Abstract
The expression of the ubiquitin-like molecule ISG15 (UCRP) and protein modification by ISG15 (ISGylation) are strongly activated by interferon, genotoxic stress, and pathogen infection, suggesting that ISG15 plays an important role in innate immune responses. Inducible nitric-oxide synthase (iNOS) is induced by the similar stimuli as ISG15 and enhances the production of nitric oxide (NO), a pleiotropic free radical with antipathogen activity. Here, we report that cysteine residues (Cys-76 and -143 in mouse, Cys-78 in human) of ISG15 can be modified by NO, and the NO modification of ISG15 decreases the dimerization of ISG15. The mutation of the cysteine residue of ISG15 to serine improves total ISGylation. The NO synthase inhibitor S-ethylisothiourea reduces endogenous ISGylation. Furthermore, ectopic expression of iNOS enhanced total ISGylation. Together, these results suggest that nitrosylation of ISG15 enhances target protein ISGylation. This is the first report of a relationship between ISGylation and nitrosylation.
Interferons (IFNs)3 are a group of related cytokines that have a role in the defense against bacterial or viral infections. IFNs are induced by several stimuli, such as LPS, lipoproteins, lipopeptides, double-stranded RNA, double-stranded DNA, single-stranded RNA, or unmethylated CpG motifs mainly through Toll-like receptors (1). ISG15 (UCRP) is one of the genes that is highly induced by IFNs (2). Furthermore, ISG15 was the first reported ubiquitin-like modifier (3). Similar to ubiquitin, ISG15 forms covalent conjugates with cellular proteins, which is similar to protein ubiquitylation (4), by utilizing the ISG15 E1 (UBE1L/UBA7), E2s (UbcH6 and UbcH8), and E3s (5–11). Besides ISG15 expression, ISG15 modification (ISGylation) and most enzymes involved in this modification are strongly activated by type I interferon IFN (4, 12). IFNs are critical cytokines involved in innate immune responses (13). These facts suggest that ISG15 modification may modulate certain immune responses related to pathogen infections and various stresses. ISG15-deficient mice show no obvious phenotype against vesicular stomatitis virus and lymphocytic choriomeningitis virus (14); however, they are more susceptible to influenza A/WSN/33 and influenza B/Lee/40 virus infections (15). ISG15-deficient mice also exhibit increased susceptibility to herpes simplex virus type 1, murine gammaherpesvirus 68, and Sindbis virus infection (15). The increased susceptibility of ISG15-deficient mice to Sindbis virus infection is rescued by expressing wild-type ISG15 but not a mutant form of ISG15 that cannot form conjugates (15), which suggests that ISGylation is quite important to resistance against the infection of these viruses.
Inducible nitric-oxide synthase (iNOS, NOS2) is a high-output NOS compared with NOS1 and NOS3. The Vmax of iNOS is ∼10-fold greater than that of NOS1 and NOS3 (16). iNOS is not expressed under normal conditions; however, it is induced in response to cytokines, microbes, or microbial products, resulting in the sustained production of nitric oxide (NO) (16). As a result, reactive nitrogen intermediates, such as NO, nitrite, and nitrate, and the products of the interaction of NO with reactive oxygen species, such as peroxynitrite and peroxynitrous acid, are accumulated and utilized for the anti-bacterial or -viral effects (16–18). One of the most important molecular mechanisms mediating anti-bacterial or -viral effects is protein S-nitrosylation, the covalent attachment of a NO moiety to the thiol of cysteine, a post-translational protein modification. So far, more than 100 proteins have been reported as substrates for protein S-nitrosylation (19). Protein S-nitrosylation regulates enzymatic activity, protein-protein interaction, and signal transduction (19).
Given that some stimuli, such as type I IFN, can induce both iNOS and ISG15, we were interested in the relationship between nitrosylation and ISGylation. Here, we report that ISG15 is able to be nitrosylated. The nitrosylation of ISG15 prevents the dimerization of ISG15, and the cysteine residue of ISG15 contributes to stable dimerization. A mutant ISG15 in which the cysteine residue is replaced by a serine shows more effective ISGylation activity than wild-type ISG15, which indicates that the cysteine residue of ISG15 is an impediment to effective ISGylation.
