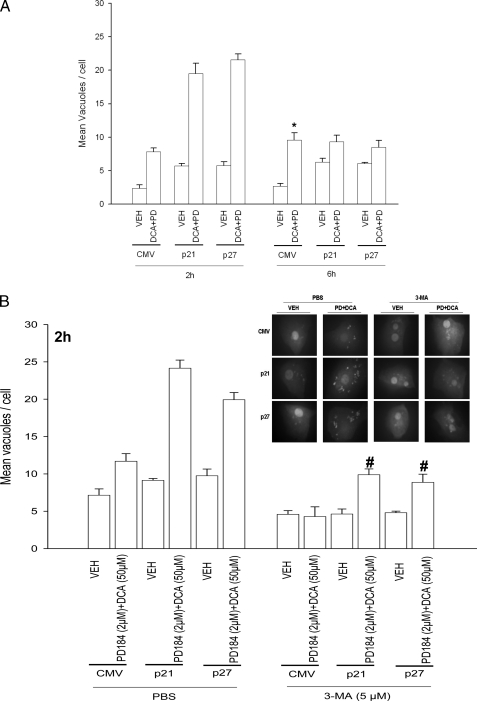
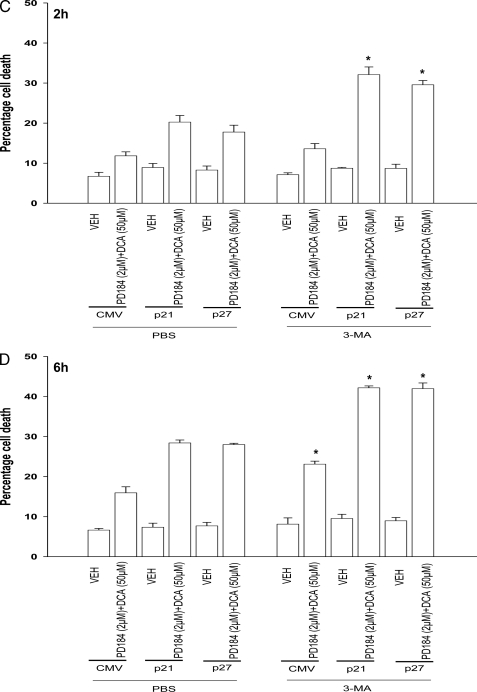
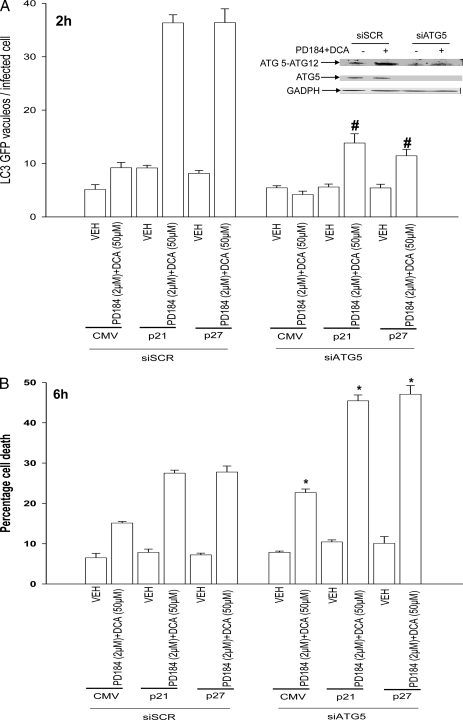
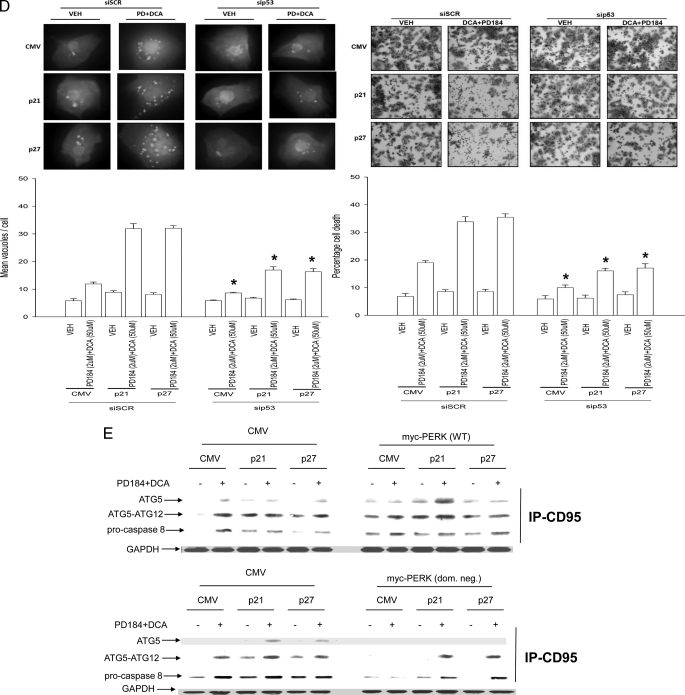
Overexpression of CDK Inhibitors Promotes Bile Acid-induced Autophagy That Is a Protective Signal—Autophagy can promote or degrade cell survival. Thus we next determined where DCA + MEK1/2 inhibitor-induced autophagy was a protective or a toxic signal. Overexpression of p21 or p27 followed by DCA + MEK1/2 inhibitor treatment increased the mean number of induced GFP-LC3 vesicles per cell 2 h after treatment, which had declined by 6 h (Fig. 4A). Treatment of cells with 3-methyladenine (3MA), a recognized small molecule inhibitor of autophagy, significantly reduced vesicle formation and simultaneously promoted DCA + MEK1/2 inhibitor-induced cell killing both 2 and 6 h after treatment (Fig. 4, B–D). In a similar manner to 3MA treatment, knockdown of ATG5 expression in hepatocytes suppressed the vesicularization of the transfected LC3-GFP construct and enhanced the lethality of DCA + MEK1/2 inhibitor treatment (Fig. 5, A and B). Thus our data demonstrate that the observed DCA + MEK1/2 inhibitor-dependent induction of autophagy was a cell survival response.
FIGURE 4.
CDK inhibitors promote bile acid-induced autophagy and apoptosis in primary hepatocytes. Primary hepatocytes were isolated as described under “Experimental Procedures.” Cells were treated with vehicle (VEH) control (DMSO), DCA (50 μm), MEK1/2 inhibitor (PD184352 (PD), 2 μm), or both agents combined, as indicated in each panel. A, primary rat hepatocytes were infected to express nothing (vector, CMV) or p21 or p27, as indicated. In parallel, cells were transfected with a construct to express LC3-GFP. Twenty four hours after plating, cells were treated with vehicle or with DCA and PD184352 for 2 or 6 h. The mean number of autophagic vesicles per cell (n = 40 cells examined in three fields) was determined at each time point ± S.E. Data are the means from three independent experiments. *, p < 0.05 differential value (vehicle cf. DCA + PD) greater than the corresponding value at 2 h. B, primary rat hepatocytes were infected to express nothing (vector, CMV) or p21 or p27, as indicated. In parallel, cells were transfected with a construct to express LC3-GFP. Twenty four hours after plating cells were pretreated for 30 min with 3-methyladenine (5 μm) then treated with vehicle or with