Abstract
TRAF6, a crucial adaptor molecule in innate and adaptive immunity, contains three distinct functional domains. The C-terminal TRAF domain facilitates oligomerization and sequence-specific interaction with receptors or other adaptor proteins. In conjunction with the dimeric E2 enzyme Ubc13-Uev1A, the N-terminal RING domain of TRAF6 functions as an E3 ubiquitin (Ub) ligase that facilitates its own site-specific ubiquitination through the generation of a Lys-63-linked poly-Ub chain. This modification does not cause its proteasomal degradation but rather serves as a scaffold to activate both the IKK and stress kinase pathways. Connecting the N-and C-terminal regions, the four internal zinc finger (ZF) motifs have yet to be functionally defined. In this study, we examined the role of the ZF domains in interleukin-1, lipopolysaccharide, and RANKL signaling by reconstitution of TRAF6-deficient cells with point mutations or deletions of these ZF motifs. Although ZF domains 2-4 are dispensable for activating IKK, p38, and JNK by interleukin-1 and lipopolysaccharide, the first ZF domain together with an intact RING domain of TRAF6 is essential for activating these pathways. Furthermore, TRAF6 autoubiquitination and its interaction with Ubc13 are dependent on ZF1 and an intact RING domain. Additionally, expression of TRAF6 lacking ZF2-4 in TRAF6-deficient monocytes rescues RANKL-mediated osteoclast differentiation and LPS-stimulated interleukin-6 production. These data provide evidence for the critical role of the Ub ligase activity of TRAF6, which is coordinated via the RING domain and ZF1 to supply the necessary elements in signaling by cytokines dependent upon TRAF6.
TRAF6, a member of the tumor necrosis factor receptor-associated factor (TRAF)2 family, is a crucial docking molecule that mediates signaling events initiated by cytokines of the tumor necrosis factor superfamily, interleukin-1 (IL-1) family, and pathogen-associated microbial patterns that are recognized by the Toll-like receptor family (1, 2). Following binding to their respective receptors, these ligands induce a cascade of signaling events leading to the activation of transcription factors, such as the nuclear factor-κB (NF-κB) and AP1 (activator protein-1) family, through activation of upstream kinases, including inhibitor of κ-B kinase (IKK) and the mitogen-activated protein kinase (MAPK) family (i.e. p38, JNK, and extracellular signal-regulated kinase) (1, 2). As a result, these ligands induce numerous genes involved in the innate immune and inflammatory responses. Generation of TRAF6-deficient mice revealed that in addition to playing a critical role in the innate and adaptive immunity, TRAF6 has a crucial role in a wide range of biological functions, such as lymph node organogenesis, formation of skin appendices, nervous system development, and bone metabolism (3-9).
TRAF6 contains three major domains: 1) the C-terminal domain, which facilitates oligomerization and interaction with receptors and other adaptor proteins in a sequence-specific manner; 2) the N-terminal domain, which possesses a RING (really interesting new gene) motif that is found in a number of E3 Ub ligases; and 3) a series of four internal ZF motifs without an ascribed function that connect the N- and C-terminal regions. Biochemical reconstitution experiments have first indicated that the RING domain of TRAF6 is a Ub ligase that in conjunction with a dimeric Ub-conjugating enzyme complex (Ubc13-UeV1A) catalyzes the formation of a unique poly-Ub chain linked through Lys-63 of Ub. That specific modification on TRAF6 triggers, by an unknown mechanism, the activation of both IKK and MKK6 (MAP kinase kinase 6) by the TAK1 (transforming growth factor β-activated kinase 1) complex (10, 11). Subsequently, we have recently demonstrated that a single point mutation in the RING domain (TRAF6-C70A) rendered TRAF6 incapable of rescuing IL-1 or RANKL (receptor activator of NF-κB ligand) signaling events in TRAF6-deficient cells (12, 13). Additionally, stable knockdown of Ubc13 in an osteoclast progenitor cell line impaired the ability of RANKL to cause TRAF6 ubiquitination, IKK and JNK activation, and osteoclast differentiation (13). Furthermore, we have also identified Lys-124 as a major Ub Lys acceptor for TRAF6 autoubiquitination, and mutation of this site abolished TRAF6-mediated NEMO ubiquitination, TAK1 and IKK activation, NF-κB activation, and spontaneous osteoclast differentiation (12). These data support the essential requirement of the RING-dependent Ub ligase activity for TRAF6 to function in cells.
Although our studies have clearly demonstrated the critical requirement of the RING domain of TRAF6 and its Ub ligase activity for RANKL and to some extent for IL-1 signaling (12, 13), a previous report concluded that the RING domain is not required for its function (14). However, in this report, large deletion mutants of the RING domain (and its ZF motifs) were employed to ascertain their function, whereas our reports used a single point mutation in the RING domain. Since these studies show conflicting results for the function of the TRAF6 RING and ZF domains in IL-1, LPS, and RANKL signaling, we decided to reevaluate the role of these domains.
To circumvent the anomalies associated with large deletions, we generated point mutations in each of the four ZF motifs separately. Transient expression of these mutants in HEK293 cells indicated that an intact ZF2, -3, or -4, was not required for activating IKK and JNK. Moreover, in TRAF6-deficient MEFs stably expressing each of the ZF mutants, only a defective ZF1, like a defective RING mutant, could not rescue IKK activation and phosphorylation of IκBα after IL-1 stimulation. Likewise, an intact RING and ZF1 were also required for JNK and p38 activation in response to IL-1 and LPS or for IL-6 production in response to IL-1. Consistent with these data, only intact RING and ZF1 domains of TRAF6 were necessary and sufficient for its autoubiquitination and interaction with Ubc13. Furthermore, TRAF6 carrying an internal deletion of ZF2 to -4 was able to form a complex with NEMO and to rescue IKK, JNK, and p38 activation in response to IL-1 in TRAF6-deficient MEFs. Notably, expression of this deletion mutant of TRAF6 in TRAF6-deficient monocytes could also rescue either RANKL-induced osteoclast differentiation or LPS-induced IL-6 secretion. In summary, we demonstrated that not only is the RING domain of TRAF6 required for its Ub ligase activity and biological function in the IL-1, LPS, and RANKL pathways, but also these studies highlight the significant utility of the first ZF domain of TRAF6 in its function.
