Abstract
It has been reported previously that inhibitory κB kinase (IKK) supports osteoclastogenesis through NF-κB-mediated prevention of apoptosis. This finding suggests that the ligand for receptor activator of NF-κB (RANKL), the master osteoclastogenic cytokine, induces apoptosis of osteoclast precursors (OCPs) in the absence of IKKβ/NF-κB competency. To validate this hypothesis, we sought to determine the pro-apoptotic signaling factors induced by RANKL in IKKβ-null osteoclast OCPs and to rescue osteoclast differentiation in the absence of IKKβ through their inhibition. To accomplish this, we generated mice that lack IKKβ in multiple hematopoietic lineages, including OCPs. We found that these mice possess both in vitro and in vivo defects in osteoclast generation, in concurrence with previous reports, and that this defect is a result of susceptibility to RANKL-mediated apoptosis as a result of gain-of-function of JNK activation. We demonstrate that differentiation of OCPs depends on IKKβ because reduced IKKβ mRNA expression correlates with impaired induction of osteoclast differentiation markers in response to RANKL stimulation. We further show that fine-tuned inhibition of JNK activation in these cells inhibits RANKL-induced apoptosis and restores the ability of IKKβ-null OCPs to become mature osteoclasts. Our data highlight the pro-osteoclastogenic and anti-apoptotic roles of IKKβ in OCPs and identify a pro-apoptotic mechanism activated within the RANK signalosome.
Osteoclasts develop from bone marrow macrophage precursors under the control of two cytokines, receptor activator of NF-κB ligand (RANKL)2 (1) and m-CSF (2). RANKL induces osteoclast commitment and development by signaling downstream to several transcription factors, the most important of which is NF-κB (3). NF-κB is a family of transcription factors whose activity coordinates a major component of the cellular inflammatory program, and its function is essential for osteoclastogenesis (4, 5). NF-κB signaling involves two distinct but cooperating pathways, one canonical and one alternative pathway (6), which has recently been shown to be critical in osteoclast biology (7).
NF-κB is activated by the inhibitory κB kinase (IKK) complex, which is crucial for osteoclastogenesis. The IKK complex is composed of two catalytically active members, IKKα and IKKβ, and a regulatory subunit IKKγ/NEMO. IKKα mediates activation of the alternative pathway by phosphorylation of NF-κB2/p100 (6), whereas IKKβ is important for activation of the canonical pathway through phosphorylation of IκB (8). The importance of the signaling activity of the IKK complex in osteoclasts is demonstrated by the defect in osteoclastogenesis noted in mice lacking IKKα (9) or IKKβ (10). Despite the sequence homology of these two kinases, their relative importance in osteoclastogenesis is strikingly different. For example, osteoclasts devoid of active IKKα only demonstrate an in vitro defect in osteoclastogenesis, whereas the bone phenotype of the mouse is remarkably normal. On the other hand, mice with an inducible osteoclast precursor-specific deletion of IKKβ demonstrate both in vitro and in vivo defects in osteoclastogenesis and are resistant to inflammatory osteolysis (10). Given these findings, it is evident that investigating the mechanism by which IKK2 supports osteoclastogenesis will improve our understanding of osteoclast biology and diseases attributable to overactive osteoclasts.
We and others have shown that diverse methods of IKK blockade arrest osteoclastogenesis by induction of apoptosis (10–12). We were interested in the pro-apoptotic signals downstream of RANKL in the absence of IKKβ, and we hypothesized that inhibition of these signals would be sufficient to rescue the osteoclast defect of cells lacking IKKβ. Our findings reveal that loss of IKKβ in osteoclast precursors (OCPs) results in a gain-of-function of JNK activation in response to RANKL that results in apoptosis. Furthermore, fine-tuned inhibition of this gain-of-function in JNK activation is sufficient to rescue osteoclastogenesis in OCPs lacking IKKβ. This finding demonstrates that the necessity of IKKβ for osteoclastogenesis may be evaded by inhibiting the pro-apoptotic effects of RANKL and designates JNK activation in the osteoclast as a potential means to induce cell death in OCPs.
MATERIALS AND METHODS
Reagents—Antibodies against IKKβ, IKKα, NEMO, actin, JNK, p38, Akt, MKP1, phospho-c-Jun, and c-Jun as well as horseradish peroxidase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibodies against phospho-JNK, phospho-p38, phospho-Akt, and PARP were purchased from Cell Signaling Technologies, Inc. (Danvers, MA). Antibody against MKP5 was purchased from Abcam (Cambridge, MA). Cytokines were purchased from R & D Systems (Minneapolis, MN). TAT-TI-JIP was purchased from EMD Biosciences, Inc. (La Jolla, CA). Enhanced chemiluminescence kit was purchased from Pierce. All other chemicals were purchased from Sigma unless otherwise indicated.
Animals—CD11b Cre Y-chromosomal transgenic and floxed IKKβ mice on a C57BL/6 background were reported previously (12, 13). Male Cre+ Floxed IKKβ homozygotes were generated by crossing the above mice.
Cell Culture—Osteoclast precursors were enriched from bone marrow of 2–3-week-old mice. Briefly, whole marrow was flushed from long bones into α-minimum Eagle's medium and was centrifuged at 453 relative centrifugal force. Marrow pellets were resuspended in whole media (α-minimum Eagle's medium with penicillin/streptomycin, 10% heat-inactivated fetal bovine serum) supplemented with 10 ng/ml m-CSF. Cell suspensions were plated onto Petri dishes at 37 °C in 5% CO2 for 5 days and then were plated according to experimental conditions.
Osteoclast Formation Assay—Osteoclast precursors were plated in triplicate at a density of 3.0 × 104 cells in 200 μl of whole media supplemented with 10 ng/ml m-CSF and RANKL in 96-well tissue culture plates. TAT-TI-JIP was added at the time of cell plating (day 1). TNF-α and LPS were added at day 4 of the assay. Mature osteoclasts form between day 5 and day 6 of culture, at which point the cells are fixed and stained for tartrate-resistant acid phosphatase (TRAP) to visualize osteoclasts (leukocyte acid phosphatase kit, Sigma). TRAP-positive multinucleated cells with three or more nuclei were scored as osteoclasts.
Protein Phosphorylation Assay—Osteoclast precursors were plated onto tissue culture dishes overnight in whole media supplemented with m-CSF. Cells were then serum-starved for 4–6 h and stimulated with the indicated cytokine for a planned time course. At the allotted time, cells were lysed in cell lysis buffer containing (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1% Triton X-100, 1 mm Na3VO4, 1 μg/ml leupeptin, 1 mm NaF, 1 mm phenylmethylsulfonyl fluoride, and distilled deionized H2O). Protein concentration was measured by standard BCA assay (Pierce). 10–20 μg of total cell protein was used for Western blot.
