Abstract
Mot1 is an essential, conserved TATA-binding protein (TBP)-associated factor in Saccharomyces cerevisiae and a member of the Snf2/Swi2 ATPase family. Mot1 uses ATP hydrolysis to displace TBP from DNA, an activity that can be readily reconciled with its global role in gene repression. Less well understood is how Mot1 directly activates gene expression. It has been suggested that Mot1-mediated activation can occur by displacement of inactive TBP-containing complexes from promoters, thereby permitting assembly of functional transcription complexes. Mot1 may also activate transcription by other mechanisms that have not yet been defined. A gap in our understanding has been the absence of biochemical information related to the activity of Mot1 on natural target genes. Using URA1 as a model Mot1-activated promoter, we show striking differences in the way that both TBP and Mot1 interact with DNA compared with other model DNA substrates analyzed previously. These differences are due at least in part to the propensity of TBP alone to bind to the URA1 promoter in the wrong orientation to direct appropriate assembly of the URA1 preinitiation complex. The results suggest that Mot1-mediated activation of URA1 transcription involves at least two steps, one of which is the removal of TBP bound to the promoter in the opposite orientation required for URA1 transcription.
The TATA-binding protein (TBP)2 is a central component of the RNA polymerase II (pol II) preinitiation complex (PIC) (1-3). Transcription can be regulated during many different steps in PIC assembly, but the regulation of TBP binding to promoters is a fundamentally important step exploited for regulation of a large number of genes (4, 5). TBP is a saddle-shaped protein that interacts with DNA as a monomer (6, 7). Because TBP can straddle DNA in either of two directions, its binding polarity determines the orientation of the assembled PIC and hence the direction in which transcription initiates (8). TBP recruitment to promoters is regulated by chromatin, activators, and co-activators that contact TBP directly, and general factors that modulate TBP binding and activity (1, 2, 4, 9-11).
NC2 and Mot1 are essential, conserved regulators of TBP function that cooperate to regulate gene expression on a global scale (12-14). NC2, a heterodimer of the Bur6 and Ydr1 subunits in yeast (15), interacts with the TBP-DNA complex and can block subsequent steps in PIC assembly (16). NC2 is also a direct activator of gene expression (17) that is found at promoters in vivo in proportion to the level of transcription (14). How this apparent negative regulator of transcription functions as an activator is likely related to the recent observation that NC2 binding can allow TBP to relocalize along the DNA contour (18). Such mobilization of TBP may be critical for clearance of TBP from inactive but high affinity sites, and may facilitate selection of appropriate TATA sequences among a collection of binding sites along accessible promoter DNA. Mot1 is a member of the Snf2/Swi2 ATPase family (19, 20). Like NC2, it also interacts directly with TBP and regulates TBP binding. However, Mot1 uses a completely different mechanism; ATP hydrolysis by Mot1 catalyzes TBP-DNA dissociation (21).
TBP is unusual among DNA-binding proteins that bind duplex DNA with high affinity in that it binds both defined TATA boxes as well as repetitive TpA sequences (22). Its high affinity results in a long lifetime that compensates for slow association (23). The stability of TBP binding to DNA is critical for nucleation of the pol II PIC at TATA-containing promoters, but the plethora of “TATA-like” sequences in the genome complicates the discrimination of bona fide core promoters from spurious sites. The solution to ensuring high TBP mobility in the face of the requirement for stable DNA binding apparently involves the combined action of NC2, which mobilizes TBP along DNA, and Mot1, which uses ATP to eject it. Many NC2- and Mot1-regulated promoters are repressed by these factors, but paradoxically, a great number of promoters require these factors for transcriptional activation (13, 14, 24, 25). The ability of NC2 to facilitate TBP diffusion along the DNA length can in principle explain how it activates or represses transcription, depending on the sequence and chromatin context. Although TBP-DNA dissociation activity of Mot1 can explain how it represses transcription, how it activates transcription is much less well understood.
We previously showed that a mutation in MOT1 increases TBP occupancy at Mot1-activated promoters in vivo (26). This increase in TBP occupancy was in contrast to the decrease in occupancy observed for other PIC components, including TFIIB, TAFs, and pol II (26). These results are consistent with the idea that Mot1 is required to remove a transcriptionally inactive, kinetically trapped TBP-containing complex. Removal of such complexes would stimulate transcription by allowing additional cycles of TBP binding to occur in a timely fashion, with active TBP-containing complexes being stabilized against Mot1 action by binding of other PIC components. However, the results do not rule out the possibility that Mot1 functions as a co-activator, which facilitates PIC assembly subsequent to TBP binding in some novel way.
In parallel to the in vivo studies, biochemical approaches have provided insight into the architecture of the Mot1-TBP-DNA complex and the mechanism of TBP-DNA dissociation catalyzed by the enzyme. Analyses of different DNA templates, Mot1 mutants, footprinting, and cross-linking have established that although Mot1 has no detectable DNA binding on its own, in the absence of ATP, it forms a ternary complex with TBP and DNA via interaction with residues in TBP and contact with base pairs within an ∼17-bp region upstream of the TATA box (27-29). The length of the upstream DNA region is about what would be expected if the Mot1 ATPase docks onto DNA in a manner similar to that of a highly related ATPase, SsoRad54 (30).
ATP hydrolysis by Mot1 greatly accelerates the dissociation of TBP from DNA. Combined biochemical, mutational, and model building studies suggest that the ATPase catalytic mechanism involves translocation along DNA, and displacement of TBP by Mot1 may occur by translocation through the TBP-DNA interface (31). One surprising observation was that even in the absence of ATP, almost half of the ternary complexes have stabilities that are much reduced compared with TBP-DNA alone, whereas the remaining ternary complexes were very stable, and ATP hydrolysis was required to disassemble them (31). The heterogeneity in ternary complex stability was interpreted to reflect conformational heterogeneity in the Mot1 ATPase itself, which may be related to conformational changes driven by ATP binding and hydrolysis (31).
