Abstract
p75 and the Nogo receptor form a signaling unit for myelin inhibitory molecules, with p75 being responsible for RhoA activation. Because p75 lacks the GDP/GTP exchange factor domain, it has remained unclear how p75 activates RhoA. Here, we report that Kalirin9, a dual RhoGEF, binds p75 directly and regulates p75-Nogo receptor-dependent RhoA activation and neurite inhibition in response to myelin-associated glycoprotein. The region of p75 that Kalirin9 binds includes its mastoparan-like fifth helix, which was shown to recruit RhoGDI-RhoA. As predicted from the presence of a shared binding site, we found that Kalirin9 competes with RhoGDI for p75 binding in a dose-dependent manner in vitro. In line with these data, myelin-associated glycoprotein addition to cerebellar granule neurons resulted in a reduction in the association of Kalirin9 with p75, and a simultaneous increase in the binding of RhoGDI to p75. These results reveal a mechanism by which the fifth helix of p75 regulates RhoA activation.
Central nervous system axons do not regenerate, partly because of the actions of myelin inhibitory molecules expressed among oligodendrocytes, such as Nogo (1), myelin-associated glycoprotein (MAG)2 (2), oligodendrocyte myelin glycoprotein (3), Ephrin-B3 (4), and more recently, netrin-1 (5). When the Nogo receptor (NgR) was cloned using the Nogo-66 domain, which is exposed on the surface of oligodendrocytes (6), it was evident that NgR would require a coreceptor to signal, because NgR is tethered to the membrane via glycosyl phosphatidylinositol linkage (7). p75 soon emerged as a coreceptor for NgR (8, 9), after it was shown that MAG-mediated RhoA activation was lost in p75-/- cerebellar granule neurons (CGN) (10). Since then, NgR has been shown to bind MAG (11) and oligodendrocyte myelin glycoprotein (8), both of which activate RhoA in a p75-dependent manner.
RhoA activation by p75-NgR was shown to involve a guanosine dissociation inhibitor, RhoGDI (12). RhoA is recruited to p75 via RhoGDI that binds p75 directly, and the recruited RhoA is then released and activated by unknown means. Because p75 does not contain a GDP/GTP exchange factor (GEF) domain, the mechanism by which the recruited RhoA becomes activated has remained undetermined. Here, we present data indicating that Kalirin9, a dual RhoGEF, is one of the key components of the signaling unit for RhoA activation by myelin inhibitory molecules.
Kalirins are mammalian orthologs of Trio in Drosophila and Unc73, which were shown to play a critical role in axon guidance in Drosophila and Caenorhabditis elegans (13–16). In the mammalian nervous system, several Kalirin isoforms are known to date, including Kalirin5, 7, 8, 9, and 12 (17). These isoforms all contain Rac GEF, but only Kalirin9 and 12 additionally contain RhoGEF, like Trio. Kalirin12 contains a kinase-like domain in addition to Rac and RhoA GEF domains, but its kinase activity has not yet been characterized. Kalirin9 was shown to promote axon outgrowth during development, (18), being the predominant form expressed (17). As its role in axon outgrowth suggests, Kalirin9 is localized to growth cones and neurites in neuronal cultures (18).
Although Trio dual RhoGEFs have been shown to play critical roles in axon outgrowth, the extracellular factors or receptors that regulate the actions of the Kalirins are not known. It was reported that EphrinB and its receptor, EphB2, regulates Kalirin7-dependent Rac activation at synaptic sites (19), but Kalirin7 is not a dual GEF and is localized to post-synaptic compartments, unlike the other members of the family. Recently, Kalirin5 was shown to interact with TrkA and play a role in nerve growth factor-dependent neurite extension in PC12 cells, presumably by activating Rac (20). Whether nerve growth factor signaling modulates Kalirin5-mediated Rac activation was, however, not clear. Here, we present evidence that MAG serves as a ligand that induces Kalirin9 to activate RhoA via p75-NgR. Kalirin9 binding to p75 is regulated by MAG, and knocking down endogenous Kalirin9 results in inhibition of MAG-mediated RhoA activity and its attendant effect on neurite outgrowth.
EXPERIMENTAL PROCEDURES
Constructs—The constructs carrying the m1–m6 regions in Kalirin9 were generated by placing the PCR fragment corresponding to each region into the pcDNA4/HisMax TOPO TA kit (Invitrogen). The PCR fragments were generated using pEAK-Kalirin9 (18) as the template and subsequently sequenced for any errors. The pTYB12-RhoGDI was generated by placing the PCR fragment of RhoGDI with appropriate linker sequences as directed by the vendor (NE BioLabs). The final product was sequenced for any errors. RhoGDI protein was produced in the ER2566 host and purified from the inteinchitin binding protein tag using the chitin column according the protocols provided by the vendor (NE BioLabs).
Primary Neuronal Cultures—CGN cultures were prepared as described (8). For transfection of small interfering RNA constructs, CGNs were transfected right after dissection with 3 μg of small interfering RNA plus GFP cDNA using the G13 program of Amaxa Nucleofactor. Rho-GTP assays were performed 2 days after transfection. The sequence for the RNAi for Kalirin9 was 6134AGUACCAGCUGCUACUCAA6152, that for Kalirin7 was 4902GGAUGGCAACCUUGUUCCU4920, and for the scrambled control, the sequence was CCAGGAUAACACAGAGAUG, all of which were synthesized by Dharmacon.
