Abstract
Cell migration is central to a number of normal and disease processes. Small organic molecules that inhibit cell migration have potential as both research probes and therapeutic agents. We have identified two tetrahydroisoquinoline natural product analogs with antimigratory activities on Madin-Darby canine kidney epithelial cells: a semisynthetic derivative of quinocarmycin (also known as quinocarcin), DX-52-1, and a more complex synthetic molecule, HUK-921, related to the naphthyridinomycin family. It has been assumed that the cellular effects of reactive tetrahydroisoquinolines result from the alkylation of DNA. We have reported previously that the primary target of DX-52-1 relevant to cell migration appears to be the membrane-cytoskeleton linker protein radixin. Here we extend the analysis of the protein targets of DX-52-1, reporting that the multifunctional carbohydrate-binding protein galectin-3 is a secondary target of DX-52-1 that may also be relevant to the antimigratory effects of both DX-52-1 and HUK-921. All known inhibitors of galectin-3 target its β-galactoside-binding site in the carbohydrate recognition domain. However, we found that DX-52-1 and HUK-921 bind galectin-3 outside of its β-galactoside-binding site. Intriguingly HUK-921, although a less potent inhibitor of cell migration than DX-52-1, had far greater selectivity for galectin-3 over radixin, exhibiting little binding to radixin, both in vitro and in cells. Overexpression of galectin-3 in cells led to a dramatic increase in cell adhesion on different extracellular matrix substrata as well as changes in cell-cell adhesion and cell motility. Galectin-3-overexpressing cells had greatly reduced sensitivity to DX-52-1 and HUK-921, and these compounds caused a change in localization of the overexpressed galectin-3 and reversion of the cells to a more normal morphology. The converse manipulation, RNA interference-based silencing of galectin-3 expression, resulted in reduced cell-matrix adhesion and cell migration. In aggregate, the data suggest that DX-52-1 and HUK-921 inhibit a carbohydrate binding-independent function of galectin-3 that is involved in cell migration.
The movement of cells is a basic feature of a range of normal and pathological processes, including embryonic development, tissue repair, immune cell function, inflammation, angiogenesis, and cancer cell invasion and metastasis. The actin cytoskeleton is the “engine” of cell crawling, whereas dynamic actin-linked cell attachment provides traction for movement (for reviews, see Refs. 1–6). In multicellular organisms, certain cells, such as leukocytes and fibroblasts, tend to move as single individuals, whereas others, such as epithelial and endothelial cells, generally migrate as groups, maintaining cell-cell contacts as they move.
A common way of initiating cell migration in vitro is to grow cells to high density in tissue culture dishes and then mechanically disrupt the cell monolayer. This kind of “scratch wounding” initiates cell migration in myriad cell types. Epithelial cells generally migrate in a collective fashion upon scratch wounding with contacts between cells maintained throughout the process of wound closure. Epithelial cell sheet migration following wounding of cell monolayers of Madin-Darby canine kidney (MDCK)2 cells, a model differentiated epithelial cell line, is regulated by the small GTPase Rac with active protrusive force generation distributed from the wound edge to multiple rows of cells behind it in the moving cell sheet (7, 8). Rac proteins are members of the Rho family of small GTPases, which also includes Rho isoforms and Cdc42; Rac activation leads to membrane ruffling and formation of lamellipodia, broad sheetlike membrane protrusions (for reviews, see Refs. 9 and 10). Unlike Rac, Rho and Cdc42 are not required for wound closure by MDCK cell sheets and instead appear to regulate how uniformly the wound edge advances (7). Epithelial cell sheet migration in this system also depends on phosphoinositides (7), c-Jun N-terminal kinase (11), glycogen synthase kinase 3 (12), and ADP-ribosylation factor 6 (12, 13).
Although there are many available small molecule inhibitors of actin dynamics and cell migration that target actin directly, there is a lack of specific inhibitors of actin-binding proteins and upstream regulators of actin dynamics, cell motility, and cell adhesion (for a review, see Ref. 14). We have exploited the classical scratch-wound closure assay for screening chemical libraries to discover new compounds that inhibit or accelerate cell migration. We have identified a number of new inhibitors of cell migration, including locostatin (15–17) and the tetrahydroisoquinoline DX-52-1, a semisynthetic derivative of quinocarmycin (also known as quinocarcin) (18). These small molecules target Raf kinase inhibitor protein (16) and the membrane-actin cytoskeleton linker protein radixin (18), respectively.
We discovered that radixin is the major target of DX-52-1 following preparation of a biotinylated derivative of DX-52-1, incubation of cells with this derivative, and then affinity-based target purification from cell lysates (18). DX-52-1 acts by specifically and covalently modifying the C-terminal region of radixin, thereby causing radixin to dissociate from actin and certain membrane proteins like the cell adhesion protein CD44, a receptor for hyaluronic acid (18). Overexpression of radixin makes cells less sensitive to the antimigratory activity of DX-52-1, whereas RNA interference (RNAi)-based silencing of radixin expression results in a reduced rate of cell migration (18). Although radixin is consistently the protein that is most intensely labeled by biotinylated DX-52-1 in MDCK cells, we also detected three other less intensely labeled proteins that also appeared to bind DX-52-1 in a saturable manner. In the present study, we describe the isolation of secondary biotinylated DX-52-1-labeled proteins and the evaluation of their possible roles in cell migration.
One of the three secondary targets of DX-52-1 we identified is galectin-3. This protein is a member of a family of lectins encoded by at least 15 different genes (for reviews, see Refs. 19–21). Galectin-3 is composed of an N-terminal repetitive collagen-like domain rich in proline, glycine, and tyrosine and a conserved C-terminal carbohydrate recognition domain. Galectin-3 is found in the nucleus and cytoplasm as well as extracellularly and has a range of apparent functions with roles in cell-extracellular matrix (ECM) adhesion, cell-cell adhesion, cell motility, cell differentiation, cell growth, apoptosis, and pre-mRNA splicing (for reviews, see Refs. 20–27). It has been implicated in numerous normal and disease processes, including inflammation, kidney development, angiogenesis, and cancer progression and metastasis (for reviews, see Refs. 22, 23, and 28–33).
Here we present evidence that galectin-3 is involved in epithelial cell migration during wound closure and, along with radixin, may be one of the targets accounting for the antimigratory activity of DX-52-1. Overexpression of galectin-3 in MDCK cells resulted in dramatically altered morphology, increased cell adhesion and spreading, increased rates of cell motility, and markedly decreased sensitivity of cells to the antimigratory activity of DX-52-1. In contrast, RNAi-based knockdown of galectin-3 in MDCK cells resulted in decreased cell motility and cell adhesion. DX-52-1 appeared to covalently bind to galectin-3 in a saturable and specific manner. Furthermore, alkylation of galectin-3 by biotinylated DX-52-1 was strongly competitive with binding of a more complex synthetic naphthyridinomycin-related tetrahydroisoquinoline that we designated HUK-921. However, binding of both DX-52-1 and HUK-921 to galectin-3 displayed little competition with binding of the β-galactosides N-acetyllactosamine (LacNAc) or lactose. Thus, DX-52-1 and HUK-921 apparently bind outside of the carbohydrate-binding site of galectin-3, unlike other known inhibitors of galectin-3 (for reviews, see Refs. 34, 35). Of particular note is the fact that HUK-921, which also inhibited cell migration, albeit less potently than DX-52-1, bound galectin-3 but exhibited little binding to radixin. Both DX-52-1 and HUK-921 largely reversed the morphology of galectin-3-overexpressing MDCK cells, restoring the cells to a more normal epithelial state. Sensitivity of cells to the antimigratory activity of HUK-921 was greatly diminished in galectin-3-expressing cells but only weakly diminished in radixin-overexpressing cells, unlike the case with DX-52-1 (18). HUK-921 is thus remarkably selective for galectin-3 over radixin and represents a potentially useful probe for evaluating the distinct contributions of the non-carbohydrate-binding functions of galectin-3 to cell motility. The results collectively suggest that functions independent of carbohydrate binding may be involved in the role of galactin-3 in cell migration.