Although the existing biochemical data have provided insight into the Mot1 catalytic mechanism, what is missing is a link between the biochemical results and the function of Mot1 at natural, Mot1-activated target promoters. Most of the biochemical studies of Mot1 action have been performed using variants of the adenovirus major late promoter (AdMLP), which was advantageous because it provides a unique, well characterized, high affinity TBP-binding site. However, it is clear that core promoters have different properties in vitro and in vivo (32), and the extent to which promoter sequence influences Mot1 activity, or perhaps even confers novel biochemical activity upon the enzyme, has not been investigated. Here we show that the basic biochemical understanding of Mot1 binding and catalytic activity obtained using TBP bound to AdMLP-derived templates is well supported by analyses of the activity of Mot1 on the Saccharomyces cerevisiae URA1 promoter, a natural Mot1-activated target promoter. However, the results also reveal multiple differences in Mot1 function at URA1, indicating that the core promoter sequence strongly influences Mot1 activity and suggesting that Mot1-mediated activation of URA1 transcription involves at least two separate activities of Mot1. Surprisingly, we show that TBP tends to bind to the URA1 TATA box with the wrong polarity to support URA1 PIC formation, suggesting that displacement of TBP bound to the promoter in the reverse orientation by Mot1 is one critical aspect of its activation function.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Growth Conditions—S. cerevisiae strains used for the in vivo analyses are derivatives of YPH499 (33) and were described previously (13, 34). MOT1 shuffling strains were transformed with plasmids carrying the indicated MOT1 alleles and plated on 5-FOA to select for loss of the URA3-marked MOT1 plasmid. For the experiments in Fig. 9, MOT1 shuffling strains were transformed with two plasmids, one carrying MOT1 and the other SPT15, prior to selection on 5-FOA. TBP plasmids (35) were obtained from Karen Arndt and are as follows: pJVS49 (TBP A100P), pJVS50 (TBP P191A), and pJVS54 (TBP A100P, P191A). A yeast plasmid expressing TBPm3 (36) was obtained from Kevin Struhl. For growth assays, spots are 10-fold serial dilutions of the indicated strains grown at 30 °C for 2-3 days. For Northern blotting, cells were grown at 30 °C to an OD ∼1.0. The temperature was then rapidly switched to 35 °C by addition of an equal volume of pre-warmed 40 °C media and heat-shocked at 35 °C for 45 min. Cells were then harvested for isolation of total RNA as described below.
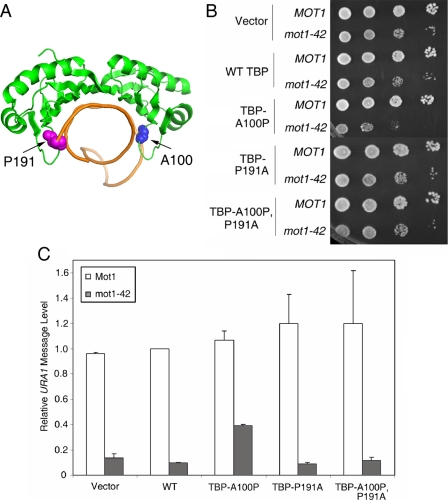
FIGURE 9.
A TBP allele with relaxed DNA binding polarity partially suppresses the requirement for Mot1 for URA1 transcription in vivo. A, schematic representation of the TBP-DNA complex (7) showing the positions of alanine 100 (blue spheres) and proline 191 (magenta spheres). These two residues disrupt the symmetry of the DNA binding surface and play a role in determining the binding orientation of TBP on promoter DNA (35). B, spot assay showing the synthetic growth defect in mot1-42 TBP-A100P cells. 10-Fold serial dilutions of the indicated strains were grown at 30 °C on synthetic media without leucine or tryptophan (for plasmid selection) for 2-3 days. C, quantitation of URA1 RNA levels, determined by Northern blotting, in MOT1+ and mot1-42 cells, with TBP alleles as indicated. Note the approximate 4-fold increase in RNA level in the mot1-42 TBP-A100P strain compared with the RNA level in mot1-42 cells only harboring wild-type TBP. Relative URA1 RNA levels were normalized to ACT1 in the same samples. Error bars are standard deviations from three sets of samples.
RNA Isolation and Northern Blotting—Total RNA was isolated using hot acid-phenol extraction (37). For Northern blots, 20 μg of total RNA was separated by electrophoresis on 1% formaldehyde gels and transferred to a nylon membrane (Nytran, Schleicher & Schuell). DNA probes were generated by random priming of PCR products amplified from a portion of the indicated open reading frames. Blots were hybridized overnight in low stringency hybridization solution (30% formamide, 4× SSC, 10 mm EDTA, 0.25 mg/ml RNA, 10% dextran sulfate, 1× Denhardt's, 5% SDS) and washed twice for 15 min, followed by 1 h with 0.1× SSC, 0.1% SDS. Bands were detected by autoradiography and visualized using a PhosphorImager. Quantitation was performed using ImageQuant software.
In Vivo Site-directed Mutagenesis—Chromosomal mutations were performed by site-directed mutagenesis using oligonucleotides as described previously (38). In brief, the CORE cassette, containing the markers URA3 and KanMX, was recombined into the URA1 promoter in place of the TATA element. Integration was scored by G418 resistance. Oligonucleotides were designed with the desired sequence elements (TATAΔ, MLP-F, MLP-R, and TGTA) and 45 bases of homologous sequence on either side to allow for efficient recombination to take place. Double-stranded, completely overlapping sequences were annealed and transformed at varying concentrations (25-125 μg) to achieve the maximum recombination efficiency. Homologous recombinants were selected by growth on 5-FOA, and integration was confirmed by colony PCR using primers that flank the URA1 promoter region, as well as sensitivity to growth on media containing G418. Total genomic DNA was harvested from positive colonies, and the URA1 promoter was sequenced to verify the sequence change. Strains were transformed with LEU2-marked plasmids carrying MOT1 or mot1-42 prior to transformation with the double-stranded oligonucleotides so that growth on 5-FOA indicated colonies that had lost both the CORE cassette and the URA3 plasmid from the original MOT1 shuffling strain.
Protein Purification—Full-length yeast Mot1 was isolated to apparent homogeneity from a yeast overexpression system as described previously (34). Full-length yeast TBP and TBP core domain (TBPc) were isolated to apparent homogeneity using bacterial overexpression systems as described previously (39). A bacterial expression plasmid for production of TBP E188C was constructed using standard procedures (40). TBP E188C was expressed and purified as described for the wild-type protein (39).
Footprinting and Electrophoretic Mobility Shift Assays—These assays were performed as described (29, 31, 41) using the radiolabeled probes shown in Table 1. The DNA concentration in each reaction was < 1 nm, and the concentrations of TBP, Mot1, and ATP are indicated in the figure legends. DNA molecules were radiolabeled by the Klenow fill-in reaction using [α-32P]dCTP or [α-32P]dATP to label the top or bottom strand and to achieve the DNA fragment lengths shown in Fig. 5. Table 1 shows the annealed oligoduplexes with their names written above and below the annealed strands. In Table 1, T or B signifies the top (written 5′ to 3′) or bottom strand (written 3′ to 5′), respectively. The probes used in Fig. 5 were named as in Table 1 without the T or B designations; the probes with the suffix C indicate labeling was performed using [α-32P]dCTP, whereas those probes with no suffix were labeled with [α-32P]dATP. Footprinting was performed using the URA1 FP1 probe or, in the case of TATA box mutants, analogous probes with the indicated sequence changes. Kinetic analysis was performed by quantifying the change in the TATA box footprint as described (30).