Apoptosis Assay—Osteoclast precursors were plated onto tissue culture dishes overnight in whole media supplemented with m-CSF. Cells were serum-starved for 6 h and stimulated with 10 ng/ml RANKL for the indicated time. At the allotted time, cells were lysed as described above; protein was normalized, and samples were analyzed by Western blot.
Rescue of Apoptosis—Osteoclast precursors were plated onto tissue culture dishes for 24 h in whole media supplemented with m-CSF. Four groups of two plates of cells were plated in this assay. Each group was treated with either sterile PBS, 0.4, 1.0, or 2.0 μm TAT-TI-JIP. Also at the time of plating, one plate from each group was stimulated with either sterile PBS or 20 ng/ml RANKL. After 24 h, cells were lysed as described above; protein was normalized, and samples were analyzed by Western blot.
Osteoclast Differentiation Assay—Osteoclast precursors were plated in whole media supplemented with m-CSF. Cells were either not stimulated or were stimulated with 10 ng/ml RANKL for 5 days. Total RNA was isolated from cells using TRIzol reagent (Invitrogen) according to the manufacturer's standard protocol.
Reverse Transcription—1.0 μg of total RNA was subjected to reverse transcription under the following conditions. 1.0 μg of RNA and 1.0 μg of random hexamer primer in 10 μl of nuclease-free deionized H2O in PCR tubes were heated to 70 °C for 5 min, cooled to 42 °C, and set on ice. The following components were then added at the indicated amounts or concentrations for a total reaction volume of 20 μl: 1× RT avian myeloblastosis virus buffer (Roche Applied Science), 40 units of RNasin (Promega, San Luis Obispo, CA), 1.25 mm dNTPs, 5 mm sodium pyruvate, 5 units of reverse transcriptase enzyme (Roche Applied Science). To produce cDNA, tubes were placed in a thermocycler programmed as follows: 42 °C for 60 min, 50 °C for 10 min, 95 °C for 5 min, and 4 °C to hold.
Bone Resorption Assay—Bone marrow osteoclast precursors were plated onto BD Biocoat osteologic tissue culture slides (BD Biosciences) in the presence of 10 ng/ml m-CSF and RANKL with or without 0.4 μm TAT-TI-JIP for the indicated times. Resorption pits were determined as clear areas in the osteologic matrix. Representative photographs were taken at ×10 Magnification.
Real Time PCR—Triplicate samples of 4 μl of cDNA product (5× diluted), 10 μl of SybrGreen PCR Master Mix (Applied Biosystems, Inc. Foster City, CA), 0.1 μl each of 10 μm forward and reverse primer stocks, and 6 μl of nuclease-free deionized H2O were subjected to real time PCR according to the following program in a 7300 AB real time PCR system: 50 °C for 2 min, 95 °C for 10 min (95 °C for 15 s, 60 °C for 1 min) for 40 cycles. Results were analyzed using AB RQ Study software. Real time PCR primers were designed using Primer Express software (Applied Biosystems, Inc.) as follows: mouse actin forward 5′-CTTCTACAATGAGCTGCGTG-3′ and mouse actin reverse 5′-TCATGAGGTAGTCTGTCAGG-3′; mouse TRAP forward 5′-CGACCATTGTTAGCCACATACG-3′ and mouse TRAP reverse 5′-CACATAGCCCACACCGTTCTC-3′; mouse calcitonin receptor forward 5′-CAAGAACCTTAGCTGCCAGAG-3′ and mouse calcitonin receptor reverse 5′-CAAGCACGCGGACAATGTTG-3′; mouse MMP9 forward 5′-CAGGGAGATGCCCATTTCG-3′ and mouse MMP9 reverse 5′-GGGCACCATTTGGAGTTTCCA-3′; mouse A20 forward 5′-CAGAAAAAAGTGGTGAAGGTGTGA-3′ and mouse A20 reverse 5′-CCAGGCTCTGACCTCTGTTACA-3′; mouse cIAP1 (birc2) forward 5′-GTGATGGTGGCTTGAGATGTTG-3′ and mouse cIAP1 (birc2) reverse 5′-CAAGAACTCACACCTTGGAAACC-3′; mouse cIAP2 (birc3) forward 5′-GAAGTGGGCTGCGGTATCA-3′ and mouse cIAP2 (birc3) reverse 5′-GCGCTGTCTTGAACCATGTTC-3′; mouse Bcl-xL forward 5′-GCGGCTGGGACACTTTTG-3′ and mouse Bcl-xL reverse 5′-CAGAACCACACCAGCCACAGT-3′; mouse XIAP forward 5′-CGGATCGTTACTTTTGGAACATG-3′ and mouse XIAP reverse 5′-CGCCTTCACCTAAAGCATAAAATC-3′; mouse cathepsin K forward 5′-GGAAGAAGACTCACCAGAAGC-3′ and mouse cathepsin K reverse 5′-GTCATATAGCCGCCTCCACAG-3′; mouse β3 integrin forward 5′-TTACCCCGTGGACATCTACTA-3′ and mouse β3 integrin reverse 5′-AGTCTTCCATCCAGGGCAATA-3′; and mouse GAPDH forward 5′-CTTCACCACCATGGAGAAGGC-3′ and mouse GAPDH reverse 5′-GACGGACACATTGGGGGTAG-3′.
Western Blot Assay—Total cell lysates were boiled in the presence of an equal volume of 2× SDS sample buffer (0.5 m Tris-HCl, pH 6.8, 10% (w/v) SDS, 10% glycerol, 0.05% (w/v) bromphenol blue, 3% β-mercaptoethanol, and distilled water) for 5 min and subjected to electrophoresis on 8–10% SDS-PAGE. The proteins were transferred to nitrocellulose membranes using a semi-dry blotter (Bio-Rad) and incubated in blocking solution (10% skim milk prepared in phosphate-buffered saline containing 0.05% Tween 20) to reduce nonspecific binding. The membranes were washed with phosphate-buffered saline/Tween buffer and exposed to primary antibodies (16 h at 4 °C), washed again four times, and incubated with the respective secondary horseradish peroxidase-conjugated antibodies (1 h at room temperature). The membranes were washed extensively (four times for 15 min), and an ECL detection assay was performed following the manufacturer's directions.