DCA and PD184352 for 2 h. The mean number of autophagic vesicles per cell (n = 40 cells examined in 3 fields) was determined at each time point ± S.E. Data are the means from 3 independent experiments. #, p < 0.05 differential value (Vehicle cf. DCA + PD) less than corresponding value in PBS treated cells. C and D, primary rat hepatocytes plated in triplicate were infected to express nothing (vector, CMV) or p21 or p27, as indicated. Twenty four hours after plating cells were pretreated for 30 min with 3-methyladenine (3-MA, 5 μm) and then treated with vehicle or with DCA and PD184352 for 2 h(C) or for 6 h (D). Cells were isolated 2 and 6 h after DCA and PD184352 (PD184) exposure, after which cells were isolated and spun onto glass slides for determination of apoptosis as described under “Experimental Procedures.” (n = 3, ±S.E.). *, p < 0.05 differential value (Vehicle cf. DCA + PD) greater than corresponding value in PBS-treated cells.
FIGURE 5.
Knockdown of ATG5 expression abolishes CDK inhibitor-enhanced autophagy and promotes DCA + MEK1/2 inhibitor-induced apoptosis. A, primary rat hepatocytes plated in triplicate were infected to express nothing (vector, CMV) or p21 or p27, as indicated. In parallel, cells were transfected with a plasmid to express LC3-GFP and with a scrambled siRNA (siSCR) or an siRNA to knock down ATG5 expression (siATG5). Twenty four hours after plating cells were treated with vehicle (VEH) or with DCA and PD184352 (PD184). The mean number of autophagic vesicles per cell (n = 40 cells examined in three fields) was determined 2 h after DCA + PD184352 exposure ± S.E. Data are the means from three independent experiments. #, p < 0.05 differential value (Vehicle cf. DCA + PD) less than corresponding value in siSCR treated cells. B, primary rat hepatocytes plated in triplicate were infected to express nothing (vector, CMV) or p21 or p27, as indicated. In parallel, cells were transfected with either a scrambled siRNA (siSCR) or an siRNA to knock down ATG5 expression (siATG5). Twenty four hours after plating cells were treated with vehicle or with DCA and PD184352. Cells were isolated 6 h after DCA and PD184352 exposure after which cells were isolated and spun onto glass slides for determination of apoptosis as described under “Experimental Procedures” (n = 3, ±S.E.). *, p < 0.05 differential value (Vehicle cf. DCA + PD) greater than corresponding value in siSCR-treated cells.