TABLE 1.
URA1 probes
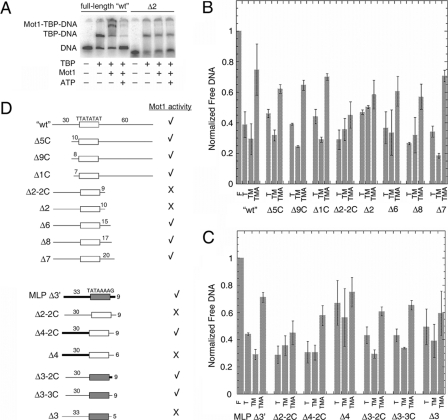
FIGURE 5.
DNA downstream of TATA is required for Mot1 binding and dissociation of TBP from URA1 DNA. A, representative electrophoretic gel mobility shift experiment performed as in Fig. 2A using either radiolabeled WT URA1 DNA or Δ2, a URA1 fragment truncated on the downstream site of the TATA box (see schematic in D). B and C, quantitation of gel mobility shift results for the indicated DNA probes. The bar height represents the relative free DNA in each reaction, ± S.D., determined from at least three independently performed experiments. F, free DNA; T, reaction containing 14 nm TBP; TM, reaction containing 14 nm TBP and 11.6 nm Mot1; TMA, reaction containing 14 nm TBP, 11.6 nm Mot1, and 25 μm ATP. D, schematic of the DNA probes used in the experiments in A-C, and summary of the results. The top set of nine probes was derived from the URA1 promoter. The open rectangle represents the TATA element sequence denoted above it, and the numbers above the flanking DNA segments indicate their lengths in base pairs. The Δ2-2C, Δ2, Δ6, Δ8, and Δ7 probes possess 30 bp of flanking DNA upstream of the TATA sequence and varying lengths of 3′-flanking DNA as indicated. The Δ5, Δ9, and Δ1 probes possess 60 bp of 3′-flanking DNA and varying lengths of 5′-flanking DNA as indicated. The bottom set of seven probes was derived by combining portions of the URA1 and AdMLP promoters, plus appropriate controls. MLP Δ3′ is derived entirely from the AdMLP promoter. In this and other probes in this series, the shaded rectangle represents the AdMLP TATA sequence, TATAAAAG, and the thick black bars indicate AdMLP upstream or downstream sequence. As in the first set of probes, the lengths of flanking DNA, in base pairs, are indicated above the DNA segments. Δ2-2C is shown twice in D simply to make comparison with other constructs easier. Δ4-2C and Δ4 probes possess the indicated lengths of AdMLP flanking DNA upstream of the URA1 TATA box and the indicated lengths of URA1 3′-flanking DNA. Probes Δ3-2C, Δ3-3C, and Δ3 possess the AdMLP TATA sequence but URA1 5′-flanking DNA. The 3′-flanking DNA for Δ3-2C was from the AdMLP promoter, whereas the 3′-flanking DNA for the Δ3-3C and Δ3 probes was derived from the URA1 promoter. A check mark indicates that TBP complexes formed on the indicated probe formed ternary complexes with Mot1 and were dissociated in the presence of Mot1 and ATP. An X indicates that Mot1 binding was poor or undetectable, and little or no catalysis of TBP-DNA dissociation was observed in reactions containing ATP. In no case was ternary complex formation observed without ATP-dependent catalytic activity.
Hydroxyl Radical Footprinting and Fe-BABE Cleavage Assays—DNA fragments of ∼100 nucleotides bearing approximately centered AdMLP or URA1 TATA elements were prepared by PCR amplification of plasmids pRW2 and pURA1 using the primers GCAGGTCGACTCTAGAGGAT, TCGCAGACAGCGATGCGGAA and GGTGGCGGCCGCTCTAGAAT, TGTAATCCAAGGAAGAATA, respectively. A single 32P end label was introduced by kinasing one of the primers in each amplification. The products were purified on 5% native acrylamide gels; the full-length DNA was electroeluted from a gel slice. Changes in the solvent accessibility of the DNA as a function of TBP binding were probed by its reactivity to hydroxyl radicals (.OH) produced by the Fenton reaction (42). Production of the .OH was initiated by simultaneous mixing of 0.9% of H2O2 and 7 mm Fe(II)-EDTA with the solutions of DNA or DNA-protein complex in 50 mm Tris-HCl, pH 8, 60 mm KCl, 5 mm MgCl2, 1 mm CaCl2, 0.01% Brij, 0.1 mm EDTA, 0.1 mm dithiothreitol. The footprinting reaction was terminated at 1 min by addition of thiourea to 30 mm. The [32P]DNA was ethanol-precipitated, and the fragments resulting from .OH cleavage were separated by denaturing PAGE. Autoradiograms of the gels were obtained by exposure to phosphor storage plates and quantitated with single band resolution using the SAFA software (52). The intensity of each band was normalized for the average intensity of resolved bands in the lane. Relative .OH reactivity was calculated as the difference in the normalized band intensities for the DNA-protein complex and the DNA alone.
TBP E188C was labeled by FeBABE using the ProFound™ protein interaction mapping kit (Pierce). The labeling procedure described in the manufacturer's manual was followed exactly except for the condition for the conjugation of the FeBABE. TBP E188C was conjugated with FeBABE at 20 °C for 3 h to prevent protein denaturation. The 32P-end-labeled DNA fragment was incubated with FeBABE-labeled TBP E188C at 25 °C for 30 min in 50 mm Tris-HCl, pH 8, 60 mm KCl, 5 mm MgCl2, 1 mm CaCl2, 0.01% Brij, 0.1 mm EDTA, 0.1 mm dithiothreitol. The cleavage reaction was initiated by addition of ascorbic acid and the stable peroxide reagent provided in the kit (Pierce). The reaction was quenched after 30 s by the addition of 2.5 volumes of absolute ethanol. The DNA fragments resulting from cleavage were separated and visualized as described above. A “DNA only” control lane contained all the reagents except for TBP E188C. Each band density was divided by an average density for all the analyzed bands in its lane. Presented is the difference between the normalized band densities in the TBP E188C-FeBABE-containing and DNA-only lanes.