Western Blot Quantification—Where indicated, protein expression was quantified using Quantity One 1-D Analysis Software (Bio-Rad).
Histology—Long bones were collected from mice and fixed in 10% buffered formalin for 24 h. Bones were then decalcified for 7 days in decalcification buffer (14% (w/v) EDTA, H4NOH, pH 7.2), dehydrated in graded ethanol (30–70%), cleared through xylene, and embedded in paraffin. Paraffin sections were stained histochemically for TRAP to visualize osteoclasts or immunohistochemically for terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) with the ApopTag peroxidase in situ apoptosis detection kit (Millipore/Chemicon International, Temecula, CA) to detect apoptotic cells.
RESULTS
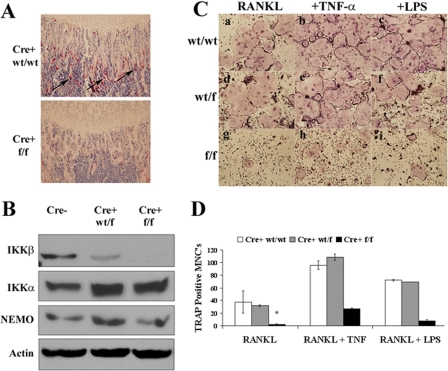
Mice with an Osteoclast Precursor IKKβ Deficiency Demonstrate in Vitro and in Vivo Defects in Osteoclastogenesis—IKKβ has been shown to be necessary for osteoclast formation (10). To define the mechanism through which IKKβ supports osteoclastogenesis, we generated mice with a deficiency of IKKβ in multiple hematopoietic lineages, including OCPs by crossing CD11b Cre recombinase transgenic mice (13) with mice possessing floxed IKKβ (14). In this study, we focus on the osteoclast phenotype, so we will refer to the resultant knock-out (Cre-positive floxed/floxed (f/f) IKKβ) mice as OCPΔIKKβ or Cre+ f/f. OCPΔIKKβ mice possess a hampered ability to generate osteoclasts in vivo as evidenced by a significantly reduced number of TRAP-positive osteoclasts compared with controls observed by histochemical staining for TRAP in long bones (Fig. 1A). This is further supported by lack of IKKβ protein in osteoclast precursors of OCPΔIKKβ mice (Fig. 1B). We demonstrate that this defect is cell-autonomous by culturing bone marrow-derived OCPs in the presence of m-CSF and RANKL. OCPΔIKKβ cells form significantly fewer multinucleated osteoclasts (Fig. 1C, panel g) compared with Cre-positive IKKβ wild-type/wild-type (WT/WT) (Fig. 1C, panel a) and Cre-positive IKKβ wild-type/floxed (WT/f) heterozygous littermate controls (Fig. 1C, panel d). Furthermore, stimulation with TNF-α or LPS (Fig. 1C, panels h and i) is insufficient to rescue the osteoclast defect of OCPΔIKKβ cells. Compared with WT cells, Cre-positive IKKβ (WT/f) OCPs also show decreased IKKβ protein expression (Fig. 1B), but this difference does not result in impaired osteoclastogenesis (Fig. 1C, panels d–f).
FIGURE 1.
Mice with IKKβ-deleted osteoclast precursors possess a defect in in vivo and in vitro osteoclastogenesis. A, histochemical TRAP stain at growth plate of femur of CD11b Cre-positive WT/WT IKKβ and CD11b Cre-positive floxed/floxed IKKβ (OCPΔIKKβ) mice to visualize osteoclasts. Arrows indicate osteoclasts. B, Western blot for indicated proteins in total cell lysates of osteoclast precursors from CD11b Cre-negative, CD11b Cre-positive WT/floxed IKKβ, and CD11b Cre-positive floxed/floxed (f/f) IKKβ mice. C and D, osteoclast precursors from CD11b Cre-positive wt/wtIKKβ, CD11b Cre-positive WT/floxed IKKβ, and CD11b Cre-positive floxed/floxed IKKβ mice were cultured in osteoclastogenic conditions. C, cells were either not stimulated (panels a, d, and g) or were further stimulated with 10 ng/ml TNF-α (panels b, e, and h) or 100 ng/ml LPS (panels c, f, and i) on day 4 of culture. Cells were fixed and histochemically stained for TRAP to visualize osteoclasts on day 6 of culture. D, quantification of C. TRAP-positive multinucleated cells (MNCs) with three or more nuclei were scored as osteoclasts. Asterisk indicates p < 0.005 for difference between number of TRAP-positive MNCs in wells represented by panels d and g.
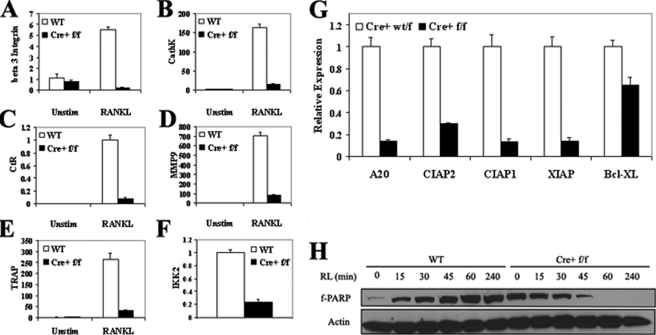
OCPΔIKKβ Are Prone to RANKL-induced Apoptosis and Display Defective Osteoclast Differentiation—IKKβ has previously been demonstrated to protect OCPs from TNF-α-mediated apoptosis (10). We sought to determine whether IKKβ-deficient OCPs are similarly susceptible to apoptosis in response to RANKL and to test whether the absence of IKKβ results in defective osteoclast differentiation. To accomplish this, we cultured OCPΔIKKβ and control OCPs in the presence of m-CSF and RANKL for 0 or 5 days to induce osteoclast differentiation. We measured by real time PCR the expression of several markers for osteoclast differentiation. We observe a significant decrease in the expression of mRNA for the osteoclast markers β3 integrin (Fig. 2A), cathepsin K (Fig. 2B), calcitonin receptor (Fig. 2C), matrix metalloproteinase 9 (Fig. 2D), and TRAP (Fig. 2E) in RANKL-stimulated OCPΔIKKβ cells compared with controls. This failure to express osteoclast markers in osteoclastogenic conditions correlates with up to an 81% reduction in expression of IKKβ mRNA in OCPΔIKKβ cells compared with controls (Fig. 2F). Interestingly, in OCPΔIKKβ cells that express higher levels of IKKβ mRNA-63% reduction compared with control OCPs, osteoclast marker expression after RANKL stimulation is not affected, yet they still fail to form multinucleated osteoclasts in in vitro culture. This finding indicates that IKKβ also serves a differentiation-independent function to support osteoclastogenesis. These observations led us to surmise that IKKβ is essential at various stages for differentiation and survival of RANKL-stimulated OCPs.