EXPERIMENTAL PROCEDURES
Cell Culture and Wound Closure Assay—MDCK cells were purchased from the American Type Culture Collection. Cells were maintained in a growth medium consisting of minimum essential medium with 10% newborn calf serum and grown at 37 °C with 5% CO2 in a humidified tissue culture incubator. Early passages of cells thawed from frozen stock cultures were used in all experiments. Wound closure experiments were conducted and analyzed as described previously (15). Images taken as a function of time postwounding were imported into ImageJ for morphometric analyses. Cell viability was determined at the end of each experiment by the Trypan blue dye exclusion assay and morphological observations, noting any rounding up of cells or cell detachment.
Cell Proliferation Assay—MDCK cells were plated onto 96-well tissue culture plates (4.0 × 104 cells/well) in the presence or absence of compounds. After culturing the cells for 72 h, cell numbers were measured with the Cell Counting Kit-8 (Dojindo), a tetrazolium salt-based assay, according to the manufacturer's protocol with an absorbance plate reader (μQuant, BioTek Instruments).
Identification of Secondary DX-52-1-binding Proteins—Purification and identification of the three secondary DX-52-1-binding proteins were performed in a manner similar to that described previously for the primary DX-52-1-binding protein, radixin, from lysates of MDCK cells that had been preincubated with biotinylated DX-52-1 (18). All steps were conducted at 4 °C. At each step, fractions containing biotinylated DX-52-1-labeled protein were identified by subjecting samples to SDS-PAGE, followed by transfer to Millipore Immobilon polyvinylidene difluoride membranes and incubation with streptavidin-horseradish peroxidase from Sigma as described previously (18). Protein visualization was by enhanced chemiluminescence (Amersham Biosciences/GE Healthcare) on a Bio-Rad ChemiDoc XRS with Bio-Rad Quantity One software. Ammonium sulfate precipitation of whole-cell lysates resulted in separation of the three secondary DX-52-1-binding proteins into the following fractions: 40% ammonium sulfate pellet for the ∼120-kDa protein and 60% ammonium sulfate pellet for the ∼31- and ∼55-kDa proteins. This was followed by dialysis of the samples against low salt buffer and separation on a 5-ml HiTrap Q-Sepharose anion exchange column (Amersham Biosciences/GE Healthcare). The ∼31- and ∼55-kDa proteins eluted around 100 mm NaCl, whereas the ∼120-kDa protein eluted around 200 mm NaCl. Following desalting, the DX-52-1-binding proteins were affinity-purified by pulldown with streptavidin-agarose beads (Prozyme) after preclearing nonspecific agarose-binding proteins with agarose beads (MP Biomedical) as described previously (18). After SDS-PAGE, specific DX-52-1-binding protein-containing bands were cut out from SYPRO Ruby dye-stained gels and subjected to in-gel digestion with trypsin. Liquid chromatography-tandem mass spectrometry was performed using a 75-μm reverse-phase microcolumn terminating in a PicoView nanoelectrospray source (New Objective) directly coupled to a Thermo Scientific LTQ Orbitrap linear quadrupole ion trap mass spectrometer. These procedures and the subsequent analyses have been described elsewhere (16, 18).
Expression and Purification of Recombinant Galectin-3—A glutathione S-transferase fusion with full-length human galectin-3 in the pGEX-2T (36) vector was expressed in BL21(DE3) Escherichia coli cells. Single bacterial colonies were picked to seed cultures in 500 ml of Luria-Bertani medium. After growing the cells overnight, isopropyl d-thiogalactopyranoside was added to a final concentration of 2 mm, followed by incubation for 4 h, and cells were then collected by centrifugation (5000 × g for 10 min). Bacterial pellets were resuspended in 25 ml of ice-cold phosphate-buffered saline (PBS) containing 2 mm EDTA, 1 mm dithiothreitol (DTT), 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 0.1 mm phenylmethylsulfonyl fluoride, and 0.1 mm benzamidine. Homogenized lysates were centrifuged at 15,000 × g for 30 min at 4 °C. The supernatants were removed and mixed with 5 ml of a 1:1 slurry of glutathione-agarose beads (Sigma) in PBS and then incubated on a rotating wheel for 2 h at 4 °C. Beads were washed four times with PBS, and glutathione S-transferase fusion proteins were eluted with 10 mm reduced glutathione in 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 2.5 mm CaCl2, and 1 mm DTT. The glutathione S-transferase moiety was removed by incubation with bovine thrombin (Sigma) for 12 h at 4 °C. The cloning, expression, and purification of both full-length and C-terminal mouse radixin have been described previously (18, 37). Each recombinant protein was ultimately used in a buffer of 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 2 mm EDTA, 2 mm DTT, 0.5% (w/v) Triton X-100, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 0.1 mm phenylmethylsulfonyl fluoride, and 0.1 mm benzamidine.
Competitive Lactose-binding Assay—The experiment was carried out by procedures similar to those described previously (38). Recombinant galectin-3 (0.5 μg/μl) was incubated for 3 h at room temperature with lactose, LacNAc, DX-52-1, HUK-921, or carrier solvent as control. The samples (30-μl volume) were mixed with 40 μl of a 1:1 lactose-conjugated agarose slurry (Sigma) and then incubated for 2 h at room temperature with gentle rocking. Following centrifugation (1000 × g for 5 min), supernatants were carefully transferred to new tubes, and beads were washed three times to remove unbound galectin-3 with centrifugation (1000 × g for 5 min) between each wash to collect the beads. Bound galectin-3 was eluted from the beads with 20 μl of 2× SDS sample buffer containing β-mercaptoethanol. For Western blot analysis, samples were boiled for 5 min and then subjected to SDS-PAGE on 12% gels. Proteins were transferred from the gels to polyvinylidene difluoride, and the resulting blots were blocked for 2 h with 5% (w/v) bovine serum albumin (BSA) in 1% (w/v) Tween 20/Tris-buffered saline. The blots were probed with an anti-galectin-3 primary antibody (Santa Cruz Biotechnology) for 2 h and horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology) for another 2 h and then washed four times (10 min each) with Tween 20/Tris-buffered saline. Proteins were visualized as described above by chemiluminescence.
Competitive Biotinylated DX-52-1-binding Assay— Recombinant full-length galectin-3, recombinant full-length radixin, and a C-terminal domain fragment of radixin were all tested under identical conditions, i.e. 50 μl of a 400 μg/ml solution in a buffer of 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 2 mm EDTA, 2 mm DTT, 0.5% Triton X-100, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 100 μm phenylmethylsulfonyl fluoride, and 100 μm benzamidine. To each sample, biotinylated DX-52-1 was added with or without simultaneous addition of a 50-fold molar excess of free, non-biotinylated DX-52-1 or 50-fold molar excess of HUK-921. After incubation at 4 °C for 4 h, an equal volume of 2× SDS sample buffer was added to each sample, followed by SDS-PAGE and transfer to polyvinylidene difluoride. Detection of biotinylated DX-52-1-labeled proteins was performed with streptavidin-horseradish peroxidase as described previously (18).