RESULTS
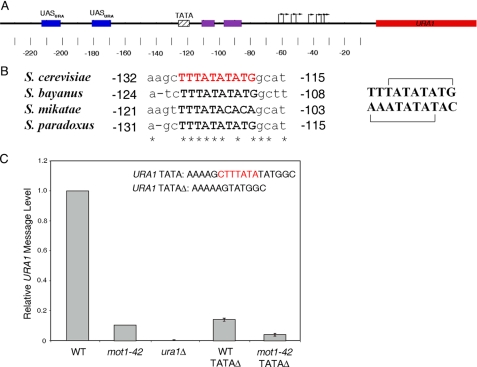
A Functional, Conserved TATA Element in the URA1 Promoter—The URA1 promoter was chosen for detailed analysis because our previous work showed that it is a direct target of Mot1-mediated activation (26). Additionally, the URA1 transcription start sites were previously mapped (43), and the promoter harbors a well defined, phylogenetically conserved TATA element (Fig. 1, A and B). The TATA element is unusual in containing two overlapping potential TBP-binding sites oriented in opposite directions. Based on prior work (22, 44), the top strand sequence TATATATG would be predicted to direct TBP binding in the appropriate orientation and distance for transcription initiation at the previously mapped URA1 start sites, whereas the overlapping bottom strand sequence, TATATAAA, would be predicted to bind TBP in the opposite orientation. The arrangement was intriguing because an inappropriately oriented TBP would interfere with PIC formation and might be a preferred substrate for Mot1-mediated displacement. To determine whether the TATA region of the URA1 promoter is functionally significant, we deleted it from the chromosomal copy of the gene. The 7-bp deletion removed both of the potential binding sites for TBP discussed above. Northern blotting showed that URA1 RNA was reduced about 10-fold in the TATAΔ strain compared with wild-type cells (WT) (Fig. 1C). The effect of TATAΔ was quantitatively similar to the decrease in URA1 RNA observed in mot1-42 cells bearing a WT URA1 promoter. The reduced expression of URA1 RNA driven by the TATAΔ promoter was further decreased in mot1-42 cells, indicating that the feeble expression driven by the TATAΔ promoter was still at least partially Mot1-dependent (Fig. 1C).
FIGURE 1.
Organization of the URA1 promoter and requirement for the URA1 TATA element in vivo. A, schematic of the URA1 locus and 5′-untranslated region showing the locations of regulatory regions identified through prior molecular studies and phylogenetic sequence alignments. The location of the TATA box is indicated by the hatched box, and the transcription start sites (43) are shown by the arrows. The URA1 promoter harbors two binding sites for the Ppr1 transcription factor (52, 53) (blue boxes, UASURA), as well as two conserved sequences (purple boxes) that may be also be involved in URA1 transcriptional control (43). The URA1 gene product is required for uracil biosynthesis (43). In our strain background, Ppr1 is required for URA1 expression; however, transcriptional activity is insensitive to the level of uracil in the media (data not shown). Numbers indicate position relative to the start of translation, ATG. B, sequence alignment showing the TATA element of URA1 in four yeast strains. Note the S. cerevisiae“conventional” TATA element TATATATG and the overlapping reverse TATA element TTTATATA (shown in red). Asterisks indicate identical base pairs, and numbers are relative to the start of translation. The top and bottom strands of the S. cerevisiae sequence are shown on the right, with the overlapping putative TATA elements marked by brackets. C, quantitation of URA1 message levels in the indicated strains, determined by Northern blotting. The 7-bp region shown in red (top right) was deleted in the TATAΔ strains. URA1 RNA levels were normalized to the levels of ACT1 RNA in the same samples. Errors are standard deviations obtained from three separate RNA samples for each strain.
In Vitro Analysis of TBP Binding to the URA1 Promoter—Having confirmed that this TATA element is critical for URA1 expression in vivo, we next tested whether TBP interacts with the TATA element in vitro and if Mot1 modulates the interaction of TBP with the promoter. Although Mot1 enzymatically displaces TBP from DNA, one possibility was that its requirement for URA1 activation was because of an alternative biochemical activity specified by the promoter sequence. For example, Mot1 might be required for delivery of TBP or a TBP-containing complex to the URA1 promoter, as suggested by in vivo studies of other Mot1-activated promoters (24). However, a unique, TBP-bound complex was readily detectable by gel mobility shift analysis using a radiolabeled URA1 promoter probe and TBP alone (Fig. 2A, lane 2). The TBP-DNA complex was further shifted to slightly slower mobility by TFIIA (Fig. 2A, lane 5 versus lane 2); a greater shift was seen with the addition of Mot1 (Fig. 2A, lane 3 versus lane 2). Note that Mot1 can also stabilize TBP binding to weaker sites on the DNA probe, which explains why addition of Mot1 can result in the formation of more than one higher order complex (27). These results show that there is a high affinity TBP-binding site on the URA1 probe, and Mot1 is not required for stable binding of TBP to the promoter. Addition of ATP resulted in Mot1-mediated clearance of TBP from the probe (Fig. 2A, lane 4 versus lanes 2 and 3), indicating that although Mot1 activates URA1 expression in vivo, the biochemical activity of Mot1 measured using the URA1 probe is similar to results obtained with other TATA sequences.
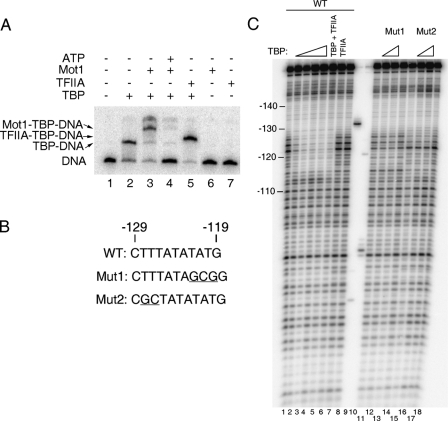
FIGURE 2.
Interaction of TBP with the URA1 promoter in vitro. A, electrophoretic mobility shift assay using radiolabeled URA1 promoter DNA. TBP (14 nm) was incubated with the probe for ∼20 min at 22 °C, followed by the addition of Mot1 (9.2 nm), TFIIA (10.5 “units” of recombinant TBP-DNA binding activity (54)), and/or ATP (25 μm) for 5 min prior to loading on the gel. Arrows indicate the positions of the TBP-DNA, Mot1-TBP-DNA, and TFIIA-TBP-DNA complexes. B, sequences of the TATA regions of the WT, Mut1, and Mut2 probes used for footprinting. Mut1 alters both putative TATA sequences; Mut2 eliminates only the reverse TATA sequence (see Fig. 1B). Deviations from the WT sequence are underlined. C, DNase I footprinting experiment in which TBP was incubated with URA1 DNA. The reactions contained 5.6, 14, 33.6, 56, and 14 nm TBP in lanes 2-6, respectively. Reactions in lanes 12 and 16 contained 14 nm TBP; reactions in lanes 13 and 17 contained 33.6 nm TBP. Reactions in lanes 7 and 8 contained 12 units of TFIIA. Lanes 9 and 10 show markers obtained by restriction enzyme digestion of the URA1 probe and were used to assign positions of the other bands on the gel. Note the TBP footprint from -127 to -115 (with respect to the start codon) that was observed using the WT probe only.