FIGURE 2.
OCPΔIKKβ are defective in osteoclast differentiation and demonstrate increased susceptibility to apoptosis. OCPΔIKKβ and control OCPs were plated in whole media supplemented with 10 ng/ml m-CSF. Cells were either not stimulated (Unstim) or were stimulated with 10 ng/ml RANKL for 5 days to induce osteoclast differentiation. mRNA was collected and analyzed by real time PCR for markers of osteoclast differentiation: A, β3 integrin; B, cathepsin K (Cath K); C, calcitonin receptor (CtR); D, matrix metalloproteinase 9 (MMP9); E, tartrate-resistant acid phosphatase (TRAP); F, IKKβ (IKK2). GAPDH served as the internal standard for cDNA normalization. Data are presented as relative quantification with WT nonstimulated levels serving as the reference point (relative expression value of 1). Values represent mean quantification plus the S.E. G, OCPΔIKKβ and control OCPs were plated in whole media supplemented with 10 ng/ml m-CSF. mRNA was collected and analyzed by real time PCR for the indicated NF-κB-regulated anti-apoptotic proteins as follows: A20, cIAP2 (cellular inhibitor of apoptosis 2); cIAP1 (cellular inhibitor of apoptosis 1); XIAP (X-linked inhibitor of apoptosis); and Bcl-xL. β-Actin served as the internal standard for cDNA normalization. Data are presented as relative quantification with control levels serving as the reference point (relative expression value of 1). Values represent mean quantification plus the S.E. Data are representative of three independent experiments. H, OCPΔIKKβ and control OCPs were serum-starved and were either not stimulated or were stimulated with 10 ng/ml RANKL for 15, 30, 45, 60, or 240 min. Total cell lysates were analyzed by Western blot for integrity of full-length PARP (f-PARP). β-Actin served as the loading control.
Therefore, we tested whether OCPΔIKKβ cells were more susceptible to apoptosis than control cells. First, we cultured OCPΔIKKβ and control OCPs in the presence of whole media supplemented with fetal bovine serum and m-CSF to promote survival and measured by real time PCR the expression of mRNA for several NF-κB-controlled anti-apoptotic proteins (15). We note significant reduction in expression of mRNA for A20, cellular inhibitor of apoptosis 2 (c-IAP2), c-IAP1, Bcl-xL, and X-linked inhibitor of apoptosis (XIAP) in OCPΔIKKβ compared with control cells (Fig. 2G).
To determine whether OCPΔIKKβ undergo apoptosis in response to RANKL, we exposed serum-starved OCPΔIKKβ and control OCPs to RANKL for a time course of 4 h. We detected the kinetics of PARP cleavage by Western blot as a molecular signature of apoptosis. We note the disappearance of full-length PARP in OCPΔIKKβ cells after 1 h, whereas in control cells, the integrity of full-length PARP is preserved over the time course of RANKL exposure (Fig. 2H), which indicates that RANKL has a pro-apoptotic effect on osteoclast precursors deficient in IKKβ. We conclude that IKKβ is necessary for osteoclast differentiation and the survival of osteoclast precursors exposed to RANKL.
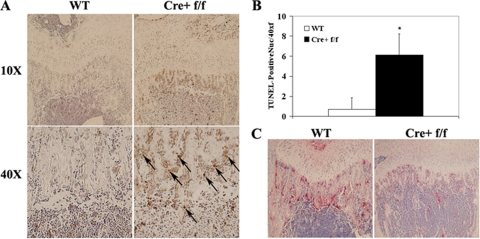
To determine whether apoptosis of osteoclasts or OCPs from OCPΔIKKβ mice contributes to the paucity of osteoclasts observed in vivo, we stained sections of long bones of OCPΔIKKβ and control mice immunohistochemically with the TUNEL method to detect apoptosis. We note a significantly greater number of TUNEL-positive peritrabecular nuclei resembling apoptotic osteoclasts and OCPs in OCPΔIKKβ compared with control long bones (Fig. 3, A and B). Based on our data, we conclude that apoptosis contributes to the osteoclast defect in OCPΔIKKβ mice.
FIGURE 3.
Apoptosis contributes to the in vivo deficiency of osteoclasts in OCPΔIKKβ mice. A, immunoperoxidase TUNEL stain and hematoxylin counterstain of histological sections of growth plate of humerus from OCPΔIKKβ and control mice to visualize apoptosis of peritrabecular osteoclasts and OCPs. Upper images were taken at ×10 magnification, and lower images are panels from upper images taken at ×40 magnification. B, graph depicting quantification of TUNEL-positive peritrabecular nuclei per ×40 field visualized by light microscopy. Arrows indicate apoptotic nuclei. Asterisk denotes p < 0.001. C, TRAP stain of sections taken from same paraffin-embedded bones used for TUNEL stain in A to demonstrate correlation between apoptosis and defective in vivo osteoclastogenesis.