EXPERIMENTAL PROCEDURES
Plasmid Construction—pcDNA3.1-HA-hUBE1L, pcDNA3.1-mUbcH8, and pCAGGS-6×His-mISG15 have been described previously (10). pFLAG-CMV2 plasmid was purchased from Sigma. iNOS (NM_010927) cDNA was amplified by reverse transcription-PCR using the mRNA of LPS-stimulated RAW264.7 cells and inserted into pcDNA3.1 (Invitrogen). Site-directed point mutation was generated by QuikChange® XL site-directed mutagenesis kit (Stratagene).
Cell Culture and Transfection—293T cells were cultured as described previously (20). HeLa cells were cultured as 293T cells. 293T cells were transfected using calcium phosphate precipitation as described previously (21). For small-scale transfection, PolyFect reagent (Qiagen) or Lipofectamine™ 2000 (Invitrogen) was used according to the manufacturer's protocols. ISG15-deficient lung fibroblasts (14) were immortalized by SV40 large T expression. HeLa cells were stimulated by 5000 units/ml of human IFNα-2a (IFNα) for 36 h as reported previously (20).
NO Donors and Inhibitors—S-Nitrosocysteine (SNOC) was produced as reported previously (22). S-Nitroso-glutathione was purchased from Sigma-Aldrich. Cell lysates or immunopurified proteins were treated with SNOC (100 μm) in the dark at room temperature for up to 1 h. S-Ethylisothiourea (ETU) was purchased from Calbiochem. Human IFNα-2a (5000 units/ml) and/or ETU (1 mm) were added to the culture medium of HeLa cells and incubated for 36 h.
Immunoprecipitation, Ni-NTA Pulldown, and Western Blot Analysis—Immunoprecipitation, Ni-NTA pulldown, and Western blot analysis were done as reported previously (20).
Antibodies—Antibodies against FLAG (Sigma), HA (Covance), iNOS (BD Transduction Laboratories™), and His6 (Clontech) were purchased from the respective manufacturers. Rabbit anti-mouse ISG15 polyclonal antibodies have been described previously (23). Rabbit anti-mUbcH8 antibodies were reported previously (7). Anti-biotin antibody and the specific second antibody were from NitroGlo nitrosylation detection kit (PerkinElmer Life Sciences).
RESULTS
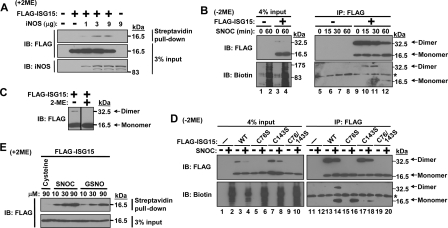
Nitrosylation of ISG15 Prevents Dimer Formation by ISG15—To test whether ISG15 is able to be nitrosylated, we expressed FLAG-tagged mouse ISG15 in 293T cells with or without iNOS. The cell lysates were subjected to the biotin switch method followed by streptavidin-agarose pulldown and immunoblotting with anti-FLAG antibody. The biotin switch method (24) was widely used to detect protein nitrosylation. FLAG-ISG15 was pulled down in the presence of iNOS expression in a dose-dependent manner (Fig. 1A), which indicates that ISG15 was nitrosylated by endogenously produced nitric oxide (NO) synthesized by iNOS. We further examined the nitrosylation of ISG15 using immunopurified ISG15. FLAG-ISG15 was expressed in 293T cells and purified by immunoprecipitation with anti-FLAG antibody and protein A-agarose. The lysates and immunoprecipitates were treated with the physiological NO donor SNOC (22, 25) for different periods to allow detection of nitrosylation of ISG15 by anti-biotin antibody. In this case nonreducing conditions are required to detect the nitrosylation, because Cys-S-NO was changed to Cys-S-S-biotin by the biotin switch method. Using this method, nitrosylation of ISG15 was detected after SNOC treatment (Fig. 1B). More interestingly, we also detected the dimerization of ISG15 in vivo (Fig. 1B). It was detected in input (lane 3) and more easily detected in immunopurified samples (lanes 9–12), which suggested that the dimerization of ISG15 depends on ISG15 concentration. In brief, the concentration of ISG15 was increased on the protein A-agarose by immunoprecipitation, and this resulted in a higher chance for interaction and dimer formation. Importantly, the dimerized form of ISG15 was decreased after SNOC treatment for 1 h (Fig. 1B, lanes 4 and 12). It has been previously reported that ISG15 forms homodimers (26). Our results confirm these findings and indicate that nitrosylation of ISG15 prevents homodimer formation. We further confirmed that dimer formation was made by disulfide bonds (Fig. 1C). Cell lysates containing FLAG-ISG15 were subjected to either nonreducing or reducing SDS-PAGE followed by anti-FLAG immunoblotting. The dimer form was detected only in the nonreducing gel. Given that cysteine residues are targeted for nitrosylation, we mutated two cysteines of mouse ISG15 and checked the effect on dimerization and nitrosylation (Fig. 1D). We found that ISG15 (C76S and C76S/C143S) mutants did not show dimerization, which means that Cys-76 but not Cys-143 contributes to stable dimer formation, which is detectable by nonreducing SDS-PAGE. The dimerization of ISG15 (wild-type (WT) and C143S) was inhibited by SNOC treatment (Fig. 1D, lanes 4, 8, 14, and 18). Nitrosylation was completely inhibited only by double mutation of two cysteines. Nitrosylation of ISG15 was also confirmed by another NO donor S-nitroso-glutathione (GSNO) in a dose-dependent manner (Fig. 1E).