Preparation of MDCK Cells Expressing Green Fluorescent Protein (GFP)-Galectin-3—A construct encoding human galectin-3 fused at its C terminus to enhanced GFP was prepared by polymerase chain reaction of galectin-3 from the pGEX-2T vector with 5′-CCGCTCGAGATGGCAGACAATTTTTCGCTC-3′ and 5′-CTGGGATCCTTATATCATGGTATATGAAGCACT-3′ primers to introduce XhoI and BamHI restriction sites, followed by cloning into the pEGFP-C1 mammalian expression vector (Clontech). Success of cloning was confirmed by evaluating restriction enzyme digestion patterns and by DNA sequencing. Transfection of the pEGFP-C1-galectin-3 construct or the pEGFP-C1 vector alone was mediated by Lipofectamine (Invitrogen) according to the manufacturer's instructions. Stable MDCK transfectants expressing GFP-galectin-3 or GFP alone were selected and maintained in growth medium with 500 μg/ml G418 sulfate (Geneticin; Invitrogen). GFP-positive MDCK cells were then isolated by fluorescence-activated cell sorting on a FACSCalibur flow cytometer (BD Biosciences), and expression was confirmed by fluorescence microscopy and Western blot analysis. For the latter, cells were grown to 80% confluence, and whole-cell lysates were prepared in ice-cold lysis buffer consisting of 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 2 mm EDTA, 2 mm DTT, 0.5% (w/v) Triton X-100, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 0.1 mm phenylmethylsulfonyl fluoride, and 0.1 mm benzamidine. After 30 min on ice, the plates were scraped with cell scrapers. Samples were homogenized by passing through a 27-gauge needle, transferred to new tubes, and centrifuged at 15,000 × g for 30 min. Supernatants were concentrated in Amicon centrifugal concentrators (Millipore) with a molecular mass cutoff of 5,000 Da at 4,000 × g at 4 °C. Total protein concentration was determined by the Bradford assay (39). Western blot analyses were then carried out as described earlier with anti-GFP and anti-galectin-3 antibodies (both from Santa Cruz Biotechnology).
Fluorescence Microscopy—MDCK cells were plated onto glass coverslips placed in 12-well tissue culture plates at 2 × 105 cells/well and grown to confluence. Cell monolayers were wounded by manual scratching with ultramicropipette tips. For experiments involving sparsely seeded cultures, cells were plated at low density onto fibronectin-coated glass coverslips in 12-well plates (4 × 104 cells/well) that had been blocked with 2% BSA. After incubation at 37 °C in 5% CO2 for 3 h in the case of the wounded monolayers and 48 h in the case of the sparsely seeded cultures, cells were fixed with 3.7% formaldehyde and permeabilized with 0.5% Triton X-100 for 15 min. In some experiments, cells were also stained for filamentous actin with 50 nm tetramethylrhodamine isothiocyanate-labeled phalloidin (Sigma). Coverslips were mounted onto glass slides in Mowiol 4-88 (0.1 g/ml) mounting medium (Calbiochem) containing 15 mg/ml antifade agent 1,4-diazabicyclo[2,2,2]octane (Sigma). Slides were examined on an inverted fluorescence microscope (Leica DMI 6000B), and images were captured using a Hamamatsu Orca AG cooled charge-coupled device camera.
Small Interfering RNA (siRNA)-mediated Silencing of Galectin-3 Expression—Four potentially effective siRNA target sequences corresponding to different parts of the coding region of canine galectin-3 and not found elsewhere in the canine genome, were identified by using Ambion's siRNA Target Finder algorithm. The siRNA target sequences were synthesized as short hairpin RNA (shRNA) molecules and ligated into an shRNA expression vector under the direction of the U6 promoter. This shRNA expression vector has been described previously (12); it contains a GFP expression cassette to aid in isolation of stable transfectants and was modified from an earlier vector we designed (16). The constructs were transfected into MDCK cells with Lipofectamine (Invitrogen) according to the manufacturer's protocol. GFP-positive stable transfectants were isolated by fluorescence-activated cell sorting after selection in growth medium with 100 μg/ml hygromycin B (A. G. Scientific). The efficiency of knockdown was evaluated by Western blot analysis of whole-cell lysates prepared as described earlier. The most efficient knockdown was achieved with an siRNA corresponding to nucleotides 708–728 of canine galectin-3 (GenBank™ accession number XM 848582), although nucleotides 589–609 were also highly effective as an siRNA sequence for knockdown. As a control, we used a cell line expressing an siRNA corresponding to nucleotides 631–651 of canine galectin-3 that we found to cause no change in galectin-3 protein levels by Western blot analysis. Each cell line was maintained in growth medium containing 100 μg/ml hygromycin B.
Cell Adhesion Assay—Cell adhesion experiments were performed in 96-well plates coated with human fibronectin, human laminin, rat tail collagen I, or gelatin (BD BioCoat, BD Biosciences). As a negative control, BSA-coated 96-well plates were used. Prior to plating cells for these experiments, the wells were washed three times with PBS, and nonspecific binding sites were blocked with 1% BSA in PBS at 37 °C for 1 h. Wells were then washed again three times with PBS. Experiments were initiated by plating MDCK cells in serum-free minimum essential medium containing 0.5% (w/v) BSA in the presence or absence of DX-52-1 onto the different precoated surfaces of the 96-well plates at 5 × 104 cells/well. Cells were allowed to adhere at 37 °C for 2 h. Non-adherent cells were removed by gently washing the plates three times with cold PBS, and adherent cells were fixed with 150 μl of methanol for 10 min. The methanol was removed by aspiration, and the plates were allowed to air dry. The relative number of adherent cells was determined as described previously (40). Adherent cells were stained for 30 min with 100 μl of filtered 1% (w/v) methylene blue in 10 mm borate buffer (pH 8.5) and then washed extensively with 10 mm borate buffer (pH 8.5). The stain from each well was eluted with 100 μl of a 1:1 (v/v) solution of ethanol and 0.1 m HCl. The level of staining was determined by measuring absorbance at 650 nm on an absorbance plate reader. Control values from BSA-coated plates were subtracted from values from ECM-coated plates. Each experiment was performed in triplicate.
RESULTS
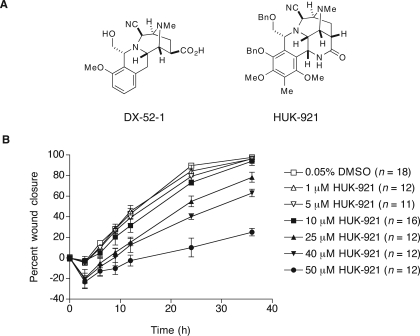
Antimigratory Tetrahydroisoquinolines and Their Targets—Given that DX-52-1 (Fig. 1A) potently inhibits epithelial cell migration (18), we evaluated the biological activity of a more complex tetrahydroisoquinoline (Fig. 1A), herein designated HUK-921, as part of an effort to investigate whether structurally related tetrahydroisoquinolines might have activity similar to that of DX-52-1. HUK-921 is an advanced intermediate in a recent synthesis of cyanocycline A and bioxalomycin β2 (41), compounds of the naphthyridinomycin family. We found that HUK-921 also inhibits cell migration during wound closure in MDCK cell monolayers (Fig. 1B). We calculated the half-maximal inhibitory concentration (IC50) for inhibition of cell sheet migration in the wound closure assay at 24 h postwounding by HUK-921 as 32.5 μm. The antimigratory activity of HUK-921 is thus considerably less than that of DX-52-1, whose IC50 for inhibition of wound closure at 24 h after wounding is 140 nm (18). HUK-921 did not display any cytotoxic or antiproliferative effects at the concentrations that inhibited cell migration based on the Trypan blue dye exclusion assay, the tetrazolium salt-based assay, and observations of cell morphology (data not shown).
FIGURE 1.
Structures and antimigratory activities of analogs of reactive tetrahydroisoquinoline natural products. A, chemical structures of DX-52-1 and HUK-921. Me, methyl; Bn, benzyl. B, HUK-921 inhibits cell sheet migration during wound closure in MDCK cell monolayers. Final dimethyl sulfoxide (DMSO) carrier solvent concentration was 0.05% for each treatment (compound was added 30 min prior to wounding). Error bars represent S.E. for the indicated number of wounds. The IC50 for inhibition of wound closure at 24 h by HUK-921 is 32.5 μm as calculated from the data presented in this figure, whereas the corresponding value for DX-52-1 is 140 nm (18).
We previously reported the preparation of biotinylated DX-52-1 and the finding that it specifically and covalently modifies four proteins in MDCK cells (18). The binding to all the DX-52-1-binding proteins is largely competed away when a 50-fold molar excess of free, non-biotinylated DX-52-1 is added simultaneously with biotinylated DX-52-1. Although radixin, which migrates as an ∼80-kDa protein by SDS-PAGE, is by far the most intensely labeled DX-52-1-binding protein and was confirmed as a molecular target of DX-52-1 relevant to the antimigratory activity of the compound in our previous work (18), we wished to identify and assess the possible roles of the secondary DX-52-1-binding proteins in cell migration. To determine their identity, the secondary DX-52-1-binding proteins were each purified to apparent homogeneity following lysis of biotinylated DX-52-1-treated MDCK cells. After SDS-PAGE, the bands corresponding to each of the DX-52-1-binding proteins were excised from the gels and subjected to in-gel digestion with trypsin. The identity of each protein was revealed by tandem mass spectrometry as galectin-3 (∼31 kDa), ATP synthase β-chain (∼55 kDa), and heterogeneous nuclear ribonucleoprotein U isoform b (∼120 kDa).