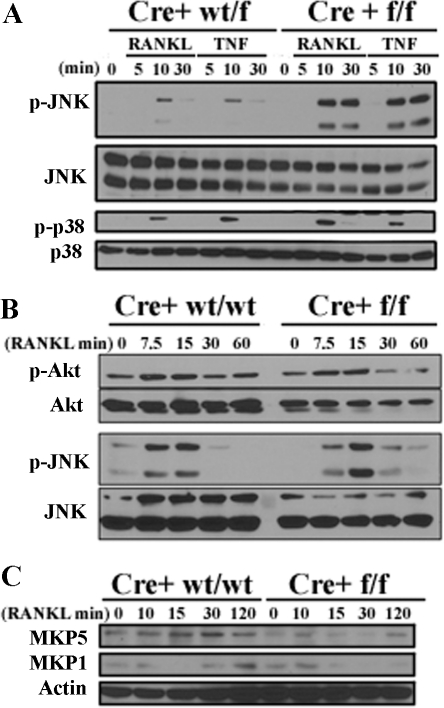
OCPΔIKKβ Possesses a Gain-of-Function in JNK Activation—We were interested in potential pro-apoptotic signals induced by RANKL in differentiating IKKβ-deficient OCPs. To address this, we performed a phosphoprotein screen by Western blot analysis in OCPΔIKKβ and control OCPs after stimulation with a time course of RANKL or TNF-α. We postulated that OCPΔIKKβ kinase signaling would possess a signature that would favor apoptosis. We noted several aberrations in the pattern of protein phosphorylation in OCPΔIKKβ compared with control cells. Of particular interest was an increase and prolongation of JNK phosphorylation in response to RANKL (Fig. 4, A and B) and TNF-α (Fig. 4A) in OCPΔIKKβ compared with control OCPs. Interestingly, p38 (Fig. 4A) and Akt (Fig. 4B) phosphorylation remain unaffected in the absence of IKKβ, suggesting that JNK down-regulation is a specific function of IKKβ. We observe that MAP kinase phosphatase 1 (MKP1) protein resynthesis after degradation and MKP5 protein synthesis are dampened after RANKL stimulation of OCPΔIKKβ cells compared with controls. In particular, MKP1 and MKP5 protein levels maximize after 30 min of RANKL stimulation of control OCPs (Fig. 4C). This time point correlates with down-regulation of JNK phosphorylation after RANKL stimulation (Fig. 4, A and B). In OCPΔIKKβ cells, JNK phosphorylation is sustained at 30 min of RANKL stimulation (Fig. 4, A and B), which correlates with the absence of detectable MKP1 and MKP5 protein at this time point in OCPΔIKKβ cells (Fig. 4C). Because MKP1 (16) and MKP5 (17) serve as JNK phosphatases, MKP induction may serve as an IKKβ-dependent mechanism for JNK down-regulation after RANKL stimulation. We postulated that the gain-of-function of JNK activation may result in apoptosis of OCPΔIKKβ cells.
FIGURE 4.
Loss of IKKβ in OCPs results in a gain-of-function in JNK phosphorylation. OCPΔIKKβ and control OCPs were serum-starved for 4–6 h. A, cells were either not stimulated or were stimulated with 10 ng/ml RANKL or TNF-α for 5, 10, or 30 min. B, cells were either not stimulated or were stimulated with 10 ng/ml RANKL for 7.5, 15, 30, or 60 min. Total cell lysates were then analyzed by Western blot for the indicated phosphorylated proteins and whole proteins. Equal loading for phosphorylated proteins was determined by stripping the membrane and re-probing for the respective whole protein (A and B). C, cells were stimulated with 10 ng/ml RANKL for 10, 15, 30, or 120 min. Total cell lysates were then analyzed by Western blot for MKP5 and MKP1. β-Actin served as the loading control.
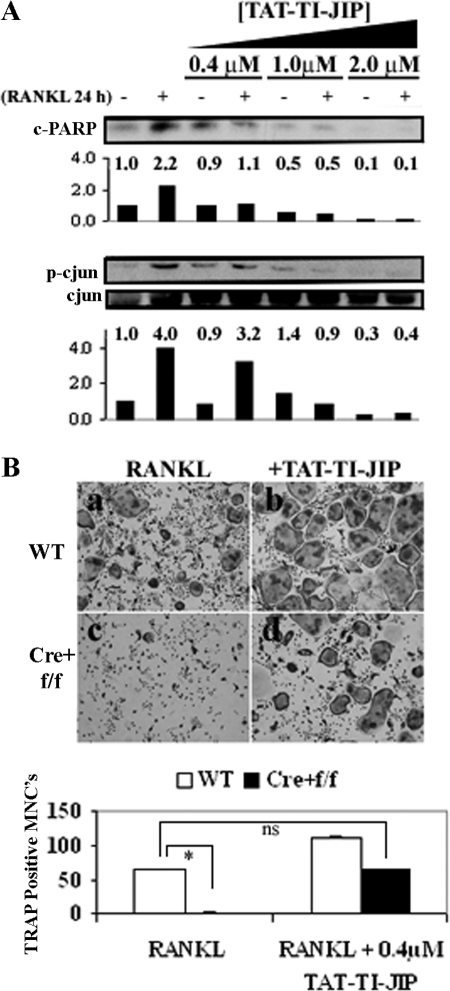
Inhibition of JNK Blocks RANKL-induced Apoptosis of Osteoclast Precursors and Rescues Osteoclastogenesis in IKKβ-deficient Osteoclast Precursors—JNK activation has been linked to RANKL-induced apoptosis of differentiating osteoclasts (18). We hypothesized that because OCPΔIKKβ cells are susceptible to RANKL-induced apoptosis, inhibition of RANKL-mediated JNK activation in these cells would rescue osteoclastogenesis. We took advantage of a cell-permeable peptide (TAT-TI-JIP) to specifically inhibit JNK activation (19) after RANKL stimulation. In WT OCPs, TAT-TI-JIP peptides inhibit osteoclastogenesis at concentrations above 1 μm (not shown). This finding is expected given the established importance of c-Jun in osteoclast differentiation (20, 21). Surprisingly, TAT-TI-JIP peptides enhance osteoclastogenesis in WT cells at a concentration of 0.4 μm (Fig. 5B, panel b). We hypothesized that this concentration of TAT-TI-JIP is sufficient to block the pro-apoptotic action of JNK without affecting the activity of JNK toward c-Jun after RANKL stimulation. Indeed, 0.4 μm of TAT-TI-JIP blocks RANKL-induced PARP cleavage without dramatically altering RANKL-induced phosphorylation of c-Jun (Fig. 5A). At a concentration of 1 μm, TAT-TI-JIP inhibits RANKL-induced PARP cleavage in OCPs, but it also inhibits c-Jun phosphorylation (Fig. 5A), which explains the inhibitory effect of this concentration on osteoclastogenesis. These results suggest that JNK serves two distinct functions in osteoclast differentiation and survival. Because low dose TAT-TI-JIP inhibits apoptosis of OCPs induced by RANKL stimulation without affecting c-Jun activation, we tested this concentration for its potential to rescue osteoclastogenesis of OCPs in the absence of IKKβ. Indeed, in the in vitro osteoclastogenesis assay, 0.4 μm TAT-TI-JIP peptide rescues osteoclastogenesis of OCPΔIKKβ cells (Fig. 5B). For example, OCPΔIKKβ cells (Fig. 5B, panel c) treated with RANKL produce less than 5% of the number of TRAP-positive osteoclasts produced by control OCPs (Fig. 5B, panel a). However, TAT-TI-JIP treatment of OCPΔIKKβ cells (Fig. 5B, panel d) results in slightly, but statistically not significant, higher number of TRAP-positive osteoclasts compared with non-TAT-TI-JIP-treated controls (Fig. 5B, panel a). Importantly, JIP peptide-treated control OCPs (Fig. 5B, panel b) produced more osteoclasts in in vitro culture than JIP peptide-treated IKKβ-deficient OCPs (Fig. 5B, panel d), indicating that OCPΔIKKβ possess defects in osteoclast differentiation that are independent of JNK-mediated apoptosis.