FIGURE 1.
Nitrosylation of ISG15. A, nitrosylation of ISG15 by endogenously produced nitric oxide. FLAG-tagged mouse ISG15 was expressed in 293T cells with or without mouse iNOS followed by use of the biotin switch method to substitute biotin for nitric oxide. The amounts of expression plasmid for iNOS were indicated. Biotin-labeled samples were pulled-down by streptavidin-agarose followed by SDS-PAGE (+2ME means reducing conditions) and Western blotting (IB) with anti-FLAG or iNOS antibody. B, inhibition of dimerization of ISG15 by nitrosylation. FLAG-ISG15 was expressed in 293T cells and immunoprecipitated (IP) by anti-FLAG antibody. Immunopurified FLAG-ISG15 as well as lysates (input) were subjected to the biotin switch method followed by SDS-PAGE (-2ME means nonreducing conditions) and Western blotting with anti-FLAG or biotin antibody. The asterisk indicates a nonspecific band. C, disulfide bond-based dimerization of ISG15. FLAG-ISG15 was expressed in 293T cells, and then lysates were subjected to either nonreducing or reducing SDS-PAGE followed by anti-FLAG antibody. D, Cys-76 and -143 are nitrosylated, and the stable dimerization of mouse ISG15 is dependent on Cys-76. FLAG-tagged wild-type, C76S, C143S, or C76S/C143S mutant ISG15 was expressed in 293T cells and immunoprecipitated by anti-FLAG antibody. Immunopurified FLAG-ISG15 as well as lysates (input) were subjected to the biotin switch method followed by SDS-PAGE (-2ME means nonreducing conditions) and Western blotting with anti-FLAG or biotin antibody. The asterisk indicates the nonspecific band. E, in vitro nitrosylation of ISG15 by SNOC or S-nitroso-glutathione (GSNO). FLAG-ISG15 was expressed in 293T cells, and then lysates were subjected to in vitro nitrosylation by SNOC or S-nitroso-glutathione in different concentrations. After biotin switch, nitrosylated FLAG-ISG15 was detected by streptavidin pulldown and anti-FLAG immunoblotting.
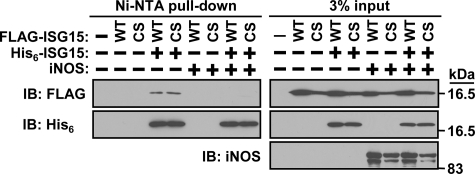
iNOS Prevents ISG15-ISG15 Interaction—Next we investigated if ISG15 molecules interact with each other, as interaction is necessary for dimerization. Either FLAG-tagged wild-type or C76S/C143S mutant ISG15 was expressed with or without His6-wild-type ISG15 followed by Ni-NTA pulldown to check the interaction (Fig. 2). Both wild-type and C76S/C143S mutant ISG15 could interact with wild-type ISG15, which demonstrates that cysteine residues are not necessary for the interaction between individual ISG15 monomers. Interestingly, co-expression of iNOS prevented the interaction. Because we did not detect interaction between ISG15 and iNOS (data not shown), nitric oxide (NO) itself or nitrosylation of another protein might regulate ISG15-ISG15 interaction. Taken together, cysteine residues are required for disulfide bond-based dimerization but not for just interaction.