EXPERIMENTAL PROCEDURES
Cell Lines, Reagents, and Antibodies—The HEK293 cells were obtained from the ATCC (Manassas, VA) and cultured as previously described (15-17). Retroviral packaging cell line GP2-293 was purchased from Clontech (Palo Alto, CA) and cultured as previously described (12). Mouse embryonic fibroblasts (MEFs) from wild type and TRAF6-/- mice were kindly provided by Dr. Tak Mak (University of Toronto). MEFs from wild type and NEMO-/- mice were kindly provided by Dr. Marc-Schmidt-Supprian (CBR Institute for Biomedical Research). Monoclonal antibodies to phospho-IκBα, phosphop38, and p38 were purchased from Cell Signaling Technology (Beverly, MA); rabbit polyclonal antibodies against JNK1 and IκBα and monoclonal antibodies for Ub and NFATc1 (nuclear factor of activated T cells) were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); goat anti-rabbit IgG-conjugated horseradish peroxidase was from Bio-Rad; goat anti-mouse IgG-conjugated horseradish peroxidase was from BD Biosciences. Rabbit polyclonal antibodies against TRAF6 and β-actin were purchased respectively, from Calbiochem and Cytoskeleton (Denver, CO). Monoclonal antibody to HA was a generous gift from Dr. G. B. Mills (University of Texas M. D. Anderson Cancer Center). Monoclonal antibodies for IKKβ and NEMO were a generous gift from Dr. S. Singh (Imgenex, CA). Monoclonal anti-FLAG, N-ethylmaleimide (NEM), LPS, and tartrate-resistant acid phosphatase (TRAP) kit were purchased from Sigma. Yeast E1 and Ub were purchased from BostonBiochem Inc. (Cambridge, MA). Recombinant GST-c-Jun-(1-79), GST-IκBα-(1-54), GST-TRAF6, and His-tagged proteins Ubc13 and Uev1A were purified essentially as described (12, 15, 17, 18). Recombinant IL-1 was purchased from PeproTech (Rocky Hill, NJ).
Plasmids—Expression vectors for FLAG-TRAF6, GST-TRAF6, HA-Ub, HA-JNK, HA-IKKβ, GST-TAB2 (TAK1-binding protein 2)-(574-693), His-Ubc13, and His-UeV1A were described previously (12, 13, 17-19). All TRAF6 ZF mutations were generated by site-directed mutagenesis using the QuikChange kit (Stratagene, La Jolla, CA) and verified by DNA sequencing. To generate the internal ZF mutants, first a region of mouse TRAF6 (residues 239-530) was amplified by PCR, digested with EcoRI and XbaI, and inserted into pCR3FLAG. Second, a region of mouse TRAF6 (residues 1-158 for TRAF6-ΔZF2-4; residues 1-185 for TRAF6-ΔZF3-4) was amplified by PCR and cloned into the EcoRI site of pCR3FLAG-TRAF6-(239-530). All TRAF6 constructs were subcloned into pGEX-4T1, pGADT7, and pMX-IRES-GFP-Puro (12, 20) by standard techniques and verified by DNA sequencing. Full-length NEMO cloned in-frame with an N-terminal FLAG tag in pcDNA3 was a gift from Dr. Tetsu Kamitani (University of Texas M. D. Anderson Cancer Center) and subsequently cloned into pMX-IRES-GFP-Puro. NEMO-UBD mutant (UBDm; L329P/D311N) was generated by site-directed mutagenesis using the QuikChange kit (Stratagene, La Jolla, CA) and verified by DNA sequencing. Residues 242-419 of NEMO and NEMO-UBDm were cloned into pGEX-4T1 by standard techniques and verified by DNA sequencing.
Measurement of IL-6 Production—TRAF6-deficient MEFs stably expressing various TRAF6 proteins were seeded at a density of 3.0 × 104 in a 24-well plates. Primary mouse bone marrow-derived monocytes (BMM) were isolated from 5-7-day-old femurs of TRAF6-deficient mice, expended, lifted with versene buffer, counted, and seeded at a density of 4.0 × 104 in a 48-well plate. The next day, the cells were infected with various TRAF6 proteins for 2 days and then changed with fresh medium for 2 days. In each case, cells were starved (in serum-free medium or 0.5% FBS) overnight and then stimulated with IL-1 (5 ng/ml) or LPS (30 ng/ml) for 12 h. Culture supernatants were collected, and IL-6 was measured with a mouse IL-6 enzyme-linked immunosorbent assay kit (BD Biosciences, San Diego, CA).
Transfection and Retroviral Infection—Transfection of HEK293 cells was performed essentially as described (12, 17, 20). Production of retroviral supernatants from GP2-293 cells and retroviral infection were performed as previously described (12, 20).
In Vitro Ubiquitination Assay and Poly-Ub Chain Synthesis—In vitro ubiquitination assays were performed as previously described (12).
Western Blotting, Immunoprecipitation, and in Vitro Kinase Assays—Cells were left unstimulated or stimulated as indicated in the figure legends and washed two times with cold phosphate-buffered saline. Cells were lysed in Buffer A (20 mm Tris, pH 7.4, 250 mm NaCl, 1 mm 1, 4-dithiothreitol, 1 mm sodium orthovanadate, 2 mm EDTA, 1% Triton X-100, 2 μg/ml leupeptin, and 2 μg/ml aprotinin) or Buffer B (50 mm HEPES, pH 7.4, 150 mm NaCl, 1 mm 1, 4-dithiothreitol, 1 mm sodium orthovanadate, 2 mm EDTA, 0.5-0.1% Nonidet P-40, 2 μg/ml leupeptin, and 2 μg/ml aprotinin) for 30 min on ice and centrifuged at 13,000 rpm for 15 min. Protein was measured on the clarified lysates, and equal protein was then processed for Western blotting, immunoprecipitation, pull-down, or kinases assays as previously described (12, 15-17, 20). The incorporation of 32P into the substrate was quantitated with a Phosphor-Imager and represented as -fold activation compared with vector or time 0. Immunoprecipitation was carried out at 4 °C for 2 h or overnight with end-over-end rotation and washed three times in Buffer A or B followed by two washes in low salt buffer (20 mm Tris, pH 7.4, 25 mm NaCl, and 1 mm 1, 4-dithiothreitol). In some experiments, possible protein-protein interactions were prevented by immunoprecipitating in Buffer C (Buffer A with the addition of 0.1% SDS and 0.5% deoxycholate) or, where indicated, boiled in 1% SDS (50 μl) for 5 min followed by the addition of 1 ml of Buffer A and then centrifuged and reimmunoprecipitated with the appropriate antibody. To prevent the deubiquitination of some proteins, cells were lysed in Buffer A or B containing 10 mm NEM.
Primary Mouse Monocyte Culture and Osteoclast Differentiation Assays—Primary BMM were isolated from 5-7-day-old femurs of TRAF6-deficient mice and cultured in α-minimal essential medium (10% serum) with 5% L929-M-CSF conditioned medium as previously described (12). When the density of the monocytes was sufficient for the experiment, the cells were washed in phosphate-buffered saline, lifted with versene buffer, counted, and seeded at a density of 2-6.103 in 96-well plates or in 12-well plates containing a dentine slice for bone resorption assays. The next day, the cells were infected with the indicated retrovirus for 2 days and then stimulated in fresh medium containing RANKL (100 ng/ml) and 5% L929-M-CSF until osteoclasts appeared (4-5 days) and then stained for TRAP. The mice were housed in a temperature-controlled environment with free access to food and water. All of the protocols and procedures were approved by the Institutional Animal Care and Use Committee at The University of Texas M.D. Anderson Cancer Center.