Galectin-3 is an attractive candidate as a DX-52-1-binding protein also relevant to the antimigratory activity of DX-52-1 because it has already been implicated in cell motility, tumor invasion, and metastasis. On the other hand, both ATP synthase and heterogeneous nuclear ribonucleoprotein U are housekeeping proteins with no known or immediately plausible direct role in cell motility. We nonetheless tested whether inhibition of ATP synthase had any effect on cell migration by treating cells with oligomycin, an inhibitor of ATP synthase, and found no subtoxic antimigratory activity of oligomycin (data not shown). There are currently no available small molecule inhibitors of heterogeneous nuclear ribonucleoproteins, which share weak sequence homology with the N-termnal domain of galectin-3 (42) and, like galectin-3, have roles in pre-mRNA splicing (for a review, see Ref. 43). We do not yet know whether DX-52-1 binds in the region of homology or affects pre-mRNA splicing.
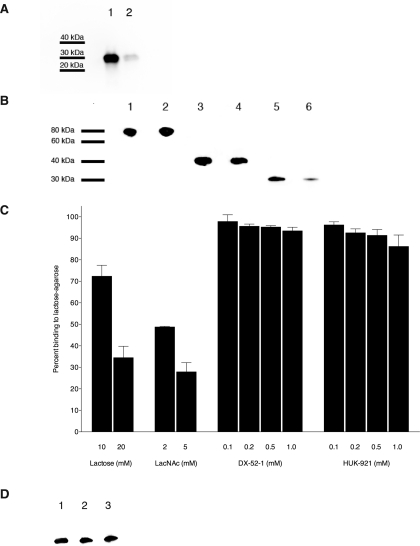
We investigated the binding of DX-52-1 and HUK-921 to pure recombinant galectin-3 in vitro. The binding of biotinylated DX-52-1 to recombinant human galectin-3 was largely competed away when a 50-fold molar excess of free, non-biotinylated DX-52-1 was added simultaneously with the biotinylated compound (Fig. 2A). We calculated that the rate of alkylation of galectin-3 by biotinylated DX-52-1 is 5-fold slower than that of radixin based on the kinetics of binding in vitro. Moreover, although HUK-921 had much lower antimigratory activity than DX-52-1, competitive binding experiments revealed that HUK-921 competed strongly with biotinylated DX-52-1 for covalent binding to galectin-3, but there was little competition between biotinylated DX-52-1 and HUK-921 for binding to full-length or a C-terminal domain fragment of radixin (Fig. 2B), the latter containing the putative DX-52-1-binding site on radixin (18).
FIGURE 2.
DX-52-1 and HUK-921 bind galectin-3 non-competitively with known carbohydrate ligands of galectin-3. A, DX-52-1 covalently and specifically binds galectin-3. Recombinant galectin-3 was incubated with 10 μm biotinylated DX-52-1 (lane 1) or 10 μm biotinylated DX-52-1 plus 500 μm free, non-biotinylated DX-52-1 competitor added simultaneously (lane 2). Following SDS-PAGE and transfer to polyvinylidene difluoride, the blot was probed with streptavidin-horseradish peroxidase. The degree of competition of biotinylated DX-52-1 by free DX-52-1 was calculated as 84.3 ± 3.5% (mean ± S.E.) from three independent experiments. B, HUK-921 competes with biotinylated DX-52-1 for binding to galectin-3 but not radixin. Lane 1, full-length radixin with 10 μm biotinylated DX-52-1; lane 2, full-length radixin with 10 μm biotinylated DX-52-1 plus 500 μm HUK-921; lane 3, C-terminal domain fragment of radixin with 10 μm biotinylated DX-52-1; lane 4, C-terminal domain fragment of radixin with 10 μm biotinylated DX-52-1 plus 500 μm HUK-921; lane 5, galectin-3 with 10 μm biotinylated DX-52-1; lane 6, galectin-3 with 10 μm biotinylated DX-52-1 plus 500 μm HUK-921. This experiment was done similarly to that shown in A except that potential competition of the binding of biotinylated DX-52-1 to galectin-3 and radixin by HUK-921 added simultaneously was evaluated. Based on quantitation of the degree of binding from the intensity of the bands from three independent experiments, we calculated the percent competition between biotinylated DX-52-1 and HUK-921 as 16.5 ± 6.8% for full-length radixin, 29.5 ± 6.9% for the C-terminal domain fragment of radixin, and 84.3 ± 4.2% for galectin-3 (means ± S.E. in all cases). C, percent binding of galectin-3 to lactose-agarose beads in the presence of the indicated concentrations of lactose, LacNAc, DX-52-1, or HUK-921. The extent of galectin-3 binding to lactose-conjugated agarose in the presence of DMSO is defined as 100% binding. (Every individual experiment was performed along with a parallel DMSO control, and each value was normalized to its parallel control.) Error bars represent S.E. for four to seven independent experiments, quantitated from Western blots, as described under “Experimental Procedures.” D, lactose and LacNAc do not compete with biotinylated DX-52-1 for binding to galectin-3. Lane 1, galectin-3 with 20 μm biotinylated DX-52-1; lane 2, galectin-3 with 20 μm biotinylated DX-52-1 plus 20 mm lactose; lane 3, galectin-3 with 20 μm biotinylated DX-52-1 plus 5 mm LacNAc. Based on quantitation of the intensity of bands from three independent experiments, we found only very weak competition between 20 μm biotinylated DX-52-1 and 20 mm lactose or 5 mm LacNAc and calculated the percent competition values as 25.0 ± 5.2 and 20.4 ± 2.4% (means ± S.E.), respectively.
DX-52-1 and HUK-921 Bind Outside of the Carbohydrate-binding Site of Galectin-3—We investigated whether DX-52-1 or HUK-921 could compete with binding of galectin-3 to lactose-agarose beads. We found that pretreatment of galectin-3 with DX-52-1 or HUK-921 prior to pulling down lactose-binding proteins had no significant effect at any concentration of the compounds on the binding of galectin-3 to the lactose-agarose beads compared with the solvent carrier control (Fig. 2C). In contrast, free lactose and LacNAc competed with binding of galectin-3 to the lactose-agarose beads as expected. We then examined whether lactose or LacNAc could compete with biotinylated DX-52-1 for binding to galectin-3. We observed little competition between DX-52-1 and lactose and LacNAc for binding galectin-3 (Fig. 2D).