Next, DNase I footprinting was used to locate bound TBP along the URA1 probe. As shown in Fig. 2C, the TBP footprint was centered over the TATA box, but the location of TBP could not be determined precisely because the A/T-rich DNA just upstream of the TATA element was refractory to DNase I cutting, and because DNase I typically generates footprints that are larger than the actual protein-binding sites (45). We observed no change in the DNase I footprint when TFIIA was added to the reaction even though TFIIA extends the TBP footprint asymmetrically on some templates (46) (Fig. 2C, lane 6 versus lanes 2-5). These data do not distinguish between the oppositely oriented overlapping TATA box sequences.
To better define the location of TBP on the TATA region, two TATA substitution mutants were analyzed (Fig. 2, B and C). Substitution of 3 bp in TATA mutant 1 disrupts both of the potential TBP-binding sites, so its failure to support TBP binding was not surprising. TATA mutant 2 alters the potential TBP-binding sequence in the orientation opposite to the direction of transcription, whereas the top strand TATA sequence TATATATG, an apparently quite reasonable TBP-binding site, remains intact. The remarkable absence of TBP binding to the mutant 2 template suggests that TBP alone cannot bind to the predicted high affinity site appropriately oriented for transcription. Moreover, the importance of base pairs at the upstream edge of the TATA element suggests that TBP preferentially binds to the URA1 promoter in the opposite direction of transcription. This reverse orientation is a potential molecular explanation for the “inactive” TBP-containing complex that accrues at Mot1-activated promoters when Mot1 is depleted in vivo (26).
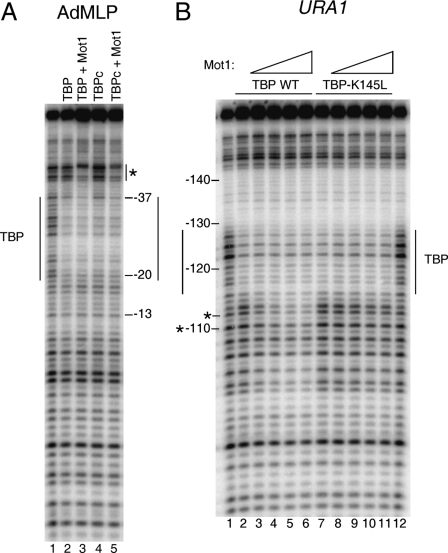
Using a DNA probe containing the high affinity TATA sequence from the adenovirus major late promoter (AdMLP; TATAAAAG), we have shown that Mot1 interacts with DNA upstream of the TATA box (31). This interaction was observed for both TBP and its C-terminal core domain (TBPc) (Fig. 3A). This experiment was performed with both TBP and TBPc to permit comparison of the new URA1 studies described below with the previously published work (31).
FIGURE 3.
Organization of the Mot1-TBP-URA1 DNA complex. A, DNase I footprinting analysis of TBP and Mot1 binding to the radiolabeled AdMLP probe containing the TATA element TATAAAAG. The reactions contained no added protein (1st lane), full-length TBP (14 nm, 2nd and 3rd lanes), TBP core domain (TBPc, 8 nm, 4th and 5th lanes), and 11.6 nm Mot1 (3rd and 5th lanes). Reactions were incubated at 22 °C for ∼20 min and then processed as described previously (31). The TBP footprint is indicated by the long vertical lines, and upstream protection induced by Mot1 binding is shown by the asterisk. B, DNase I footprinting experiment in which Mot1 was added to TBP-URA1 DNA complexes. DNA and 14 nm TBP (WT or K145L) were incubated together for ∼20 min at 22 °C, followed by the addition of Mot1 as follows: 1.2 nm to lanes 3 and 8; 3.6 nm to lanes 4 and 9; 7.2 nm to lanes 5 and 10; and 10.8 nm to lanes 6 and 11. After ∼5 min of incubation, reactions were processed as described (31). Lanes 1 and 12 show free DNA. Vertical lines indicate the TBP footprint, and asterisks indicate downstream positions where digestion was inhibited by Mot1 below the level of digestion seen in DNA alone (see Fig. 4).
In contrast, titration of Mot1 to pre-formed TBP-URA1 DNA complexes resulted in changes in the footprint downstream of the TATA element (Fig. 3B). The Mot1-induced modulation of DNase I cleavage was unusual in that extension of the footprint was because of suppression of TBP-induced enhanced cleavage (Fig. 4). Mot1 suppressed DNase I cleavage in the downstream DNA below that observed in the naked DNA control at only two positions (Fig. 4, asterisks). Thus, it was unclear if Mot1 protects the DNA downstream of TATA or if it alters the conformation of the TBP-DNA complex so as to alleviate enhanced downstream DNase I cutting.
FIGURE 4.
Quantitation of URA1 DNase I footprinting results. The graph shows the relative band intensities measured from the gel image shown in Fig. 3B, lanes 1-6, obtained by single band peak fitting (49, 55). Note that the addition of Mot1 resulted in suppression of DNase I digestion at downstream positions, but at only two positions (asterisks) was the digestion reduced below the level of digestion seen in reactions with DNA alone. Thus, Mot1 reduced the enhanced downstream digestion attributable to TBP, rather than generating a true extension of the TBP footprint.
The Mot1-TBP interaction on the URA1 promoter was further probed using the TBP mutant K145L. Lys-145 is located on the “top” surface of TBP opposite the DNA binding surface, and this residue is critical for Mot1 binding to TBP bound to the AdMLP (29). The DNase I footprint on URA1 was unchanged when Mot1 was added to TBP-K145L-URA1 complexes (Fig. 3B), indicating that this residue is important for Mot1 binding to TBP on the URA1 promoter as well as the AdMLP. A similar result was obtained using TBP-K138L, another mutant that has been shown previously to be defective in interaction with Mot1 (29; data not shown). The results show that although Mot1 interacts similarly with TBP on both the AdMLP and URA1 probes, the organization of the DNA-bound proteins differs.
