Abstract
The polyguanine-rich DNA sequences commonly found at telomeres and in rDNA arrays have been shown to assemble into structures known as G quadruplexes, or G4 DNA, stabilized by base-stacked G quartets, an arrangement of four hydrogen-bonded guanines. G4 DNA structures are resistant to the many helicases and nucleases that process intermediates arising in the course of DNA replication and repair. The lagging strand DNA replication protein, Dna2, has demonstrated a unique localization to telomeres and a role in de novo telomere biogenesis, prompting us to study the activities of Dna2 on G4 DNA-containing substrates. We find that yeast Dna2 binds with 25-fold higher affinity to G4 DNA formed from yeast telomere repeats than to single-stranded DNA of the same sequence. Human Dna2 also binds G4 DNAs. The helicase activities of both yeast and human Dna2 are effective in unwinding G4 DNAs. On the other hand, the nuclease activities of both yeast and human Dna2 are attenuated by the formation of G4 DNA, with the extent of inhibition depending on the topology of the G4 structure. This inhibition can be overcome by replication protein A. Replication protein A is known to stimulate the 5′- to 3′-nuclease activity of Dna2; however, we go on to show that this same protein inhibits the 3′- to 5′-exo/endonuclease activity of Dna2. These observations are discussed in terms of possible roles for Dna2 in resolving G4 secondary structures that arise during Okazaki fragment processing and telomere lengthening.
The Dna2 protein, which is involved in the maintenance of genomic stability, is a multifaceted enzyme, with 5′–3′ DNA helicase, DNA-dependent ATPase, 3′-exo/endonuclease, 5′-exo/endonuclease, single strand annealing, and strand exchange activities (1–3). A global synthetic lethal screen in yeast has identified a network of pathways consisting of at least 322 Dna2-interacting genes, which are involved in DNA replication, DNA repair, telomere maintenance, and chromatin dynamics, consistent with the complex role of Dna2 in preserving genome integrity (4). The best characterized function of Dna2 is that in Okazaki fragment processing (OFP)2 during DNA replication. Biochemical and genetic evidence suggests that Dna2 assists FEN1 (flap endonuclease) in RNA/DNA primer removal during processing of a subset of Okazaki fragments with long 5′ flaps and flaps with secondary structures arising because of excessive strand displacement by polymerase δ (5). Such flaps cannot be processed efficiently by the primary nuclease FEN1 alone.
Dna2 also appears to play an important role in telomere maintenance, and there is strong evidence that Dna2 is associated with telomeres. Overexpression of Dna2 leads to derepression of genes embedded in telomeres (6, 7). Dna2 is localized to telomeres in G1 phase, when the telomere is transcriptionally silenced. In S phase Dna2 is redistributed from telomeres to sites throughout the chromosomes and is found again at telomeres in late S phase through G2 phase (7). This dynamic localization of Dna2 suggests roles for Dna2 in telomere capping (G1) and in telomere replication (S/G2). DNA damage also induces Dna2 to dissociate from telomeres. Dna2 is required for de novo telomere synthesis and is involved in telomere lengthening in telomerase-deficient mutants. The lethality of dna2 est2 (EST2 encodes the reverse transcriptase subunit of telomerase) double mutants suggests involvement of Dna2 in the resolution of the end replication problem at the telomeres (7). Also, the suppression of lethality in dna2 mutants by deletion of PIF1, a helicase interacting with telomerase, and the telomere phenotypes of the dna2Δ pif1Δ mutants points to a role of Dna2 at telomeres (8). By the same token, the Dna2/Pif1 interaction may suggest a role for Pif1 in OFP.
The molecular function of Dna2 at telomeres remains elusive. Telomeres are structures at the ends of chromosomes, providing for genomic stability by ensuring complete DNA replication and end protection. Yeast telomeres consist of roughly 300 bp of heterogeneous C1–3A/TG1–3 repetitive sequences at the chromosome termini. Telomere length is maintained by telomerase, which elongates the 3′ G-rich strand. Various stable structures may form on the 3′ G-rich strand, based on interactions between G residues that can form G quartets. Since 1991, when Zahler et al. (9) showed that G quadruplex-folded telomeric DNA inhibited telomerase action in vitro, a rapidly growing list of proteins has been found to bind to and resolve or bind to and favor the formation of specific G quadruplex topologies. The human RecQ helicases BLM and WRN have been shown to have far greater unwinding activity with parallel G quadruplex tetramers than their equivalent double-stranded DNA counterparts in vitro. Their yeast ortholog, Sgs1, has also shown a preference for these substrates, as well as for antiparallel dimeric G quadruplex structures (10–14). In addition, some endonucleases have been shown to bind to G quadruplexes, but in the absence of unwinding activity, they can only cleave outside of the higher order structure, leaving it intact (15–18). All this, as well as the ability of the human shelterin complex components such as hsPot1 and hsRap1 to respectively disrupt and promote G quadruplexes, suggests that these structures interfere with telomere elongation and DNA replication by virtue of their resistance to nucleases and telomerase (19, 20). Resolution of such G quadruplex DNA structures may therefore be indispensable in preserving genome integrity.
As mentioned above, RecQ helicases (BLM, WRN, and Sgs1) are known to bind and unwind G quadruplex DNA (12, 14, 21). Because human BLM helicase can suppress Dna2 mutants, it is reasonable to hypothesize that both proteins similarly contribute to the resolution of secondary structures during replication. Here we show that Dna2 can recognize structures forming G4 DNA, bind to such structures, and unwind G4 DNA or cleave it in the presence of replication protein A (RPA). Remarkably, RPA seems to act as a specificity factor, directing Dna2 to 5′ ends for subsequent nuclease cleavage, while completely inhibiting the 3′ end cleavage. The functional significance of the nuclease and the helicase activities of Dna2 on G quadruplex DNA in OFP and telomere maintenance are discussed. Thus, in both OFP and telomere maintenance, Dna2 may be viewed as an accessory protein important in helping to prevent replication fork stalling by providing an unstructured DNA template for polymerase elongation and telomere lengthening or for processing by nucleases.
EXPERIMENTAL PROCEDURES
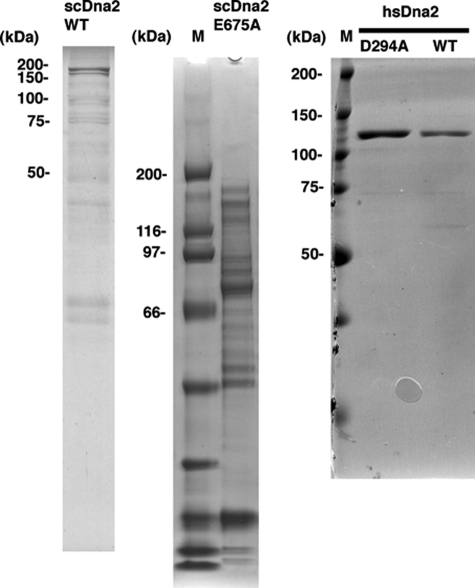
Recombinant Proteins—Recombinant yeast and human Dna2 were prepared as described previously. hsRPA was the gift of Marc Wold (University of Iowa, Iowa City). scRPA was prepared as described previously. The purity of the proteins is shown in Fig. 1.
FIGURE 1.
Protein preparations used in this study. Proteins were subjected to electrophoresis, and gels were stained with Coomassie Blue. M indicates molecular mass markers.
Oligonucleotides—The oligonucleotides used are as follows: scGQ1, 5′-CTA ACT TCA TAA TGA TGT GTG GGT GTG TGG GTG TGT GGG AGT AAT ACT TCA ATC-3′; OX1, 5′-ACT GTC GTA CTT GAT ATT TTG GGG TTT GGGG-3′; hsGQ, 5′-TTA GGG TTA GGG TTA GGG TTA GGG-3′; hsGQ-aT8, 5′-TTA GGG TTA GTG TTA GGG TTA GGG-3′; hsGQ1, 5′-CTA ACT TCA TAA TGA GGG TTA GGG TTA GGG TTA GGG ATT CAA TAA CGT ATA-3′; hsGQ1-aT8, 5′-CTA ACT TCA TAA TGA GGG TTA GTG TTA GGG TTA GGG ATT CAA TAA CGT ATA-3′; hsGQ2, 5′-CTA ACT TCA TAA TTA GGG TTA GGG TTA GGG TTA GGG-3′; hsGQ2-aT8, 5′-CTA ACT TCA TAA TTA GGG TTA GTG TTA GGG TTA GGG-3′; hsGQ3, 5′-CTA ACT TCA TAA TGA GGG TTA GGG TTA GGG TTA GGG ATT CAA TAA CGT ATA CAC CAT-3′; hsGQ3-aT8 5′-CTA ACT TCA TAA TGA GGG TTA GTG TTA GGG TTA GGG ATT CAA TAA CGT ATA CAC CAT-3′; hsGQ4, 5′-CTA CTC CAT TCG GCA TCA CAT ACT GCT AAT GAG GGT TAG GGT TAG GGT TAG GGA TTC AAT AAC GTA TA-3′; hsGQ5, 5′-CTA ACT TCA TAA TGA GGG TTA GGG TTA GGG TTA GGG ATT CAA CTA CTC CAT TCGGCA TCA CAT ACT GC-3′; C (complement to 5′ tail of hsGQ4 and 3′ tail of hsGQ5) 5′-GCA GTA TGT GAT GCC GAA TGG AGT AG-3′. Flap substrate (22) is as follows: D(55-mer), AGG TCT CGA CTA ACT CTA GTC GTT GTT CCA CCC GTC CAC CCG ACG CCA CCT CCT G; T(51-mer), GCA GGA GGT GGC GTC GGG TGG ACG GGA TTG AAA TTT AGG CTG GCA CGG TCG; U(26-mer), CGA CCG TGC CAG CCT AAA TTT CAA TA.
Preparation of G4 Substrates—All oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). To prepare yeast G4 DNA, scGQ1 DNA (3 μg/μl) was boiled in TE (10 mm Tris-HCl, 1 mm EDTA) for 4 min, and then NaCl was added to a final concentration of 1 m. The mixture was incubated at 37 °C for 4–7 days, and formation of intermolecular G4 was confirmed by native gel electrophoresis. The ciliate G4 DNA (OX1) (3 μg/μl) was prepared in 1 m KCl, 10 mm Tris-HCl, 1 mm EDTA buffer. Intermolecular G4 was 5′-radiolabeled with [γ-32P]ATP by T4 polynucleotide kinase according to manufacturer's instructions, except that the reaction mixture contained 5 mm KCl to stabilize the G quadruplex structure. For 3′ end labeling, 10 pmol of intermolecular G4 was incubated with terminal transferase (25 units; Roche Applied Science) in buffer containing 1× terminal transferase reaction buffer, 2.5 mm CoCl2, and 50 μCi [α-32P]ddATP (>3000 Ci/mmol). After labeling for 1 h at 37 °C, unincorporated nucleotides were removed by Micro Bio-Spin 30 chromatography column (Bio-Rad) containing 10 mm KCl in TE. Substrates were further purified by native gel electrophoresis (8% polyacrylamide, 37.5:1, 0.5× TBE, 10 mm KCl) at 4 °C, and gel slices containing bands corresponding to G4 DNA were excised. Substrates in the gel were eluted in TE in the presence of 50 mm NaCl (scGQ1) or in the presence of 50 mm KCl (OX1), purified by ethanol precipitation, and suspended in 50 mm NaCl in TE (scGQ1) or 50 mm KCl in TE (OX1). To prepare intramolecular hsG4, DNA oligonucleotides were 5′- or 3′-labeled as described above and purified by denaturing gel electrophoresis. Intramolecular hsG4 DNA substrates were diluted in 10 mm sodium cacodylate, pH 7.4, and 0.1 m KCl and heated at 95 °C for 5 min and immediately placed on ice to favor intramolecular folding. For hsGQ4 and hsGQ5, substrates were heated and slowly cooled in the presence of C (complementary strand) at a molar ratio of 1 to 4.