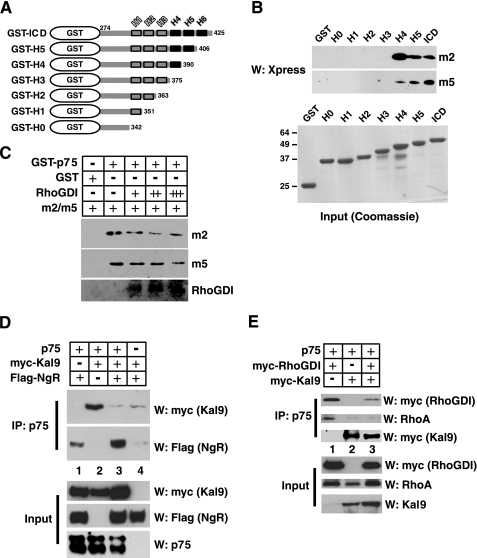
GST Pulldown Assays—The m1–m6 proteins were harvested from 293T cells following transfection. For pulldown analyses, the 293T lysates containing m1–m6 fragments were incubated with GST or GST-p75ICD proteins, and the bound m1–m6 fragments were detected with Xpress antibody in Western assays. For in vitro competition with RhoGDI, m1–m5 fragments were purified from 293T cells using nickel-nitrilotriacetic acid magnetic agarose beads (Qiagen). The purified m1–m5 fragments were incubated with GST or GST-p75ICD in the presence of increasing amounts of RhoGDI, which was obtained following dithiothreitol treatment of intein-RhoGDI that was loaded onto a chitin column. For competition assays shown in Fig. 3C, 1 μg of m2 and m5 fragments were bound to 10 μg of GST-p75ICD in the presence of 0, 1.8, 5.4, and 16.2 μg of purified RhoGDI. For GST binding of m2 and m5 fragments, 10 μg of GST was used as well. The kalirin fragments (m1 to m6) that had bound to GST or GST-p75ICD were detected using Xpress antibody.
FIGURE 3.
Kalirin9 and RhoGDI compete for p75 binding in vitro. A and B, the helix 4 in p75 is necessary for Kalirin9 binding. A, a diagram of helix deletion mutants of p75 in GST. The helices are indicated as boxes with respective positions, H1–H6, above the boxes. The numbers in each mutant indicate the position of the last amino acid included. The black helices indicate those helices that are involved in Kalirin9 binding. The deletion mutants and GST control are shown as Coomassie stains. B, helix 4 in p75 is necessary for binding of the m2 and m5. The 293 lysates containing m2 and m5 were subjected to pulldown assays with GST or GST-p75ICD; helix deletion mutants are shown in the diagram. Note that upon deletion of the fourth helix, both the m2 and m5 fail to bind p75ICD. C, RhoGDI competes with Kalirin9 for binding of p75 in vitro. The m2 and m5 constructs were transfected into 293T cells and subsequently purified using a nickel column prior to being subjected to pull-down assays with GST-p75ICD in the absence or presence of increasing amounts of purified RhoGDI. RhoGDI was purified following dithiothreitol treatment of intein-RhoGDI from a chitin column (NE BioLabs). The specific amount of each protein used in competition is described under “Experimental Procedures.” D, the full-length Kalirin9 dissociates from p75 when NgR forms a complex with p75. The lysates from transfected 293T cells were subjected to immunoprecipitation (IP) with p75 antibody, and the immune complexes were probed with Myc antibody to detect the full-length recombinant Kalirin9 using the upper part of the transferred membrane, and FLAG antibody to detect NgR in the lower part of the membrane. The control Western assays (W) are also shown as Input. E, the full-length Kalirin9 competes with RhoGDI for p75 binding, which coincides with the loss of RhoA from p75. The lysates from transfected 293T cells were divided into three equal parts, and each was subjected to immunoprecipitation with p75 antibody. The immunoprecipitate was probed for the presence of Myc-RhoGDI, RhoA, or Myc-Kalirin9 in Western assay. The control Western assays are also shown as Input.
Rho-GTP Measurements—To measure Rho-GTP, the CGN cultures were treated for 15 min with 0.3 μg/ml of conjugated MAG-Fc (Sigma) and 8 μg/ml conjugated AP-Nogo-66. The pulldown reactions with GST-Rhotekin beads were performed according to the manufacturer's protocol (Upstate Biotech Inc.).
Neurite Outgrowth Assay—Neurite outgrowth assays were performed as described previously (3, 8). For quantification of average neurite length, the lengths of GFP+ neurites were measured on at least 150 neurons/condition, from three independent experiments.
Immunoprecipitation/Western Analyses—The procedures for immunoprecipitation and Western analyses were identical to what was described (21). The antibodies used in these assays include p75, hemagglutinin (Covance), Nogo receptor (Chemicon), Myc (Santa Cruz), pan-Kalirin antibody (18), RhoA, RhoGDI (Sigma), and Xpress (Invitrogen).
RESULTS
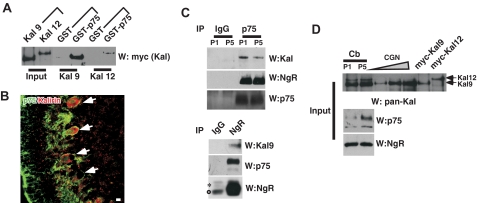
Kalirin9 and 12 Bind p75 in Vitro—Several GEF molecules have been implicated in nerve growth factor-dependent regulation of small G proteins, such as Trio (22), Kalirin5 (20), Rap1-PDZ-GEF1 (23), and p85 betaPIX (24). Of these, the Kalirin family of dual RhoGEFs was of special interest for p75-mediated, and TrkA-independent regulation of Rac1 and RhoA, because p75 can activate these GTPases depending on with which receptor it has become associated. We therefore tested whether Kalirin9 and 12 that contain both RhoGEF and RacGEF domains (17) can interact with p75 in vitro using pull-down assays with GST-p75ICD. The 293T lysates that contained the full-length, Myc-tagged Kalirin9 or 12 were incubated with the GST or GST-p75 intracellular domain (GST-p75ICD). Both Kalirin9 and 12 were pulled down with the GST-p75ICD but not with the GST (Fig. 1A), suggesting that both kalirins can bind the cytoplasmic domain of p75 in vitro.