DNA Downstream of TATA Is Required for Mot1 Binding to TBP-URA1 DNA—Gel mobility shift assays were performed using probes of decreasing length to determine the DNA that is required for Mot1 activity on the URA1 promoter. Representative primary data are shown in Fig. 5A, the quantified results from multiple experiments in Fig. 5, B and C, and the overall conclusions in Fig. 5D. On the AdMLP (27), DNA upstream of the TATA box was required for Mot1 binding and ATP-dependent TBP-DNA dissociation, whereas downstream DNA was not required (MLP Δ3′ probe, Fig. 5, C and D). This behavior strikingly contrasts with the URA1 probes. Downstream URA1 DNA was required for Mot1 binding and catalysis, whereas upstream DNA was not (e.g. compare probe Δ2 versus WT in Fig. 5A; see also results with Δ1C and Δ2-2Cin Fig. 5, B and D). A URA1 probe with 15 bp of 3′-flanking DNA (probe Δ6) was the shortest probe tested that supported Mot1 action. Taken together, the results show that between 11 and 15 bp of downstream flanking DNA were required for Mot1 to interact with TBP bound to the URA1 promoter and to catalyze displacement of TBP in the presence of ATP. As this DNA length requirement is similar to the length of upstream DNA required for Mot1 binding to TBP positioned on the AdMLP TATA, we suggest that the URA1 and AdMLP Mot1-TBP complexes are bound in opposite orientations.
Chimeric probes that combine AdMLP and URA1 sequences were used to reveal the features of DNA sequence responsible for the different ternary complexes formed on the parent promoters. Mot1 binding and catalytic activity were supported by a probe consisting of AdMLP TATA and URA1 upstream flanking DNA (Fig. 5, C and D, compare Δ3-2C or Δ3-3C to Δ2-2C) demonstrating that the TATA element can dictate the upstream DNA requirement. Swapping the upstream DNA from the URA1 promoter with the upstream DNA from the AdMLP also switched the requirement for upstream DNA (Fig. 5, C and D; compare Δ4-2C to Δ2-2C). Thus, the sequences flanking the TATA box can dictate the architecture and functionality of Mot1-TBP-DNA ternary complex in addition to the identity of the TATA box.
It is important to note that regardless of the probe used, there was a requirement for at least a short sequence of duplex DNA abutting the TATA sequence. For example, although the AdMLP TATA element had the capacity to switch the flanking DNA requirement of the URA1 probe, between 6 and 9 bp of flanking DNA were required on the opposite side of the TATA box. This requirement can be seen in the activity of the Δ3-3C probe, which possessed 9 bp of downstream flanking DNA and supported Mot1 activity, versus the Δ3 probe which possessed a long upstream DNA tail but was inactive with only 6 bp of downstream flanking DNA.
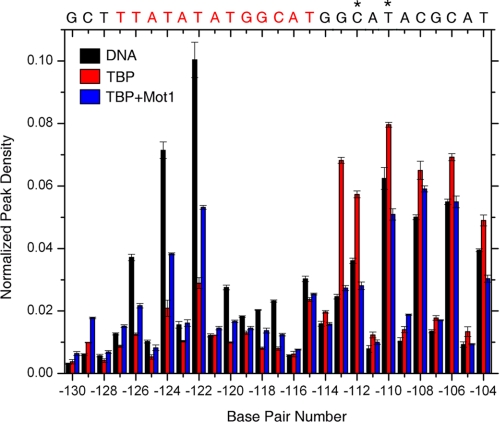
Preferential Reversed Orientation of TBP Binding to the URA1 Promoter—TBP occupancy of the oppositely oriented URA1 TATA box (Fig. 2), the Mot1 downstream footprint, and the requirement for downstream DNA (Figs. 3 and 5) suggest that the orientation of TBP and Mot1 is reversed relative to the AdMLP. To precisely localize the binding of TBP to the URA1 promoter, we turned to hydroxyl radical footprinting. Because of its small size and relative insensitivity to base sequence, the hydroxyl radical provides higher resolution information about protein-DNA complexes compared with DNase I (47). Full-length TBP and TBP core domain (TBPc) behaved similarly in these experiments; both proteins were analyzed to facilitate comparison with published work (31).
The hydroxyl radical footprints of TBPc and TBP binding to the AdMLP TATA box (Fig. 6A) were consistent with previous studies (47). The top strand protection was asymmetric with the greatest protection at T3; the bottom strand protection was more evenly distributed over the first six nucleotides. Only when the orientation of the URA1 probe was inverted (Fig. 6B) did the strand-specific patterns of hydroxyl radical protection mimic the AdMLP. The deepest protection on the top was on the consensus half of the indicated TATA box; the protection on the bottom strand was more evenly distributed. These similarities (and the mutational results shown in Fig. 2C) suggest that it is the reverse TATA element defined by the bottom strand sequence that binds TBP at the URA1 promoter.
FIGURE 6.
Hydroxyl radical footprinting of the TBP-URA1 DNA complex. The graphs show the extent of hydroxyl radical DNA cleavage of TBPc-DNA (open circles) and TBP-DNA (closed circles) complexes formed on the AdMLP (A) and URA1 (B) DNAs. The blue box in A indicates the AdMLP DNA sequence bound by TBPc in the co-crystal structure (6, 22). The blue box in B shows the DNA sequence contacted by TBP and TBPc in the URA1 promoter inferred by the effects of the TATA mutants and the similarities with the AdMLP footprinting patterns. Note that compared with the AdMLP, the URA1 sequence is inverted with respect to the direction of transcription.
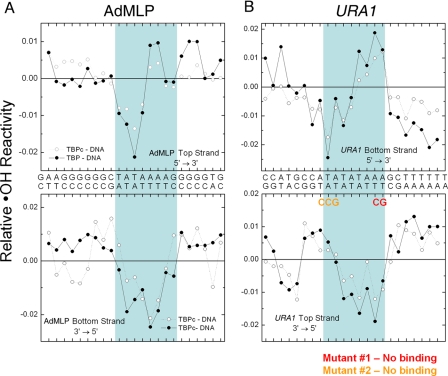
Although the hydroxyl radical footprinting patterns suggest that only the reverse TATA box binds TBP, these data do not directly address the orientation of the bound protein. FeBABE was conjugated to TBP carrying a cysteine residue at the tip of the C-terminal stirrup (Fig. 7A) to directly test if TBP binds to the URA1 promoter in a unique orientation opposite that of the AdMLP. The FeBABE moiety generates a localized “spray” of hydroxyl radicals that cleave DNA in the vicinity of the tethered reagent.
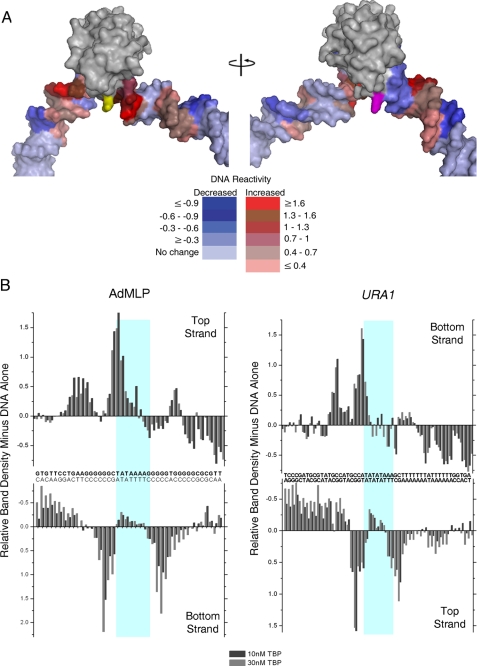
FIGURE 7.