Snake Venom Phosphodiesterase I (SVPI) Nuclease Assay—SVPI nuclease (Worthington) was resuspended in stock solution (100 mm Tris-HCl, pH 8.9, 100 mm NaCl, 15 mm MgCl2) at 1 mg/ml. Reaction mixtures (20 μl) contained 50 mm Tris-HCl, pH 8.0, 50 mm KCl, 1 mm MgCl2, 15 mm NaCl, 5 mm 2-mercaptoethanol, 1 mm spermidine, 50 fmol of 5′ end-labeled DNA substrates, and indicated amounts of SVPI nuclease (75, 150, and 300 ng). Reactions were incubated for 5 min at 30 °C and stopped by addition of 2 μl of 100 mm EDTA and heat inactivation for 2 min. The products were resolved on a 12% polyacrylamide, 7 m urea denaturing gel.
DMS Protection Assay—Standard Maxam-Gilbert G reaction was performed (23) except 50 mm KCl or 50 mm NaCl was included in the DMS buffer (10 mm sodium cacodylate, pH 8, 1 mm EDTA). Reaction times were 5 and 10 min, as indicated in figure legend.
DNA Binding Assay—The G4 binding activity of recombinant Dna2 was measured using a gel shift assay. Recombinant Dna2 was incubated with G4 substrate in a reaction mixture containing 50 mm Tris-HCl, pH 8.0, 0.25 mg/ml BSA, 55 mm NaCl, 1 mm EDTA, 2 mm DTT, 10% glycerol. If necessary, enzymes were diluted to appropriate concentrations just prior to use in Dna2 dilution buffer containing 500 mm NaCl, 50 mm Tris-HCl, pH 8.0, 2 mm DTT, 0.5 mg/ml BSA, 10% glycerol (v/v), and 0.02% Nonidet P-40. After incubation on ice for 30 min, reaction mixtures were directly loaded onto 5% native polyacrylamide gel (0.5× TBE, 10 mm KCl) and separated at 4 °C for 90 min, and gels were analyzed using a Storm 860 PhosphorImager. For the competition experiments, labeled G4 substrate was mixed with the indicated amounts of unlabeled G4 DNA or boiled G4 DNA and incubated with recombinant Dna2. For quantitation of DNA binding, the intensity of bands corresponding to free DNA and Dna2-DNA complex was determined using ImageQuant software (GE Healthcare); free DNA (%) = free DNA/(free DNA +Dna2-DNA) × 100.
Nuclease Assay—Nuclease activity of Dna2 was measured using a reaction mixture (20 μl) containing 50 mm Tris-HCl, pH 7.5, 2 mm DTT, 0.25 mg/ml BSA, 50 mm NaCl, 100 mm KCl, 32P-labeled DNA substrate, and various concentrations of MgCl2 and ATP as indicated in the figure legends. The reaction was started by adding 1 μl of Dna2 protein or Dna2 dilution buffer. After incubation at 37 °C, reactions were stopped with 2× denaturing termination dye (95% deionized formamide, 10 mm EDTA, 0.1% bromphenol blue, and 0.1% xylene cyanol) and analyzed as described previously (22). When RPA was included, reaction mixtures were preincubated without Dna2 at 4 °C for 10 min to allow binding of RPA to the substrate. If necessary, RPA was diluted with 30 mm Hepes-KOH, pH 7.8, 1 mm DTT, 0.25 mm EDTA, 0.5% inositol, 0.01% Nonidet P-40, 300 mm KCl. A 32P-labeled 10-bp ladder was used as marker to indicate the length of cleaved product.
Helicase Assay—Helicase assays were performed with the nuclease-deficient mutant of hsDna2(D294A) and scDna2(E675A). The standard reaction mixtures contained 50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 2 mm DTT, 0.25 mg/ml BSA, 32P-labeled scGQ1 G4 DNA, and various concentrations of ATP, AMP-PNP, and MgCl2 as described in the legends. After incubation, reactions were stopped by the addition of an equal volume of 2× G4 loading buffer (20% glycerol, 20 mm EDTA, 0.2% SDS, 20 mm KCl). Reaction products were then separated using 8% native PAGE (0.1% SDS, 0.5× TBE, 10 mm KCl) and detected with a Storm PhosphorImager.
Affinity purification of scDna2(E675A). The scDna2(E675A) shown in Fig. 1 was further purified with anti-HA antibody (12CA5). 9 pmol of scDna2 was mixed with 19 μl of ascites and incubated at 4 °C. 30 μl of Protein G beads (GE Healthcare) was added after 1 h and incubated another 1 h. The matrix was then thoroughly washed with 1 mg/ml BSA in TBSG-Triton (25 mm Tris-HCl, pH 7.6, 0.1% Triton X-100, 150 mm NaCl, 10% glycerol), followed by washes with Dna2 dilution buffer. Proteins bound to beads were then eluted with 60 μl of 1 mg/ml HA peptide at 4°C for 1 h.
Preparation of Experimental Data as Computer Images—Images were edited using the levels command of Adobe Photoshop. Specifically, images were enhanced by dragging the black and white input levels sliders to the edge of the first group of pixels on either end of the histogram.
RESULTS
In Vitro Formation of Radiolabeled G-Quadruplex Structures—G-rich telomeric repeat sequences of eukaryotes from humans to yeast to protozoans are capable of interacting with monovalent cations to form both intramolecular and intermolecular G quadruplex structures in vitro. (The terminology used is as follows: G quadruplex, a four-stranded structure found in sequences rich in runs of guanines that is stabilized by formation and stacking of G quartets and can be formed from one, two, or four G-rich strands; G4 DNA, synonymous with G quadruplex; G quartet, square, planar arrangement of four guanines in a tetrad stabilized by Hoogsteen pairing and a monovalent cation.) We have used a yeast telomeric G-rich sequence that has been previously shown to form intermolecular, tetramolecular, parallel G4 DNA, Oxytrichia telomeric sequences that form bimolecular intermolecular G4 DNA, and human telomeric sequences that form unimolecular, intramolecular G4 DNAs (19, 24–26). These DNAs differ from those used previously only in that the G-rich repeats are flanked by 15 bp of random sequence. These tails were added because Dna2 nuclease tracks along single strands from the ends and blocking of the end inhibits nuclease and helicase activities (22, 27, 28). In our hands, repeats of the yeast G-rich telomere sequence flanked by 15 bp of random sequence assembled into tetrameric G4 structures after incubation in 1 m NaCl over several days at 37 °C, based on electrophoretic mobility shown below and the results of previous studies (18, 24). DMS footprinting analysis (see “Experimental Procedures”) was performed to confirm formation of G quartets in our substrates. Compared with guanines in single-stranded DNA, guanines in G4 DNA are relatively resistant to DMS modification and cleavage. As shown in Fig. 2, the guanine tracts, most prominently the central triplet in the yeast sequences used, scGQ1, are clearly protected from DMS cleavage in the presence of either Na+ or K+ ions. The protection of the two runs of four guanines in Oxytrichia DNA is even more evident than in the yeast sequences, most likely due to the enhanced stability of a four-residue guanine tract. The DMS protection analysis establishes the existence of G quartets in both yeast and Oxytrichia sequences used in our work.
FIGURE 2.
Substrates for G4 DNA formation and DMS protection analysis. DNA substrates were used for intermolecular G4 formation; brackets indicate guanine tracts that are thought to be involved in G4 formation. DMS footprint analysis of 32P end-labeled single-stranded (ss) or G4 DNA (G4) structures formed from oligonucleotides scGQ1 and OX1. Substrates, as indicated in the figure, were treated with DMS for 5 or 10 min before piperidine cleavage. Salts added to stabilize G4 structure are also indicated. Brackets denote the region of protection from DMS treatment.
Binding of Yeast Dna2 to Yeast and Oxytrichia G4 DNA—The protein preparations used in this study are shown in Fig. 1.
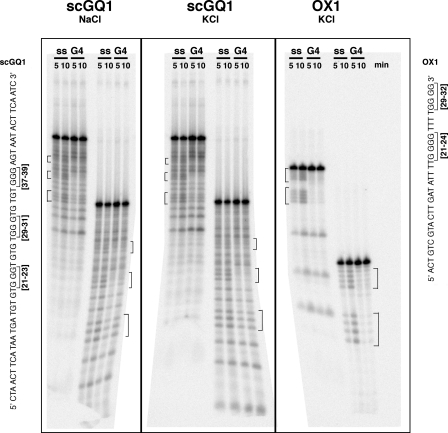
Gel shift assays indicated that scDna2 bound with high affinity to the yeast G4 DNA and bound G4 DNA preferentially over the single-stranded DNA of the same sequence or a 5′ flap structure, a preferred substrate for scDna2 nuclease activity (Fig. 3A). Even at the lowest protein level tested (3.75 fmol; Fig. 3A, lane 7), nearly all of the free G4 DNA formed a complex with Dna2. By comparison, single-stranded DNA and the 5′ flap failed to show Dna2 binding at even the highest protein level tested (30 fmol; Fig. 3A, lanes 5 and 15). To more directly compare relative binding affinities, scDna2 was incubated with radiolabeled G4 DNA in the presence of competing unlabeled G4 or single-stranded DNA of the same sequence. As shown in Fig. 3B, at a 600-fold excess of G4 DNA (lane 8), only a small fraction of labeled G4 DNA-Dna2 protein complex was observed, whereas even a 6000-fold excess of single-stranded DNA failed to titrate scDna2 away from forming complexes with G4 DNA (Fig. 3B, lane 14). Taking into account that a single G4 structure may consist of four individual G-rich strands, and binding of Dna2 to one molecule may cause a shift of the G4 DNA, we conclude that Dna2 has at least a 25-fold higher affinity for G4-containing DNA than for ssDNA of the same sequence. These results strongly suggest that binding of scDna2 to G4 DNA is structure-specific.