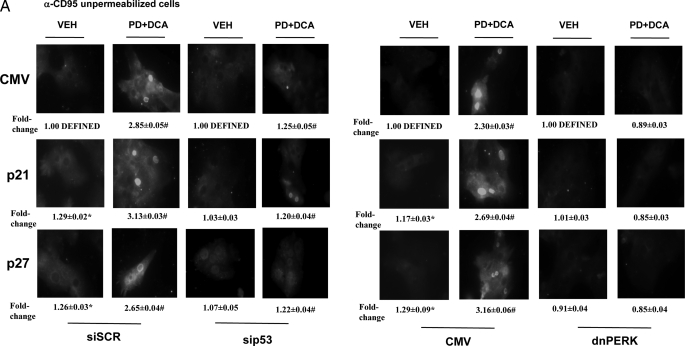
Overexpression of CDK Inhibitors Promotes p53-dependent Increases in ER Stress and Autophagy Proteins That Regulate Hepatocyte Survival after Bile Acid Exposure—The data in Figs. 2, 3, 4, 5 represented the phenomenology of cell killing and autophagy in our system using hepatocytes derived from genetically modified animals. We next attempted to place the sequential regulation of these processes in molecular terms. As noted previously, overexpression of p21 or p27 modestly enhanced basal levels of plasma membrane immunostaining for CD95 but did not profoundly enhance DCA + MEK1/2 inhibitor treatment-induced activation of CD95. Knockdown of p53 expression suppressed, but did not abolish, DCA + MEK1/2 inhibitor treatment-induced activation CD95 (Fig. 6A). Prior studies demonstrated that PERK was not activated by DCA + MEK1/2 inhibitor treatment in CD95–/– hepatocytes (Fig. 3D). However, to our surprise, we noted that expression of a dominant negative form of PERK abolished CD95 activation after DCA + MEK1/2 inhibitor treatment (Fig. 6A). This finding demonstrates that PERK is both an upstream and a downstream effector of CD95 signaling in our cell system.
FIGURE 6.
Knock-out of CD95 expression abolishes CDK inhibitor-enhanced autophagy and suppresses DCA + MEK1/2 inhibitor-induced apoptosis. A, primary rat hepatocytes plated in triplicate on glass slides were infected to express nothing (vector, CMV) or p21 or p27, as indicated. In parallel, cells, as indicated in the figure, were transfected either with empty vector plasmid (CMV) or a plasmid to express dominant negative PERK (dnPERK) or transfected with a scrambled siRNA (siSCR) or an siRNA to knock down p53 expression (sip53). Twenty four hours after plating/transfection, cells were treated with vehicle or with DCA and PD184352. Cells were fixed in situ but not permeabilized 30 min after DCA + PD184352 exposure and the surface immunofluorescence of CD95 was determined ± S.E. (n = 2 studies). #, p < 0.01 value greater than the corresponding value in vehicle-treated cells. *, p < 0.01 value greater than corresponding value in vector control (CMV)-infected cells. B, upper immunoblot, primary rat hepatocytes were infected to express nothing (vector, CMV) or p21 or p27, as indicated. In parallel, cells were transfected with either a vector control plasmid (CMV) or a plasmid to express dominant negative PERK (dnPERK). Twenty four hours after plating cells were treated with vehicle or with DCA and PD184352 (PD184). Cells were isolated 2 h after DCA and PD184352 exposure, after which cells were isolated and subjected to SDS-PAGE, and immunoblotting was performed to determine the expression of ATG5, ATG5-ATG12 conjugate, P-eIF2α, P-PERK, and GRP78 (n = 2, a representative study is shown). Lower immunoblot, primary rat hepatocytes were infected to express nothing (vector, CMV) or p21 or p27, as indicated. In parallel, cells were transfected with either a scrambled siRNA (siSCR) or an siRNA to knock down p53 expression (sip53). Twenty four hours after plating cells were treated with vehicle or with DCA and PD184352. Cells were isolated 2 h after DCA and PD184352 exposure after which cells were isolated and subjected to SDS-PAGE and immunoblotting performed to determine the expression of CD95, p53, ATG5, ATG5-ATG12 conjugate, P-eIF2α, P-PERK, and GRP78 (n = 2, a representative study is shown). C, left graph, primary rat hepatocytes were infected to express nothing (vector, CMV) or p21 or p27, as indicated. In parallel, cells were transfected with either a vector control plasmid (CMV) or a plasmid to express dominant negative PERK (dnPERK) together with a plasmid to express LC3-GFP. Twenty four hours after plating cells were treated with vehicle or with DCA and PD184352. The mean number of autophagic vesicles per cell (n = 40 cells examined in three fields) was determined 2 h after DCA + PD184352 exposure ± S.E. Data are the means from three independent experiments. Right graph, primary rat hepatocytes were infected to express nothing (vector, CMV) or p21 or p27, as indicated. In parallel, cells were transfected with either a vector control plasmid (CMV) or a plasmid to express dominant negative PERK (dnPERK). Twenty four hours after plating cells were treated with vehicle or with DCA and PD184352. Cells were isolated 2 h after DCA and PD184352 exposure, after which cells were isolated and spun onto glass slides for determination of apoptosis as described under “Experimental Procedures” (n = 3, ±S.E.). *, p < 0.05 value less than corresponding parallel value in CMV transfected treated cells. D, left graph, primary rat hepatocytes were infected to express nothing (vector, CMV) or p21 or p27, as indicated. In parallel, cells were transfected with either a scrambled siRNA (siSCR) or an siRNA to knock down p53 expression (sip53) together with a plasmid to express LC3-GFP. Twenty four hours after plating cells were treated with vehicle or with DCA and PD184352. The mean number of autophagic vesicles per cell (n = 40 cells examined in three fields) was determined 2 h after DCA + PD184352 exposure ± S.E. Data are the means from three independent experiments. Right graph, primary rat hepatocytes were infected to express nothing (vector, CMV) or p21 or p27, as indicated. In parallel, cells were transfected with either a scrambled siRNA (siSCR) or an siRNA to knock down p53 expression (sip53). Twenty four hours after plating cells were treated with vehicle or with DCA and PD184352. Cells were isolated 2 h after DCA and PD184352 exposure, after which cells were isolated and spun onto glass slides for determination of apoptosis as described under “Experimental Procedures” (n = 3, ±S.E.). *, p < 0.05 value less than corresponding parallel value in CMV transfected treated cells. E, primary rat hepatocytes were transfected to express MYC-tagged wild type or dominant negative PERK as indicated. Cells were treated 24 h after transfection with vehicle or with DCA and PD184352. After 30 min exposure, cells were isolated, and CD95 was immunoprecipitated (IP), and the formation of a DISC complex containing pro-caspase 8, ATG5, or PERK (via MYC tag) was determined. The cytosolic fraction was analyzed for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression to determine equal protein loading (n = 2).
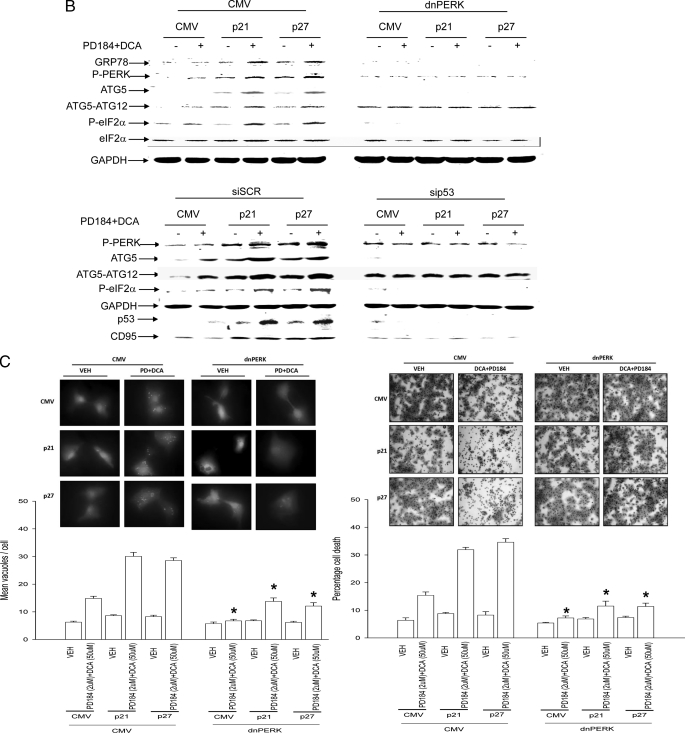
Regardless of p21/p27 overexpression, expression of dominant negative PERK blocked DCA + MEK1/2 inhibitor-induced PERK and eIF2α phosphorylation and alterations in ATG5 and GRP78/BiP expression as well as increased levels of LC3-GFP vesicularization and apoptosis (Fig. 6, B and C). Thus, the DCA + MEK1/2 inhibitor-induced stimulation of autophagy is PERK-dependent.