Yeast Two-hybrid Analysis—Saccharomyces cerevisiae strain AH109 was a generous gift from Dr. Shrikanth Reddy (University of Texas M.D. Anderson Cancer Center) and used for protein-protein interaction studies. Yeast cells were transformed using the lithium acetate method as described by the manufacturer (Clontech). The yeast cells were co-transformed with the indicated genes in pGAD-T7- and pGBK-T7-based plasmids. Following double selection on synthetic dextrose (SD) lacking Trp and Leu (SD-Trp-Leu), at least three separate colonies were then picked and spotted onto both SD-Trp-Leu and SD-Trp-Leu-His (with 5 mm 1, 2, 4-amino triazole) plates. The plates were incubated at 30 °C until growth appeared on the master plate (SD-Trp-Leu), typically within 2-3 days, and positive interaction was then determined by equivalent growth of colonies on SD-Trp-Leu-His plates.
RESULTS AND DISCUSSION
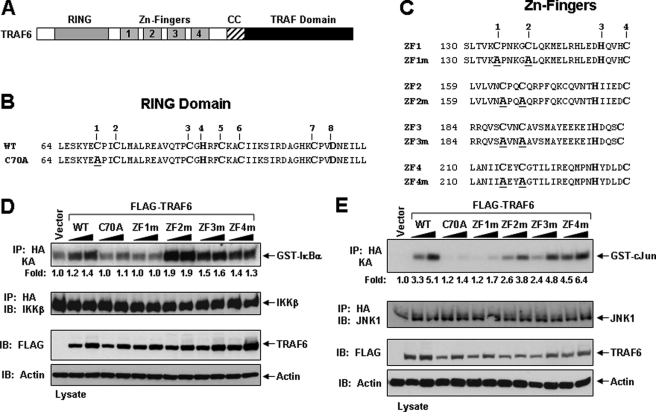
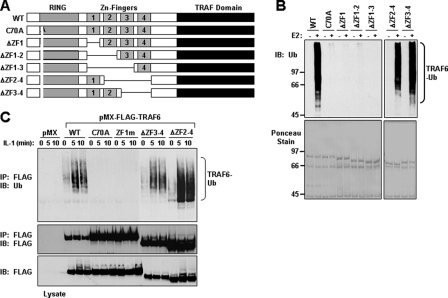
An Intact RING Domain and ZF1 of TRAF6 Are Required for IKKβ and JNK Activation in HEK293 Cells—TRAF6 is composed of an N-terminal RING domain followed by a series of ZF motifs and a C-terminal TRAF homology domain (Fig. 1A). Although considerable characterization of the C-terminal TRAF domain has been elucidated, recent biochemical and biological data have revealed that the N-terminal RING domain of TRAF6 belongs to a family of Ub ligases, also known as E3s. RING-type E3s contain an octet of Cys and His residues distinctly separated to constitute a novel zinc-binding domain, which coordinates two zinc ions in a unique “cross-brace” arrangement (Fig. 1B). Following the RING domain of TRAF6, there are five predicted classical ZF domains, of which four conform to the Cys2-His-Cys (C2HC) motif (Fig. 1C).
FIGURE 1.
Activation of IKKβ and JNK by TRAF6 and its RING and ZF mutants in HEK293 cells. A, schematic diagram of TRAF6 with the indicated RING, ZF, coiled-coil (CC), and TRAF-C domains. B, the primary amino acid sequence of the RING domain consisting of the conserved Cys and His residues (numbered on top) of mouse TRAF6. The TRAF6-C70A mutation is shown below with the Ala substitution underlined. C, the primary amino acid sequence of the four ZF motifs consisting of the conserved Cys and His residues (numbered on top) of mouse TRAF6. The TRAF6 ZF mutants are shown below with the Ala substitution underlined. D-E, TRAF6-ZF1 is important for IKKβ and JNK activation. HEK293 cells were co-transfected with empty vector (vector) or increasing amounts of the indicated FLAG-tagged TRAF6 constructs together with an HA-tagged IKKβ (D) or JNK (E). Thirty-six hours after transfection, cells were harvested, lysed in Buffer A, and subjected to immunoprecipitation (IP) with an anti-HA antibody followed by an in vitro kinase assay using GST-IκBα (D) or GST-c-Jun (E), respectively, as a substrate. The experiment was performed at least three times with similar results each time. The relative incorporation of 32P into the substrates was quantified by a PhosphorImager and represented as -fold activation as compared with vector. The immunoprecipitates were then immunoblotted (IB) with an IKK (D) or JNK (E) antibody, respectively. Expression of the various TRAF6 proteins in the lysates was detected by immunoblotting with anti-FLAG, and then the membrane was stripped and reprobed with anti-actin.
We have previously shown that the Ub ligase activity of TRAF6 requires an intact RING domain by generating a mutant of TRAF6 in which the first Cys residue in its RING domain was mutated to Ala (TRAF6-C70A) (Fig. 1B) (12, 13). To investigate the functional role of the ZF domains of TRAF6 to activate signaling in cells, we respectively mutated the first two Cys residues to Ala in ZF1 (C135A/C140A; TRAF6-ZF1m), ZF2 (C164A/C167A; TRAF6-ZF2m), ZF3 (C189A/C192A; TRAF6-ZF3m), and ZF4 (C215A/C218A; TRAF6-ZF4m) (Fig. 1C). As an initial characterization of these mutations, we examined whether they could activate IKKβ and JNK by transient expression in HEK293 cells. Co-transfection of the FLAG-tagged TRAF6 mutants with HA-tagged IKKβ or HA-tagged JNK indicated that TRAF6-WT, TRAF6-ZF2m, TRAF6-ZF3m, and TRAF6-ZF4m, but not TRAF6-C70A or TRAF6-ZF1m, activated IKKβ (Fig. 1D) and JNK (Fig. 1E). These results suggest that an intact RING domain and ZF1 domain are critical for TRAF6 to activate IKK and JNK.
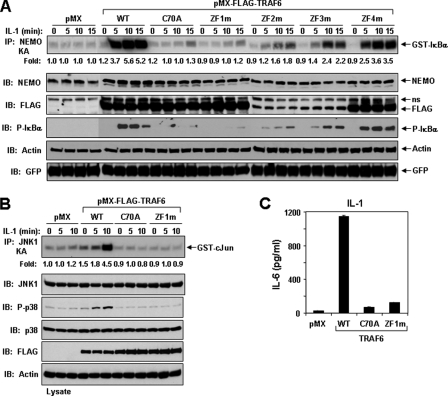
An Intact ZF1 and RING Domain Are Critical for IL-1 and LPS Signaling in TRAF6-deficient MEFs—TRAF6 plays a critical role in IL-1 and LPS signaling, as indicated by loss of function in TRAF6-deficient MEFs (7, 12, 21). By establishing stable cell lines using a retroviral expression system (12, 20), we next examined whether the TRAF6 ZF mutants could rescue IL-1-mediated IKK activation in TRAF6-deficient MEFs. Following IL-1 stimulation, only expression of TRAF6-WT, TRAF6-ZF2m, TRAF6-ZF3m, and TRAF6-ZF4m, but not TRAF6-C70A and TRAF6-ZF1m, were able to activate IKK, resulting in the phosphorylation of IκBα (Fig. 2A). Immunoblotting with an anti-FLAG antibody revealed that protein expression of TRAF6-ZF2m and TRAF6-ZF3m and to a lesser extent TRAF6-ZF4m was less than TRAF6-WT or the other mutants (Fig. 2A) but sufficient to induce significant IKK activation or phosphorylation of IκBα as compared with TRAF6-C70A or TRAF6-ZF1m. This difference in protein expression suggests that mutations in ZF domains 2-4 might affect the stability of the protein in the cells, since the level of GFP, which is expressed from the same transcript, was similar in each cell line (Fig. 2A). Furthermore, IL-1-induced JNK activation and phosphorylation of p38 was not rescued in cells expressing TRAF6-C70A or TRAF6-ZF1m as compared with TRAF6-WT (Fig. 2B). To examine whether the failure of TRAF6-C70A and TRAF6-ZF1m to rescue IKK and stress kinase activation would be translated to their inability to activate target genes, we measured IL-6 secretion by TRAF6-deficient MEFs expressing these mutants after IL-1 stimulation. Although expression of TRAF6-WT rescued IL-6 production by IL-1, TRAF6-C70A- and TRAF6-ZF1m-expressing cells showed impaired induction of IL-6 (Fig. 2C).