FIGURE 3.
scDna2 displays higher affinity for yeast G4 DNA over single-stranded DNA. A, direct binding assays. Reaction mixtures contained 15 fmol of scGQ1 single-stranded DNA (lanes 1–5, ss), scGQ1 G4 DNA (lanes 6–10, G4), and flap (lanes 11–15, FLAP) substrates and were incubated with scDna2 at 0, 3.75, 7.5, 15, and 30 fmol (lanes 1–5, 6–10, and 11–15). Formation of the protein-DNA complex was analyzed by gel shift assay as described under”Experimental Procedures.“DNA substrates used in the assay are shown at the top of the figure (star, 32P-labeled ends). Single-stranded substrate was prepared by boiling the G4 DNA just before use. B, competition binding assays show scDna2 displays higher affinity for yeast G4 DNA than for single-stranded DNA. Binding of scDna2 (15 fmol) to scGQ1 G4 (2.35 fmol) was assayed in the presence of indicated amounts of unlabeled scGQ1 G4 DNA (lanes 1–2 and 5–9) or scGQ1 single-stranded DNA (lanes 3–4 and 10–14) as competitors. Lanes 1–4 show”no protein“control. Formation of protein-DNA complex was analyzed by gel shift assay as described under”Experimental Procedures.“Oligonucleotide used in the assay is shown at the top of the figure (star, 32P-labeled ends). Single-stranded substrate was prepared by boiling the G4 substrate just before use. C, scDna2 displays higher affinity for Oxytrichia G4 DNA over single-stranded DNA. Reaction mixtures contained 15 fmol of OX1 G4 (lanes 1–7, G4), and single-stranded (lanes 8–14, ss) substrates and were incubated with scDna2 at 0, 5, 15, 45, 100, 300, and 600 fmol. Formation of protein-DNA complex was analyzed by gel shift assay as under”Experimental Procedures“and in the legend to Fig. 3A. D, competition assay. scDna2 displays higher affinity for Oxytrichia G4 DNA over single-stranded DNA. Binding of scDna2 (100 fmol) to OX1 intermolecular G4 (15 fmol) was assayed in the presence of indicated amounts of unlabeled OX1 G4 (lanes 1–2 and 5–9) or OX-1 single-stranded (lanes 3–4 and 10–14) as competitors. Lanes 1–4 show no protein control. Formation of protein-DNA complex was analyzed by gel shift assay as under”Experimental Procedures“and in the legend to Fig. 3A. Free DNA (%) = free DNA/(free DNA +Dna2-DNA) × 100.
We also tested an Oxytrichia G4 DNA, using the oligonucleotide containing two repeats of Oxytrichia telomere sequence flanked on the 5′ end by single-stranded DNA of random sequence (see OX1, Fig. 2). Judging by electrophoretic mobility, Fig. 3C, and previous studies of the Oxytrichia structure, the predominant Oxytrichia G4 species is bimolecular (25, 26). As in the case of the yeast sequences, scDna2 seems to have higher affinity for the Oxytrichia G4 DNA versus single-stranded DNA; however, the preference for the G4 DNA is less pronounced than for the yeast G4 construct (Fig. 3C). The Oxytrichia G4 is only marginally more efficient (∼5-fold) than linear single-stranded forms of the same sequence in competing for scDna2 binding (Fig. 3D). About 10 times more scDna2 protein is required to fully bind Oxytrichia than the yeast DNA (compare Fig. 3, A and C). This suggests that scDna2 binds structure specifically, with higher affinity to tetrameric G4 DNA than to bimolecular hairpin forms, although further work would be required to establish precisely the structures of these substrates. This might, in turn, suggest that scDna2 has higher affinity for yeast than for Oxytrichia telomeric DNA.
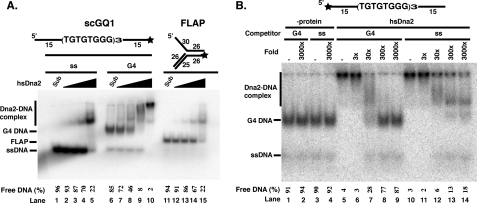
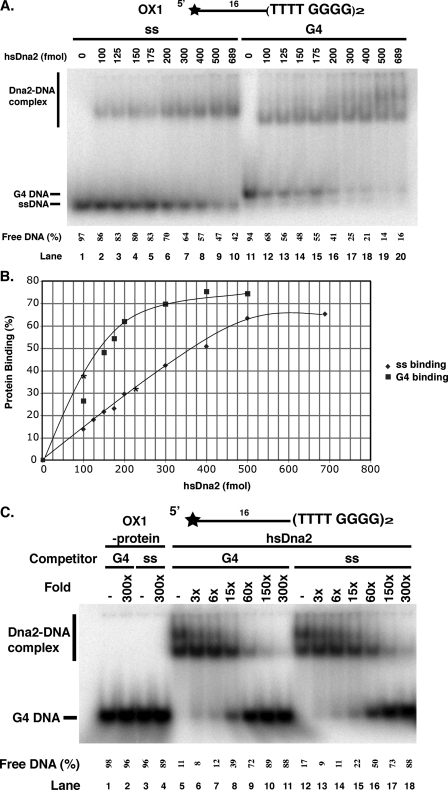
Human Dna2 Binds to G4 DNA but with Lower Differential Affinity for Single-stranded Versus G4 DNA than scDna2—We next tested the binding of hsDna2 to G4 DNA. As shown in Fig. 4A, hsDna2 binds to yeast G4 DNA. It is notable that, in contrast to the yeast protein, hsDna2 gives rise to multiple retarded bands, indicating the formation of multiple complexes containing different molar ratios of protein to DNA (Fig. 4A, lanes 7–10). The most rapidly migrating complexes in the gel retardation experiment most likely arise from 1:1 binding of hsDna2 and G4 DNA, whereas the lower mobility complexes are likely the result of multiple molecules of Dna2 binding. Comparison of free DNA remaining in Fig. 4A, lanes 4 and 5 (single-stranded DNA) or lanes 14 and 15 (flap DNA) with that remaining in lanes 9 and 10 (G4 DNA), shows a quantitative preference for G4 structure versus single-stranded DNA, although preference for G4 is not as strong as in scDna2. To further investigate whether hsDna2 has higher affinity for G4 DNA than for single-stranded DNA, we performed competition experiments. As shown in Fig. 4B (lane 8), 300-fold excess of G4 DNA can almost completely compete the binding of hsDna2 to the G4 DNA. Both the slowly and rapidly migrating Dna2-G4 DNA complexes are competed by the G4 DNA. In contrast, single-stranded competitor cannot completely compete with binding of labeled G4 to hsDna2, even at 3000-fold excess, confirming that hsDna2 does have a different affinity for G4 and single-stranded DNA (Fig. 4B, lanes 10–14). Furthermore, the single-stranded DNA competitor affects the various hsDna2-G4 DNA complexes differently. The lower mobility hsDna2/G4 DNA bands disappeared at a relatively low molar excess of cold single-stranded DNA (Fig. 4B, lane 12, 30-fold), consistent with these species representing Dna2 bound to the single-stranded tails of the G4 DNA molecule. However, the faster moving complex is still detectable when even a 3000-fold molar excess of single-stranded DNA is added (Fig. 4B, lane 14, and compare lanes 8 and 9).
FIGURE 4.
hsDna2 displays slightly higher affinity for yeast G4 DNA than single-stranded DNA. A, direct binding assay. Reaction mixtures contained 15 fmol of scGQ1 single-stranded DNA (lanes 1–5, ss) and scGQ1 intermolecular G4 DNA (lanes 6–10, G4) and flap DNA (lanes 11–15, FLAP) substrates, and were incubated with hsDna2 at 0, 5, 25, 100, and 690 fmol (lanes 1–5, 6–10, and 11–15). Formation of protein-DNA complex was analyzed by gel shift assay as under”Experimental Procedures“and in the legend to Fig. 3A. Free DNA (%) = free DNA/(free DNA +Dna2-DNA) × 100. B, competition assay shows hsDna2 displays higher affinity for yeast G4 DNA over single-stranded DNA. Binding of hsDna2 (689 fmol) to scGQ1 intermolecular G4 (15 fmol) was assayed in the presence of indicated amounts of unlabeled scGQ1 G4 (lanes 1–2 and 5–9) or scGQ-1 single-stranded (lanes 3–4 and 10–14) as competitors. Lanes 1–4 show no protein control. Amounts of competitor are indicated, and free DNA (%) = free DNA/(free DNA +Dna2-DNA) × 100.
We also investigated the interaction between Oxytrichia G4 DNA and hsDna2. hsDna2 showed a 2-fold difference in equilibrium dissociation constant, KD, between single-stranded (KD ∼12 nm) and G4 DNA complexes (KD ∼ 6 nm), as determined by the hsDna2 concentration that bound 50% of the respective DNA substrate (Fig. 5, A and B). Because Oxytrichia G4 is suggested to be a dimer, the 2-fold difference may not be significant. To clarify this point, we performed competition experiments using single-stranded and Oxytrichia G4 DNA as competitors (Fig. 5C). 60-Fold excess G4 competitor inhibits 70% of the G4/hsDna2 interaction, but single-stranded competitor showed only 50% inhibition. This difference in affinity is much lower than the difference found for binding yeast G4 versus yeast single-stranded DNA (Fig. 5C, lanes 9 and 16, compare with Fig. 4, A or B). We conclude that human Dna2 protein exhibited lower specificity for Oxytrichia G4 DNA versus Oxytrichia single-stranded DNA than did yeast Dna2 (compare Fig. 5C and Fig. 3, C or D). Interestingly, multiple complexes were observed between Oxytrichia G4 DNA and both hsDna2 and scDna2 proteins.
FIGURE 5.
hsDna2 binds Oxytrichia G4 DNA, but with only slight preference over single-stranded DNA. A, direct binding. Binding of the indicated amounts of hsDna2 to OX1 single-stranded DNA (15 fmol, lanes 1–10) or OX1 DNA G4 (15 fmol, lanes 11–20) was assayed by gel shift assay as described under”Experimental Procedures.“ B, quantification of the results of six experiments like those in Fig. 5A; stars represent protein level where half of DNA is bound. C, competition binding. Binding of hsDna2 (689 fmol) to OX1 intermolecular G4 (15 fmol) was assayed in the presence of the indicated amounts of unlabeled OX1 G4 (lanes 1–2 and 5–11) or OX-1 single-stranded (lanes 3–4 and 12–18) as competitors. Lanes 1–4 show no protein control. Amounts of competitor are indicated, and the % free DNA as determined by ImageQuant is shown below the lanes.