FIGURE 2.
ISG15-ISG15 interaction. FLAG-tagged mouse ISG15 (WT or C76S/C143S mutant (CS)) was expressed in 293T cells with or without His6-tagged wild-type mouse ISG15 and iNOS. The resulting cell lysates were subjected to Ni-NTA-agarose pulldown to purify His6-tagged wild-type ISG15. The resulting agarose beads were subjected to SDS-PAGE and Western blotting (IB) with anti-FLAG, iNOS, or His6-tag antibody.
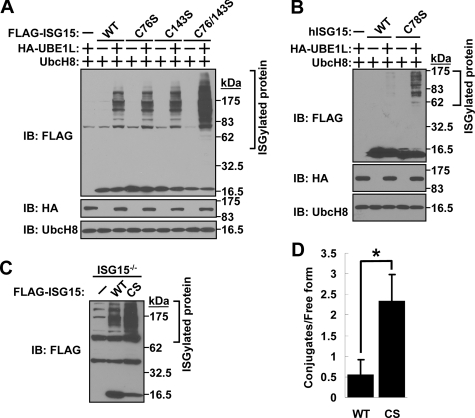
Cysteine Residues of ISG15 Prevent ISGylation—Because dimerized ISG15 might have a different physiological effect from monomer ISG15, we investigated the effect of a Cys to Ser mutation of ISG15 on ISGylation. Wild-type, C76S, C143S, or C76S/C143S mutant mouse ISG15 was expressed with or without ISG15 activating enzyme UBE1L and ISG15 conjugating enzyme UbcH8 in 293T cells to assess ISGylation (Fig. 3A). Interestingly, ISG15(C76S) showed no difference in the level of ISGylation, although the C76S mutation inhibited the formation of stable ISG15 dimers (Fig. 1D). More importantly, double mutation of both cysteine residues of mouse ISG15 enhanced ISGylation (Fig. 3A), which suggests that both cysteine residues inhibit ISGylation. Cys-143 might result in unstable dimerization, which is not detectable by Western blotting in Fig. 1D. Together, these data suggest that Cys-76 and Cys-143 contributed to dimer formation (Figs. 1D and 3A) and decreased the availability of monomer ISG15 to be used for ISGylation. We also confirmed this result with human ISG15, which has only one Cys in mature form. Human ISG15(C78S) has higher ISGylation activity compared with wild-type ISG15 (Fig. 3B). We further checked this idea with ISG15-deficient lung fibroblasts, which have been immortalized with SV40 large T antigen (Fig. 3C). This cell line shows high endogenous ISGylation activity, which means that transfection to express exogenous ISG15-activating and -conjugating enzymes is not necessary to get ISGylation. This cell line was reconstituted with either FLAG-tagged wild-type mouse ISG15 or mutant mouse ISG15(C76S/C143S) followed by Western blotting with anti-FLAG antibody, and the resulting ratio of ISG15 conjugates and free ISG15 were compared (Fig. 3, C and D). As a result, we found again that ISG15(C76S/C143S) has greater effective ISGylation activity than wild-type ISG15.
FIGURE 3.
Cysteine residues prevent effective ISGylation. A, cysteine residues of mouse ISG15 inhibit ISGylation. FLAG-tagged wild-type, C76S, C143S, or C76S/C143S mutant ISG15 was expressed in 293T cells with or without HA-UBE1L and mUbcH8 followed by SDS-PAGE and Western blotting (IB) with anti-FLAG, HA, or mUbcH8 antibody. B, the cysteine residue of human ISG15 inhibits ISGylation. FLAG-tagged wild-type or C78S mutant human ISG15 was expressed in 293T cells with or without HA-UBE1L and mUbcH8 followed by SDS-PAGE and Western blotting with anti-FLAG, HA, or mUbcH8 antibody. C, effective ISGylation of FLAG-mouse ISG15(C76S/C143S) in ISG15-deficient lung fibroblasts. ISG15-deficient lung fibroblasts were reconstituted by FLAG-tagged wild-type or C76S/C143S mutant mouse ISG15 followed by SDS-PAGE and Western blotting with anti-FLAG antibody. D, the quantification of the ratio between conjugated and free form of ISG15. The amount of ISG15 in either conjugated or free form was determined by scanning densitometry of the immunoblot of Fig. 3C. Data are expressed as means ± S.D. from three independent experiments. *, p < 0.01 for the indicated comparison (Student's t test).