FIGURE 2.
IL-1 signaling in TRAF6-deficient MEFs expressing TRAF6 or various TRAF6 mutants. A, an intact RING and ZF1 are required for IKK activation by IL-1. TRAF6-deficient MEFs stably expressing empty vector (pMX) or the indicated FLAG-tagged TRAF6 construct were left unstimulated (time 0) or stimulated with IL-1 (1 ng/ml) for the indicated times. Lysates from the indicated cell lines were prepared in Buffer A, and equivalent protein was immunoprecipitated (IP) with anti-NEMO followed by in vitro kinase assay with GST-IκBα. The experiment was performed at least three times with similar results each time. Cell lysates were immunoblotted (IB) with the indicated antibodies. B, an intact RING and ZF1 are required for JNK and p38 activation by IL-1. Lysates from the indicated cell lines were prepared as in A and immunoprecipitated with anti-JNK1, followed by in vitro kinase assay with GST-c-Jun. The experiment was performed at least three times with similar results each time. Cell lysates were also immunoblotted with the indicated antibodies. C, an intact RING and ZF1 domain are required for IL-6 production induced by IL-1. TRAF6-deficient MEFs stably expressing empty vector (pMX) or the indicated FLAG-tagged TRAF6 constructs were treated with IL-1 (5 ng/ml) for 12 h. The cultured supernatant was collected and assayed for IL-6 using a standard enzyme-linked immunosorbent assay kit. The experiment was performed at least three times with similar results each time.
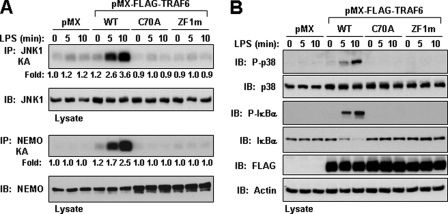
Having established that an intact RING domain and ZF1 are required for IL-1 responsiveness, we next addressed whether these domains are also important for LPS signaling. Under LPS stimulation, stable cell lines expressing TRAF6-WT, but not TRAF6-C70A or TRAF6-ZF1m, were able to activate IKK, resulting in the phosphorylation of IκBα (Fig. 3A). Similar to our data with IL-1, TRAF6-C70A and TRAF6-ZF1m were unable to rescue JNK activation and phosphorylation of p38 following LPS stimulation (Fig. 3, A and B). Collectively, these data establish that a functional RING domain and first ZF motif of TRAF6 are required for IKK, JNK, and p38 activation by IL-1 and LPS.
FIGURE 3.
An intact RING and ZF1 domain of TRAF6 is required for LPS signaling. A and B, TRAF6-deficient MEFs stably expressing empty vector (pMX) or the indicated FLAG-tagged TRAF6 construct were left unstimulated (time 0) or stimulated with LPS (5 μg/ml) for the indicated times. Cell were lysed in Buffer A and immunoprecipitated (IP) with anti-JNK1 or anti-NEMO followed by an in vitro kinase assay using GST-c-Jun (A, top) or GST-IκBα (A, bottom), respectively, as a substrate. The experiment was performed at least three times with similar results each time. Cell lysates were immunoblotted (IB) with the indicated antibodies (A and B).
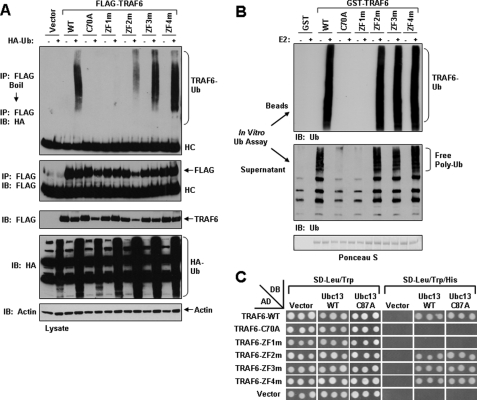
An Intact RING Domain and ZF1 Are Required for TRAF6 Autoubiquitination and Ligase Activity—The RING domain of TRAF6 is essential for its autoubiquitination, which is critical for activation of downstream signaling (12, 13). Since expression of TRAF6, TRAF6-ZF2m, TRAF6-ZF3m, and TRAF6-ZF4m, but not TRAF6-C70A or TRAF6-ZF1m, activated both IKKβ and JNK, we next examined whether they are ubiquitinated in HEK293 cells. The FLAG-tagged TRAF6 constructs were transfected into HEK293 together with HA-Ub, immunopurified by anti-FLAG antibody, boiled in the presence of SDS, and again immunoprecipitated with anti-FLAG followed by SDS-PAGE and immunoblotted with anti-HA. Similar to our previous results (12), when expressed in HEK293 cells along with HA-Ub, FLAG-tagged TRAF6 immunoprecipitates were highly ubiquitinated, as indicated by immunoblotting with anti-HA, whereas TRAF6-C70A was not ubiquitinated (Fig. 4A). Of the ZF mutants tested, only TRAF6-ZF1m was not ubiquitinated (Fig. 4A). To further examine the Ub ligase activity of each of these TRAF6 mutants and to eliminate the possibility of contaminating Ub ligases when expressed in mammalian cells, we expressed and purified TRAF6 and its mutants as GST fusion proteins from bacteria. We tested their ability to become autoubiquitinated and to catalyze free Ub chain synthesis in the presence of Ubc13-UeV1A by performing an in vitro ubiquitination assay. TRAF6, but not TRAF6-C70A, was highly ubiquitinated and promoted free Ub chain synthesis in presence of Ubc13-UeV1A (Fig. 4B). Of the ZF mutants tested, only TRAF6-ZF1m was incapable of serving as an E3 Ub ligase with Ubc13-UeV1A (Fig. 4B). Taken together, these results indicate that not only is an intact RING domain of TRAF6 necessary for its Ub ligase activity, as we have previously reported (12), but also the first ZF domain appears to provide structural integrity to the RING domain.
FIGURE 4.