FIGURE 5.
Inhibition of RANKL-mediated JNK-induced apoptosis rescues osteoclastogenesis defect in OCPs deficient in IKKβ. A, OCPs from Cre+ wild-type/floxed IKKβ mice were plated in whole media supplemented with 10 ng/ml m-CSF. Four groups of cells were treated at the time of plating with either no TAT-TI-JIP or with 0.4, 1.0, or 2.0 μm TAT-TI-JIP. Also at the time of plating, one sample in each group was stimulated with 20 ng/ml RANKL. Cells were lysed after 24 h of stimulation, and total cell lysates were analyzed by Western blot for cleaved PARP (c-PARP), phosphorylated c-Jun (p-c jun), and total c-Jun. Cleaved PARP and phospho-c-Jun quantification in the different conditions is shown in numerical and graph form under the corresponding blot image. B, OCPΔIKKβ and control OCPs were plated in osteoclastogenic conditions. At the time of plating, one group of cells from each population was either left untreated (panels a and c) or treated with 0.4 μm TAT-TI-JIP (panels b and d). Cells were fixed and histochemically stained for TRAP to visualize osteoclasts on day 6 of culture. Quantification is shown in graph below. TRAP-positive multinucleated cells (MNCs) with three or more nuclei were scored as osteoclasts. Data are representative of three independent experiments, and error bars represent S.E. Asterisk indicates p < 0.0001 for difference between number of TRAP-positive MNCs in wells represented by panels a and c. No significant difference exists between panels a and d.
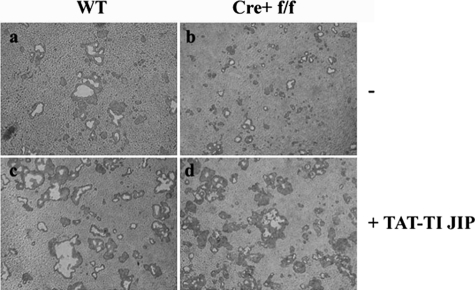
Finally, we sought to determine whether rescue of osteoclastogenesis in OCPΔIKKβ through JNK inhibition results in a concomitant rescue of bone resorption. To accomplish this, we plated WT and OCPΔIKKβ osteoclast precursors on an artificial bone substrate in osteoclastogenic conditions in the presence and absence of 0.4 μm TAT-TI-JIP. Resorption pits created by RANKL-treated OCPΔIKKβ cells (Fig. 6b) were significantly smaller (22 ± 3% resorption area compared with controls) than those created by control osteoclasts (Fig. 6a), indicating that OCPΔIKKβ are defective in resorbing bone. However, when we treated OCPΔIKKβ cells with TAT-TI-JIP (Fig. 6d), we restored resorption pit size over that noted in WT non-TAT-TI-JIP treated cells (165 ± 7% of control) (Fig. 6a). Consistent with our in vitro osteoclastogenesis assay data shown in Fig. 5, resorption pit size of TAT-TI-JIP-treated WT osteoclasts (Fig. 6c) is 2-fold larger than that of TAT-TI-JIP-treated OCPΔIKKβ (210 ± 14%) indicating that IKKβ also acts through mechanisms independent of JNK inhibition to support osteoclastogenesis.
FIGURE 6.
Inhibition of JNK in OCPΔIKKβ cells rescues bone resorption. Control (a and c) and OCPΔIKKβ (b and d) osteoclast precursors were plated onto BD Biocoat Osteologic tissue culture slides in osteoclastogenic conditions in the absence (a and b) or presence (c and d) of 0.4 μm TAT-TI-JIP. Cells were removed from the slides with deionized water, and resorption pits were noted as clear areas. Images were taken at ×10 magnification.
DISCUSSION
In previous studies, we and others have shown that IKK-NF-κB function is necessary for osteoclastogenesis (4, 10, 12). The diverse activities of the individual IKK and NF-κB members suggest that each molecule plays a unique role in the overall program of osteoclast development. It has been demonstrated that IKKβ protects OCPs from apoptosis in response to TNF-α (10). Although this finding is consistent with previous reports regarding the function of IKKβ in other settings (22), its precise role downstream of RANK during osteoclastogenesis has remained unclear. The most likely possibility is that IKKβ serves some capacity in osteoclastogenesis that is dependent upon NF-κB-p65 action. However, the exact function of p65 in the osteoclast remains to be elucidated. It has been suggested that IKKβ may be important for the survival and differentiation of OCPs (10). We show that OCPs deficient in IKKβ display a defect in osteoclast differentiation, which is consistent with impaired induction of mRNA for the following osteoclast markers: β3 integrin, cathepsin K, calcitonin receptor, MMP9, and TRAP after RANKL stimulation.
We also show that IKKβ-deficient OCPs are susceptible to apoptosis in response to RANKL stimulation. Based on our real time PCR data, this phenomenon is likely to partially result from impaired NF-κB-mediated transcription of anti-apoptotic genes. However, the pro-apoptotic function of RANKL in osteoclast precursors has not been fully described. Importantly, in OCPΔIKKβ cells, which expressed higher levels of IKKβ mRNA, induction of osteoclast differentiation markers was not affected, although impaired in vitro osteoclastogenesis was still observed. We examined the expression of mRNA for dendritic cell-specific transmembrane protein (DC-STAMP) (23) and the d2 isoform of the vacuolar ATPase V0 domain (ATP6v0d2) (24), two gene products known to be important in osteoclast fusion, in these cells to determine whether a fusion deficiency was responsible for the defect. Induction of these markers was equivalent to controls in this population of OCPΔIKKβ cells (data not shown). Although induction of other unknown osteoclast fusogenic genes may be impaired in RANKL-treated OCPΔIKKβ cells, our data suggest that RANKL-induced apoptosis is a major contributor to the osteoclast defect in these cells.
We hypothesized that the kinase signaling environment downstream of RANK in OCPΔIKKβ cells would reveal pro-apoptotic signaling changes. Among several observed signaling aberrations, we noted in particular that JNK displayed a more robust and prolonged phosphorylation profile after RANKL stimulation in OCPs that lack IKKβ. Because JNK activation has been correlated with RANKL-induced apoptosis previously (18), we postulated that OCPΔIKKβ cells undergo apoptosis as a result of RANKL-mediated JNK activation. Indeed, by inhibiting JNK-dependent apoptosis, we rescued osteoclastogenesis in OCPΔIKKβ cells.