Regardless of p21/p27 overexpression, knockdown of p53 levels reduced the ability of CDK inhibitors to promote CD95 expression as well as to increase PERK and eIF2α phosphorylation, to elevate ATG5 or GRP78/BiP expression, and to enhance LC3-GFP vesicularization (Fig. 6, B and D). Thus, DCA + MEK1/2 inhibitor-induced PERK activation and the increased expression of proteins that facilitate autophagy and the elevation of their protein levels by CDK inhibitor overexpression were dependent on hepatocytes expressing p53.
It has been demonstrated that ATG5 can associate with the CD95-binding protein FADD (40). DCA + MEK1/2 inhibitor treatment promoted both ATG5 and pro-caspase 8 association with CD95 (Fig. 6E). Overexpression of PERK caused constitutive association of procaspase 8 and ATG5 with CD95. Expression of dominant negative PERK profoundly reduced procaspase 8 and ATG5 association with CD95 after DCA + MEK1/2 inhibitor treatment (Fig. 6E). Thus, PERK regulates CD95 and DISC complex formation.
DISCUSSION
Previous studies by this group in primary hepatocytes have linked bile acid-induced activation of CD95 to pro-apoptotic signaling via the extrinsic apoptosis pathway. Other studies have shown that overexpression of the CDK inhibitor p21 promoted bile acid-induced apoptosis that was p53-dependent. This study attempted to extend our analyses to determine whether other CDK inhibitors promoted bile acid-induced apoptosis and determine the downstream molecular mechanisms by which the promotion of cell killing occurred.
We discovered that in addition to p21, the related CDK inhibitor family member p27 also promoted bile acid-induced apoptosis. Of note, as primary hepatocytes are not proliferating and were in early G1 phase during our analyses, the mechanism by which this event occurred will in all likelihood not have been due to inhibition of CDK activity(ies). Increased expression of p21 or p27 resulted in enhanced p53 levels that correlated with reduced expression of MDM2 and reduced ubiquitination of p53. In cells overexpressing p21, p53 was predominantly localized in the nucleus, and treatment of cells with bile acid caused p53 to become localized in the nucleus. Increased expression/nuclear localization of p53 correlated with increased expression of multiple pro-apoptotic gene products, including BAX, NOXA, PUMA, and CD95. Knockdown of BAX, NOXA, and PUMA significantly reduced the enhancement in cell killing caused by p21 and p27 overexpression. However, in the absence of CDK inhibitor overexpression, the relative role of p53 in bile acid-induced apoptosis was more modest. BAX, NOXA, and PUMA are well recognized transcriptional effectors of p53, and in multiple cell systems activation of p53 leads to elevation of their expression and enhancement of cell death (29, 30).
Prior studies examining p21-promoted bile acid apoptosis in hepatocytes had not observed any significant alteration in CD95 expression levels that we believe were because of antibody specificity/quality issues in our prior work. In the present studies using a newer antibody reagent, we observed increased expression of CD95 in hepatocytes overexpressing p21 or p27. Overexpression of p21 or p27 in a p53-dependent fashion enhanced basal plasma membrane levels of CD95 but did not further enhance bile acid stimulated CD95 activation. These findings suggest that the major mechanism by which p21/p27 enhance bile acid toxicity in a p53-dependent fashion is by promoting expression of BAX, NOXA, and PUMA rather than by increasing CD95 activation.
The alternate splice product of the CDK inhibitor p16INK4a, p19ARF, has been shown by many groups to inhibit MDM2 association with, and E3 ligase activity against, p53. This acts to promote p53 protein levels (31, 32). One of the transcriptional targets of p53, p21, has been shown to increase its expression upon elevation of p19ARF levels that can result in cell cycle arrest. The present findings argue that overexpression of p21 itself is competent to suppress p53 ubiquitination and enhance p53 levels, as well as promote increased expression of p53 target genes. That p27 also enhances p53 expression and the apoptotic response of bile acid-treated hepatocytes suggests that p21 and p27 may have some form of potential to act in a similar manner against MDM2 as does p19ARF. Structure-function relationships examining how these proteins reduce p53 ubiquitination and reduce MDM2 expression will require considerable further analyses beyond the scope of this study.