Requirement of the RING and ZF1 domains of TRAF6 for Ub ligase activity. A, overexpressed TRAF6-C70A and TRAF6-ZF1m are not ubiquitinated in HEK293 cells. HEK293 cells were co-transfected with empty vector (vector) or the indicated FLAG-tagged TRAF6 constructs in the absence (-) or presence (+) of HA-tagged Ub. Thirty-six hours after transfection, cells were harvested, and cell lysates were subjected to immunoprecipitation (IP) with an anti-FLAG antibody followed by boiling in 1% SDS and reimmunoprecipitated with anti-FLAG. Finally, the eluted protein was subjected to SDS-PAGE and immunoblotted (IB) with anti-Ub. The membrane was then stripped and reprobed with anti-FLAG. The experiment was performed at least two times with similar results each time. Cell lysates were immunoblotted with the indicated antibodies (bottom). B, bacterially expressed TRAF6-C70A and TRAF6-ZF1m lack ubiquitin ligase activity. The indicated GST fusion proteins bound to glutathione-agarose beads were subjected to an in vitro ubiquitination assay in the absence (-) or presence (+) of Ubc13-Uev1A (E2). After the ubiquitination assay, a portion of the supernatant (middle) was subjected to SDS-PAGE and immunoblotted with anti-Ub. The GST fusion proteins bound to glutathione-agarose beads were washed and then subjected to SDS-PAGE and immunoblotted with anti-Ub (top). The membrane was stained with Ponceau S (bottom). C, TRAF6-C70A and TRAF6-ZF1m do not interact with Ubc13 in yeast. S. cerevisiae strain AH109 was co-transformed with Ubc13 expressed in pGBK-T7 (DB) and the indicated TRAF6 constructs expressed in pGAD-T7 (AD). Following double selection on SD-Trp-Leu, three separate colonies were then picked and spotted onto both SD-Trp-Leu and SD-Trp-Leu-His plates. The plates were incubated at 30 °C until growth appeared on the master plate (SD-Trp-Leu), typically within 2-3 days, and positive interaction was then determined by equivalent growth of colonies on SD-Trp-Leu-His plates.
Since the destruction of the first ZF domain of TRAF6 presents similar characteristics as a mutation in the RING domain, we next investigated whether both mutations perturb the capacity of TRAF6 to interact with Ubc13. To address this hypothesis, we evaluated the interaction of TRAF6 and its different mutants with Ubc13 in a yeast two-hybrid analysis. TRAF6-WT, but not TRAF6-C70A, was able to interact with Ubc13 or a catalytic mutant of Ubc13 (Ubc13-C87A) (Fig. 4C). Of the four ZF mutants, only TRAF6-ZF2m, TRAF6-ZF3m, and TRAF6-ZF4m were able to interact with Ubc13 or Ubc13C87A (Fig. 4C). Comparable with TRAF6-C70A, TRAF6-ZF1m was incapable of interacting with Ubc13 (Fig. 4C). These results clearly demonstrate that not only is an intact RING domain of TRAF6 required for its interaction with Ubc13, but also the ZF1 participates in maintaining an integral conformation of the E2-binding site. These results also provide insight into our previous findings that ZF1 is required for both IL-1 and LPS signaling.
The RING and ZF1 Domains Are Necessary and Sufficient for TRAF6 Ub Ligase Activity—We next asked whether the RING and ZF1 are necessary and sufficient for TRAF6 Ub ligase activity in vitro. To test this hypothesis, we generated a series of GST fusion proteins containing various perturbations of the RING and ZF domains (Fig. 5A). An in vitro ubiquitination assay using bacterial purified protein confirmed that TRAF6 mutants without an intact RING domain or mutants lacking ZF1 (TRAF6-ΔZF1, TRAF6-ΔZF1-2, and TRAF6-ΔZF1-3) were deficient in TRAF6 autoubiquitination (Fig. 5B). Alternatively, TRAF6 mutants lacking ZF domains 2-4 (TRAF6-ΔZF2-4) or 3 and 4 (TRAF6-ΔZF3-4) possessed significant Ub ligase activity in vitro (Fig. 5B). Furthermore, the absence of TRAF6 autoubiquitination was not due to the loss of its Ub acceptor Lys, since all of the constructs contained the critical Lys that has been mapped to Lys124 (12). Taken together, these results clearly demonstrate that ZF domains 2-4 of TRAF6 are not essential for its Ub ligase activity.
FIGURE 5.
ZF Domains 2-4 are dispensable for TRAF6 autoubiquitination. A, schematic diagram of TRAF6-WT and the internal deletion mutants. B, bacterially expressed TRAF6 mutants lacking ZF1 are not ubiquitinated in vitro. The indicated GST fusion proteins bound to glutathione-agarose beads were subjected to an in vitro ubiquitination assay in the absence (-) or presence (+) of Ubc13-Uev1A (E2). After the ubiquitination assay, the GST fusion proteins bound to glutathione-agarose beads were washed and then subjected to SDS-PAGE and immunoblotted (IB) with anti-Ub (top). The membrane was stained with Ponceau S (bottom). C, TRAF6-C70A and TRAF6-ZF1m are not ubiquitinated following IL-1 stimulation. TRAF6-deficient MEFs stably expressing empty vector (pMX) or the indicated FLAG-tagged TRAF6 constructs were left unstimulated (time 0) or stimulated with IL-1 (10 ng/ml) for the indicated times. Cell lysates were prepared in Buffer A and immunoprecipitated (IP) with anti-FLAG, followed by boiling in 1% SDS, and again immunoprecipitated with anti-FLAG. The samples were then subjected to SDS-PAGE and immunoblotted with anti-Ub (top). The membrane was stripped and reprobed with anti-FLAG (middle). Expression of the TRAF6 proteins in the cell lysates was detected by immunoblotting with anti-FLAG (bottom). The experiment was performed at least two times with similar results each time.
ZF Domains 2-4 Are Dispensable for IL-1-induced TRAF6 Ubiquitination—The experiments described so far demonstrate that the RING and first ZF domains of TRAF6 are critical for IL-1 and LPS signaling and for the ability of TRAF6 to activate these signaling pathways through its Ub ligase activity. Furthermore, in an in vitro ubiquitination assay, TRAF6 lacking ZF2-4 was still capable of autoubiquitination in the presence of Ubc13-Uev1A. Since TRAF6 ubiquitination plays a critical role in its ability to activate downstream pathways (12, 13), we next examined whether TRAF6 mutants lacking ZF2-4 or ZF3-4 expressed in TRAF6-deficient MEFs are ubiquitinated in the presence of IL-1. Stimulation with IL-1 for various times and immunoprecipitation with anti-FLAG revealed that TRAF6-WT, TRAF6-ΔZF2-4, and TRAF6-ΔZF3-4, but not TRAF6-C70A or TRAF6-ZF1m, were highly ubiquitinated in response to IL-1 (Fig. 5C). These results reveal that ZF2 to -4 domains of TRAF6 are not essential for its ability to become ubiquitinated in an IL-1-dependent manner.
ZF Domains 2-4 Are Dispensable for Interacting with NEMO—In view of the fact that TRAF6-ΔZF2-4 has E3 ligase activity and becomes ubiquitinated after IL-1 stimulation, we next examined whether this internal deletion of TRAF6 would interfere with its capacity to form a complex with downstream signaling molecules, in particular with NF-κB essential modifier (NEMO), a critical regulatory subunit of the IKK complex. Recent reports have demonstrated that NEMO preferentially binds Lys-63-linked poly-Ub chains through a novel Ub-binding domain (UBD) that resides within residues 246-346 (22, 23), and mutation within this region, namely Asp-311 (D311N) or Leu-329 (L329P), prevented its binding to Lys-63-linked poly-Ub chains (23). Moreover, mutation of the NEMO UBD prevented its recruitment to the ubiquitinated RIP-1 (receptor-interacting protein-1) complex followed by IKK activation after tumor necrosis factor stimulation (22).