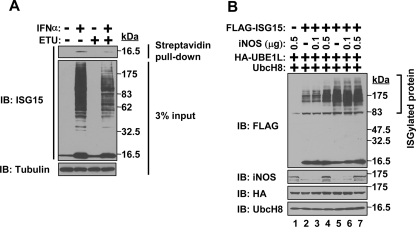
Nitrosylation Contributes to Effective ISGylation—We next analyzed the relationship between ISGylation and nitrosylation. HeLa cells were stimulated with IFNα in the presence or absence of NOS inhibitor ETU. Endogenous ISG15 nitrosylation and total protein ISGylation were examined (Fig. 4A). The presence of ETU decreased ISG15 nitrosylation, which is shown in a biotin switch-streptavidin pulldown assay. Furthermore, total protein ISGylation was also substantially decreased in the presence of NOS inhibitor ETU. These results further support that nitrosylation promotes effective ISGylation. We also checked ISGylation by WT or C76S/C143S mutant ISG15 in the presence or absence of the expression of iNOS. As a result, we found that the expression of iNOS substantially increased ISGylation of WT ISG15 (Fig. 4B, lane 4). Although the expression of iNOS inhibited ISG15-ISG15 interaction of both WT and C76S/C143S mutant ISG15 (Fig. 2), iNOS expression did not increase ISGylation of C76S/C143S mutant ISG15 (Fig. 4B, lane 7). Overall these data indicate that nitrosylation enhances effective ISGylation.
FIGURE 4.
Effective ISGylation in the presence of nitrosylation. A, decreased endogenous ISGylation by NOS inhibitor. HeLa cells were stimulated by IFNα in the presence or absence of NOS inhibitor ETU. The resulting cell lysates were subjected to either biotin switch method followed by streptavidin pulldown to detect nitrosylation of ISG15 or Western blotting (IB) with anti-ISG15 antibody to detect ISGylation. Anti-tubulin immunoblotting was shown as a loading control. B, enhancement of ISGylation by iNOS expression. FLAG-mouse ISG15 (WT (lanes 2–4) or C76S/C143S mutant (lanes 5–7)), iNOS, HA-UBE1L, and mUbcH8 were co-expressed in 293T cells in the indicated combinations followed by SDS-PAGE and Western blotting with anti-FLAG, iNOS, HA, or mUbcH8 antibody. The amounts of expression plasmid for iNOS were indicated.
DISCUSSION
NO has been shown to act as an antibacterial and antiviral agent (16). iNOS-derived NO contributes to both early and late phases of antibacterial activity (27). ISG15 itself and ISG15 modification also have been reported to have antiviral functions (15, 28), although the details of the molecular mechanism are not yet clear. Virus infection is one of the common stimuli which induce nitrosylation and ISGylation (Fig. 5), which suggests that there might be a correlation between nitrosylation and ISGylation to generate the maximum immune response. Here, we reported that nitrosylation of ISG15 prevented the dimerization of ISG15 and increased the availability of free ISG15 to be used for ISGylation. Given that ISG15 is able to form a homodimer through a disulfide bond (26), it is also possible that ISG15 may form disulfide bonds with other unidentified proteins. As a result, the availability of free ISG15 to be used for ISGylation will be decreased. Furthermore, ISG15 activating enzyme Ube1L null mice, which lack protein ISGylation (29), showed the same antiviral defects as ISG15 knock-out mice.4 Taken together, these data suggest that nitrosylation might contribute to the ISGylation-dependent anti-viral pathway. In fact, NO has been found to affect infection by more than a dozen viruses (30). For example, iNOS-deficient mice were significantly more susceptible to herpes simplex virus-1 infection, displayed a delayed clearance of virus, and exhibited an increase in the frequency of virus reactivation in dorsal root ganglia compared with similarly infected heterozygous mice (31). Another example is that iNOS-deficient mice, which were infected by murine cytomegalovirus (MCMV), developed significantly higher titers of infectious MCMV compared with the wild-type mice (32). Coxsackie B3 virus is also regulated by NO (33, 34). Coxsackievirus replicates to higher titers in iNOS-deficient mice, and the clearance of virus is delayed in these mice compared with wild-type mice (33, 35). Mice lacking NOS have a severe, necrotizing pancreatitis, with elevated pancreatic enzymes in the blood and necrotic acinar cells (35).