FIGURE 1.
Kalirin9 binds p75. A, the recombinant Kalirin9 and 12 bind the cytoplasmic domain of p75 in vitro. GST or GST-p75ICD was incubated with the lysates from 293T cells that were transfected with the Myc-tagged wild type Kalirin9 or 12. The bound Kalirins were detected by Myc Western analysis (W). For input, 5% of the lysates that were used for GST pulldown were loaded as controls. B, Kalirin is expressed among p75+ Purkinje neurons in the P5 cerebellum. P5 rat sagittal sections were stained with an anti-p75 antibody, 192, and pan-Kalirin antibodies (28). The arrows point to doubly stained Purkinje neurons. Scale bar, 12.5 μm. C, Kalirin9, and not Kalirin12, associates with p75 and NgR in the developing cerebellum. Upper panel, the cerebellar lysates from P5 rat pups were subjected to immunoprecipitation (IP) with a p75 antibody and a control IgG. The bound proteins were simultaneously probed with pan-Kalirin and NgR antibodies using a divided membrane. For detection of p75, the same membrane was stripped and reprobed. *, nonspecific bands. Lower panel, detection of Kalirin9 and p75 in reciprocal immunoprecipitation with NgR antibody. The bound proteins were simultaneously probed with pan-Kalirin and NgR antibodies using a divided membrane. As a control, the same blot was reprobed for NgR following membrane stripping. *, a nonspecific band; ○, IgG heavy chain. D, input controls for C. Note that Kalirin9 is the major isoform present in CGN cultures, whereas Kalirin12 is not expressed at detectable level.
Kalirin9 Is in Complex with p75-NgR in Vivo—We next asked whether Kalirins are expressed in p75+ neurons in vivo. Immunohistochemical analyses of postnatal day 5 (P5) rat cerebellum with pan-Kalirin antibody revealed that Kalirin is expressed among developing Purkinje cells, including their axons and dendrites (Fig. 1B). The positive immunoreactive signals in the cerebellum are likely to represent Kalirin9 and 12, because they are the predominant isoforms detected in the cerebellar lysates from P1 and P5, based on the expected size, 350 kDa for Kalirin9 and 450 kDa for Kalirin12 (Fig. 1D and supplemental Fig. S1). The specificity of the Kalirin antibody is shown in supplemental Fig. S1. More importantly, both Kalirins were expressed in cerebellar granule neurons (see CGN in Fig. 1D), in which p75 was shown to regulate RhoA activation in response to myelin inhibitory molecules (8, 10–12). It should be pointed out, however, that Kalirin12 expression is extremely low in CGN cultures compared with that of Kalirin9, suggesting that Kalirin9, and not Kalirin12, is likely to be the GEF that is involved in RhoA activation by myelin inhibitory molecules.
We next asked whether both Kalirin9 and 12 interact with p75-NgR complex in vivo. The cerebellar lysates from P5 rats were subjected to immunoprecipitation with the control IgG or MC192, an anti-p75 antibody. The resulting p75 immune complexes were separated and probed for both Kalirin and NgR by dividing the membrane into two parts and then using one for Kalirin and the other for NgR in Western analyses. As a control, the same blot was stripped and reprobed for p75. A Kalirin-immunoreactive band was detected in the p75 immune complex at the size corresponding to Kalirin9 and not Kalirin12 (Fig. 1C). Also, in reciprocal immunoprecipitation with NgR antibody, p75 was present along with Kalirin9, but not with Kalirin12 (Fig. 1C). Neither Kalirin9 nor NgR was detected in the control IgG immunoprecipitation. These results suggest that Kalirin9 is the isoform that forms a complex with p75-NgR in vivo and may play a role in p75-NgR signaling.
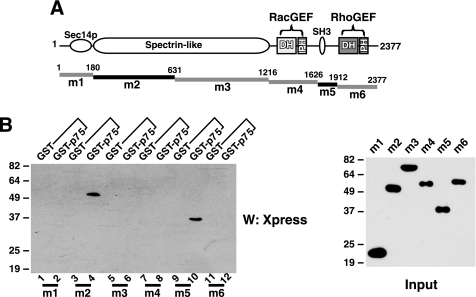
Two Distinct Domains in Kalirin9 Are Involved in Binding p75—We next determined which domains in Kalirin9 are involved in the interaction with p75. For these experiments, the constructs containing the m1–m6 regions of Kalirin9 depicted in Fig. 2A were introduced into 293T cells, and the resulting lysates were subjected to pulldown assays with GST or GST-p75ICD. Of the six regions tested, two domains interacted with p75ICD; m2, which contains the most of the spectrin-like repeats of Kalirin9; and m5, which contains the SH3 domain plus the sequences before the Dbl homology (DH) and pleckstrin homology (PH) domains begin (Fig. 2). The binding of the m2 and m5 domains to p75 was direct (Fig. 3C), because the purified m2 and m5 from 293T lysates also interacted with p75 in vitro, suggesting that it is likely that both domains are involved in forming a complex with p75. This may be regarded as atypical, in that two widely separated domains in Kalirin9 are jointly interacting with p75. It is highly plausible, however, that the m2 and m5 domains are structurally juxtaposed in folded, native proteins, and thereby concurrently accessible to the C termini of p75. It should also be pointed out that the same region in hTrio, that is, the N-terminal half of the spectrin-like repeats in hTrio, was shown to be critical in eliciting neurite outgrowth in PC12 cells (22).