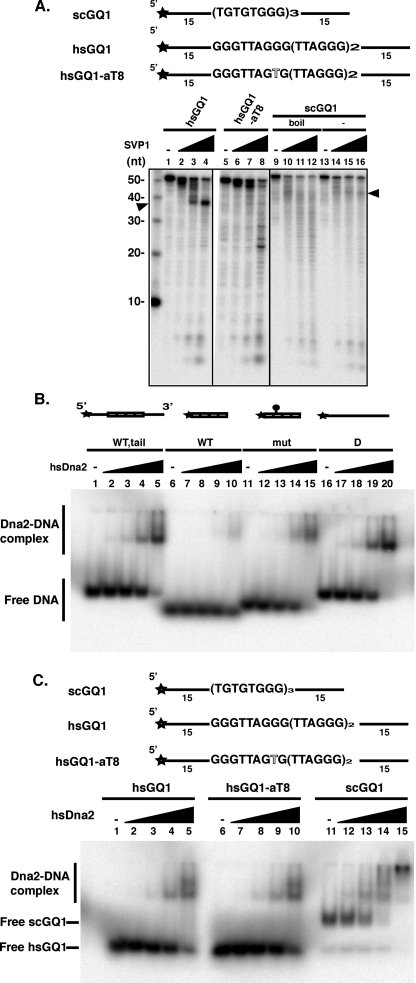
Binding of hsDna2 to Human G4-forming DNA Oligonucleotides—Because hsDna2 showed only marginally higher affinity for the yeast and Oxytrichia G4 putative telomeric structures versus single-stranded DNA, we next asked if hsDna2 showed a greater affinity for human G4 DNA than for single-stranded DNA of the same sequence. Oligonucleotides composed of human telomeric sequence repeats readily form an intramolecular G4 DNA, with a propensity toward antiparallel chair-type topologies in the presence of K+ and toward antiparallel basket-type conformations in the presence of Na+ (29–31). Because the tailed structures we used showed indistinguishable electrophoretic mobility from single-stranded DNA in neutral polyacrylamide gels (not shown), formation of G4 from the human sequence was confirmed by treatment with SVPI, a 3′-exonuclease unable to efficiently cleave DNA past a single-stranded/G4 DNA junction (19). As shown in Fig. 6A, SVPI removes the 15-bp 3′ tail from the hsGQ1 G4 DNA but is inhibited by the guanine repeat tracts (Fig. 6A, lanes 1–4). In contrast, a base pair alteration at position 8 (G to A) of hsGQ1, which disrupts the hydrogen bonding of the G quartets, shows a reduced pause at 39 nt and allows SVPI to cut all the way through the G-rich sequence (Fig. 6A, lanes 5–8). The yeast G4 substrate described above was also resistant to SVPI at the position of the single-stranded/G-rich junction (Fig. 6A, lanes 13–16) compared with the same sequence in unfolded, single-stranded form (Fig. 6A, lanes 9–12). Although the inhibition is less pronounced than in hsGQ1, this experiment also provides further evidence for the existence of G4 DNA in the yeast sequence (see also Fig. 2).
FIGURE 6.
Characterization of G4 DNA formed by hsGQ1, mutant hsGQ1-aT8, and scGQ1 determined by sensitivity to SVPI. A, formation of G4 DNA is inhibitory to SVPI nuclease. 50 fmol of 5′ end-labeled substrates of human wild type (hsGQ1) and mutant (hsGQ1-aT8) telomeric substrates were treated with SVPI nuclease as described under”Experimental Procedures.“Nuclease products were analyzed by denaturing gel electrophoresis. Substrates were preincubated in the absence (lanes 1 and 5) or the presence of increasing amounts of SVPI nuclease as follows: 75 ng (lanes 2 and 6), 150 ng (lanes 3 and 7), and 300 ng (lanes 4 and 8) of G4 DNA was formed from the hsGQ1 oligonucleotides by boiling and slow cooling. hsGQ1-aT8 contains a G4-disrupting mutation in the second G-tract. Wedges represent increasing amounts of SVPI nuclease; – represents no enzyme addition. Arrowhead points to the band generated by removal of the 15-nucleotide 3′ tail by SVPI and the block at the junction with G4 DNA. Left lane shows length markers (nt). Lanes 9–16, scGQ1 G4 DNA was formed as described under”Experimental Procedures.“Single-stranded scGQ1 was prepared by boiling the G4 DNA. Lanes 13–16 show pausing of SVPI as the nuclease encounters the G4 DNA from the 3′ direction. Lanes 9–12 show cutting in the G-rich sequences (bands less than 40 nt in length). B, hsDna2 needs single-stranded region for efficient binding to hsGQ1 G4 DNA. Reaction mixtures contained 15 fmol of hsGQ1 wild type with tail (lanes 1–5), hsGQ wild type without tail (lanes 6–10), hsGQ mutant without tail (lanes 11–15), and single-stranded DNA (lanes 16–20) substrates and were incubated with hsDna2 at 0, 5, 25, 100, and 690 fmol (represented as triangles above lanes 1–5, 6–10, 11–15, and 16–20). Formation of protein-DNA complex was analyzed by gel shift assay. C, comparison of hsDna2 binding to yeast (intermolecular) and human (intramolecular) G4. Reaction mixtures contained 15 fmol of hsGQ1 wild type (lanes 1–5), hsGQ1-aT8 mutant (lanes 6–10, G4), and scGQ1 (lanes 11–15) substrates and were incubated with hsDna2 at 0, 5, 25, 100, and 690 fmol (represented as triangles above lanes 1–5, 6–10, and 11–15). Formation of protein-DNA complex was analyzed by gel shift assay.
Gel shift assays indicate that the presumed intramolecular G4 DNA containing human telomeric repeats flanked by single-stranded 5′ and 3′ mixed flanking sequences are bound by hsDna2 (Fig. 6B, lanes 1–5). G4 DNA lacking 5′ and 3′ mixed flanking sequences serve as poor ligands for hsDna2 binding (Fig. 6B, lanes 6–10). At the highest concentration of hsDna2 tested, less than 50% of tailless G4 DNA formed a complex with the protein (Fig. 6B, lane 10). The G4-disrupting mutation increased hsDna2 binding affinity to the tailless G4 DNA (Fig. 6B, lanes 11–15) almost to the levels seen with random sequence single-stranded DNA (Fig. 6B, lanes 16–20). Fig. 6C, like Fig. 6A, shows that hsDna2 binds hsGQ1 G4 DNA, but that a single nucleotide, G4-disrupting alteration in one of the telomeric repeats, does not seem to affect binding by hsDna2 to the hsGQ1 (Fig. 6C, compare lanes 1–5 with 6–10); thus affinity for single-stranded DNA seems to dominate hsDna2 binding to these molecules rather than the existence of G4 DNA. Binding of hsDna2 to yeast G4 DNA is shown here as a positive control. We conclude that hsDna2 does not bind these human G4 sequences with significantly higher preference than it binds DNA of random sequence. In general, we conclude that, whereas hsDna2 does bind to G4 DNA, it does not discriminate as strongly between single-stranded DNA and G4 DNA as does scDna2.
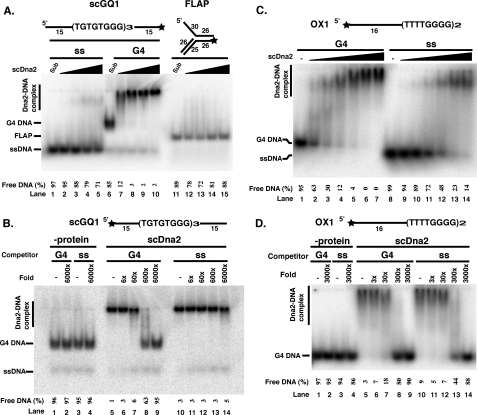
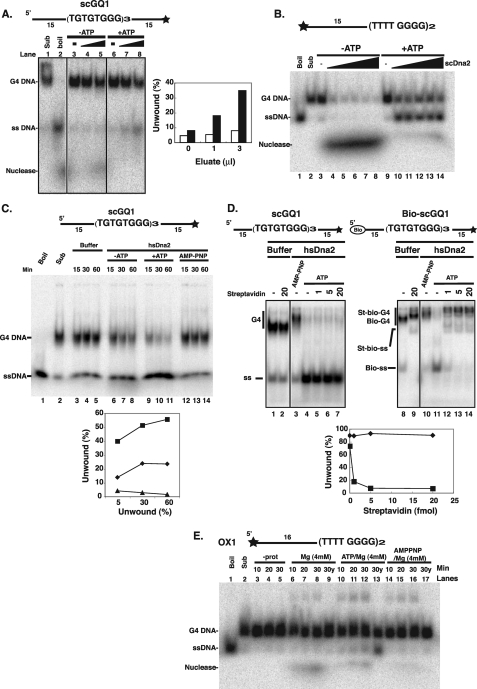
scDna2 Unwinds G4 DNA—We next asked if any of the enzymatic functions of Dna2 are active on the G4 DNA substrates and, if so, whether the activities are conserved. Because loss of Dna2 function can be partially rescued by expression of the human Bloom syndrome RecQ helicase known to resolve intermolecular G4 structures, it seemed possible that Dna2 helicase activity might also carry out the unusual reaction of unwinding G4 DNA (Fig. 7). We first used the yeast G4 DNA substrate that binds to scDna2 with high affinity and specificity. Both yeast and human Dna2 have potent endo- and exonuclease activities and a relatively weak ATP-dependent helicase activity whose single-stranded unwinding products are obscured from detection by the ensuing cleavage activity of its nuclease once ATP has been depleted (refer to Fig. 7B). We therefore used scDna2 containing a point mutation (E675A) in the nuclease domain that significantly attenuated its cleavage activity. We were able to detect ATP-dependent unwinding of yeast G4 DNA by scDna2 (Fig. 7A). Unwinding of Oxytrichia G4 DNA by scDna2 was also observed (Fig. 7B). In the case of the Oxytricha G4 DNA, only 50% of the input DNA was unwound, even at the highest protein concentration (Fig. 7B, lane 14 and under “Discussion”). We also note that the Oxytrichia G4 DNA was degraded by Dna2 in the absence of ATP and do not know if this is due to Dna2 or to a contaminating nuclease. hsDna2 Also Unwinds Yeast G4 DNA—Although we did not detect strong preferential binding of hsDna2 to G4 DNA in experiments described above, we wished to see if, like scDna2, hsDna2 could unwind G4 DNA. Helicase activity was indeed observed when helicase assays were repeated with nuclease-defective human Dna2(D294A). As shown in Fig. 7C, human Dna2 unwinds yeast G4 DNA in an ATP-dependent manner, and even though some spontaneous unwinding can be observed in the absence of protein or the absence of ATP, the stimulatory effect of ATP is clearly visible at shorter incubation times (Fig. 7C, compare lanes 6 and 9, for instance). To increase confidence that the observed unwinding is because of the intrinsic helicase activity of hsDna2 and not a contaminating activity of another protein, we prepared G4 substrates with biotinylated 5′ ends. Previous work has shown that streptavidin binding to the 5′-biotinylated ssDNA arm of a forked substrate inhibits unwinding by hsDna2, an unusual specificity for DNA helicases, which generally bind any single-stranded DNA (22). Suggesting that hsDna2 is doing the unwinding, hsDna2 appeared to bind to biotin-free G4 DNA substrate (Fig. 7D, lane 3, slightly retarded band), and in the presence of ATP was able to unwind it in the presence of streptavidin (Fig. 7D, lane 4–7). Streptavidin, however, inhibited unwinding of the 5′-biotinylated G4 DNA (Fig. 7D). These data suggest that Dna2 may potentially load onto G4 structures with a threading and tracking mechanism similar to its mode of action in Okazaki fragment processing. They also say that Dna2 cannot unwind G4 DNA translocating on a free 3′ tail, similar to its specificity for unwinding normal B DNA.