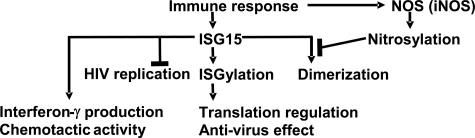
FIGURE 5.
A model of the relationship between ISGylation and nitrosylation. Immune response induces the expression of ISG15 and iNOS. Induced ISG15 is used for ISG15 modification (ISGylation) of many substrates and contributes to protein translation regulation and anti-virus effect. ISG15 itself can prevent the replication of human immunodeficiency virus. ISG15 also induces interferon-γ production and has chemotactic activity. Some of the induced ISG15 forms homodimer, which is an inactive form and cannot be used for ISGylation. The nitric oxide produced mainly by iNOS contributes to the nitrosylation of ISG15 and prevents the dimerization of ISG15, which means more free monomer ISG15 is available for ISGylation.
Recently, we have found that the mRNA cap structure binding protein eIF4E cognate 4EHP is modified by ISG15, resulting in higher cap structure binding activity and repression of protein translation (20). Given that Drosophila 4EHP selectively inhibits Caudal and hunchback protein translation (36, 37), mammalian 4EHP is believed to inhibit mRNA-specific protein translation. These data suggest that ISGylation probably controls protein translation under particular conditions, for example during the immune response to bacterial or viral infection (Fig. 5). Given that the homodimerization of ISG15 or the formation of disulfide bonds with other proteins seems to inhibit ISGylation, it is possible that some factors which can inhibit the formation of disulfide bonds by ISG15 would enhance total ISGylation. Reducing agents, for example glutathione (GSH), are the other factors which might enhance ISGylation. On the other hand, it has been reported that hepatic glutathione synthesis is decreased 24 h after injection of LPS (38). They also showed that the ratio of GSH (reduced form)/GSSG (oxidative form) was decreased by LPS treatment, although there was no increase in absolute amount of GSSG, which suggests that LPS treatment generates oxidative conditions. In this case, ISG15 should easily form disulfide bonds with itself to form homodimers or with other proteins, and nitrosylation should prevent disulfide bond formation by ISG15 under these conditions. This evidence suggests that nitrosylation of ISG15 may be an emergency pathway to achieve effective ISGylation to generate a full immune response.
ISG15 has been reported to inhibit human immunodeficiency virus replication (28) (Fig. 5). Interestingly, ISG15 has also been reported to be released from cells into the culture medium (39, 40). Furthermore, the addition of ISG15 into the culture medium of CD3+ cells induces the secretion of interferon-γ (41). It also has been reported that ISG15 has neutrophil chemotactic activity (42). This evidence suggests that ISG15 has at least two physiological roles; one is post-translational protein modification under particular conditions to enhance the immune response; another is the use of ISG15 itself for the immune response as shown in Fig. 5. However, it is not yet clear whether monomer or dimer forms of ISG15 are important for these activities. Nitrosylation of ISG15 could be one of the important regulatory steps during the ISG15 and ISGylation-dependent immune responses. However, current available assays to study ISG15 function are limited. It is necessary to develop new approaches to further elucidate the biological function of ISG15.
Acknowledgments
We thank Dr. Tomohiro Nakamura (Burnham Institute for Biomedical Research) for great discussions and Dr. Ernest Borden (Cleveland Clinical Foundation, Cleveland, OH) for human ISG15 antibody. We also thank members of the Zhang laboratory for valuable discussions and Dr. Joseph Biggs for critical editing of this manuscript. The Stein Endowment Fund has partially supported the departmental molecular biology service laboratory for DNA sequencing and oligonucleotide synthesis.
This work was supported, in whole or in part, by National Institutes of Health Grants GM66955 and CA102625 (to D.-E. Z.). This work was also supported by a Uehara Memorial Foundation postdoctoral fellowship (to F. O.). This is manuscript 19193 from The Scripps Research Institute. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: IFN, interferons; iNOS, inducible nitric-oxide synthase; LPS, lipopolysaccharide; SNOC, S-nitrosocysteine; ETU, S-ethylisothiourea; Ni-NTA, nickel-nitrilotriacetic acid; HA, hemagglutinin; WT, wild type.