FIGURE 2.
Two distinct domains in Kalirin9 are involved in binding p75 through its C-terminal helices. Two domains encompassing the SH3 motif and spectrin-like repeats are involved in binding p75. A, the regions in Kalirin9 tested for binding p75 are depicted as m1–m6. The m2 and m5 that bind p75 are indicated as black lines. The number above each deletion construct refers to the deleted amino acid position in Kalirin9. Each mutant was introduced into the pcDNA4/HisMax-TOPO TA vector (Invitrogen) and detected with an Xpress Western analysis (W). B, the spectrin-like repeat domain, m2, and the region containing SH3 motif, m5, bind p75. The m1–m6 regions were transfected into 293T cells, and the resulting lysates were subjected to pulldown assays with GST or GST-p75ICD.
A recent study reported that the SH3 domain of Kalirin9 interacts with Crk (25). When bound to Crk, Kalirin9 failed to activate Rac, suggesting that Crk binding to Kalirin9 via its SH3 domain inhibits its GEF activity. It may then be reasonable to extrapolate that the binding of p75 through the SH3 domain of Kalirin9 is also likely to result in inhibition of its GEF activity. Concordantly, when released, Kalirin9 would be likely to regain its GEF activity.
Kalirin9 and RhoGDI Compete for p75ICD Binding in Vitro—It was reported that RhoGDI-RhoA binds p75 via its mastoparan-like fifth helix, which has the ability to facilitate the release of RhoA when it is bound to RhoGDI (12). If Kalirin9 is the GEF responsible for the subsequent GDP/GTP exchange reaction, we may expect that Kalirin9 will bind near helix 5 of p75. We therefore tested where the m2 and m5 domains bind p75 using a series of nested deletion constructs (Fig. 3A). Upon deleting the fourth helix, both the m2 and m5 fragments failed to bind p75, suggesting that helix 4 is necessary for m2 and m5 to bind p75 (Fig. 3B). The modes of binding appear somewhat different between m2 and m5, however; helices 5 and 6 appear to augment the binding of m5 to p75 significantly, but they had only a minor effect on the binding of m2 to p75 (Fig. 3B).
The result that Kalirin9 binding to p75 is enhanced by helices 5 and 6, at least with the m5 domain, suggests that Kalirin9 may compete with RhoGDI-RhoA for p75 binding. This hypothesis was next tested entirely in vitro, using the purified m2 and m5 and GST-p75ICD in the presence of increasing amounts of RhoGDI that was purified from bacteria. In these assays, the binding of m2 and m5 to p75ICD was competed by the presence of RhoGDI in a dose-dependent manner (Fig. 3C). These in vitro results thus suggest that Kalirin9 and RhoGDI may not bind p75 at the same time and that their binding may be mutually exclusive.
The reciprocal interaction between Kalirin9 and RhoGDI in p75 binding was further tested using the full-length recombinant Kalirin9 construct in 293T cells. FLAG-NgR was detected in the p75 immune complex when cells were transfected with hemagglutinin-p75 and FLAG-NgR (Fig. 3D, lane 1), which is in agreement with previous reports (8, 9). Similarly, Kalirin9 interacted with p75 (Fig. 3D, lane 2) as in Fig. 1. When NgR was cotransfected along with p75 and Kalirin9, however, Kalirin9 dissociated from p75 to the background level (Fig. 3D, lanes 3 and 4). These results suggest that association between p75 and NgR facilitates the dissociation of Kalirin9 from p75.
We next asked whether the presence of RhoGDI modulates the full-length Kalirin9 binding to p75 in 293T cells. For these experiments, the lysates were equally divided to produce three identical immunoprecipitation reactions with a p75 antibody, and then each was probed in Western assay for Myc-RhoGDI, RhoA, or Myc-Kalirin9. In the absence of Kalirin9, RhoGDI interacted with p75, and this interaction brought RhoA to p75 (Fig. 3E, lane 1), which is in agreement with the previous report (12). When Kalirin9 was introduced, on the other hand, the amount of Myc-RhoGDI bound to p75 was significantly reduced compared with that without Kalirin9, which also correlated with the loss of RhoA from p75 (Fig. 3E, lane 3). Together, these results suggest that Kalirin9 and RhoGDI compete for p75 binding. Such mutually exclusive binding behavior can be interpreted as suggesting that Kalirin9 may participate in the release of RhoA from p75 after RhoA is recruited to p75 via RhoGDI.