TBP-tethered FeBABE cleavage of URA1- and AdMLP DNA. A, model of the TBPc-DNA complex, based on the yeast TBP-DNA co-crystal (7) to which extensions of duplex were appended to each end of the DNA visible in the structure. TBP is shown in gray. The stirrup residue Glu-188, which was converted to cysteine for conjugation to FeBABE, is colored yellow. Residue Lys-97 on the opposite stirrup that corresponds to the position of E188 is colored magenta. The relative reactivity of the AdMLP DNA to FeBABE cleavage (B) is color-coded in PyMOL as follows: increased reactivity is colored in shades of red ranging from red (≥1.6×), chocolate (1.3-1.6), firebrick (1-1.3), raspberry (0.7-1), dark salmon (0.4-0.7), to salmon (≤0.4). Decreased relative reactivity is colored in shades of blue ranging from density (≤ -0.9), blue (-0.6 to -0.9), tv blue (-0.3 to -0.6), to slate (≥ -0.3). The coincidence of the regions of increased reactivity in shades of red with the site of FeBABE conjugation (shown in yellow) indicates TBP is bound in a single orientation on both promoters. B, graphs show the relative FeBABE-directed DNA cleavage on both strands and at each position of the AdMLP and URA1 promoters as indicated. The light blue boxes mark the sequences contacted by TBP (Fig. 6). The URA1 and AdMLP sequences are oriented in opposite directions with respect to one another as in Fig. 6. Experiments were performed in reactions containing either 10 or 30 nm TBP as indicated.
Because TBP stirrups bind at the ends of the TATA box sequence, the location of DNA cleavage reveals the orientation of the bound protein (8). If TBP binds with a preferred orientation appropriate for PIC assembly, FeBABE tethered to the C-terminal stirrup is predicted to preferentially cleave the DNA at the upstream edge of the TATA box and farther upstream with one helical turn periodicity (7, 8). Indeed, FeBABE cleavage was seen for the top strand of the AdMLP at the upstream end of the TATA box and one helical turn farther up the DNA (Fig. 7B). Upstream cleavage of the bottom strand was also observed in helical phase with the FeBABE moiety.
The agreement of the observed FeBABE reactivity pattern with TBP bound in a single orientation is seen when the reactivity pattern is mapped onto the structure of the TBPc-DNA complex; the highest reactivity was observed in the DNA immediately adjacent to the FeBABE moiety followed by high reactivity in more distant regions within a “clear line of sight” (Fig. 7A, the site of coupling of the FeBABE moiety is colored yellow; also see supplemental Fig. S1). Protection from FeBABE cleavage was clearly evident in regions of the DNA obscured from the moiety by either TBP itself or the helical phasing of the DNA. This interpretation was reinforced by inspection of the reactivity in relation to the symmetric residue Lys-97 on the opposite stirrup (Fig. 7A, colored magenta). No DNA reactivity was observed adjacent to Lys-97 or in-phase on the downstream DNA. We consider this interpretation under “Discussion” in relation to published TBP-DNA-directed cleavage patterns (8, 48).
The FeBABE cleavage patterns of the URA1 and AdMLP promoters are strikingly similar when the orientation of the URA1 sequence is inverted (Fig. 7B) as was observed for the hydroxyl radical cleavage patterns (Fig. 6B). We conclude from the results presented in Figs. 6 and 7 that TBP binds to the TATA box defined by the bottom strand URA1 sequence in a preferred orientation opposite to its orientation on the AdMLP. This opposite orientation to the start of transcription is thus predicted to be responsible for the opposite orientation and flanking DNA requirement of Mot1 bound to the URA1 promoter. The results suggest that TBP does not on its own interact with the URA1 promoter in an orientation that supports transcription of the URA1 open reading frame.
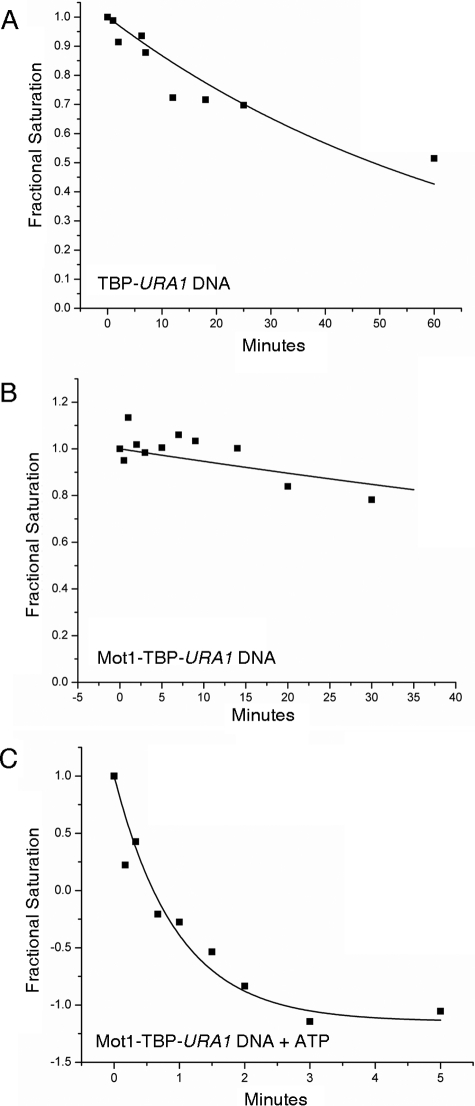
Kinetic Analysis of Mot1 Catalytic Activity—The unexpected organization of the TBP-DNA and Mot1-TBP-DNA complexes formed on the URA1 template prompted us to consider that quantitatively different complex stabilities might contribute to the differential sensitivity of the URA1 promoter to Mot1 action. TBP bound to URA1 DNA with a Kd of 5.4 ± 0.3 nm, as determined by DNase I footprint titration (not shown), comparable with that measured for the AdMLP. However, the kinetics of TBP binding to these two promoters differ (see supplemental Table S1). TBP bound to the URA1 promoter was about 4-fold more stable compared with the AdMLP (Fig. 8A; supplemental Table S1; t½ = 70.4 ± 8.3 versus ∼18 min, respectively (31)). The Mot1-TBP-URA1 DNA complex was also distinguishably different. On URA1 DNA, the ternary complex behaved as a single, highly stable species with a dissociation half-time of 181.9 ± 67.2 min (Fig. 8B). On the AdMLP TATA, the ternary complex dissociation was biphasic, with about half the complexes displaying very short half-lives and about half the complexes much longer lives (31) (supplemental Table S1). Thus, the effect of Mot1 on TBP-DNA complex behavior was affected by DNA sequence.