Recently, Fujiwara et al. (33) noted in mouse embryonic fibroblasts that p21 also played a pivotal role in regulating ceramide-induced apoptosis and autophagy. In these studies, expression of p21 was obligate for ceramide to cause cell killing in fibroblasts, whereas a lack of p21 expression in this cell type resulted in ceramide-promoting autophagy instead of apoptosis. Previous studies from our laboratories have demonstrated that bile acid-induced activation of CD95 is dependent on ASMase and the generation of endogenous ceramide (27, 28). In agreement with our CD95 data and the fact that loss of ASMase expression abolishes CD95 activation, we also noted that loss of ASMase expression abolished the induction of autophagic vesicles and abolished the promotion of apoptosis by DCA + MEK1/2 inhibitor treatment, regardless of CDK inhibitor overexpression.
Our present studies in primary hepatocytes thus provide a slightly more nuanced series of observations concerning endogenous ceramide signaling, and the roles that CD95 and p21 play in these processes, than those of Fujiwara et al. (33) In agreement with Fujiwara et al. (33), at times where apoptosis had become strongly manifested, 6 h after DCA+MEK1/2 inhibitor exposure, the induction of autophagy was suppressed, relative to vector control, by expression of p21 or p27; and apoptosis was strongly promoted relative to vector control by expression of p21 or p27. However, our data 2 h after DCA + MEK1/2 inhibitor exposure demonstrate that apoptosis and autophagy are more closely interrelated rather than simply being the opposite sides of a cell survival coin. For example, at the 2-h exposure point, relative to vector control, expression of p21 or p27 enhanced LC3-GFP vesicularization, increased the protein levels of multiple protein markers of autophagy, and also enhanced apoptosis. Inhibition of LC3-GFP vesicularization/autophagy, either by using 3MA or siATG5, further enhanced cell killing. Thus the overall conclusion in our cell system is that p21 or p27 strongly and simultaneously promotes both apoptosis and autophagy, which is manifest 2 h after exposure, and that the loss of an autophagy response increases the amount of cell killing. Studies beyond the scope of this study will be required to define in precise detail how and why the CDK inhibitor-stimulated apoptotic/pro-death signal overcomes the CDK inhibitor-stimulated autophagic/pro-survival signal in primary hepatocytes.
In primary hepatocytes, bile acid-induced activation of PERK enhanced LC3-GFP vesicularization, and its potentiation by expression of p21 or p27 was dependent on expression of CD95. Thus not only the promotion of apoptosis but also of autophagy was dependent on ASMase/ceramide-induced activation of a death receptor. Previously we have shown that CD95 signaling, per se, does not always result in cell death in primary hepatocytes, and we have shown that bile acid-induced activation of a CD95-JNK pathway was important in the regulation of cholesterol 7α-hydroxylase expression (27, 34). The present studies demonstrate that CDK inhibitors, by indirectly elevating p53 levels, which in turn promote CD95 expression, act to facilitate both toxic apoptotic and protective autophagic signaling.
A recent study by Abida and Gu (35) demonstrated that p14/p19ARF can promote p53-dependent and p53-independent autophagy in transformed cell types. Several studies in a variety of cell types have shown that p53 can induce autophagy (36, 37). Our present findings, in general agreement with those of Abida and Gu(35), also demonstrate that p53 plays a key role in promoting CDK inhibitor-stimulated autophagy after DCA + MEK1/2 inhibitor treatment. Others have noted that damage-regulated autophagy modulator can have its expression increased at the transcriptional level by both p53 and the related factor p73, and this protein can also regulate the fate of cells exposed to amino acid starvation and DNA damage in terms of the balance between their apoptotic and autophagy responses (37, 38). It will be of interest in the future to determine whether damage-regulated autophagy modulator expression or localization represents one of the downstream factors involved in the decision-making process that hepatocytes utilize in terms of how and why the CDK inhibitor-stimulated apoptotic/pro-death signal overcomes the CDK inhibitor-stimulated autophagic/pro-survival signal in primary hepatocytes.
In conclusion, this study demonstrates that CDK inhibitors via CD95 promote bile acid-induced apoptosis (a toxic response) and autophagy (a survival response). This implies that both p21 and p27 have functions other than those as CDK inhibitors (39). Whether elevated CDK inhibitor expression has true biological relevance for apoptosis and autophagy signaling, such as in the livers of older individuals that constitutively overexpress p21 and lack regenerative capacity, will need to be addressed in future studies.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-DK52825, P01-CA104177, and R01-CA108520 (to P. D.) and R01-CA63753 and R01-CA77141 (to S. G.) from the USPHS, Department of Defense Award DAMD17-03-1-0262, The V Foundation, and a Leukemia Society of America Grant 6405-97. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: DCA, deoxycholic acid; DMSO, dimethyl sulfoxide; CMV, cytomegalovirus; siRNA, short interfering RNA; PBS, phosphate-buffered saline; siSCR, scrambled siRNA; ASMase, acidic sphingomyelinase; CDK, cyclin-dependent kinase; ER, endoplasmic reticulum; PERK, PKR-like endoplasmic reticulum kinase; E3, ubiquitin-protein isopeptide ligase.