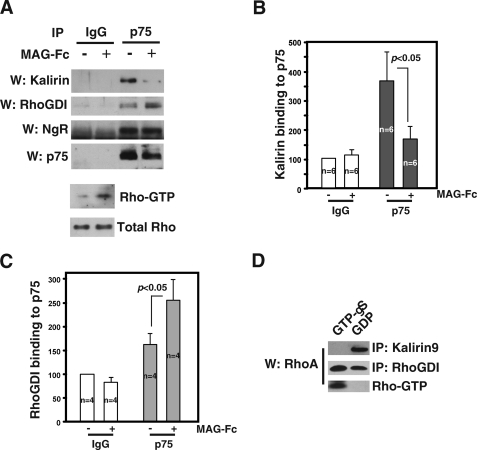
Kalirin9 Dissociates from p75-NgR following MAG Activation—Yamashita and Tohyama (12) reported that MAG-Fc treatment facilitates RhoGDI-RhoA recruitment to p75-NgR complex in CGN cultures. We therefore tested whether RhoGDI indeed competes for p75 binding with Kalirin9 after MAG activation using CGN cultures. Neonatal CGN cultures are known to respond to all known NgR ligands that activate RhoA in a manner dependent on p75-NgR (8–10, 26). Also in our P5–6 neonatal rat CGN cultures, the addition of MAG-Fc resulted in RhoA activation (Fig. 4A, lower panels). After treating CGN cultures with or without MAG, the resulting lysates were subjected to immunoprecipitation reactions using anti-p75 antibody or the control IgG. Following Western analysis, the blot was divided into two parts, and each was probed for the endogenous Kalirin9 or RhoGDI. As a control, the blot was stripped and reprobed for NgR and p75. We found that Kalirin9 was bound to p75 in the basal state, but the amount of Kalirin9 that associated with p75 decreased by 60% after MAG-Fc treatment (Fig. 4, A, upper panels, and B, for quantification). The amount of RhoGDI that associated with p75 increased with MAG-Fc, as reported (12) (Fig. 4, A and C, for quantification). The control IgG did not bring down any proteins. These results together suggest that MAG-dependent recruitment of RhoGDI-RhoA to p75 coincides with dissociation of Kalirin9 in a reciprocal manner.
FIGURE 4.
Reciprocal interaction between Kalirin9 and RhoGDI with p75 in response to MAG-Fc. A, with MAG activation, Kalirin9 binding to p75 decreased, whereas RhoGDI binding to p75 increased in CGN cultures. CGN cultures were treated with 0.3 μg/ml conjugated MAG-Fc for 15 min, and the lysates were subjected to immunoprecipitation (IP) with IgG control or p75 antibody. The presence of Kalirin9 and RhoGDI in the immune complexes was probed in Western assays (W). The same blot was stripped and reprobed for the presence of NgR and p75. In parallel, MAG-Fc-treated CGN lysates were subjected to RhoA activity assays using Rhotekin binding proteins in pulldown assays (lower panels). B, quantification of the amount of Kalirin9 bound to p75 in immunoprecipitation shown in B. The amount of Kalirin9 bound to p75 decreased by 60% following MAG-Fc treatments (Student's t test, p < 0.05). C, quantification of the amount of RhoGDI bound to p75 in immunoprecipitation shown in B. The amount of RhoGDI bound to p75 increased by 1.5-fold after MAG-Fc treatments (Student's t test, p < 0.05). D, Kalirin9 binds Rho-GDP and not Rho-GTP. 293T lysates were loaded with GDP or unhydrolyzable GTPγS and subjected to immunoprecipitation with Kalirin or RhoGDI antibodies. The bound RhoA was detected in a RhoA Western assay. As a control, the same lysates were subjected to GST-Rhotekin pulldown assays to measure the extent of GTP/GDP loading.
In this reciprocal interaction between Kalirin9 and RhoGDI-RhoA, we envision that the dissociating Kalirin9 binds RhoA that was brought to p75 by RhoGDI following MAG activation. Because RhoA binding to RhoGEF is likely to be too fast and transient to detect in immunoprecipitation assays, we opted instead to ask whether Kalirin9 binds GDP or GTP-bound form of RhoA. If Kalirin9 were to activate RhoA by facilitating GDP to GTP exchange reaction on the bound RhoA after it is released from RhoGDI, Kalirin9 will bind the GDP-bound form specifically. Indeed, Kalirin9 bound Rho-GDP and not Rho-GTP, whereas RhoGDI bound both GTP and the GDP-bound form of RhoA (Fig. 4D). In these experiments, we preloaded 293T lysates with either GDP or unhydrolyzable GTP analog GTPγS and subjected the lysates to immunoprecipitation reactions with Kalirin9 or RhoGDI antibodies. The control Rho-GTP assay of the same lysates shows that the preloading was complete, because Rho-GTP was detected only in GTPγS samples and not in GDP samples. These results together suggest that activation of RhoA by p75-NgR includes Kalirin9, whose action is likely to bind RhoA-GDP after it is released from the activated p75-NgR complex and facilitate the GDP to GTP exchange reactions.
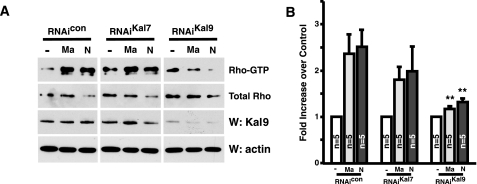
Kalirin9 Is Involved in RhoA Activation by p75-NgR—We next examined whether Kalirin9 is necessary for RhoA activation by myelin inhibitory molecules using RNAi to knock down the endogenous Kalirin9. As controls, an RNAi with scrambled sequences of Kalirin9 RNAi was used as well as an RNAi for Kalirin7, an alternatively spliced isoform of Kalirin9. In scrambled-RNAi and Kalirin7-RNAi transfected cultures, both MAG (Ma) and Nogo-66 (N) activated RhoA, whereas they failed to do so when the cultures were transfected with Kalirin9-RNAi (Fig. 5; p < 0.01). In Kalirin9-RNAi cultures, the overall Kalirin9 protein levels were reduced significantly, whereas in the control-RNAi and Kalirin7-RNAi cultures, they were not (Fig. 5A). These results suggest that Kalirin9 is necessary for ligand-dependent RhoA activation in CGN cultures.
FIGURE 5.