FIGURE 7.
Unwinding of G4 DNA by scDna2 and by hsDna2. A, unwinding of yeast telomeric G4 DNA by scDna2. 3′ end-labeled scGQ1 intermolecular G4 (5 fmol) was incubated with scDna2(E675A) purified by an additional HA immunoprecipitation as described under”Experimental Procedures“in the absence (lanes 3–5) and presence (lanes 6–8) of 2 mm ATP for 1 h at 37°C, and helicase products were analyzed by native gel electrophoresis. scDna2 was omitted from the reactions shown in lanes 3 and 6. Positions of scGQ1 G4 (G4 DNA), scGQ1 single-stranded (ss DNA), and nuclease products of scGQ1 (Nuclease) are indicated on the left. Lanes 4 and 7 contain 1 μl of protein; lanes 5 and 8 contain 3 μl of protein. Data from lanes 3 to 8 are quantified in the histogram. B, unwinding of Oxytrichia telomeric G4 DNA by scDna2. 5′ end-labeled OX1 intermolecular G4 was incubated with 0, 3, 6, 12, 23, and 58 fmol of scDna2(E675A) (lanes 3–8 and 9–14) in the absence (lanes 3–8) and presence (lanes 9–14) of 4 mm ATP for 45 min at 30 °C, and helicase products were analyzed by native gel electrophoresis. Substrate only (Sub) and boiled substrate only (Boil) are also shown. Positions of OX1 G4 (G4 DNA), OX1 single-stranded (ss DNA), and nuclease products of OX1 (Nuclease) are indicated on the left. C, unwinding of yeast G4 DNA by recombinant hsDna2 protein. 3′ end-labeled scGQ1 intermolecular G4 (star, 32P-labeled end) was incubated with nuclease-defective hsDna2(D294A) in the presence of 4 mm ATP (lanes 9–11, +ATP) or nonhydrolyzable AMP-PNP (lanes 12–14, AMP-PNP) as indicated. As a control, ATP was omitted from the reaction (lanes 6–8,–ATP). hsDna2 was omitted from the reaction mixture in lanes 3–5, Buffer. Reactions were stopped at indicated time points by adding 2× stop solution. Products were then separated using native gel electrophoresis and detected by autoradiography as described under”Experimental Procedures.“ Sub (lane 2) and Boil (lane 1) denote the position of G4 substrate and single-stranded forms, respectively. Positions of G4 substrate (G4 DNA) and helicase products (ssDNA) are as indicated on the left. Data are quantified on the graph. D, unwinding of G4 DNA by hsDna2 needs free 5′ end. 3′ end-labeled scGQ1 intermolecular G4 (scGQ1, 5 fmol, lanes 1–7) or 5′ biotin modified scGQ1 (Bio-scGQ1, 5 fmol, lanes 8–14) were preincubated with indicated amounts (fmol) of streptavidin for 15 min at 4 °C. Biotinylated oligonucleotides were prepared as described previously (22). After incubation, substrates were incubated with hsDna2(D294A) for 1 h at 37 °C, and products were analyzed by native gel electrophoresis. Reaction mixtures contained 4 mm ATP (lanes 4–7 and 11–14) or 4 mm AMP-PNP (lanes 3 and 10). hsDna2p was omitted from the reaction mixture in lanes 1–2 and 8–9. Positions of scGQ1 G4 (G4), scGQ1 single-stranded (ss), Bio-scGQ1 G4 (Bio-G4), Bio-scGQ1 single-stranded (Bio-ss), bio-scGQ1 G4 bound to streptavidin (St-bio-G4), and bio-scGQ1 single-stranded bound to streptavidin (St-bio-ss) are indicated. Data are quantified on the graph. E, unwinding of Oxytrichia telomeric G4 DNA by scDna2 and hsDna2. 5′ end-labeled OX1 intermolecular G4 was incubated with HA-purified scDna2(E675A) (lanes 9, 13, and 17) or hsDna2(D294A) (1000 fmol, lanes 6–8, 10–12, and 14–16) in the presence and absence of Mg2+ (4 mm), ATP (4 mm), and AMP-PNP (4 mm) as indicated in the figure. After incubation at 30 °C, reactions were stopped at indicated time points, and helicase products were analyzed by native gel electrophoresis. Enzyme was omitted from the reaction in lanes 3–5. G4 substrate, without incubation, is shown in lane 2 (Sub) and single-stranded OX1 prepared by boiling the G4 form is shown in lane 1 (Boil). Positions of OX1 G4 (G4 DNA), OX1 single-stranded (ssDNA), and nuclease products of OX1 (Nuclease) are indicated on the left.
Oxytrichia—G4 DNA was also a substrate for the helicase activity of hsDna2, but some of the unwinding products were then subjected to cleavage by the Dna2 nuclease (Fig. 7E). A parallel assay with scDna2 suggests that scDna2 is more active on this substrate than is hsDna2 (Fig. 7E, lane 13), which is also true for helicase activity on B DNA (22, 32).
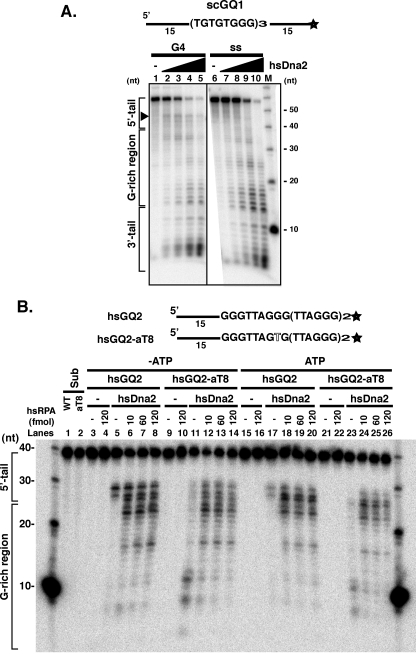
The Nuclease Activity of Dna2 Is Inhibited by G4 DNA Unless Assisted by RPA—G4 DNA is resistant to many nucleases, just as the putative intramolecular human sequence G4 DNA is resistant to cleavage by SVPI. Therefore, it was of interest to test whether Dna2 nuclease activity could act upon G4 DNA structures and whether the nuclease and helicase could act in concert in the presence of ATP. As shown in Fig. 8A, hsDna2 nuclease appears to pause upon encountering a single strand/G4 DNA junction in the yeast G4 substrate (Fig. 8A, lanes 1–5, G4 DNA region is marked by a line at the left and pause is visible as a band at about 44 nt; small products are due to 3′-nuclease, as shown below). hsDna2 nuclease shows a much less pronounced, if any, pause in the single-stranded yeast DNA and cleaves through the G-rich DNA region yielding primarily short products (Fig. 8A, compare lanes 5 and 10). The inhibition of Dna2 in this yeast sequence was not complete, and therefore we investigated additional G4 substrates.
FIGURE 8.
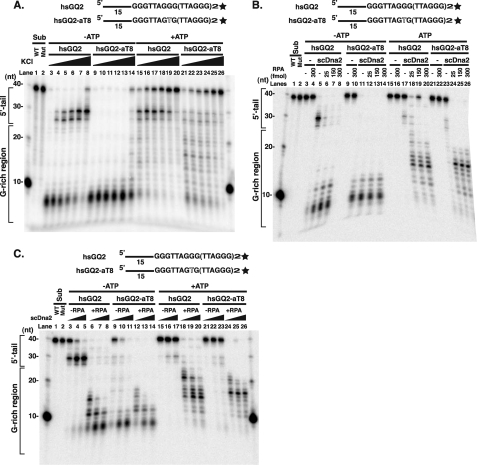
hsRPA stimulates hsDna2 on G4 DNA substrates. A, cleavage of intermolecular yeast telomeric G4 by hsDna2. 3′ end-labeled scGQ1 intermolecular G4 (15 fmol, G4) and single-stranded scGQ1 (15 fmol, ss) were incubated with 0, 3, 15, 60, and 300 fmol of hsDna2 (lanes 1–5 and 6–10) in the presence 4 mm ATP for 5 min at 37 °C, and nuclease products were analyzed by denaturing gel electrophoresis. The reaction mixture (20 μl) contained 50 mm Tris-HCl, pH 7.5, 2 mm DTT, 0.25 mg/ml BSA, 50 mm NaCl, 4 mm MgCl2, 4 mm ATP, and 32P-labeled DNA. Reactions were started by adding Dna2 protein or Dna2 dilution buffer. M = markers. B, hsRPA stimulates hsDna2 nuclease against intramolecular human telomeric G4. 3′ end-labeled hsGQ2 intramolecular G4 wild type (5 fmol) and hsGQ2-aT8 mutant (5 fmol) were preincubated on ice with indicated amounts of RPA in the absence (lanes 3–14) and presence (lanes 15–26) of 4 mm ATP. Nuclease reactions were started by adding hsDna2 (100 fmol), and reaction mixtures were incubated for 60 min at 37 °C. Nuclease products were then analyzed by high resolution denaturing gel electrophoresis. hsDna2 was omitted from the reaction in lanes 3–4, 9–10, 15–16, and 21–22. hsGQ2 (lane 1) and hsGQ2-aT8 (lane 2), without incubation, are also shown.
When human telomeric DNA (hsGQ2) was used, hsDna2 removed most of the 5′ single-stranded tail but showed a more distinct pause near the 5′ tail/G4 DNA junction than was observed in the yeast G4 (Fig. 7B, lane 5). There are no short products that might arise from cleavage by the 3′-nuclease activity. The block to 5′-nuclease activity was not overcome by the addition of ATP (Fig. 8B, lanes 5 and 17). However, if a single nucleotide change is introduced into the second G run, the propensity of the oligonucleotide to form G4 structures is disrupted (hsGQ2-aT8), and hsDna2 is able to cleave the substrates within the guanine tracts (Fig. 8B, lanes 11 and 23). Pausing at the position of the 5′ single-strand/G4 DNA junction in the presence of ATP suggests that Dna2 is incapable of coupling its helicase and nuclease activities to process such structures under these conditions.
To determine whether Dna2 activity on G4 DNA could be stimulated by RPA, we tested the effect of scRPA on scDna2 and of hsRPA on hsDna2. Addition of RPA into the reaction mixture strongly stimulated hsDna2 5′-nuclease activity on human G4 DNA (Fig. 8B, lanes 5–8 and lanes 17–20). Even at the lowest levels of RPA tested, stimulation was saturating. Products derived from hsGQ2 cleavage in the G-rich region are clearly visible and form the same spectrum as that for hsGQ2-aT8 in the presence of RPA (Fig. 8B, compare lanes 6–8 with 12–14). Interestingly, but unexpectedly, when RPA is added to hsGQ2-aT8, a single-stranded form, the pattern of product lengths is different from cleavage in the absence of RPA, revealing the predominance of much longer products (Fig. 8B, compare lane 11 with lanes 12–14 or lane 23 with lanes 24–26). We will show below that the difference in the cleavage of single-stranded DNA hsGQ2-aT8 in the absence and presence of RPA likely arises because cleavage of the hsGQ2-aT8 is occurring from both the 5′ and 3′ ends, and cleavage from the 3′ end is inhibited by RPA.