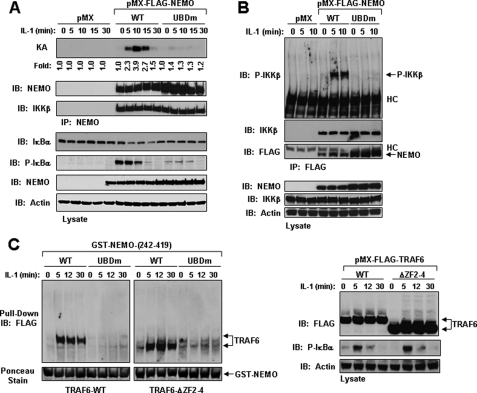
Since RIP1 is not involved in the IL-1 signaling pathway, we decided to first examine the importance of the UBD of NEMO in IL-1 signaling. We generated stable cell lines expressing empty vector (pMX) or FLAG-tagged NEMO-WT or NEMO-UBDm (D311N/L329P) in NEMO-deficient MEFs. Following stimulation with IL-1, expression of NEMO-UBDm did not result in IKKβ activation and phosphorylation leading to phosphorylation and degradation of IκBα, as compared with NEMO-deficient MEFs expressing NEMO-WT (Fig. 6, A and B). Notably, mutation of the UBD of NEMO did not disrupt its interaction with IKKβ, suggesting that the UBD of NEMO is required for recruitment to a proximal activated complex that can phosphorylate IKKβ (Fig. 6B). In agreement with two recent reports (24, 25), our results confirm the critical role of the UBD of NEMO for activation of the IKK complex leading to the phosphorylation of IκBα in IL-1 signaling.
FIGURE 6.
The UBD of NEMO is required for IL-1-mediated IKK activation and its interaction with TRAF6. A, the UBD of NEMO is required for IL-1-induced IKK activation. NEMO-deficient MEFs stably expressing empty vector (pMX), NEMO-WT (WT), or NEMO-UBDm (UBDm) were left unstimulated (time 0) or stimulated with IL-1 (10 ng/ml) for the indicated times. Lysates from the specified cell lines were prepared in Buffer A, and equivalent protein was immunoprecipitated (IP) with anti-NEMO followed by an in vitro kinase assay with GST-IκBα. The membrane was probed with anti-NEMO and then stripped and probed with IKKβ. Cell lysates were immunoblotted (IB) with the indicated antibodies. The experiment was performed at least three times with similar results each time. B, the NEMO-UBD is required for the phosphorylation of IKKβ. The indicated cell lines were treated as in A, and the cell lysates were immunoprecipitated with anti-FLAG. The membrane was probed with phospho-IKKβ antibody and subsequently stripped and immunoblotted with anti-IKKβ and anti-FLAG, respectively. Cell lysates were immunoblotted with the indicated antibodies. The experiment was performed at least two times with similar results each time. C, GST-NEMO through its UBD forms a complex with TRAF6-WT or TRAF6-ΔZF2-4 following IL-1 stimulation. TRAF6-deficient MEFs stably expressing TRAF6-WT or TRAF6-ΔZF2-4 were left unstimulated (time 0) or stimulated with IL-1 (50 ng/ml) for the indicated times. Cell lysates were prepared in Buffer B and precleared with GST bound to glutathione-agarose beads for 1 h at 4°C with rotation. The samples were then centrifuged, and the supernatant was mixed with GST-NEMOorGST-NEMO-UBDm bound to glutathione-agarose beads and incubated overnight at 4 °C with rotation. The samples were washed and subjected to SDS-PAGE and immunoblotted with anti-FLAG (left). The GST fusion proteins were detected by staining the membrane with Ponceau S (bottom). The cell lysates were immunoblotted with the indicated antibodies (right). The experiment was performed at least three times with similar results each time.
Given that these two reports (24, 25) disagree on a potential interaction between NEMO and TRAF6, we decided to examine if IL-1 stimulation could induce the formation of a complex containing TRAF6 and NEMO by using a GST fusion protein consisting of the C-terminal domain of NEMO-WT or UBD mutant (residues 242-419). TRAF6-deficient MEFs stably expressing TRAF6-WT or TRAF6-ΔZF2-4 were treated for various times with IL-1, and the lysates were used in a GST pull-down assay with either GST-NEMO-WT or GST-NEMOUBDm. GST-NEMO-WT formed a complex with both TRAF6-WT and TRAF6-ΔZF2-4 in an IL-1-dependent manner, which occurred after 5 min and persisted for 30 min; however, the type of TRAF6 associating with NEMO appeared to be mostly unmodified (Fig. 6C). These results indicate that IL-1 induced a complex formation between TRAF6 and NEMO, which was not affected by deletion of ZF2 to -4 in TRAF6. In contrast, GST-NEMO-UBDm failed to interact with either TRAF6-WT or TRAF6-ΔZF2-4 (Fig. 6C). These data appear to agree with the model proposed earlier demonstrating that NEMO preferentially binds to Lys63-linked polyubiquitinated IRAK1, which itself interacts with TRAF6 (24, 25).
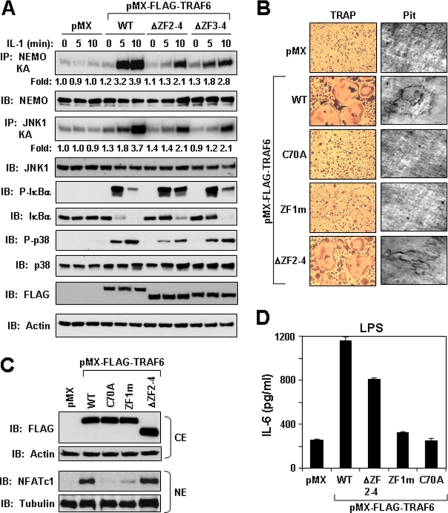
ZF Domains 2-4 Are Dispensable for IL-1, LPS, and RANKL Signaling—We have demonstrated above that TRAF6 lacking the internal ZF domains 2-4 is ubiquitinated in response to IL-1 and that these domains are nonessential for forming a complex with NEMO. We next examined signaling properties of TRAF6 mutants lacking either ZF2-4 or ZF3-4 in TRAF6-deficient cells. Expression of either deletion mutant of TRAF6 was able to rescue IKK, JNK, and p38 activation as compared with pMX in response to IL-1 (Fig. 7A). Similar to IL-1, these mutants also rescued the ability of LPS to activate these signaling pathways (data not shown).
FIGURE 7.