Kalirin9 is necessary for ligand-dependent RhoA activation in CGN cultures. A, knockdown of Kalirin9 inhibits RhoA activation by myelin inhibitory molecules in CGN cultures. Note that the Kalirin9 protein level is significantly reduced with Kalirin9-RNAi, but not with Kalirin7-RNAi or the control RNAi. The RNAi duplexes were introduced to CGN cultures by transfection using the Amaxa kit. Ma, MAG-Fc; N, Nogo66. B, quantification of the RhoA activities shown in A. The inhibition in RhoA activity by Kalirin9-RNAi is significant compared with the control RNAi (**, p < 0.05, Student's t test). The reduction in RhoA activity with Kalirin7-RNAi is not significantly different from the control (p > 0.1). W, Western analysis.
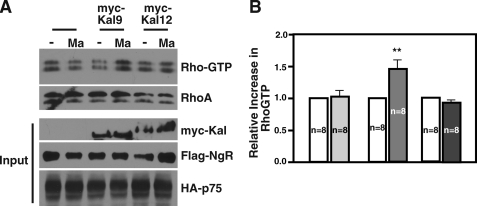
It should be pointed out that the sequences that we used to knock down Kalirin9 in Fig. 5 are derived from the RhoGEF domain (6134–6152 in nucleotide sequences), suggesting that the Kalirin9-RNAi can potentially target Kalirin12 that contains the same sequences. Although Kalirin12 did not interact with p75 in the cerebellum, making it unlikely that it plays a role, we thought it was important to determine whether it was Kalirin9 or Kalirin12 that is involved in RhoA activation by MAG-Fc in these cultures. Unfortunately, Kalirin12 levels are extremely low in CGN cultures (Fig. 1D), rendering it very difficult for us to use RNAi-based knockdown approaches effectively, even with several different sets of RNAi that were unique to Kalirin12 (data not shown). We therefore decided to test whether Kalirin9 or Kalirin12 is sufficient to allow RhoA activation in response to myelin inhibitory molecules using an ectopic system, such as 293T cells, as was reported (26). 293T cells were therefore transfected with p75/NgR, p75/NgR/Kalirin9, or p75/NgR/Kalirin12. Following 30-min treatments with MAG-Fc (Ma), there was 1.5-fold increase in RhoA-GTP levels when Kalirin9 was introduced, but not when Kalirin12 was (Fig. 6). The relative expression levels of transfected constructs were similar in each set of transfections, based on control Western assay (Fig. 6A, Input). Because the introduction of p75 with NgR alone failed to activate RhoA in these cells, these data suggest that Kalirin9 and not Kalirin12 constitutes the missing component in the RhoA activation machinery of the p75-NgR complex in response to MAG-Fc.
FIGURE 6.
Kalirin9 and not Kalirin12 is involved in p75-NgR-mediated RhoA activation. A, introduction of Kalirin9 and not Kalirin12 reconstitutes a ligand-dependent RhoA activation by p75-NgR in 293T cells. The transfected 293T cells were untreated or treated with 0.3 μg/ml conjugated MAG-Fc (Ma) for 30 min, and the resulting lysates were subjected to RhoA activity assays using Rhotekin-binding protein pulldown assays. The control Western assays are also shown for each construct introduced as input. B, quantification of the RhoA-GTP levels. The RhoA-GTP levels were adjusted to the total RhoA. The increase in RhoA-GTP levels with Kalirin9 is significant compared with the control and Kalirin12 (**, p < 0.05, Student's t test).
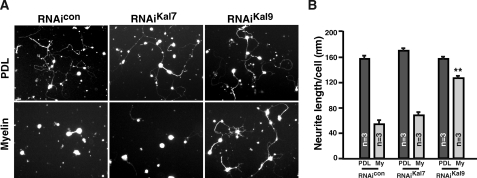
Kalirin9 Is Required for Neurite Inhibition by Myelin Inhibitory Molecules in CGN Cultures—If Kalirin9 activates RhoA in response to myelin inhibitory molecules, knocking down Kalirin9 should allow neurons to continue extending processes even when they are plated onto myelin. To address this question, the RNAi duplexes were introduced along with a GFP construct into CGN cultures, and the lengths of the subsequently grown neurites were measured among GFP+ neurons. All of the transfected neurons sent out long neurites on a permissive substrate, poly-d-lysine, regardless of the RNAi introduced, indicating that RNAi duplexes did not perturb the normal process outgrowth on permissive substrates (Fig. 7A). On myelin substrates, however, both the control-RNAi and Kalirin7-RNAi expressing neurons failed to extend neurites, whereas Kalirin9-RNAi expressing neurons continued to send out processes as they did on poly-d-lysine (Fig. 7; **, p < 0.001). These data suggest that Kalirin9 is indeed involved in p75-NgR-dependent RhoA activation and inhibition of neurite outgrowth.
FIGURE 7.
Kalirin9 is necessary for ligand-dependent RhoA activation and inhibition of neurite out-growth in CGN cultures. A, Kalirin9 is necessary for inhibitory action of myelin in CGN neurite outgrowth. The images are of representative cultures that were transfected with RNAi duplexes plus GFP plasmid using the Amaxa transfection kit. B, quantification of neurite outgrowth per cell is shown in D (**, p < 0.001, one-way analysis of variance). The number of cells whose neurite length was measured was 100–150/each group in individual experiment. PDL, poly-d-lysine.
DISCUSSION
In this report, we present data that support a functional role for Kalirin9, a Trio family of a dual Rac and RhoGEF in p75-NgR signaling; Kalirin9 binds p75 through its helices 4–6, and this binding appears to exclude RhoGDI binding to p75 via its helix 5 region. Functionally, knocking down Kalirin9 protein levels in CGN cultures resulted in inhibition of RhoA activation and continued neurite outgrowth on myelin substrates. Together, these data suggest that Kalirin9 is a GEF that plays a role in the activation of RhoA by the p75-NgR complex.