Similar results were obtained with yeast Dna2. Fig. 9A shows that scDna2 nuclease removes the 5′ single-stranded tails on the human G4 DNA substrate but is inhibited when it encounters the human G4 DNA. Increasing the stability of the G4 DNA by increasing the K+ concentration further inhibits (Fig. 9A, lanes 1–8 and 15–20). However, Dna2 cleaves throughout both the random sequence tail and the G-rich DNA, even with increasing KCl concentration, when the G4 structure formation is disrupted in the hsGQ2-aT8 mutant sequence (Fig. 9A, lanes 9–14 and 21–26). As with hsDna2, scRPA overcomes the inhibition of cutting of the hGQ2 G4 DNA, and in its presence scDna2 cleaves the G-rich DNA in a dose-dependent manner (Fig. 9B, lanes 5–8 and 17–20). The intermediates arising from cuts in the G-rich DNA are best seen in Fig. 9B, lanes 17–20, where ATP inhibits overall nuclease activity. The pattern of cleavage in the presence of RPA resembles the cleavage pattern obtained on the substrate containing the G4-disruption mutation, hsGQ2-aT8 (Fig. 9B). Fig. 9C shows a similar experiment, except that the levels of RPA are held constant, and the levels of Dna2 are varied. This experiment shows that even low levels of scDna2 are stimulated to cleave at the single strand/G-rich DNA junction by scRPA, and give the same products as observed on the substrate with G4-disrupting point mutation in the DNA (Fig. 9C). RPA may unwind the G4 DNA, exposing it to cleavage by Dna2, or RPA may stimulate Dna2 as it does on random sequence DNA.
FIGURE 9.
scRPA stimulates scDna2 cleavage of G4 DNA. A, cleavage of intramolecular human telomeric G4 by scDna2. 3′ end-labeled hsGQ2 intramolecular G4 wild type hsGQ2 (15 fmol) and mutant hsGQ2-aT8 (30 fmol) were incubated with scDna2 (100 fmol) at KCl concentrations of 5, 15, 25, 35, 55, 105 mm (lanes 3–8, 9–14, 15–20, and 21–26) in the absence (lanes 3–14) and presence (lanes 15–26) of 2 mm ATP for 15 min at 37 °C, and nuclease products were analyzed by denaturing gel electrophoresis. Wild type hsGQ2 (lane 1) and mutant hsGQ2-aT8 (lane 2) substrate, without incubation, are also shown. Numbers shown on the left of the figure indicate the size of the markers. B, scRPA titrations show stimulation of scDna2 nuclease against intramolecular human telomeric G4. 3′ end-labeled hsGQ2 intramolecular G4 (5 fmol) and mutant hsGQ-aT8 (5 fmol) were preincubated on ice with indicated amounts of RPA in the absence (lanes 3–14) and presence (lanes 15–26) of 2 mm ATP. The nuclease reaction was started by adding scDna2 (100 fmol), and the reaction mixture was kept at 37 °C for 60 min. Nuclease products were then analyzed by denaturing gel electrophoresis. As a control, yDna2 was omitted from the reaction (lanes 3–4, 9–10, 15–16, and 21–22). Wild type (lane 1, WT) and mutant (lane 2, Mut) substrate were also loaded. C, scDna2 titration shows scRPA stimulates scDna2 nuclease against intramolecular human telomeric G4. 3′ end-labeled hsGQ2 intramolecular G4 wild type (5 fmol) and mutant hsGQ2-aT8 (5 fmol) were preincubated on ice in the absence (lanes 3–5, 9–11, 15–17, and 21–23) and presence (150 fmol, lanes 6–8, 12–14, 18–20, and 24–26) of scRPA. Increasing amounts of scDna2 (10, 50, and 100 fmol, as indicated by triangles) were then added to start the reaction, and the reaction mixture was incubated at 37 °C for 60 min. Nuclease products were analyzed by denaturing gel electrophoresis. hsGQ2 (lane 1) and hsGQ2-aT8 (lane 2) substrates, without incubation, are also shown.
RPA Directs Yeast and Human Dna2 to Cleave at the 5′ End and Inhibits 3′ End Cleavage—In further investigating how RPA affected Dna2 cleavage of these substrates, we gained insight into one of the most poorly understood aspects of Dna2 enzymology, the general mechanism of stimulation of Dna2 by RPA. To investigate the effect of RPA on hsDna2 cleavage, we used human G4 DNA substrate with both 3′ and 5′ tails and the same mutant version of this sequence used above (G-DNA disrupting point mutation in the second G-run). In the absence of hsRPA, the tails, labeled at the 3′ end, are cleaved, but very little cleavage can be observed in the G4 DNA region (Fig. 10A, lanes 3 and 4). Cleavage appears to occur both from the 5′ end (long products) and from the 3′ end (short products) and pauses or arrests at the point where it encounters the G4 structure, giving no intermediates from cleavage in the G-rich region. In the mutant, non-G4 forming sequence, cleavage occurs throughout the fragment. Addition of 5 fmol of RPA allows hsDna2 to cleave into the G4 DNA (Fig. 10A, lanes 7–8, products of intermediate length), and the cleavage pattern looks similar to that of the non-G4 DNA (mutant) sequence (Fig. 10A, lanes 9–10). Remarkably, at higher RPA concentrations, there is inhibition of overall cutting, and we suggest this to be due to cleavage from the 3′ end being dramatically inhibited, as demonstrated by an increase in long products but the disappearance of short products (less than 12 nt) most likely derived from cleavage from the 3′ end. We suggest that RPA prevents Dna2 from cleaving the 3′ end, perhaps because Dna2 cannot track along RPA-coated DNA in the 3′–5′ direction. The opposite is true at the 5′ end, where RPA unfolds the G4 DNA allowing Dna2 to track and cleave, and high RPA concentrations have a stimulatory rather than an inhibitory effect.
FIGURE 10.
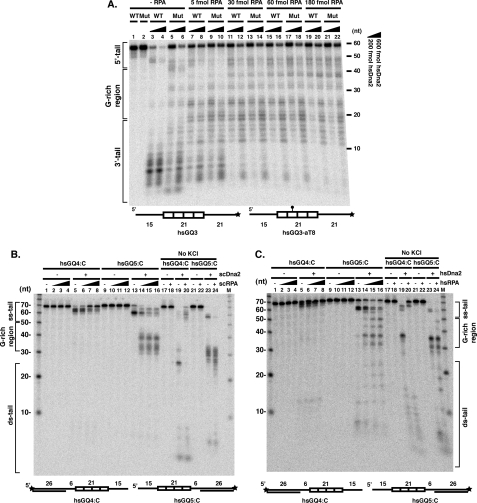
RPA allows Dna2 cleavage of human G4 DNA from the 5′ end but protects the 3′ end from cleavage. A, effect of hsRPA on hsDna2 cleavage. 5 fmol of wild type (hsGQ3, WT) and mutant (hsGQ3-aT8, Mut) labeled substrates were incubated without hsRPA (lanes 1–6) or increasing amounts of hsRPA (5 fmol, lanes 7–10; 30 fmol, lanes 11–14; 60 fmol, lanes 15–18; 180 fmol, lanes 19–22). Except for lanes 1 and 2, hsDna2 was included at 200 (odd-numbered lanes) or 600 fmol (even-numbered lanes). Products of the reactions were resolved on a 12% polyacrylamide, 7 m urea denaturing gel. B, scRPA stimulates scDna2 5′-nuclease and inhibits 3′-nuclease against G4 DNA. 5′ end-labeled hsGQ4:C (lanes 1–8 and 17–20) or 3′ end-labeled hsGQ5:C (lanes 9–16 and 21–24) partially double-stranded human telomeric intramolecular G4 DNAs (5 fmol) were preincubated on ice in the presence of increasing amounts of scRPA (0, 25, 150, and 300 fmol, as indicated by triangles). scDna2 (100 fmol for 3′ end-labeled substrate and 300 fmol for 5′ end-labeled substrate) was then added to start nuclease reactions, and reaction mixtures were incubated at 37 °C for 60 min. Nuclease products were analyzed by denaturing gel electrophoresis. scDna2 was omitted from the reaction in lanes 1–4, 9–12, 17–18, and 21–22. In lanes 17–24, KCl was omitted from the reaction, and nuclease assay was performed in the absence (lanes 17, 19, 21, and 23) and presence (300 fmol, lanes 18, 20, 22, and 24) of scRPA. Reaction mixtures contained 1 mm MgCl2 and no ATP. C, hsRPA stimulates hsDna2 5′-nuclease and inhibits 3′-nuclease against G4 DNA. 5′ end-labeled hsGQ4:C (lanes 1–8, and 17–20) or 3′ end-labeled hsGQ5:C (lanes 9–16and21–24) partially double-stranded human telomeric G4 DNAs (5 fmol) were preincubated on ice in the presence of increasing amounts of hsRPA (0, 5, 25, and 100 fmol, as indicated by triangles). hsDna2 (200 fmol) was then added to start the nuclease reaction, and the reaction mixture was incubated at 37 °C for 60 min. Nuclease products were analyzed by denaturing gel electrophoresis. hsDna2 was omitted from the reaction in lanes 1–4, 9–12, 17–18, and 21–22. In lanes 17–24, KCl was omitted from the reaction, and nuclease assay was performed in the absence (lanes 17, 19, 21, and 23) and presence (lanes 18, 20, 22, and 24) of hsRPA (100 fmol). Reaction mixtures contained 4 mm MgCl2 and no ATP.
To obtain direct evidence for the above interpretation, we used G4 DNA substrates that allowed us to monitor 5′ and 3′ end cleavage independently. The 5′ and 3′ single-stranded tails were converted to Dna2-resistant forms by hybridizing with complementary 5′ and 3′ oligonucleotides, respectively (22, 27, 28). As shown in Fig. 10B, on the substrate with a 5′ end blocked by the duplex configuration, scDna2, in the absence of RPA, can remove several nucleotides from the single strand 3′ tail to give a product of about 65 nucleotides (label is at the 5′ end). However, scDna2 cannot cleave past the single strand/G4 DNA junctions (Fig. 10B, lane 5). Addition of RPA nearly completely inhibits 3′ end cleavage (Fig. 10B, lanes 6–8, increase in full-length remaining and reduction of cleaved fragments). On the other hand, when the 3′ end is blocked by an oligonucleotide, scDna2 cleaves from the 5′ end, to give a product about 58 nt long (label at the 3′ end). When Dna2 enters from the 5′ end, as from the 3′ end, it also stops upon encountering the single-strand/G4 DNA junction (Fig. 10B, lane 13). This product is reminiscent of the product of cleavage of a 5′ flap substrate, where the flap is cleaved to within 5–7 nucleotides of the junction between single- and double-stranded DNA. However, addition of RPA, rather than inhibiting scDna2 tracking from a 5′ end, both allows scDna2 to cleave into the G4 DNA and also significantly stimulates the overall Dna2 nuclease activity (Fig. 10B, lanes 14–16). In the absence of monovalent cation and therefore of G4 structure, Fig. 10B, lanes 17–24, cleavage from the 3′ end proceeded into the G-rich region, lane 19, and was still inhibited by RPA (lane 20). Cleavage from the 5′ end also extended into the G-rich region (Fig. 10B, lane 23) but was stimulated by RPA (Fig. 10B, lane 24). Similar results were obtained with hsDna2 on the same substrates (Fig. 10C). Cleavage from the 3′ end was completely inhibited by hsRPA, but cleavage from the 5′ end was stimulated, and hsRPA allowed hsDna2 to cleave into the G4 DNA from the 5′ end but not from the 3′ end (Fig. 10C). Thus, regardless of the details of the structure of the G4 DNA, it is more susceptible to Dna2 cleavage in the presence of RPA than is duplex DNA. (This is probably because RPA destabilizes the G4 structure but does not unwind duplex B DNA to significantly extend cutting of a 5′ random DNA flap beyond the single-stranded/duplex junction.) We conclude that RPA, in general, inhibits Dna2 cleavage from 3′ ends and directs Dna2 to 5′ ends where it stimulates cleavage.