C. Lai, J. Schneider, J. J. Struckhoff, L. Martinez-Sobrido, A. Garcia-Sastre, T. Wolff, D.-E. Zhang and D. J. Lenschow, manuscript submitted.
References
- 1.Kaisho, T., and Akira, S. (2006) J. Allergy Clin. Immunol. 117979 –987 [DOI] [PubMed] [Google Scholar]
- 2.Korant, B. D., Blomstrom, D. C., Jonak, G. J., and Knight, E., Jr. (1984) J. Biol. Chem. 25914835 –14839 [PubMed] [Google Scholar]
- 3.Haas, A. L., Ahrens, P., Bright, P. M., and Ankel, H. (1987) J. Biol. Chem. 26211315 –11323 [PubMed] [Google Scholar]
- 4.Loeb, K. R., and Haas, A. L. (1992) J. Biol. Chem. 2677806 –7813 [PubMed] [Google Scholar]
- 5.Kok, K., Hofstra, R., Pilz, A., van den, B. A., Terpstra, P., Buys, C. H., and Carritt, B. (1993) Proc. Natl. Acad. Sci. U. S. A. 906071 –6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao, C., Beaudenon, S. L., Kelley, M. L., Waddell, M. B., Yuan, W., Schulman, B. A., Huibregtse, J. M., and Krug, R. M. (2004) Proc. Natl. Acad. Sci. U. S. A. 1017578 –7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim, K. I., Giannakopoulos, N. V., Virgin, H. W., and Zhang, D. E. (2004) Mol. Cell. Biol. 249592 –9600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dastur, A., Beaudenon, S., Kelley, M., Krug, R. M., and Huibregtse, J. M. (2006) J. Biol. Chem. 2814334 –4338 [DOI] [PubMed] [Google Scholar]
- 9.Takeuchi, T., Iwahara, S., Saeki, Y., Sasajima, H., and Yokosawa, H. (2005) J. Biochem. (Tokyo) 138711 –719 [DOI] [PubMed] [Google Scholar]
- 10.Zou, W., and Zhang, D. E. (2006) J. Biol. Chem. 2813989 –3994 [DOI] [PubMed] [Google Scholar]
- 11.Wong, J. J., Pung, Y. F., Sze, N. S., and Chin, K. C. (2006) Proc. Natl. Acad. Sci. U. S. A. 10310735 –10740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farrell, P. J., Broeze, R. J., and Lengyel, P. (1979) Nature 279523 –525 [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Sastre, A., and Biron, C. A. (2006) Science 312879 –882 [DOI] [PubMed] [Google Scholar]
- 14.Osiak, A., Utermohlen, O., Niendorf, S., Horak, I., and Knobeloch, K. P. (2005) Mol. Cell. Biol. 256338 –6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenschow, D. J., Lai, C., Frias-Staheli, N., Giannakopoulos, N. V., Lutz, A., Wolff, T., Osiak, A., Levine, B., Schmidt, R. E., Garcia-Sastre, A., Leib, D. A., Pekosz, A., Knobeloch, K. P., Horak, I., and Virgin, H. W. (2007) Proc. Natl. Acad. Sci. U. S. A. 1041371 –1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowenstein, C. J., and Padalko, E. (2004) J. Cell Sci. 1172865 –2867 [DOI] [PubMed] [Google Scholar]
- 17.Fang, F. C. (1997) J. Clin. Investig. 992818 –2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nathan, C., and Shiloh, M. U. (2000) Proc. Natl. Acad. Sci. U. S. A. 978841 –8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hess, D. T., Matsumoto, A., Kim, S. O., Marshall, H. E., and Stamler, J. S. (2005) Nat. Rev. Mol. Cell Biol. 6150 –166 [DOI] [PubMed] [Google Scholar]
- 20.Okumura, F., Zou, W., and Zhang, D. E. (2007) Genes Dev. 21255 –260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okumura, F., Hatakeyama, S., Matsumoto, M., Kamura, T., and Nakayama, K. I. (2004) J. Biol. Chem. 27953533 –53543 [DOI] [PubMed] [Google Scholar]