We believe that Kalirin9 dissociates from p75 as a result of competition with RhoGDI-RhoA that is being recruited to p75 upon MAG activation. It is possible, however, that dissociation is simply a result of p75 being cleaved by γ-secretase after MAG treatment. Cleavage of p75 by γ-secretase was recently shown to be necessary for RhoA activation by MAG (27). We believe this latter scenario is highly unlikely based on two observations. One, Kalirin9 can bind GST-p75ICD. Two, the proportion of cleaved p75ICD to the full-length form is relatively small, whereas the extent of reduction in Kalirin9 bound to p75 is much greater after MAG treatment in CGN cultures. Alternatively, it is possible that dissociation of 350 kDa Kalirin9 from p75 allows γ-secretase complex to access p75-NgR.
Kalirin9 is a dual RhoGEF, containing both RacGEF and RhoGEF domains, suggesting that it can potentially activate Rac1 and RhoA simultaneously. When overexpressed in fibroblasts, the RacGEF domain alone induced lamellipodia and membrane ruffles, whereas overexpression of the full-length Kalirin9 failed to exhibit prominent membrane ruffles, although lamellipodia production increased along with increased production of stress fibers (18). These results suggest that the RacGEF domain is functional, but its activity may be modulated in the context of the full-length protein when appropriate signals are received. Overexpression of Kalirin9 in neuronal cultures led to a similar conclusion, suggesting that although each GEF domain in isolation acts as a RacGEF and RhoGEF, there may be cross-talk between the two GEF domains in the context of a full-length protein, thereby eliciting a coordinated response that differs from an arithmetic sum of individual activities (18). Determination of whether both GEF domains are responsible for RhoA activation will thus have to rely on mutant constructs that bear deletion of each GEF domain.
In regard to myelin inhibitory molecules that signal through p75-NgR coreceptors, we believe that Rac1 activation is unlikely to play critical roles. A major reason is that our unpublished data indicate that neurotrophin-mediated Rac1 activation by p75 is dependent on Kalirin7, and not Kalirin9. In addition, we found that RacGTP levels are concentrated in membrane-rich compartments of the cerebellum, whereas RhoGTP levels are not.3 These results suggest that the site of Rac1 activation is distinct from RhoA activation inside the cell. Indeed, Kalilrin7 is localized to membrane-rich compartments, such as postsynaptic densities (19), whereas Kalirin9 is distributed widely.4 Based on these observations, we believe that Kalirin9 is not likely to be involved in activating Rac1, at least not in response to myelin inhibitory molecules.
Our data suggest that Kalirin9, and not Kalirin12, is involved in RhoA activation by p75-NgR. The primary difference between the two proteins is an additional kinase domain present in Kalirin12. In hTrio, it was shown that its kinase domain was dispensable for Trio to promote neurite outgrowth (22). We are not certain what prevents Kalirin12 from activating RhoA in 293T cells, but aside from the difference in kinase domains, it seems possible that a difference in cellular localization plays a role. Kalirin9 is expressed in neuritic processes in neuronal cultures, whereas Kalirin12 expression is mostly confined to the cell body (18). p75 is also highly expressed in the dendritic layers as well as in developing axons of Purkinje neurons (Fig. 1). This suggests that Kalirin9 may be sequestered in the particular cellular compartment where it can interact with p75 in the neuritic processes and grow axonal endings to activate RhoA.
Besides Kalirin9, Lingo-1, was previously reported to confer RhoA activation in an ectopic Cos cells, suggesting that Lingo-1 is also involved in Nogo signaling (26). Lingo-1 does not bind myelin inhibitory molecules but associates with p75 and NgR in Cos cells, although in vivo interaction has not been demonstrated. The mechanism by which Lingo-1 activates RhoA has remained unclear, especially because it is a glycosyl phosphatidylinositol-linked protein without an obvious RhoGEF motif. It is possible that Lingo-1 recruits a RhoGEF that can substitute the function of Kalirin9 in ectopic cells, but in the presence of Kalirin9, Lingo-1 utilizes Kalirin9.
In conclusion, we have documented that Kalirin9 is a necessary, functional component for p75-NgR signaling, thereby facilitating RhoA activation. We also identify myelin inhibitory molecules as the first known group of ligands that regulate the function of Kalirin9 in process outgrowth.
Supplementary Material
Acknowledgments
We thank Dr. Betty Eipper, who generously provided Kalirin constructs and antibodies. We also thank Dr. Y. Takai for the pEF-BOS-Myc-Rho-GDI construct and Dr. J. Twiss for the 192 antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants NS 39472 (to S. O. Y.) and NS045758. This work was also supported by the Christopher and Dana Reeve Paralysis Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: MAG, myelin-associated glycoprotein; GEF, GDP/GTP exchange factor; NgR, Nogo receptor; CGN, cerebellar granule neuron(s); GST, glutathione S-transferase; Pn, postnatal day n; GDI, GDP dissociation inhibitor; GTPγS, guanosine 5′-3-O-(thio)triphosphate; RNAi, RNA interference; GFP, green fluorescent protein; SH3, Src homology 3.