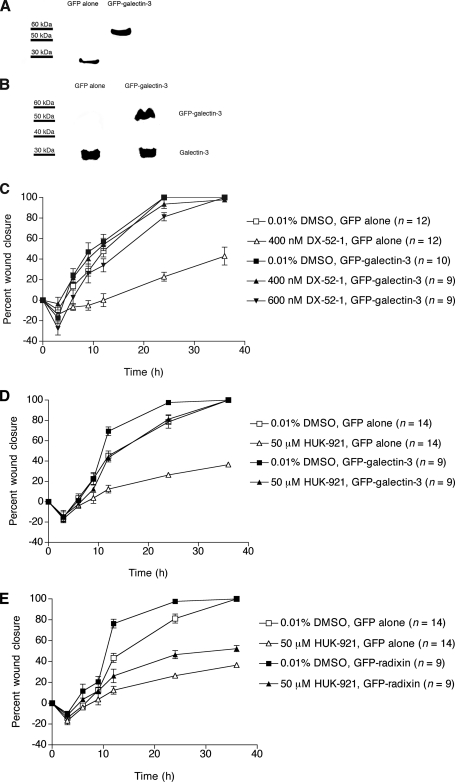
Overexpression of Galectin-3 in MDCK Cells Reduces Their Sensitivity to the Antimigratory Activities of DX-52-1 and HUK-921—We generated MDCK cells stably expressing GFP-galectin-3 to investigate the role of galectin-3 in cell migration in MDCK cell monolayers as well as the sensitivity of cells to DX-52-1 with overexpression of galectin-3. The same GFP-galectin-3 fusion protein (44) and a galectin-3 fusion with the cyan variant of GFP (45) have been reported previously and appear to have normal galectin-3 function. In the GFP-galectin-3-expressing cells we prepared, the GFP-galectin-3 fusion protein appeared stable (Fig. 3A), and the level of expression of GFP-galectin-3, as calculated from the intensity of bands on Western blots, was ∼126% that of endogenous galectin-3 (Fig. 3B). We performed wound closure assays in confluent monolayers of galectin-3-overexpressing cells. Galectin-3-overexpressing cells migrated at rates similar to that of control cells (Fig. 3C). However, overexpression of galectin-3 did result in greatly decreased sensitivity of the cells to the antimigratory activity of DX-52-1. There was little or no effect on the migration of galectin-3-overexpressing cells with treatment of DX-52-1 at a subtoxic concentration (400 nm) that strongly inhibited wound closure in control MDCK cells expressing GFP alone (Fig. 3C). At high concentrations (≥600 nm), DX-52-1 became cytotoxic in control MDCK cells with long term exposure as in the wound closure experiments (where compound was added 30 min before wounding, and then the experiment proceeded for 36 h after wounding in the presence of compound). However, the compound at 600 nm displayed no cytotoxic activity and possibly only weak antimigratory activity in galectin-3-overexpressing cells, although the decrease in rate of migration is not statistically significant. At ≥1 μm, DX-52-1 became cytotoxic in galectin-3-overexpressing cells as with long term treatment.
FIGURE 3.
Overexpression of galectin-3 results in strongly reduced sensitivity to the antimigratory activities of DX-52-1 and HUK-921, whereas overexpression of radixin only weakly reduces sensitivity to HUK-921. A, Western blot analysis with an anti-GFP antibody to probe whole-cell lysates from MDCK cells stably expressing GFP alone or GFP-galectin-3. B, Western blot analysis with an anti-galectin-3 antibody showing levels of GFP-galectin-3 and endogenous galectin-3 in MDCK cells stably expressing GFP alone or GFP-galectin-3. C, overexpression of galectin-3 markedly decreases the sensitivity of MDCK cells to the inhibitory effect of DX-52-1 on cell sheet migration in the wound closure assay. D, overexpression of galectin-3 markedly decreases the sensitivity of MDCK cells to the antimigratory activity of HUK-921. E, overexpression of radixin only mildly decreases the sensitivity of MDCK cells to the antimigratory activity of HUK-921 in contrast to previous results with DX-52-1 (18). Error bars represent S.E. for the indicated number of wounds in C–E.
Overexpression of galectin-3 also strongly reduced the sensitivity of cells to the antimigratory activity of HUK-921 (Fig. 3D). In contrast, overexpression of radixin resulted in only a mild decrease in the sensitivity of the cells to the antimigratory activity of HUK-921 (Fig. 3E), unlike our previously reported findings with DX-52-1 (18).
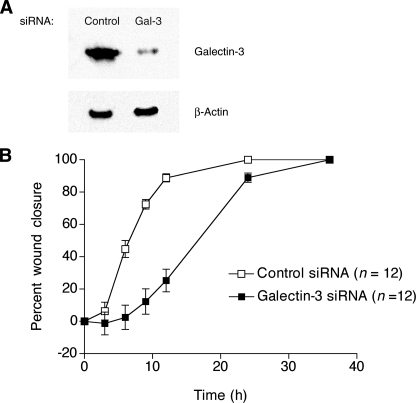
RNAi-based Knockdown of Galectin-3 Results in a Lower Rate of Cell Migration—Inhibition of the β-galactoside-binding function of galectin-3 with 5 mm LacNAc had little or no effect on wound closure in MDCK cell sheets (data not shown). However, because DX-52-1 and HUK-921 did inhibit cell migration and bound galectin-3 non-competitively with LacNAc, we wished to silence the expression of galectin-3 by RNAi to investigate the overall role of galectin-3 in cell migration. We therefore stably transfected MDCK cells with shRNA expression constructs encoding galectin-3-specific siRNAs. The efficiency of silencing of galectin-3 expression for the most effective of four siRNAs we designed and tested was 88% as determined by Western blot analysis (Fig. 4A). Galectin-3 knockdown cells exhibited reduced rates of wound closure compared with control cells expressing an inert siRNA (Fig. 4B).
FIGURE 4.
RNAi-based silencing of galectin-3 expression results in a reduced rate of cell migration. A, Western blot analysis of whole-cell lysates from MDCK cells stably expressing either an inert, control siRNA or an effective galectin-3-specific (Gal-3) siRNA prepared as described under “Experimental Procedures.” B, knockdown of galectin-3 results in a decreased rate of cell sheet migration during wound closure in MDCK cell monolayers. Error bars represent S.E. for the indicated number of wounds. Note that, unlike knockdown of galectin-3 or treatment with DX-52-1 or HUK-921, treatment with LacNAc had little or no effect on cell migration in this system.
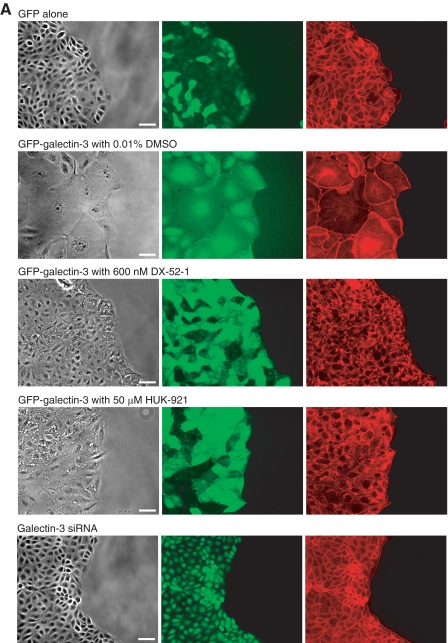
Effects of Galectin-3 Overexpression and Knockdown on Cell-ECM Adhesion—Striking morphological changes occurred in MDCK cells with overexpression of galectin-3: unlike normal MDCK cells, which have a cuboidal morphology, galectin-3-overexpressing cells were extremely spread and often had asymmetric shapes (Fig. 5A). There was an apparent decrease in strength and/or extent of cell-cell contacts because small gaps were often visible between cells. In galectin-3-overexpressing cells, GFP-galectin-3 appeared to be concentrated in and around the nucleus and at the periphery of the cell (Fig. 5A). Treatment with DX-52-1 or HUK-921 resulted in the cells adopting a much less spread shape more like the cuboidal cells expressing GFP alone (Fig. 5A). Treatment with DX-52-1 or HUK-921 also appeared to cause a change in the localization of GFP-galectin-3. The amount of GFP-galectin-3 localized to the nucleus appeared to decrease upon treatment with DX-52-1 or HUK-921 with GFP-galectin-3 having a more diffuse distribution throughout the cell. Treatment of cells with DX-52-1 or HUK-921 had little or no effect on the overall levels of galectin-3 or radixin in the cell based on Western blot analysis (data not shown).
FIGURE 5.