DISCUSSION
The existence of G4 DNA in living cells has been met until recently with some degree of skepticism. Although it was clear that a large number of G-rich sequences have a potential for G4 formation in vitro, the question remained whether such structures exist in vivo. More questions concerned the biological function of G4 DNA and proteins interacting with such higher order structures. The demonstration of the existence of specialized proteins with the ability to resolve G4 DNA structures nevertheless provided compelling, although circumstantial, evidence for the importance of G4 DNA in various biological processes. Recent advances provide more and more convincing experimental data for a role of G4 DNA in many regulatory mechanisms (33). The potential to form G4 DNA exists at G-rich, highly repetitive sequences characteristic of telomeres, the rDNA, and the immunoglobulin heavy-chain switch regions. What prompted us to study Dna2 and its possible interactions with G4 DNA? The rationales for such studies were 4-fold. One, Dna2 is localized to telomeres in both the G1 and G2 phases of the cell cycle (7). Two, yeast dna2 mutant phenotypes are suppressed by mammalian BLM and WRN, RecQ helicases known to interact with G4 DNA (34, 35). Three, scDna2 is involved in rDNA stability (36). Four, Dna2 was shown to process long, structured flaps that might arise during OFP (5), suggesting that it is also involved in processing of G-rich Okazaki intermediates with potential for G4 DNA formation.
In this work we demonstrate that scDna2 binds G4-containing DNAs flanked by single-stranded tails and has especially high specificity for G4 DNA carrying TGTGGG repeats, characteristic of yeast telomeres, compared with single-stranded DNA of the same sequence. scDna2 also binds ciliate G4 DNA, but the difference in affinity for tailed ciliate G4 DNA and for single-stranded DNA of the same sequence is less pronounced. Because the G4 DNA structures formed with yeast and ciliate DNA may differ (e.g. yeast G-rich repeats most likely form a parallel tetramer and the ciliate repeats form hairpin dimers), our results suggest that scDNA2 binding is structure-specific, although some sequence specificity cannot be completely ruled out. We also found that hsDna2 binds to G4 DNAs, although the differential affinity between G4 and single-stranded DNA found for the scDna2 was not true of hsDna2. One interesting aspect of hsDna2 binding to G4 DNA appeared in the experiment with scG4 DNA. Two different complexes formed, one that was competed efficiently by single-stranded DNA and another that was not competed by single-stranded DNA (Fig. 4B). The latter complex might represent structure-specific binding, set up by the G4 DNA. We propose that the difference in the competition by single-stranded DNA and G4 DNA shows that hsDna2 binds in different modes to yeast G4 DNA and to single-stranded DNA, respectively. In one mode, the Dna2 may bind the single-stranded tails (slower moving complexes). In an alternative mode, represented by the faster moving complex that is competed efficiently only by tailed G4 DNA and not by single-stranded DNA, it is likely, although there are other possibilities, that Dna2 is bound at or near the junction of the G4 structure and the single-stranded tail (Fig. 4B, compare lanes 8 and 9 with 13 and 14). Such junction binding has been demonstrated on flaps of random sequence (37).
Perhaps more important regarding the function of Dna2 is our observation of the efficient unwinding of both yeast and Oxytrichia G4 DNAs by Dna2 helicase activity and the conservation of this helicase activity in both scDna2 and hsDna2. The RecQ helicases have also been shown to unwind G4 DNA and have been implicated in resolution of G4 DNA that may arise in the process of replication and recombination. The unwinding of short G4 DNA substrates by BLM and WRN helicases requires a 3′ single-stranded tail adjacent to the G4 DNA structure, but single-stranded tails do not seem to be required for binding to bubble and Holliday junction intermediates (21). It was recently shown that RecQ helicases have high specificity for direct binding to G4 DNA structures per se, rather than single-stranded DNA/G4 junctions, in the context of long stretches of DNA, as opposed to short oligonucleotides. It is not known if RecQ helicases can resolve such structures, however (11). If so, they could prevent replication arrest on a template that otherwise may act as a roadblock for polymerase. In the case of Dna2, unwinding requires an unblocked 5′ terminus on the single-stranded DNA tail proximal to the G4 DNA, and we found no evidence that Dna2 binds directly to G4 DNA, but rather it seems to bind at or near a junction between single-stranded and G4 DNA, as deduced from competition assays.
Another striking reaction we observed is that G4 DNA does not seem to be susceptible to Dna2 cleavage but that RPA promotes Dna2 nuclease cleavage through runs of guanines in G4 structures, probably through RPA-mediated resolution of the G quartets. This reaction may be physiologically significant, because Dna2 is tightly associated with RPA in the cell (38) and DNA2 and RFA genes interact (38). Also, it has been shown that Dna2/FEN1/RPA function in a coordinated fashion in Okazaki fragment processing (5, 27, 39). RPA has previously been shown to stimulate Dna2 exo-endonuclease on 5′ flap structures such as that shown in Fig. 4. The single-stranded telomere-specific binding protein hPOT1 is known to disrupt G4 structures and stimulate ATP-dependent unwinding by BLM (and other RecQ helicases). RPA and Dna2 seem to behave in a slightly different manner from hPOT1 and BLM; namely, RPA, which is capable of unfolding the G4 DNA by itself (40), appears only to be required to stimulate the nuclease activity, and not the helicase, because the stimulation of nuclease activity occurs even in the absence of ATP and therefore in the absence of Dna2 helicase activity.
The fourth important observation is unrelated to the G4 structure of the DNA and reveals a more general property of the Dna2 mechanism. The Dna2 3′-exo-endonuclease activity is strongly inhibited by RPA, unlike the 5′-nuclease activity. Dna2 interacts with the three subunit RPA through the RPA1 subunit (41), and this interaction has been shown to stimulate helicase and 5′-nuclease (28, 38, 41). The primary interaction domains are in the C-terminal three-quarters of each protein, although N-N-terminal interactions appear to increase the stability of complexes (41). RPA1 binds to DNA with a specific polarity; the N terminus interacts strongly with 5′ sequences of its binding site and the C terminus weakly with 3′ sequences (42). Our results imply that this polarity may load Dna2 with the proper orientation to translocate in the 5′ to 3′ direction. RPA has been proposed to play a similar role in positioning XPG nuclease and ERCC1/XPF nuclease appropriately during excision repair (42) and in directing the DNA damage checkpoint Rad24-RFC clamp loader to 5′ single-strand/duplex junctions rather than 3′ junctions (43). In contrast to our results, previous work revealed a marked stimulation of a feeble 3′-nuclease activity in scDna2 in the presence of RPA (38). The difference may be due to differences in substrates studied or reaction conditions used and/or in the Dna2 protein preparations.
The function of the potent 3′-nuclease activity of yeast and human Dna2 we observe is not known. During OFP, there may be branch migration of the flaps created by polymerase δ, generating both 3′ and 5′ flaps. Nucleolytic attack on the 3′-equilibrating flap could potentially retard the rate of removal of the RNA/DNA primer by competing with productive binding and degradation of the 5′ flap. The switch in 3′-nuclease specificity governed by RPA might prevent this. The switch we observe may mimic the switch in nuclease activity in the RecBCD nuclease helicase observed upon encountering a χ site, in which case the 5′-nuclease activity is stimulated and the 3′-nuclease activity is inhibited.
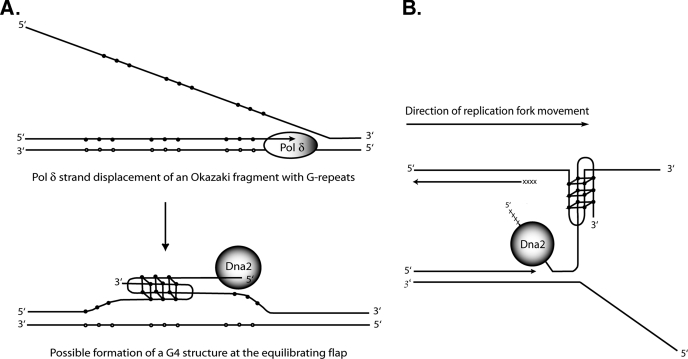
It is easy to propose a role for the G4 binding and resolving activities of Dna2 on OFs with G-rich flaps, which would exist in the rDNA, for instance. One can envision the formation of G4 DNA on G-rich flaps that would inhibit FEN1. G4 formation on G-rich flaps has been demonstrated on the human immunodeficiency virus, type 1, central flap (44). Dna2 would remove G4 DNA, either through helicase activity or by nuclease in conjunction with RPA and prepare a substrate for FEN1 (Fig. 11A).
FIGURE 11.
Models for functional interactions of Dna2 with G4 DNA. A, hypothetical role for Dna2 in resolving G4 structures during Okazaki fragment processing. Closed circles, guanines; open circles, cytosines. Flaps generated by polymerase δ (Pol δ) strand displacement might form either intramolecular G4 DNA on the 5′ flap (not shown) or intermolecular G4 structures on equilibrating 5′ and 3′ flaps. For simplicity, only an antiparallel version of the intermolecular form is shown. B, hypothetical role for Dna2 in resolving G4 structures during the replication of telomeres. The direction of DNA replication fork migration is from left (centromere proximal) to right (toward the telomere). See text for details. xxxxx, RNA primer; solid line, DNA; closed circles, guanines.