We believe that OCPΔIKKβ cells undergo RANKL-induced JNK-dependent apoptosis early after RANKL stimulation, at a stage prior to the mature osteoclast, for two reasons. First, the number of multinucleated osteoclasts of RANKL-stimulated OCPΔIKKβ cells never approaches that of controls cells.3 Second, TAT-TI-JIP only rescues the osteoclast defect of OCPΔIKKβ if added to the culture at the same time as RANKL stimulation. When treated with TAT-TI-JIP 48 h after RANKL stimulation, JNK inhibition does not fully rescue osteoclastogenesis of OCPΔIKKβ cells.
Whether the absence of IKKβ protein is essential for the gain-of-function of JNK in response to RANKL is unknown. For example, it is probable that a downstream effector of IKKβ signaling and not IKKβ itself is responsible for inhibition of JNK after RANKL stimulation. In any case, we demonstrate that fine-tuned inhibition of JNK in OCPΔIKKβ cells rescues osteoclastogenesis. Our data suggest a model whereby RANKL stimulation of OCPs leads to activation and eventual down-regulation of JNK through MAP kinase phosphatase synthesis. In the absence of IKKβ, RANKL-mediated MKP1 and MKP5 synthesis are defective leading to enhancement and prolongation of JNK phosphorylation and activation resulting in apoptosis. One may postulate that blockade of JNK down-regulation through inhibition of the JNK phosphatases will result in enhanced and prolonged RANKL-induced JNK activity that is anti-osteoclastogenic.
The mechanism of JNK-mediated apoptosis in response to RANKL is not well defined. It has been demonstrated previously that in response to TNF-α in NF-κB-deficient cells, JNK activation leads to caspase-8-independent cleavage of the proapoptotic protein, BID, resulting in mitochondrial release of Smac and apoptosis (25). Because RANKL stimulation of OCPs does not result in caspase-8 activation, it is likely that enhanced JNK activity in the absence of IKKβ directly leads to apoptosis of OCPs after RANKL stimulation. Because lower expression of IKKβ in OCPΔIKKβ cells prevents osteoclast differentiation, it is unlikely that inhibition of JNK-mediated apoptosis will rescue osteoclastogenesis in the complete absence of IKKβ. Observing the rescuing effect of a low dose JNK inhibitor on osteoclastogenesis in OCPΔIKKβ therefore requires a permissive level of IKKβ expression that allows differentiation to occur but does not inhibit apoptosis. We believe our CD11b Cre-mediated deletion of IKKβ was a successful tool in this regard.
In addition to MAP kinase phosphatases, several potential connections between the absence of IKKβ and enhanced JNK activation exist. For example, known target genes of NF-κB serve to down-regulate JNK activation such as Gadd45β, which specifically inhibits TNF-α-mediated MKK7 activation of JNK (26). Additionally, XIAP is a target of NF-κB that down-regulates JNK activation in response to TNF-α (27). Furthermore, A20 has been postulated to play a role in NF-κB-mediated inhibition of JNK activation by down-regulating TRAF2 (28), although this hypothesis has never been validated. Because OCPΔIKKβ display reduced XIAP and A20 expression, it will be interesting to test whether these mechanisms of cross-talk between IKKβ and JNK hold true in RANKL signaling during osteoclastogenesis.
It is important to note that JNK-mediated c-Jun activation is required for efficient osteoclastogenesis (20). c-Jun activation leads to a partnership between AP-1 and NFAT1, which induces expression of NFAT2 and differentiation of osteoclasts (21). Therefore, inhibition of the RANKL-RANK-JNK pathway is a candidate for treatment of osteoporosis (29). In light of the opposing effects of the two arms of JNK activation in the osteoclast, it will be critical to sort out the pro- and anti-osteoclastogenic means of RANKL-mediated JNK activation.
Our results highlight the necessity of IKKβ in osteoclastogenesis. We demonstrate that IKKβ is important for both differentiation and survival of osteoclasts. Given that we are able to rescue osteoclastogenesis in OCPΔIKKβ cells through inhibition of JNK-induced apoptosis, we conclude that IKKβ acts, at least partially, through down-modulation of JNK activity to support cell survival during osteoclastogenesis. Our results therefore suggest that hyperactivation of JNK and inhibition of IKKβ in OCPs are potential means to treat osteoclast-mediated disease.
This work was supported, in whole or in part, by National Institutes of Health Grants AR049192 and AR054326 (to Y. A.-A.). This work was also supported by Ruth L. Kirschstein Predoctoral National Research Service Award AR055392-01 (to J. E. O.) and Shriners Hospital for Children Grants 8510 and 8570 (to Y. A.-A). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: RANKL, receptor activator of NF-κB ligand; m-CSF, macrophage colony-stimulating factor; OCP, osteoclast precursor; WT, wild type; TRAP, tartrate-resistant acid phosphatase; NFAT, nuclear factor and activator of T-cells; MMP, matrix metalloproteinase; cIAP, cellular inhibitor of apoptosis; XIAP, X-linked inhibitor of apoptosis; MKP, MAPK phosphatase; PARP, poly(ADP-ribose) polymerase; TUNEL, TdT-mediated dUTP nick end labeling; JIP, JNK-interacting protein; OCP, osteoclast precursor; TNF, tumor necrosis factor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MAP, mitogen-activated protein; LPS, lipopolysaccharide; IKK, inhibitory κB kinase.