G. Zhang, M. A. Park, and P. Dent, unpublished observations.
References
- 1.Benage, D., and O'Connor, K. W. (1990) J. Clin. Gastroenterol. 12 192–194 [PubMed] [Google Scholar]
- 2.Holt, P. R. (1972) Arch. Intern. Med. 130 574–583 [PubMed] [Google Scholar]
- 3.Roberts, M. S., Magnusson, B. M., Burczynski, F. J., and Weiss, M. (2002) Clin. Pharmacokinet. 41 751–790 [DOI] [PubMed] [Google Scholar]
- 4.Faubion, W. A., Guicciardi, M. E., Miyoshi, H., Bronk, S. F., Roberts, P. J., Svingen, P. A., Kaufmann, S. H., and Gores, G. J. (1999) J. Clin. Investig. 103 137–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noto, H., Matsushita, M., Koike, M., Takahashi, M., Matsue, H., Kimura, J., and Todo, S. (1998) Artif. Organs 22 300–307 [DOI] [PubMed] [Google Scholar]
- 6.Bloomer, J. R., Allen, R. M., and Klatskin, G. (1976) Arch. Intern. Med. 136 57–61 [PubMed] [Google Scholar]
- 7.Koeppel, T. A., Trauner, M., Baas, J. C., Thies, J. C., Schlosser, S. F., Post, S., Gebhard, M. M., Herfarth, C., Boyer, J. L., and Otto, G. (1997) Hepatology 26 1085–1091 [DOI] [PubMed] [Google Scholar]
- 8.Poupon, R., Chazouilleres, O., and Poupon, R. E. (2000) J. Hepatol. 32 129–140 [DOI] [PubMed] [Google Scholar]
- 9.Qiao, L., McKinstry, R., Gupta, S., Gilfor, D., Windle, J. J., Hylemon, P. B., Grant, S., Fisher, P. B., and Dent, P. (2002) Hepatology 36 39–48 [DOI] [PubMed] [Google Scholar]
- 10.Qiao, L., Yacoub, A., Studer, E., Gupta, S., Pei, X. Y., Grant, S., Hylemon, P. B., and Dent, P. (2002) Hepatology 35 779–789 [DOI] [PubMed] [Google Scholar]
- 11.Qiao, L., Studer, E., Leach, K., McKinstry, R., Gupta, S., Decker, R., Kukreja, R., Valerie, K., Nagarkatti, P., El Deiry, W., Molkentin, J., Schmidt-Ullrich, R., Fisher, P. B., Grant, S., Hylemon, P. B., and Dent, P. (2001) Mol. Biol. Cell 12 2629–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Werneburg, N. W., Yoon, J. H., Higuchi, H., and Gores, G. J. (2003) Am. J. Physiol. 285 G31–G36 [DOI] [PubMed] [Google Scholar]
- 13.Qiao, D., Stratagouleas, E. D., and Martinez, J. D. (2001) Carcinogenesis 22 35–41 [DOI] [PubMed] [Google Scholar]
- 14.Haupt, Y., Maya, R., Kazaz, A., and Oren, M. (1997) Nature 387 296–299 [DOI] [PubMed] [Google Scholar]
- 15.Honda, R., Tanaka, H., and Yasuda, H. (1997) FEBS Lett. 420 25–27 [DOI] [PubMed] [Google Scholar]
- 16.Kubbutat, M. H., Jones, S. N., and Vousden, K. H. (1997) Nature 387 299–303 [DOI] [PubMed] [Google Scholar]
- 17.Tao, W., and Levine, A. J. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 3077–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, Z., Wang, H., Prasad, G., Li, M., Yu, D., Bonner, J. A., Agarwal, S., and Zhang, R. (2004) Clin. Cancer Res. 10 1263–1273 [DOI] [PubMed] [Google Scholar]
- 19.Shieh, S. Y., Ikeda, M., Taya, Y., and Prives, C. (1997) Cell 91 325–334 [DOI] [PubMed] [Google Scholar]
- 20.Maki, C. G. (1999) J. Biol. Chem. 274 16531–16535 [DOI] [PubMed] [Google Scholar]