At the telomere, a role for the G4 activities of Dna2 is harder to envision. The binding of Dna2 to G4 might recruit Dna2 at G1 and G2 phases of the cell cycle. This is unlikely, however, and it is more likely that protein/protein interactions are involved in localization of Dna2 to telomeres, given the effects of overexpression of Dna2 on silencing and implied effects on chromatin organization (6, 7). It is also not straightforward to envision a role for Dna2-mediated G4 DNA resolution at the telomere because of a polarity problem. It is the newly synthesized leading strand that is G-rich at telomeres, whereas the Okazaki fragments on the lagging strand at the telomere are C-rich. Thus, the role of Dna2 cannot be removal of G4 DNA from flaps during OFP. The template for the lagging strand, which is also the strand elongated by telomerase, is G-rich, and one might need to resolve either intrachromosomal or interchromosomal G4 DNAs arising in those sequences. This strand terminates at a 3′ end, however, and Dna2 appears to prefer to enter DNA from a 5′ end. Although Dna2 can bind a 3′ terminus, it does not appear to be able to track in the 3′ to 5′ direction, as it cannot unwind DNA that has only 3′ single-stranded tails (1, 2, 22). To account for a role for Dna2 involving G4 DNA resolution at telomere, we therefore propose that replication forks may stall as they approach the end of the linear chromosome, perhaps as the replicative helicase unwinds the final turns of the helix. The stalled fork can then rearrange as depicted in Fig. 11B. The newly synthesized G-rich strand can become dissociated from its leading strand template and form G4 DNA with the single-stranded G-rich lagging strand template. This G4 DNA would prevent completion of replication on the leading strand and inhibit telomerase on the lagging strand, leading to loss of telomeres on both strands. We propose that telomere loss is prevented by Dna2 as follows. Evidence is accumulating that replication forks stall within telomeric repeats in yeasts (45, 46). It has also recently been shown that long single-stranded regions, likely on the leading strand, form at fork-blocking lesions in yeast and that gaps remain on both strands behind the forks that traverse lesions (47). Such gaps might arise by repriming downstream of fork blocks during replication restart, and Dna2 might remove 5′ RNA/DNA primers on the reinitiated on the leading as well as the lagging strand. Dna2 loaded on the 5′ end of a newly synthesized G-rich strand could resolve the interstrand G4 DNA using its helicase activity or nucleolytically remove the G4 allowing for completion of leading strand synthesis and providing a free G-rich substrate for telomerase extension on the lagging strand. In a dna2 mutant, the stalled fork and resulting G4 DNA might be resolved by RecQ helicases, accounting for the suppression of dna2 mutant phenotypes by overexpression of RecQ helicase. It should be possible in yeast to demonstrate that the leading strand has discontinuities at the telomere. One would cleave genomic DNA with XhoI, which normally gives a telomeric fragment of about 1.3 kb in a native gel. In a denaturing gel, if synthesis on the leading strand is discontinuous, one would expect a shorter G-rich leading strand fragment. Use of a DNA ligase mutant might facilitate detection. In human cells, where cytological investigation of telomeres of metaphase chromosomes is feasible, this model predicts that upon depletion of human Dna2 using short hairpin RNAs, for instance, one should observe loss of both leading and lagging strand telomeres.
In conclusion, we have presented reactions catalyzed by Dna2 that may lead to an understanding of its function at telomeres. In a more general mechanistic observation, we have shown that Dna2 needs to be loaded in a fashion that is determined by the polarity of binding of RPA to be active as a nuclease.
Acknowledgments
We are grateful to Marc Wold for the gift of highly purified hsRPA and to Robert Bambara and members of his laboratory for discussions of the experiments. Proteins were expressed and partially purified in the Protein Expression Center at Caltech.
This work was supported, in whole or in part, by National Institutes of Health Grant GM087666 from the USPHS and a research grant from the Philip Morris External Research Program. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: OFP, Okazaki fragment processing; SVPI, snake venom phosphodiesterase I; DMS, dimethyl sulfate; BSA, bovine serum albumin; DTT, dithiothreitol; HA, hemagglutinin; AMP-PNP, adenosine 5′-(β, γ-imino) triphosphate; ssDNA, single-stranded DNA; nt, nucleotide; sc, Saccharomyces cerevisiae; hs, Homo sapiens.
References
- 1.Budd, M. E., and Campbell, J. L. (1995) Proc. Natl. Acad. Sci. U. S. A. 927642 –7646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Budd, M. E., Choe, W.-C., and Campbell, J. L. (1995) J. Biol. Chem. 27026766 –26769 [DOI] [PubMed] [Google Scholar]
- 3.Masuda-Sasa, T., Polaczek, P., and Campbell, J. L. (2006) J. Biol. Chem. 28138555 –38564 [DOI] [PubMed] [Google Scholar]
- 4.Budd, M. E., Tong, A. H., Polaczek, P., Peng, X., Boone, C., and Campbell, J. L. (2005) PLoS Genet. 1634 –650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kao, H. I., Veeraraghavan, J., Polaczek, P., Campbell, J. L., and Bambara, R. A. (2004) J. Biol. Chem. 27915014 –15024 [DOI] [PubMed] [Google Scholar]
- 6.Singer, M. S., Kahana, A., Wolf, A. J., Meisinger, L. L., Peterson, S. E., Goggin, C., Nahowald, M., and Gottschling, D. E. (1998) Genetics 150613 –632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choe, W., Budd, M., Imamura, O., Hoopes, L., and Campbell, J. L. (2002) Mol. Cell Biol. 222002 –2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Budd, M. E., Reis, C. C., Smith, S., Myung, K., and Campbell, J. L. (2006) Mol. Cell Biol. 262490 –2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zahler, A. M., Williamson, J. R., Cech, T. R., and Prescott, D. M. (1991) Nature 350718 –720 [DOI] [PubMed] [Google Scholar]
- 10.Fry, M., and Loeb, L. A. (1999) J. Biol. Chem. 27412797 –12802 [DOI] [PubMed] [Google Scholar]
- 11.Huber, M. D., Duquette, M. L., Shiels, J. C., and Maizels, N. (2006) J. Mol. Biol. 3581071 –1080 [DOI] [PubMed] [Google Scholar]
- 12.Huber, M. D., Lee, D. C., and Maizels, N. (2002) Nucleic Acids Res. 303954 –3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun, H., Bennett, R., and Maizels, N. (1999) Nucleic Acids Res. 271978 –1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun, H., Karow, J. K., Hickson, I. D., and Maizels, N. (1998) J. Biol. Chem. 27327587 –27592 [DOI] [PubMed] [Google Scholar]
- 15.Sun, H., Yabuki, A., and Maizels, N. (2001) Proc. Natl. Acad. Sci. U. S. A. 9812444 –12449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, Z. P., Frantz, J. D., Gilbert, W., and Tye, B. K. (1993) Proc. Natl. Acad. Sci. U. S. A. 903157 –316117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, Z. Z., and Gilbert, W. W. (1994) Cell 771083 –1092 [DOI] [PubMed] [Google Scholar]
- 18.Ghosal, G., and Muniyappa, K. (2005) Nucleic Acids Res. 334692 –4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaug, A. J., Podell, E. R., and Cech, T. R. (2005) Proc. Natl. Acad. Sci. U. S. A. 10210864 –10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giraldo, R., and Rhodes, D. (1994) EMBO J. 132411 –2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohaghegh, P., Karow, J. K., Brosh, R. M., Jr., Bohr, V. A., and Hickson, I. D. (2001) Nucleic Acids Res. 292843 –2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masuda-Sasa, T., Imamura, O., and Campbell, J. L. (2006) Nucleic Acids Res. 341865 –1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., pp.13.78 –13.104, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 24.Muniyappa, K., Anuradha, S., and Byers, B. (2000) Mol. Cell Biol. 201361 –1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sen, D., and Gilbert, W. (1992) Biochemistry 3165 –70 [DOI] [PubMed] [Google Scholar]
- 26.Sen, D., and Gilbert, W. (1990) Nature 344410 –414 [DOI] [PubMed] [Google Scholar]
- 27.Kao, H.-I., Campbell, J. L., and Bambara, R. A. (2004) J. Biol. Chem. 27950840 –50849 [DOI] [PubMed] [Google Scholar]
- 28.Bae, S.-H., and Seo, Y.-S. (2000) J. Biol. Chem. 27538022 –38031 [DOI] [PubMed] [Google Scholar]
- 29.Wang, Y., and Patel, D. J. (1993) Structure (Lond.) 1263 –282 [DOI] [PubMed] [Google Scholar]
- 30.Parkinson, G. N., Lee, M. P., and Neidle, S. (2002) Nature 417876 –880 [DOI] [PubMed] [Google Scholar]
- 31.Li, J., Correia, J. J., Wang, L., Trent, J. O., and Chaires, J. B. (2005) Nucleic Acids Res. 334649 –4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, J. H., Kim, H. D., Ryu, G. H., Kim, D. H., Hurwitz, J., and Seo, Y. S. (2006) Nucleic Acids Res. 341854 –1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maizels, N. (2006) Nat. Struct. Mol. Biol. 131055 –1059 [DOI] [PubMed] [Google Scholar]
- 34.Imamura, O., and Campbell, J. L. (2003) Proc. Natl. Acad. Sci. U. S. A. 1008193 –8198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma, S., Sommers, J. A., and Brosh, R. M., Jr. (2004) Hum. Mol. Genet. 132247 –2261 [DOI] [PubMed] [Google Scholar]
- 36.Weitao, T., Budd, M., and Campbell, J. L. (2003) Mutat. Res. 532157 –172 [DOI] [PubMed] [Google Scholar]
- 37.Stewart, J. A., Campbell, J. L., and Bambara, R. A. (2006) J. Biol. Chem. 28138565 –38572 [DOI] [PubMed] [Google Scholar]
- 38.Bae, S. H., Bae, K.-H., Kim, J. A., and Seo, Y. S. (2001) Nature 412456 –461 [DOI] [PubMed] [Google Scholar]
- 39.Kao, H. I., Henricksen, L. A., Liu, Y., and Bambara, R. A. (2002) J. Biol. Chem. 27714379 –14389 [DOI] [PubMed] [Google Scholar]
- 40.Salas, T. R., Petruseva, I., Lavrik, O., Bourdoncle, A., Mergny, J. L., Favre, A., and Saintome, C. (2006) Nucleic Acids Res. 344857 –4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bae, K.-H., Kim, H.-S., Bae, S.-H., Kang, H.-Y., Brill, S., and Seo, Y.-S. (2003) Nucleic Acids Res. 313006 –3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Laat, W. L., Appledoorn, E., Sugasawa, K., Weterings, E., Jaspers, N. G. J., and Hoeijmakers, J. H. J. (1998) Genes Dev. 122598 –2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Majka, J., Binz, S. K., Wold, M. S., and Burgers, P. M. (2006) J. Biol. Chem. 28127855 –27861 [DOI] [PubMed] [Google Scholar]
- 44.Lyonnais, S., Hounsou, C., Teulade-Fichou, M. P., Jeusset, J., Le Cam, E., and Mirambeau, G. (2002) Nucleic Acids Res. 305276 –5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivessa, A. S., Zhou, J.-Q., Schulz, V. P., Monson, E. K., and Zakian, V. A. (2002) Genes Dev. 161383 –1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller, K. M., Rog, O., and Cooper, J. P. (2006) Nature 440824 –828 [DOI] [PubMed] [Google Scholar]
- 47.Lopes, M., Foiani, M., and Sogo, J. M. (2006) Mol. Cell 2115 –27 [DOI] [PubMed] [Google Scholar]