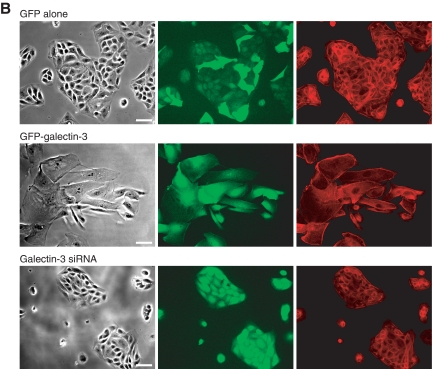
Galectin-3-overexpressing cells are highly spread and display reduced cell-cell contact, and treatment with DX-52-1 or HUK-921 causes a change in localization of GFP-galectin-3 and reversion of cells to a more normal morphology, whereas galectin-3 knockdown cells display reduced membrane protrusion. A, images of cells expressing GFP alone, GFP-galectin-3 in the presence or absence of DX-52-1 or HUK-921, or a galectin-3-specific siRNA, all 4 h after wounding of MDCK cell monolayer treatment (compounds were added 30 min prior to wounding). From left to right, each row consists of a phase-contrast image, a GFP fluorescence image, and a second fluorescence image showing TRITC-phalloidin staining of filamentous actin. Scale bars, 50 μm. B, images of cells expressing GFP alone, GFP-galectin-3, or a galectin-3-specific siRNA plated at low density on fibronectin-coated glass coverslips and then imaged 48 h later. From left to right, each row consists of a phase-contrast image, a GFP fluorescence image, and a second fluorescence image showing TRITC-phalloidin staining of filamentous actin. Scale bars, 50 μm. (Note that the shRNA expression vector harbors a GFP cassette, as described under “Experimental Procedures,” so the galectin-3 knockdown cells also display GFP fluorescence.)
In contrast to the dramatically altered morphology of galectin-3-overexpressing cells, galectin-3 knockdown cells appeared more normal in morphology (Fig. 5A). However, lamellipodial protrusion at the wound edge appeared limited, consistent with the slower observed rate of migration of the galectin-3 knockdown cells compared with controls (Fig. 4B).
Lower density cultures revealed that galectin-3-overexpressing cells do not grow in tightly cell-cell adherent islands, as do normal MDCK cells, but as collectives of highly spread cells without a uniformly high degree of cell-cell contact (Fig. 5B). As in the confluent cultures, galectin-3 knockdown cells at lower density displayed a more normal morphology but with less apparent spreading of cells at the edge of the islands.
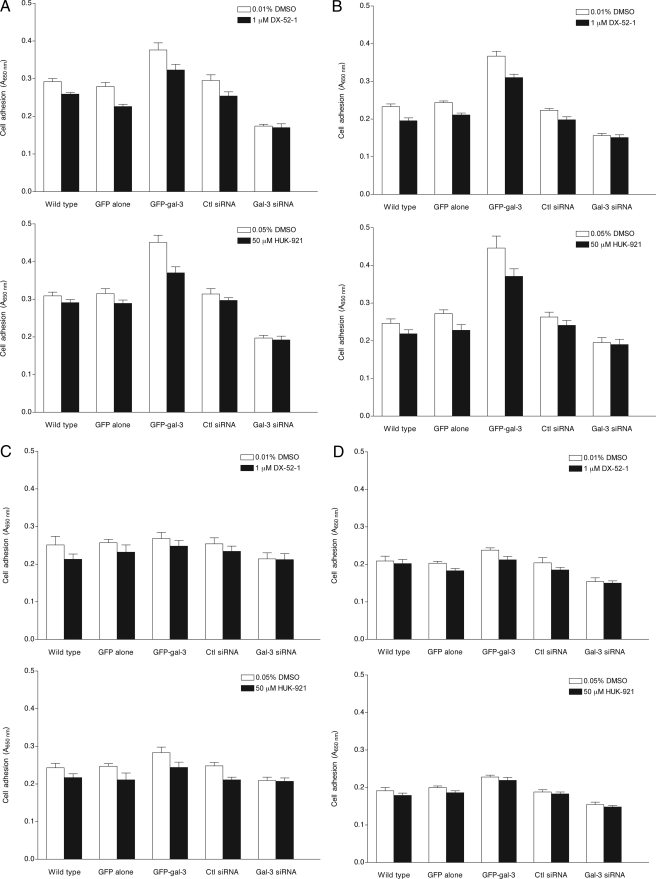
We next compared the adhesion of galectin-3-overexpressing cells, galectin-3 knockdown cells, and corresponding control cells, in the presence or absence of DX-52-1 or HUK-921, to various ECM proteins. MDCK cells were harvested and incubated in 96-well plates coated with fibronectin, laminin, collagen I, or gelatin (hydrolyzed collagen) in serum-free minimum essential medium. After 2-h incubation, the wells were washed, and adherent cells were quantified by methylene blue staining. The effects of galectin-3 overexpression and knockdown on cell-ECM adhesion were evaluated in comparison with the adhesion of control MDCK cells stably expressing GFP or a control siRNA. We found that overexpression of galectin-3 in MDCK cells results in stronger attachment to both fibronectin and laminin (Fig. 6, A and B). The difference between galectin-3-overexpressing and control cells was less marked on collagen I and gelatin (Fig. 6, C and D). When control and galectin-3-overexpressing cells were incubated in the presence of DX-52-1 or HUK-921 on the different ECM-coated plates, the numbers of strongly adherent cells were mildly reduced, particularly on fibronectin and laminin (Fig. 6, A and B). Knockdown of galectin-3 resulted in decreased adhesion of cells to fibronectin, laminin, and collagen I, but not to gelatin, as compared with controls (Fig. 6, A–D). No further reduction in adhesion of galectin-3 knockdown cells was observed upon treatment with DX-52-1 or HUK-921.
FIGURE 6.
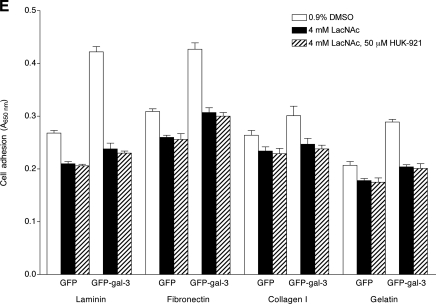
Effects of galectin-3 overexpression, galectin-3 knockdown, or treatment with DX-52-1, HUK-921, or LacNAc on adhesion of cells to different ECM proteins. Shown are the levels of attachment of MDCK cells that were not manipulated (“wild type”) and cells expressing GFP alone, GFP-galectin-3 (GFP-gal-3), a control (Ctl) siRNA, or a galectin-3 specific (Gal-3) siRNA to fibronectin (A), laminin (B), collagen I (C), and gelatin (hydrolyzed collagen; D) in the presence or absence of DX-52-1 or HUK-921, all 2 h after plating. E, levels of attachment of MDCK cells expressing GFP alone or GFP-galectin-3 (GFP-gal-3) in the presence or absence of LacNAc, with or without co-treatment with HUK-921, 2 h after plating. Error bars represent S.E. for three independent experiments in each case.
Treatment of cells expressing GFP alone and those expressing GFP-galectin-3 with LacNAc resulted in statistically significantly decreased adhesion of both cells expressing GFP alone and those expressing GFP-galectin-3 to all the ECM substrata, and co-treatment with HUK-921 led to no further reduction in cell adhesion (Fig. 6E). In contrast, as mentioned above, Lac-NAc had little or no effect on cell migration during wound closure in MDCK cell monolayers.
DISCUSSION
Despite its relatively modest size, galectin-3 is a multifunctional protein that is found in the nucleus, cytoplasm, and extracellular environment (for reviews, see Refs. 20–27). In addition, a splice variant of galectin-3 with a putative transmembrane domain has been identified (46). The extracellular fraction of galectin-3 is secreted by a non-classical secretory pathway not involving the endoplasmic reticulum and Golgi apparatus, which may be significant in that it prevents galectin-3 from binding to nascent glycoconjugates; extracellular galectin-3 binds to various glycoproteins, including ECM proteins, such as fibronectin and laminin, and cell adhesion proteins, such as integrins and neural cell adhesion molecule (for reviews, see Refs. 20, 21, 24, 25).
The fact that DX-52-1 and HUK-921 inhibited cell migration (Fig. 1B and Ref. 18) and targeted galectin-3 outside of its carbohydrate-binding site of galectin-3 (Fig. 2, C and D) raises the possibility that these compounds inhibit another galectin-3 function(s) that may also be important in cell migration. Because Lac-NAc did not inhibit migration of MDCK cells, the function of galectin-3 involved in cell motility in this system appears to be independent of its β-galactoside-binding activity.