ZF Domains 2-4 are dispensable for IL-1 signaling, RANKL-mediated osteoclast differentiation, and LPS-induced IL-6 production. A, ZF domains 2-4 are dispensable for IL-1-induced IKK, JNK, and p38 activation. TRAF6-deficient MEFs stably expressing empty vector (pMX) or the indicated FLAG-tagged TRAF6 constructs were left unstimulated (time 0) or stimulated with IL-1 (1 ng/ml) for the times indicated. Equivalent protein from the indicated cell lines was immunoprecipitated (IP) with anti-NEMO or anti-JNK1, followed by an in vitro kinase assay using GST-IκBα or GST-c-Jun, respectively, as a substrate. Cell lysates were immunoblotted (IB) with the indicated antibodies. The experiment was performed at least three times with similar results each time. B, TRAF6-ΔZF2-4 is able to rescue osteoclast differentiation and function in TRAF6-deficient monocytes. TRAF6-deficient monocytes isolated from bone marrow of TRAF6-deficient mice were plated in 96-well plates for the osteoclast assay (left) or in 12-well plates containing dentine slide for bone resorption assay (right). The cells were infected with empty virus (pMX) or the indicated TRAF6 retrovirus in septuplet for the osteoclast assay and then stimulated with RANKL (100 ng/ml). The cells were fixed and stained for TRAP (left), and the dentine slices were examined for pit formation (right). Similar results were obtained in at least three independent experiments. C, TRAF6 lacking ZF domains 2-4 is able to induce nuclear accumulation of NFATc1 following RANKL stimulation. The monocytes infected as described above were seeded in 12-well plates and stimulated with RANKL (100 ng/ml). After 48 h, the cells were harvested, and cytoplasmic and nuclear fractions were prepared. Protein from the cytoplasmic extracts (CE) and nuclear extracts (NE) were immunoblotted with anti-FLAG and anti-NFATc1, respectively. The membranes were then stripped and reprobed with anti-actin and anti-tubulin, respectively. D, TRAF6-ΔZF2-4 is able to rescue LPS-induced IL-6 production in TRAF6-deficient monocytes. TRAF6-deficient monocytes isolated from bone marrow of TRAF6-deficient mice were plated in 48-well plates and then infected in triplicate with the indicated retrovirus followed by LPS treatment (30 ng/ml) for 12 h. The cultured supernatant was collected and assayed for IL-6 using a standard enzyme-linked immunosorbent assay kit. The experiment was performed at least three times with similar results each time.
We next evaluated whether these domains are involved in the regulation of other biological endpoints by TRAF6 signaling. First, TRAF6 is an essential mediator of the differentiation of monocytes to mature bone-resorbing osteoclasts following RANKL stimulation (26). We have previously demonstrated that an intact RING domain of TRAF6, and thus its Ub ligase activity, are essential for RANKL-mediated osteoclast differentiation (12, 13). To examine whether the first ZF domain of TRAF6 is necessary and sufficient for osteoclast differentiation, we isolated primary BMM from TRAF6-deficient mice and infected them with a retrovirus programmed to express TRAF6-WT, TRAF6-C70A, TRAF6-ZF1m, or TRAF6-ΔZF2-4 and tested their ability to differentiate into functional TRAP-positive multinucleated osteoclasts after RANKL treatment. TRAF6-deficient monocytes expressing TRAF6-WT and TRAF6-ΔZF2-4, but not TRAF6-C70A or TRAF6-ZF1m, formed multinucleated osteoclasts (Fig. 7B, left) and caused bone resorption pits when differentiated on dentine slices after RANKL stimulation (Fig. 7B, right). Furthermore, RANKL treatment of TRAF6-deficient BMM infected with TRAF6-WT and TRAF6-ΔZF2-4 stimulated the nuclear accumulation of NFATc1, a critical transcription factor for osteoclast differentiation (27, 28), whereas cells expressing either TRAF6-C70A or TRAF6-ZF1m did not (Fig. 7C).
Second, cytokine production by LPS has been shown to be impaired in TRAF6-deficient macrophages (29). Reconstitution with TRAF6 or TRAF6-ΔZF2-4, but not TRAF6-C70A or TRAF6-ZF1m in TRAF6-deficient monocytes rescued LPS-induced IL-6 (Fig. 7D). Collectively, these findings support the necessary requirement of the RING and ZF1 domains of TRAF6 for the biological activity induced by IL-1, LPS, and RANKL and provide further support for the nonessential requirement of ZF domains 2-4 in TRAF6 signaling.
In conclusion, in this report, we have investigated the functional role of the ZF domains of TRAF6 in IL-1, LPS, and RANKL signaling. We have taken advantage of using cells lacking endogenous TRAF6, either TRAF6-deficient MEFs or primary BMM from TRAF6-deficient mice, and introducing various TRAF6 mutants by retroviral transfer to study their effect on signaling by these cytokines. Therefore, the results can be solely attributed to the mutation introduced on TRAF6 providing a better structure-function relationship than overexpression in a cell line containing endogenous TRAF6.
By investigating the functional role of each TRAF6 ZF motifs, we surprisingly uncovered a significant function of the first ZF motif. Previous structural studies using NMR revealed that the RING domain (residues 67-124) of TRAF6 presents a topology similar to those of other RING domains, indicating that the RING adopts a correctly folded structure in the absence of ZF1 (30). Interestingly, the NMR structure of TRAF6 RING revealed a very weak affinity for Ubc13 in the low millimolar range (30), which is too weak to be biologically relevant. This result suggests that other regions contiguous to the RING and possibly the first ZF domain may serve to increase the affinity of TRAF6 for Ubc13. From our mutagenesis studies, the exact role of the ZF1 in TRAF6 function is unclear. It is possible that ZF1 interacts directly with Ubc13 or indirectly by stabilizing some structural elements other than the RING itself. In fact, the crystal structure of the TRAF6 RING plus ZF1 alone or in complex with Ubc13 was solved.3 These data reveal a close interaction between the sequence preceding the RING and the sequences located just before and inside the ZF1. The structural analysis provides support that additional contacts from residues preceding the RING are important for TRAF6/Ubc13 interaction and that ZF1 probably plays a structural role by stabilizing RING/Ubc13 interaction for increased affinity. Nonetheless, our data reveal a close connection between the RING and the ZF1 and that the integrity of both domains is required to have a functional Ub ligase of TRAF6 able to activate downstream signaling events.
Acknowledgments
We thank Dr. Kitamura for providing the pMX vectors, Dr. Mak for providing the wild type and TRAF6-deficient MEFs, Dr. Reddy for providing the yeast two-hybrid reagents, Dr. Marc-Schmidt-Supprian for the wild type and NEMO-deficient MEFs, Dr. Hao Wu for helpful discussions, Dr. Singh for providing NEMO and TRAF6 antibodies, and Dr. Mills for providing the anti-HA antibody.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 AR053540 (to B. G. D.). The DNA Sequencing Core facility is funded in part through NCI, National Institutes of Health, Grant CA16672DAF. This work was also supported in part by institutional start-up funds from the University of Texas M.D. Anderson Cancer Center. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TRAF, tumor necrosis factor receptor-associated factor; IL, interleukin; LPS, lipopolysaccharide; NF-κB, nuclear factor-κB; JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; GST, glutathione S-transferase; GFP, green fluorescent protein; IKK, IκB kinase; MEF, mouse embryonic fibroblast; WT, wild type; NF-κB, nuclear factor-κB; TRAP tartrate-resistant acid phosphatase; UBD, ubiquitin binding domain; ZF, zinc finger; BMM, bone marrow-derived monocytes;NEMO, NF-κB essential modifier; Ub, ubiquitin; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; SD, synthetic dextrose.