Y. Abu-Amer and J. Otero, unpublished observations.
References
- 1.Kong, Y. Y., Yoshida, H., Sarosi, I., Tan, H. L., Timms, E., Capparelli, C., Morony, S., Oliveira-dos-Santos, A. J., Van, G., Itie, A., Khoo, W., Wakeham, A., Dunstan, C. R., Lacey, D. L., Mak, T. W., Boyle, W. J., and Penninger, J. M. (1999) Nature 397 315–323 [DOI] [PubMed] [Google Scholar]
- 2.Yoshida, H., Hayashi, S.-I., Kunisada, T., Ogawa, M., Nishikawa, S., Okamura, H., Sudo, T., Shultz, L. D., and Nishikawa, S.-I. (1990) Nature 345 442– 443 [DOI] [PubMed] [Google Scholar]
- 3.Anderson, D. M., Maraskovsky, E., Billingsley, W. L., Dougall, W. C., Tometsko, M. E., Roux, E. R., Teepe, M. C., DuBose, R. F., Cosman, D., and Galibert, L. (1997) Nature 390 175–179 [DOI] [PubMed] [Google Scholar]
- 4.Iotsova, V., Caamano, J., Loy, J., Young, Y., Lewin, A., and Bravo, R. (1997) Nat. Med. 3 1285–1289 [DOI] [PubMed] [Google Scholar]
- 5.Yamashita, T., Yao, Z., Li, F., Zhang, Q., Badell, I. R., Schwarz, E. M., Takeshita, S., Wagner, E. F., Noda, M., Matsuo, K., Xing, L., and Boyce, B. F. (2007) J. Biol. Chem. 282 18245–18253 [DOI] [PubMed] [Google Scholar]
- 6.Senftleben, U., Cao, Y., Xiao, G., Greten, F. R., Krahn, G., Bonizzi, G., Chen, Y., Hu, Y., Fong, A., Sun, S. C., and Karin, M. (2001) Science 293 1495–1499 [DOI] [PubMed] [Google Scholar]
- 7.Vaira, S., Johnson, T., Hirbe, A. C., Alhawagri, M., Anwisye, I., Sammut, B., O'Neal, J., Zou, W., Weilbaecher, K. N., Faccio, R., and Novack, D. V. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 3897–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zandi, E., Rothwarf, D. M., Delhase, M., Hayakawa, M., and Karin, M. (1997) Cell 91 243–252 [DOI] [PubMed] [Google Scholar]
- 9.Chaisson, M. L., Branstetter, D. G., Derry, J. M., Armstrong, A. P., Tometsko, M. E., Takeda, K., Akira, S., and Dougall, W. C. (2004) J. Biol. Chem. 279 54841–54848 [DOI] [PubMed] [Google Scholar]
- 10.Ruocco, M. G., Maeda, S., Park, J. M., Lawrence, T., Hsu, L.-C., Cao, Y., Schett, G., Wagner, E. F., and Karin, M. (2005) J. Exp. Med. 201 1677–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbas, S., and Abu-Amer, Y. (2003) J. Biol. Chem. 278 20077–20082 [DOI] [PubMed] [Google Scholar]
- 12.Dai, S., Hirayama, T., Abbas, S., and Abu-Amer, Y. (2004) J. Biol. Chem. 279 37219–37222 [DOI] [PubMed] [Google Scholar]
- 13.Ferron, M., and Vacher, J. (2005) Genesis 41 138–145 [DOI] [PubMed] [Google Scholar]
- 14.Pasparakis, M., Courtois, G., Hafner, M., Schmidt-Supprian, M., Nenci, A., Toksoy, A., Krampert, M., Goebeler, M., Gillitzer, R., Israel, A., Krieg, T., Rajewsky, K., and Haase, I. (2002) Nature 417 861– 866 [DOI] [PubMed] [Google Scholar]
- 15.Shishodia, S., and Aggarwal, B. B. (2002) J. Biochem. Mol. Biol. 35 28–40 [DOI] [PubMed] [Google Scholar]
- 16.Liu, Y., Gorospe, M., Yang, C., and Holbrook, N. J. (1995) J. Biol. Chem. 270 8377– 8380 [DOI] [PubMed] [Google Scholar]
- 17.Theodosiou, A., Smith, A., Gillieron, C., Arkinstall, S., and Ashworth, A. (1999) Oncogene 18 6981– 6988 [DOI] [PubMed] [Google Scholar]
- 18.Bharti, A. C., Takada, Y., Shishodia, S., and Aggarwal, B. B. (2004) J. Biol. Chem. 279 6065– 6076 [DOI] [PubMed] [Google Scholar]
- 19.Barr, R. K., Kendrick, T. S., and Bogoyevitch, M. A. (2002) J. Biol. Chem. 277 10987–10997 [DOI] [PubMed] [Google Scholar]
- 20.David, J. P., Sabapathy, K., Hoffmann, O., Idarraga, M. H., and Wagner, E. F. (2002) J. Cell Sci. 115 4317– 4325 [DOI] [PubMed] [Google Scholar]
- 21.Ikeda, F., Nishimura, R., Matsubara, T., Tanaka, S., Inoue, J. I., Reddy, S. V., Hata, K., Yamashita, K., Hiraga, T., Watanabe, T., Kukita, T., Yoshioka, K., Rao, A., and Yoneda, T. (2004) J. Clin. Investig. 114 475– 484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Z. W., Chu, W., Hu, Y., Delhase, M., Deerinck, T., Ellisman, M., Johnson, R., and Karin, M. (1999) J. Exp. Med. 189 1839–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yagi, M., Miyamoto, T., Sawatani, Y., Iwamoto, K., Hosogane, N., Fujita, N., Morita, K., Ninomiya, K., Suzuki, T., Miyamoto, K., Oike, Y., Takeya, M., Toyama, Y., and Suda, T. (2005) J. Exp. Med. 202 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, S. H., Rho, J., Jeong, D., Sul, J. Y., Kim, T., Kim, N., Kang, J. S., Miyamoto, T., Suda, T., Lee, S. K., Pignolo, R. J., Koczon-Jaremko, B., Lorenzo, J., and Choi, Y. (2006) Nat. Med. 12 1403–1409 [DOI] [PubMed] [Google Scholar]
- 25.Deng, Y., Ren, X., Yang, L., Lin, Y., and Wu, X. (2003) Cell 115 61–70 [DOI] [PubMed] [Google Scholar]
- 26.Papa, S., Zazzeroni, F., Bubici, C., Jayawardena, S., Alvarez, K., Matsuda, S., Nguyen, D. U., Pham, C. G., Nelsbach, A. H., Melis, T., De Smaele, E., Tang, W. J., D'Adamio, L., and Franzoso, G. (2004) Nat. Cell Biol. 6 146–153 [DOI] [PubMed] [Google Scholar]
- 27.Tang, G., Minemoto, Y., Dibling, B., Purcell, N. H., Li, Z., Karin, M., and Lin, A. (2001) Nature 414 313–317 [DOI] [PubMed] [Google Scholar]
- 28.Perkins, N. D. (2007) Nat. Rev. Mol. Cell Biol. 8 49–62 [DOI] [PubMed] [Google Scholar]
- 29.Teitelbaum, S. L. (2004) J. Clin. Invest. 114 463– 465 [DOI] [PMC free article] [PubMed] [Google Scholar]