Because of the role of galectin-3 in myriad disease-relevant processes, there is considerable interest in the development of new galectin-3 inhibitors. Existing galectin-3 inhibitors are carbohydrate- or peptide-based and target the carbohydrate-binding site of the C-terminal carbohydrate recognition domain of the protein (for reviews, see Refs. 34 and 35). DX-52-1 and HUK-921 are thus unique in that they specifically bind galectin-3 outside of its carbohydrate-binding site.
Although less potent than DX-52-1, the remarkable selectivity of HUK-921 for galectin-3 over radixin (Fig. 2B) suggests that target selectivity is derived from specific recognition interactions between the protein and small molecule prior to alkylation. It is likely that further development of related molecules would yield compounds with yet different selectivities, such as those with greater selectivity for radixin over galectin-3.
The mechanism of covalent modification of galectin-3 may involve elimination of the nitrile group of DX-52-1 or HUK-921 and attack of a nucleophilic amino acid side chain of galectin-3 on the resulting iminium ion as proposed for alkylation of radixin by DX-52-1 (18). Quinocarmycin, of which DX-52-1 is a derivative resulting from opening of the oxazolidine ring of the natural product by hydrocyanation, has DNA-damaging activity through two proposed mechanisms, one involving DNA alkylation and the other involving oxidative cleavage of DNA through the generation of superoxide (47–54). DX-52-1, unlike its parent natural product, is incapable of mediating oxidative scission of DNA (53, 54). However, the mechanism proposed for alkylation of DNA by quinocarmycin (48), also involving formation of an iminium ion, would be accessible to DX-52-1. Nevertheless, such activity alone would be unlikely to account for the antimigratory activity of DX-52-1 or HUK-921 because other DNA-alkylating agents such as saframycin C (18), another reactive tetrahydroisoquinoline, and mitomycin C (8) do not inhibit cell migration during wound closure by MDCK cell sheets. Interestingly, covalent DNA-saframycin C adducts bind glyceraldehyde-3-phosphate dehydrogenase, and this appears to be involved in the mechanism of the antiproliferative activity of saframycin C (55). The data presented here extend the notion that reactive tetrahydroisoquinolines possess important protein targets and that their cellular activities may not be confined to or even principally result from any targeting of DNA.
The sensitivity of MDCK cells to the antimigratory activity of DX-52-1 is reduced when either radixin (18) or galectin-3 (Fig. 3C) is expressed constitutively from the cytomegalovirus promoter in stably transfected cells. This is most likely due to the fact that because both of them are specific DX-52-1-binding proteins, overexpression of either protein covalently and irreversibly “titrates” out a large proportion of the soluble DX-52-1, reducing its concentration in solution and thus its ability to inhibit either protein completely. Because of continual biosynthesis of each transgenic protein and protein turnover, the observed loss of sensitivity would not even require an instantaneously high concentration of either protein.
Overexpression of galectin-3 also led to a dramatic reduction in sensitivity of cells to the antimigratory activity of HUK-921 (Fig. 3D). However, there was only weakly reduced sensitivity to the antimigratory activity of HUK-921 when radixin was overexpressed (Fig. 3F), unlike the case with DX-52-1 (18). These results are consistent with the in vitro binding data (Fig. 2, A and B, and Ref. 18). The data thus suggest that DX-52-1 binds both radixin and galectin-3 in vitro and in cells, whereas HUK-921 predominantly binds galectin-3 over radixin in vitro and in cells.
Treatment of galectin-3-overexpressing MDCK cells with DX-52-1 or HUK-921 resulted in a change in localization of GFP-galectin-3 and reversion of the galectin-3-overexpressing cells from a highly spread state to a more normal cuboidal morphology (Fig. 5A), strongly suggesting that these compounds inhibit an important function of galectin-3. This function is not purely related to cell adhesion because DX-52-1 and HUK-921 only moderately inhibited cell adhesion and did so more on certain ECM substrata (Fig. 6, A and B) than others (Fig. 6, C and D), unlike LacNAc, which inhibited cell adhesion more strongly and in a less substratum-dependent manner (Fig. 6E). DX-52-1 and HUK-921 may inhibit interactions with intracellular galectin-3-binding proteins involved in cell migration. Alternatively, these compounds may inhibit non-carbohydrate-based interactions between galectin-3 and extracellular proteins or the extracellular domains of transmembrane proteins with roles in cell adhesion and cell migration, such as ECM proteins or integrins. Interactions between galectin-3 and glycoproteins likely involve some degree of direct protein-protein contact in addition to protein-carbohydrate contact, and the protein-protein component of such an interaction could be affected by DX-52-1 and HUK-921. Efforts are ongoing to identify the precise binding site of these tetrahydroisoquinolines on galectin-3 and to define the functional interactions of galectin-3 that are disrupted by these compounds.
DX-52-1 and HUK-921 inhibited cell migration, bound galectin-3 outside of its carbohydrate-binding site, and largely reverted galectin-3-overexpressing MDCK cells to a more normal epithelial morphology. Furthermore, although less potent an inhibitor of cell migration than DX-52-1, HUK-921 displayed remarkable selectively for galectin-3 over radixin. These data suggest that carbohydrate binding-independent functions of galectin-3 are important for cell motility and validate the use of privileged natural product scaffolds for the exploration and delineation of specific protein function.
Acknowledgments
We thank William S. Lane (Harvard Microchemistry and Proteomics Analysis Facility, Harvard University) for protein identification. We thank William B. Stallcup (Burnham Institute) and Walter Nickel (University of Heidelberg) for human galectin-3 cDNA. We also thank Carol E. Norris (University of Connecticut) for technical assistance with sorting cells by fluorescence-activated cell sorting. We are grateful to the National Cancer Institute for providing quinocarmycin analog DX-52-1 (NSC 607097).
This work was supported, in whole or in part, by National Institutes of Health Grant GM077622 (to G. F.). This work was also supported by National Science Foundation Grant CHE-0553313 (to P. P. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: MDCK, Madin-Darby canine kidney; RNAi, RNA interference; ECM, extracellular matrix; LacNAc, N-acetyllactosamine; PBS, phosphate-buffered saline; DTT, dithiothreitol; BSA, bovine serum albumin; GFP, green fluorescent protein; TRITC, tetramethylrhodamine isothiocyanate; siRNA, small interfering RNA; shRNA, short hairpin RNA; IC50, half-maximal inhibitory concentration; DMSO, dimethyl sulfoxide.