B. G. Darnay and H. Wu, submitted for publication.
References
- 1.Chung, J. Y., Park, Y. C., Ye, H., and Wu, H. (2002) J. Cell Sci. 115 679-688 [DOI] [PubMed] [Google Scholar]
- 2.Wu, H., and Arron, J. R. (2003) BioEssays 25 1096-1105 [DOI] [PubMed] [Google Scholar]
- 3.Choi, Y. (2005) Adv. Exp. Med. Biol. 560 77-82 [DOI] [PubMed] [Google Scholar]
- 4.King, C. G., Buckler, J. L., Kobayashi, T., Hannah, J. R., Bassett, G., Kim, T., Pearce, E. L., Kim, G. G., Turka, L. A., and Choi, Y. (2008) J. Immunol. 180 34-38 [DOI] [PubMed] [Google Scholar]
- 5.King, C. G., Kobayashi, T., Cejas, P. J., Kim, T., Yoon, K., Kim, G. K., Chiffoleau, E., Hickman, S. P., Walsh, P. T., Turka, L. A., and Choi, Y. (2006) Nat. Med. 12 1088-1092 [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi, T., Walsh, P. T., Walsh, M. C., Speirs, K. M., Chiffoleau, E., King, C. G., Hancock, W. W., Caamano, J. H., Hunter, C. A., Scott, P., Turka, L. A., and Choi, Y. (2003) Immunity 19 353-363 [DOI] [PubMed] [Google Scholar]
- 7.Lomaga, M. A., Yeh, W. C., Sarosi, I., Duncan, G. S., Furlonger, C., Ho, A., Morony, S., Capparelli, C., Van, G., Kaufman, S., van der Heiden, A., Itie, A., Wakeham, A., Khoo, W., Sasaki, T., Cao, Z., Penninger, J. M., Paige, C. J., Lacey, D. L., Dunstan, C. R., Boyle, W. J., Goeddel, D. V., and Mak, T. W. (1999) Genes Dev. 13 1015-1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naito, A., Azuma, S., Tanaka, S., Miyazaki, T., Takaki, S., Takatsu, K., Nakao, K., Nakamura, K., Katsuki, M., Yamamoto, T., and Inoue, J. (1999) Genes Cells 4 353-362 [DOI] [PubMed] [Google Scholar]
- 9.Naito, A., Yoshida, H., Nishioka, E., Satoh, M., Azuma, S., Yamamoto, T., Nishikawa, S., and Inoue, J. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 8766-8771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang, C., Deng, L., Hong, M., Akkaraju, G. R., Inoue, J., and Chen, Z. J. (2001) Nature 412 346-351 [DOI] [PubMed] [Google Scholar]
- 11.Deng, L., Wang, C., Spencer, E., Yang, L., Braun, A., You, J., Slaughter, C., Pickart, C., and Chen, Z. J. (2000) Cell 103 351-361 [DOI] [PubMed] [Google Scholar]
- 12.Lamothe, B., Besse, A., Campos, A. D., Webster, W. K., Wu, H., and Darnay, B. G. (2007) J. Biol. Chem. 282 4102-4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamothe, B., Webster, W. K., Gopinathan, A., Besse, A., Campos, A. D., and Darnay, B. G. (2007) Biochem. Biophys. Res. Commun. 359 1044-1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi, N., Kadono, Y., Naito, A., Matsumoto, K., Yamamoto, T., Tanaka, S., and Inoue, J. (2001) EMBO J. 20 1271-1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polek, T. C., Talpaz, M., Darnay, B. G., and Spivak-Kroizman, T. (2003) J. Biol. Chem. 278 32317-32323 [DOI] [PubMed] [Google Scholar]
- 16.Ye, H., Arron, J. R., Lamothe, B., Cirilli, M., Kobayashi, T., Shevde, N. K., Segal, D., Dzivenu, O. K., Vologodskaia, M., Yim, M., Du, K., Singh, S., Pike, J. W., Darnay, B. G., Choi, Y., and Wu, H. (2002) Nature 418 443-447 [DOI] [PubMed] [Google Scholar]
- 17.Darnay, B. G., Ni, J., Moore, P. A., and Aggarwal, B. B. (1999) J. Biol. Chem. 274 7724-7731 [DOI] [PubMed] [Google Scholar]
- 18.Darnay, B. G., Haridas, V., Ni, J., Moore, P. A., and Aggarwal, B. B. (1998) J. Biol. Chem. 273 20551-20555 [DOI] [PubMed] [Google Scholar]
- 19.Wong, A., Lamothe, B., Lee, A., Schlessinger, J., and Lax, I. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 6684-6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Besse, A., Lamothe, B., Campos, A. D., Webster, W. K., Maddineni, U., Lin, S. C., Wu, H., and Darnay, B. G. (2007) J. Biol. Chem. 282 3918-3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi, T., Walsh, M. C., and Choi, Y. (2004) Microbes Infect. 6 1333-1338 [DOI] [PubMed] [Google Scholar]
- 22.Ea, C. K., Deng, L., Xia, Z. P., Pineda, G., and Chen, Z. J. (2006) Mol. Cell 22 245-257 [DOI] [PubMed] [Google Scholar]
- 23.Wu, C. J., Conze, D. B., Li, T., Srinivasula, S. M., and Ashwell, J. D. (2006) Nat. Cell Biol. 8 398-406 [DOI] [PubMed] [Google Scholar]
- 24.Conze, D. B., Wu, C. J., Thomas, J. A., Landstrom, A., and Ashwell, J. D. (2008) Mol. Cell. Biol. 28 3538-3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Windheim, M., Stafford, M., Peggie, M., and Cohen, P. (2008) Mol. Cell. Biol. 28 1783-1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darnay, B. G., Besse, A., Poblenz, A. T., Lamothe, B., and Jacoby, J. J. (2007) Adv. Exp. Med. Biol. 597 152-159 [DOI] [PubMed] [Google Scholar]
- 27.Ishida, N., Hayashi, K., Hoshijima, M., Ogawa, T., Koga, S., Miyatake, Y., Kumegawa, M., Kimura, T., and Takeya, T. (2002) J. Biol. Chem. 277 41147-41156 [DOI] [PubMed] [Google Scholar]
- 28.Takayanagi, H., Kim, S., Koga, T., Nishina, H., Isshiki, M., Yoshida, H., Saiura, A., Isobe, M., Yokochi, T., Inoue, J., Wagner, E. F., Mak, T. W., Kodama, T., and Taniguchi, T. (2002) Dev. Cell 3 889-901 [DOI] [PubMed] [Google Scholar]
- 29.Gohda, J., Matsumura, T., and Inoue, J. (2004) J. Immunol. 173 2913-2917 [DOI] [PubMed] [Google Scholar]
- 30.Mercier, P., Lewis, M. J., Hau, D. D., Saltibus, L. F., Xiao, W., and Spyra-copoulos, L. (2007) Protein Sci. 16 602-614 [DOI] [PMC free article] [PubMed] [Google Scholar]