- 22.Lei, S. Z., Pan, Z. H., Aggarwal, S. K., Chen, H. S., Hartman, J., Sucher, N. J., and Lipton, S. A. (1992) Neuron 81087 –1099 [DOI] [PubMed] [Google Scholar]
- 23.Malakhova, O. A., Yan, M., Malakhov, M. P., Yuan, Y., Ritchie, K. J., Kim, K. I., Peterson, L. F., Shuai, K., and Zhang, D. E. (2003) Genes Dev. 17455 –460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaffrey, S. R., and Snyder, S. H. (2001) Sci. STKE 2001,L1. [DOI] [PubMed]
- 25.Uehara, T., Nakamura, T., Yao, D., Shi, Z. Q., Gu, Z., Ma, Y., Masliah, E., Nomura, Y., and Lipton, S. A. (2006) Nature 441513 –517 [DOI] [PubMed] [Google Scholar]
- 26.Sorensen, C. M., Rempel, L. A., Nelson, S. R., Francis, B. R., Perry, D. J., Lewis, R. V., Haas, A. L., and Hansen, T. R. (2007) Biochemistry 46772 –780 [DOI] [PubMed] [Google Scholar]
- 27.Benz, D., Cadet, P., Mantione, K., Zhu, W., and Stefano, G. (2002) Med. Sci. Monit. 81 –4 [PubMed] [Google Scholar]
- 28.Okumura, A., Lu, G., Pitha-Rowe, I., and Pitha, P. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 1031440 –1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, K. I., Yan, M., Malakhova, O., Luo, J. K., Shen, M. F., Zou, W., de la Torre, J. C., and Zhang, D. E. (2006) Mol. Cell. Biol. 26472 –479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reiss, C. S., and Komatsu, T. (1998) J. Virol. 724547 –4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacLean, A., Wei, X. Q., Huang, F. P., Al-Alem, U. A., Chan, W. L., and Liew, F. Y. (1998) J. Gen. Virol. 79825 –830 [DOI] [PubMed] [Google Scholar]
- 32.Fernandez, J. A., Rodrigues, E. G., and Tsuji, M. (2000) Viral Immunol. 13287 –295 [DOI] [PubMed] [Google Scholar]
- 33.Zaragoza, C., Ocampo, C., Saura, M., Leppo, M., Wei, X. Q., Quick, R., Moncada, S., Liew, F. Y., and Lowenstein, C. J. (1998) Proc. Natl. Acad. Sci. U. S. A. 952469 –2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowenstein, C. J., Hill, S. L., Lafond-Walker, A., Wu, J., Allen, G., Landavere, M., Rose, N. R., and Herskowitz, A. (1996) J. Clin. Investig. 971837 –1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaragoza, C., Ocampo, C. J., Saura, M., Bao, C., Leppo, M., Lafond-Walker, A., Thiemann, D. R., Hruban, R., and Lowenstein, C. J. (1999) J. Immunol. 1635497 –5504 [PubMed] [Google Scholar]
- 36.Cho, P. F., Poulin, F., Cho-Park, Y. A., Cho-Park, I. B., Chicoine, J. D., Lasko, P., and Sonenberg, N. (2005) Cell 121411 –423 [DOI] [PubMed] [Google Scholar]
- 37.Cho, P. F., Gamberi, C., Cho-Park, Y. A., Cho-Park, I. B., Lasko, P., and Sonenberg, N. (2006) Curr. Biol. 162035 –2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Payabvash, S., Ghahremani, M. H., Goliaei, A., Mandegary, A., Shafaroodi, H., Amanlou, M., and Dehpour, A. R. (2006) Free Radic. Biol. Med. 411817 –1828 [DOI] [PubMed] [Google Scholar]
- 39.Knight, E., Jr., and Cordova, B. (1991) J. Immunol. 1462280 –2284 [PubMed] [Google Scholar]
- 40.D'Cunha, J., Ramanujam, S., Wagner, R. J., Witt, P. L., Knight, E., Jr., and Borden, E. C. (1996) J. Immunol. 1574100 –4108 [PubMed] [Google Scholar]
- 41.Recht, M., Borden, E. C., and Knight, E., Jr. (1991) J. Immunol. 1472617 –2623 [PubMed] [Google Scholar]
- 42.Owhashi, M., Taoka, Y., Ishii, K., Nakazawa, S., Uemura, H., and Kambara, H. (2003) Biochem. Biophys. Res. Commun. 309533 –539 [DOI] [PubMed] [Google Scholar]