References
- 1.Disanza, A., Steffen, A., Hertzog, M., Frittoli, E., Rottner, K., and Scita, G. (2005) CMLS Cell. Mol. Life Sci. 62 955–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li, S., Guan, J. L., and Chien, S. (2005) Annu. Rev. Biomed. Eng. 7 105–150 [DOI] [PubMed] [Google Scholar]
- 3.Vicente-Manzanares, M., Webb, D. J., and Horwitz, A. R. (2005) J. Cell Sci. 118 4917–4919 [DOI] [PubMed] [Google Scholar]
- 4.Stossel, T. P., Fenteany, G., and Hartwig, J. H. (2006) J. Cell Sci. 119 3261–3264 [DOI] [PubMed] [Google Scholar]
- 5.Ananthakrishnan, R., and Ehrlicher, A. (2007) Int. J. Biol. Sci. 3 303–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Clainche, C., and Carlier, M. F. (2008) Physiol. Rev. 88 489–513 [DOI] [PubMed] [Google Scholar]
- 7.Fenteany, G., Janmey, P. A., and Stossel, T. P. (2000) Curr. Biol. 10 831–838 [DOI] [PubMed] [Google Scholar]
- 8.Farooqui, R., and Fenteany, G. (2005) J. Cell Sci. 118 51–63 [DOI] [PubMed] [Google Scholar]
- 9.Hall, A. (2005) Biochem. Soc. Trans. 33 891–895 [DOI] [PubMed] [Google Scholar]
- 10.Bustelo, X. R., Sauzeau, V., and Berenjeno, I. M. (2007) BioEssays 29 356–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altan, Z. M., and Fenteany, G. (2004) Biochem. Biophys. Res. Commun. 322 56–67 [DOI] [PubMed] [Google Scholar]
- 12.Farooqui, R., Zhu, S., and Fenteany, G. (2006) Exp. Cell Res. 312 1514–1525 [DOI] [PubMed] [Google Scholar]
- 13.Santy, L. C., and Casanova, J. E. (2001) J. Cell Biol. 154 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenteany, G., and Zhu, S. (2003) Curr. Top. Med. Chem. 3 593–616 [DOI] [PubMed] [Google Scholar]
- 15.Mc Henry, K. T., Ankala, S. V., Ghosh, A. K., and Fenteany, G. (2002) ChemBioChem 11 1105–1111 [DOI] [PubMed] [Google Scholar]
- 16.Zhu, S., Mc Henry, K. T., Lane, W. S., and Fenteany, G. (2005) Chem. Biol. 12 981–991 [DOI] [PubMed] [Google Scholar]
- 17.Mc Henry, K. T., Montesano, R., Zhu, S., Beshir, A. B., Tang, H.-H., Yeung, K., and Fenteany, G. (2008) J. Cell. Biochem. 103 972–985 [DOI] [PubMed] [Google Scholar]
- 18.Kahsai, A. W., Zhu, S., Wardrop, D. J., Lane, W. S., and Fenteany, G. (2006) Chem. Biol. 13 973–983 [DOI] [PubMed] [Google Scholar]
- 19.Cooper, D. N. (2002) Biochim. Biophys. Acta 1572 209–231 [DOI] [PubMed] [Google Scholar]
- 20.Dumic, J., Dabelic, S., and Flögel, M. (2006) Biochim. Biophys. Acta 1760 616–635 [DOI] [PubMed] [Google Scholar]
- 21.Elola, M. T., Wolfenstein-Todel, C., Troncoso, M. F., Vasta, G. R., and Rabinovich, G. A. (2007) CMLS Cell. Mol. Life Sci. 64 1679–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, F. T., Patterson, R. J., and Wang, J. L. (2002) Biochim. Biophys. Acta 1572 263–273 [DOI] [PubMed] [Google Scholar]
- 23.Yang, R. Y., and Liu, F. T. (2003) CMLS Cell. Mol. Life Sci. 60 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krześlak, A., and Lipinśka, A. (2004) Cell. Mol. Biol. Lett. 9 305–328 [PubMed] [Google Scholar]
- 25.Ochieng, J., Furtak, V., and Lukyanov, P. (2004) Glycoconj. J. 19 527–535 [DOI] [PubMed] [Google Scholar]
- 26.Patterson, R. J., Wang, W., and Wang, J. L. (2004) Glycoconj. J. 19 499–506 [DOI] [PubMed] [Google Scholar]
- 27.Nangia-Makker, P., Nakahara, S., Hogan, V., and Raz, A. (2007) J. Bioenerg. Biomembr. 39 79–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Almkvist, J., and Karlsson, A. (2004) Glycoconj. J. 19 575–581 [DOI] [PubMed] [Google Scholar]
- 29.Califice, S., Castronovo, V., Bracke, M., and van den Brûle, F. (2004) Oncogene 23 7527–7536 [DOI] [PubMed] [Google Scholar]
- 30.Hughes, R. C. (2004) Glycoconj. J. 19 621–629 [DOI] [PubMed] [Google Scholar]
- 31.Takenaka, Y., Fukumori, T., and Raz, A. (2004) Glycoconj. J. 19 543–549 [DOI] [PubMed] [Google Scholar]
- 32.Chen, H. Y., Liu, F. T., and Yang, R. Y. (2005) Arch. Immunol. Ther. Exp. (Warsz.) 53 497–504 [PubMed] [Google Scholar]
- 33.Liu, F. T., and Hsu, D. K. (2007) Drug News Perspect. 20 455–460 [DOI] [PubMed] [Google Scholar]
- 34.Ingrassia, L., Camby, I., Lefranc, F., Mathieu, V., Nshimyumukiza, P., Darro, F., and Kiss, R. (2006) Curr. Med. Chem. 13 3513–3527 [DOI] [PubMed] [Google Scholar]
- 35.Pieters, R. J. (2006) ChemBioChem 7 721–728 [DOI] [PubMed] [Google Scholar]
- 36.Fukushi, J., Makagiansar, I. T., and Stallcup, W. B. (2004) Mol. Biol. Cell 15 3580–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaiskunaite, R., Adarichev, V., Furthmayr, H., Kozasa, T., Gudkov, A., and Voyno-Yasenetskaya, T. A. (2000) J. Biol. Chem. 275 26206–26212 [DOI] [PubMed] [Google Scholar]
- 38.Dyer, K. D., and Rosenberg, H. F. (1996) Life Sci. 23 2073–2082 [DOI] [PubMed] [Google Scholar]
- 39.Bradford, M. M. (1976) Anal. Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- 40.Oliver, M. H., Harrison, N. K., Bishop, J. E., Cole, P. J., and Laurent, G. J. (1989) J. Cell Sci. 92 513–518 [DOI] [PubMed] [Google Scholar]
- 41.Kaniskan, H.Ü., and Garner, P. (2007) J. Am. Chem. Soc. 129 15460–15461 [DOI] [PubMed] [Google Scholar]
- 42.Jia, S., and Wang, J. L. (1988) J. Biol. Chem. 263 6009–6011 [PubMed] [Google Scholar]
- 43.Carpenter, B., MacKay, C., Alnabulsi, A., MacKay, M., Telfer, C., Melvin, W. T., and Murray, G. I. (2006) Biochim. Biophys. Acta 1765 85–100 [DOI] [PubMed] [Google Scholar]
- 44.McFarlane, S., Glenn, J. V., Lichanska, A. M., Simpson, D. A., and Stitt, A. W. (2005) Br. J. Ophthalmol. 89 107–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delacour, D., Cramm-Behrens, C. I., Drobecq, H., Le Bivic, A., Naim, H. Y., and Jacob, R. (2006) Curr. Biol. 16 408–414 [DOI] [PubMed] [Google Scholar]
- 46.Gorski, J. P., Liu, F. T., Artigues, A., Castagna, L. F., and Osdoby, P. (2002) J. Biol. Chem. 277 18840–18848 [DOI] [PubMed] [Google Scholar]
- 47.Tomita, F., Takahashi, K., and Tamaoki, T. (1984) J. Antibiot. (Tokyo) 37 1268–1272 [DOI] [PubMed] [Google Scholar]
- 48.Hill, G. C., Wunz, T. P., and Remers, W. A. (1988) J. Comput.-Aided Mol. Des. 2 91–106 [DOI] [PubMed] [Google Scholar]
- 49.Kanamaru, R., Konishi, Y., Ishioka, C., Kakuta, H., Sato, T., Ishikawa, A., Asamura, M., and Wakui, A. (1988) Cancer Chemother. Pharmacol. 22 197–200 [DOI] [PubMed] [Google Scholar]
- 50.Flanagan, M. E., Rollins, S. B., and Williams, R. M. (1995) Chem. Biol. 2 147–156 [DOI] [PubMed] [Google Scholar]
- 51.Herberich, B., Scott, J. D., and Williams, R. M. (2000) Bioorg. Med. Chem. 8 523–532 [DOI] [PubMed] [Google Scholar]
- 52.Williams, R. M., Glinka, T., Gallegos, R., Ehrlich, P. P., Flanagan, M. E., Coffman, H., and Park, G. (1990) Tetrahedron 47 2629–2642 [Google Scholar]
- 53.Williams, R. M., Glinka, T., Flanagan, M. E., Gallegos, R., Coffman, H., and Pei, D. (1992) J. Am. Chem. Soc. 114 733–740 [Google Scholar]
- 54.Williams, R. M., Flanagan, M. E., and Tippie, T. N. (1994) Biochemistry 33 4086–4092 [DOI] [PubMed] [Google Scholar]
- 55.Xing, C., LaPorte, J. R., Barbay, J. K., and Myers, A. G. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 5862–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]