A. W. Harrington and S. O. Yoon, unpublished data.
References
- 1.Chen, M. S., Huber, A. B., van der Haar, M. E., Frank, M., Schnell, L., Spillmann, A. A., Christ, F., and Schwab, M. E. (2000) Nature 403 434-439 [DOI] [PubMed] [Google Scholar]
- 2.Mukhopadhyay, G., Doherty, P., Walsh, F. S., Crocker, P. R., and Filbin, M. T. (1994) Neuron 13 757-767 [DOI] [PubMed] [Google Scholar]
- 3.Wang, K. C., Koprivica, V., Kim, J. A., Sivasankaran, R., Guo, Y., Neve, R. L., and He, Z. (2002) Nature 417 941-944 [DOI] [PubMed] [Google Scholar]
- 4.Benson, M. D., Romero, M. I., Lush, M. E., Lu, Q. R., Henkemeyer, M., and Parada, L. F. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 10694-10699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Low, K., Culbertson, M., Bradke, F., Tessier-Lavigne, M., and Tuszynski, M. H. (2008) J. Neurosci. 28 1099-1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.GrandPre, T., Nakamura, F., Vartanian, T., and Strittmatter, S. M. (2000) Nature 403 439-444 [DOI] [PubMed] [Google Scholar]
- 7.Fournier, A. E., GrandPre, T., and Strittmatter, S. M. (2001) Nature 409 341-346 [DOI] [PubMed] [Google Scholar]
- 8.Wang, K. C., Kim, J. A., Sivasankaran, R., Segal, R., and He, Z. (2002) Nature 420 74-78 [DOI] [PubMed] [Google Scholar]
- 9.Wong, S. T., Henley, J. R., Kanning, K. C., Huang, K. H., Bothwell, M., and Poo, M. M. (2002) Nat. Neurosci. 5 1302-1308 [DOI] [PubMed] [Google Scholar]
- 10.Yamashita, T., Higuchi, H., and Tohyama, M. (2002) J. Cell Biol. 157 565-570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domeniconi, M., Cao, Z., Spencer, T., Sivasankaran, R., Wang, K., Nikulina, E., Kimura, N., Cai, H., Deng, K., Gao, Y., He, Z., and Filbin, M. (2002) Neuron 35 283-290 [DOI] [PubMed] [Google Scholar]
- 12.Yamashita, T., and Tohyama, M. (2003) Nat. Neurosci. 6 461-467 [DOI] [PubMed] [Google Scholar]
- 13.Bateman, J., Shu, H., and Van Vactor, D. (2000) Neuron 26 93-106 [DOI] [PubMed] [Google Scholar]
- 14.Liebl, E. C., Forsthoefel, D. J., Franco, L. S., Sample, S. H., Hess, J. E., Cowger, J. A., Chandler, M. P., Shupert, A. M., and Seeger, M. A. (2000) Neuron 26 107-118 [DOI] [PubMed] [Google Scholar]
- 15.Awasaki, T., Saito, M., Sone, M., Suzuki, E., Sakai, R., Ito, K., and Hama, C. (2000) Neuron 26 119-131 [DOI] [PubMed] [Google Scholar]
- 16.Newsome, T. P., Schmidt, S., Dietzl, G., Keleman, K., Asling, B., Debant, A., and Dickson, B. J. (2000) Cell 101 283-294 [DOI] [PubMed] [Google Scholar]
- 17.Hansel, D. E., Quinones, M. E., Ronnett, G. V., and Eipper, B. A. (2001) J. Histochem. Cytochem. 49 833-844 [DOI] [PubMed] [Google Scholar]
- 18.Penzes, P., Johnson, R. C., Kambampati, V., Mains, R. E., and Eipper, B. A. (2001) J. Neurosci. 21 8426-8434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Penzes, P., Beeser, A., Chernoff, J., Schiller, M. R., Eipper, B. A., Mains, R. E., and Huganir, R. L. (2003) Neuron 37 263-274 [DOI] [PubMed] [Google Scholar]
- 20.Chakrabarti, K., Lin, R., Schiller, N. I., Wang, Y., Koubi, D., Fan, Y. X., Rudkin, B. B., Johnson, G. R., and Schiller, M. R. (2005) Mol. Cell. Biol. 25 5106-5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon, S. O., Soltoff, S. P., and Chao, M. V. (1997) J. Biol. Chem. 272 23231-23238 [DOI] [PubMed] [Google Scholar]
- 22.Estrach, S., Schmidt, S., Diriong, S., Penna, A., Blangy, A., Fort, P., and Debant, A. (2002) Curr. Biol. 12 307-312 [DOI] [PubMed] [Google Scholar]
- 23.Hisata, S., Sakisaka, T., Baba, T., Yamada, T., Aoki, K., Matsuda, M., and Takai, Y. (2007) J. Cell Biol. 178 843-860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin, E. Y., Woo, K. N., Lee, C. S., Koo, S. H., Kim, Y. G., Kim, W. J., Bae, C. D., Chang, S. I., and Kim, E. G. (2004) J. Biol. Chem. 279 1994-2004 [DOI] [PubMed] [Google Scholar]
- 25.Schiller, M. R., Chakrabarti, K., King, G. F., Schiller, N. I., Eipper, B. A., and Maciejewski, M. W. (2006) J. Biol. Chem. 281 18774-18786 [DOI] [PubMed] [Google Scholar]
- 26.Mi, S., Lee, X., Shao, Z., Thill, G., Ji, B., Relton, J., Levesque, M., Allaire, N., Perrin, S., Sands, B., Crowell, T., Cate, R. L., McCoy, J. M., and Pepinsky, R. B. (2004) Nat. Neurosci. 7 221-228 [DOI] [PubMed] [Google Scholar]
- 27.Domeniconi, M., Zampieri, N., Spencer, T., Hilaire, M., Mellado, W., Chao, M. V., and Filbin, M. T. (2005) Neuron 46 849-855 [DOI] [PubMed] [Google Scholar]
- 28.Penzes, P., Johnson, R. C., Alam, M. R., Kambampati, V., Mains, R. E., and Eipper, B. A. (2000) J. Biol. Chem. 275 6395-6403 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.