- 21.Chehab, N. H., Malikzay, A., Stavridi, E. S., and Halazonetis, T. D. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 13777–13782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maya, R., Balass, M., Kim, S. T., Shkedy, D., Leal, J. F., Shifman, O., Moas, M., Buschmann, T., Ronai, Z., Shiloh, Y., Kastan, M. B., Katzir, E., and Oren, M. (2001) Genes Dev. 15 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yacoub, A., Park, M. A., Gupta, P., Rahmani, M., Zhang, G., Hamed, H., Hanna, D., Sarkar, D., Lebedeva, I. V., Emdad, L., Sauane, M., Vozhilla, N., Spiegel, S., Koumenis, C., Graf, M., Curiel, D. T., Grant, S., Fisher, P. B., and Dent, P. (2008) Mol. Cancer Ther. 7 297–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park, M., Yacoub, A., Rahmani, M., Zhang, G., Hart, L., Hagan, M., Calderwood, S., Sherman, M., Koumenis, C., Spiegel, S., Chen, C. S., Graf, M., Curiel, D., Fisher, P., Grant, S., and Dent, P. (2008) Mol. Pharmacol. 73 1168–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, G., Park, M. A., Mitchell, C., Hamed, H., Rahmani, M., Martin, A. P., Curiel, D. T., Yacoub, A., Graf, M., Lee, R., Roberts, J. D., Fisher, P. B., Grant, S., and Dent, P. (2008) Clin. Cancer Res., in press [DOI] [PMC free article] [PubMed]
- 26.Park, M. A., Zhang, Z., Mitchell, C., Hamed, H., Rahmani, M., Martin, A. P., Yacoub, A., Koumenis, C., Spiegel, S., Norris, J., Hylemon, P. B., Graf, M., Fisher, P. B., Grant, S., and Dent, P. (2008) Cancer Biol. Ther., in press [DOI] [PMC free article] [PubMed]
- 27.Gupta, S., Natarajan, R., Payne, S. G., Studer, E. J., Spiegel, S., Dent, P., and Hylemon, P. B. (2004) J. Biol. Chem. 279 5821–5828 [DOI] [PubMed] [Google Scholar]
- 28.Fang, Y., Han, S. I., Mitchell, C., Gupta, S., Studer, E., Grant, S., Hylemon, P. B., and Dent, P. (2004) Hepatology 40 961–971 [DOI] [PubMed] [Google Scholar]
- 29.Labi, V., Erlacher, M., Kiessling, S., and Villunger, A. (2006) Cell Death Differ. 13 1325–1338 [DOI] [PubMed] [Google Scholar]
- 30.Yu, J., and Zhang, L. (2003) Cancer Cell 4 248–249 [DOI] [PubMed] [Google Scholar]
- 31.Wang, W., and El-Deiry, W. S. (2008) Curr. Opin. Oncol. 20 90–96 [DOI] [PubMed] [Google Scholar]
- 32.Saporita, A. J., Maggi, L. B., Jr., Apicelli, A. J., and Weber, J. D. (2007) Curr. Med. Chem. 14 1815–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujiwara, K., Daido, S., Yamamoto, A., Kobayashi, R., Yokoyama, T., Aoki, H., Iwado, E., Shinojima, N., Kondo, Y., and Kondo, S. (2008) J. Biol. Chem. 283 388–397 [DOI] [PubMed] [Google Scholar]
- 34.Gupta, S., Stravitz, R. T., Dent, P., and Hylemon, P. B. (2001) J. Biol. Chem. 276 15816–15822 [DOI] [PubMed] [Google Scholar]
- 35.Abida, W. M., and Gu, W. (2008) Cancer Res. 68 352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng, Z., Zhang, H., Levine, A. J., and Jin, S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 8204–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crighton, D., Wilkinson, S., O'Prey, J., Syed, N., Smith, P., Harrison, P. R., Gasco, M., Garrone, O., Crook, T., and Ryan, K. M. (2006) Cell 126 121–134 [DOI] [PubMed] [Google Scholar]
- 38.Crighton, D., O'Prey, J., Bell, H. S., and Ryan, K. M. (2007) Cell Death Differ. 14 1071–1079 [DOI] [PubMed] [Google Scholar]
- 39.Coqueret, O. (2003) Trends Cell Biol. 13 65–70 [DOI] [PubMed] [Google Scholar]
- 40.Pyo, J. O., Jang, M. H., Kwon, Y. K., Lee, H. J., Jun, J. I., Woo, H. N., Cho, D. H., Choi, B., Lee, H., Kim, J. H., Mizushima, N., Oshumi, Y., and Jung, Y. K. (2005) J. Biol. Chem. 280 20722–20729 [DOI] [PubMed] [Google Scholar]