FIGURE 8.
Dissociation kinetic analysis of complexes formed on the URA1 promoter. Radiolabeled URA1 DNA probe was incubated with 14 nm TBP with or without 15 nm Mot1. At time 0, an excess of unlabeled TATA DNA was added to the reactions, and the extent of TATA box occupancy was determined by DNase I footprinting as described previously (31). A, rate of dissociation of the TBP-URA1 DNA complex. The half-time value for the best fit to a single exponential decay is 70.4 ± 8.3 min. The TBP-DNA complexes formed on the URA1 template are about 4-fold more stable than those formed on the AdMLP (31) (supplemental Table S1). B, rate of dissociation of the Mot1-TBP-URA1 DNA complex. The half-time value for the best fit to a single exponential is 181.9 ± 67.2 min. C, rate of dissociation of the Mot1-TBP-URA1 DNA complex in the presence of 25 μm ATP, which was added at time 0. The half-time value for the best fit to a single exponential is 0.96 ± 0.11 min. This represents an approximate 190-fold increase in the dissociation rate as a result of Mot1 ATP-dependent catalytic activity.
Dissociation of the Mot1-TBP-URA1 DNA complex was measured following addition of ATP. The reaction was characterized by a half-time of 0.96 ± 0.1 min (Fig. 8C) and is the same, within experimental error, as the rate measured under the same conditions for the AdMLP (31) (supplemental Table S1). This result indicates that Mot1 catalysis of TBP dissociation is not influenced by the stability of TATA box binding but is rather dictated by another rate-limiting aspect of the catalytic mechanism.
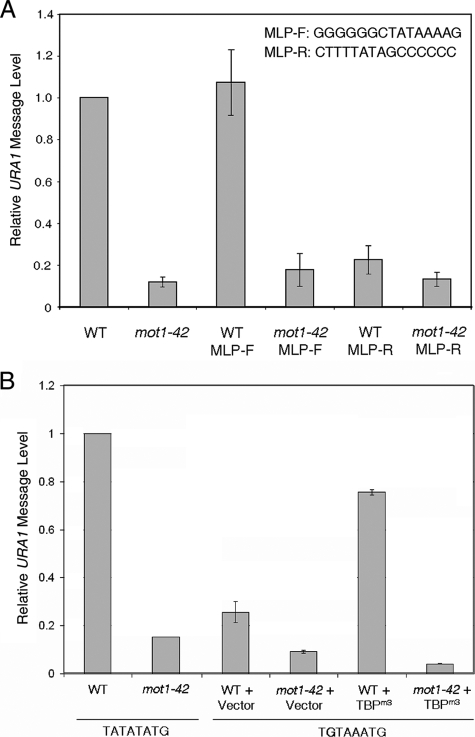
Partial Bypass of the Mot1 Requirement in Vivo by a Bidirectional TBP—The above results establish the propensity of TBP to associate with the URA1 promoter in the opposite orientation as that required for appropriate PIC assembly. Because previous chromatin mapping analysis indicates that the URA1 promoter is not occluded by nucleosomes (26), we suggest that TBP can bind in the reverse orientation to the URA1 promoter in vivo. TBP binding orientation cannot be measured directly in vivo, but by manipulating the sequence of the URA1 TATA or TBP, several experiments were performed to test the idea. First, we took advantage of a TBP allele, A100P, whose alteration on the DNA binding surface permits utilization of a reverse TATA element in vivo (35) (Fig. 9A). This allele does not support cell viability as the sole source of TBP (data not shown), so it was introduced into cells expressing wild-type TBP as well. As shown in Fig. 9B, expression of TBP A100P had no effect on growth of wild-type cells but aggravated the slow growth phenotype of mot1-42 cells. This effect on growth was specific for the A100P allele because it was not observed in cells expressing TBP P191A or TBP A100P P191A, alleles with a mutation in the symmetrically positioned residue on the DNA binding surface, Pro-191, that do not have the ability to direct transcription from a reversed TATA element in vivo (35). Interestingly, Northern blot analysis (Fig. 9C) showed that although expression of TBP A100P had no significant effect on URA1 message levels in wild-type cells, it could partially suppress the Mot1 requirement for URA1 transcription in mot1-42 cells. The effect was specific because there was no effect of the other TBP alleles on URA1 expression, and the positive effect of TBP A100P on URA1 expression was observed even though expression of TBP A100P had a negative effect on cell growth.
Replacement of the TATA Box Does Not Obviate the Requirement for Mot1 in Vivo—The observation that a TBP allele with relaxed binding polarity can partially bypass the requirement for Mot1 is consistent with the hypothesis that displacement of inappropriately oriented TBP is one function for Mot1 in gene activation in vivo. The partial suppression of the Mot1 requirement could be due to the presence of wild-type TBP in the same cells, additional roles for Mot1, or a more complex, possibly indirect mechanism underlying the activity of TBP A100P. We thus altered the sequence of the URA1 promoter itself to determine how the TATA sequence influences the Mot1 requirement in vivo. As the architectures of ternary complexes are reversed on AdMLP versus URA1 probes in vitro, we replaced the URA1 TATA with the AdMLP TATA in both orientations. As shown in Fig. 10A, the AdMLP TATA sequence directed levels of URA1 transcription comparable with the fully wild-type promoter when the TATA was inserted in the same orientation as it resides in the AdMLP promoter (“forward”). Strikingly, expression from this AdMLP-URA1 promoter was as dependent on Mot1 as expression driven by the wild-type promoter. Therefore, the identity of the TATA element alone is not sufficient to specify the Mot1 requirement in vivo. This TATA box orientation was important because the AdMLP TATA inserted in the opposite orientation (“reverse”) did not restore comparable levels of URA1 transcription, and the transcription that was detectable was only weakly dependent on Mot1. These results are consistent with results shown in Fig. 5 demonstrating that ternary complex orientation can be influenced by flanking DNA sequence as well as TATA sequence, and indicate that simple ejection of TBP bound to the promoter in the reverse orientation is not sufficient to explain how Mot1 activates this promoter (although limitations of this experiment are discussed below).
FIGURE 10.
Substitution of the URA1 TATA box with other TATA sequences does not bypass the requirement for Mot1 in vivo. A, URA1 message levels in WT and mot1-42 strains with the AdMLP TATA box inserted in the forward (MLP-F) or reverse (MLP-R) orientation. The exact sequences of the MLP-F and MLP-R constructs are shown (upper left). URA1 RNA levels were determined by Northern blotting and were normalized to the level of ACT1 RNA in the same samples. B, quantitation of URA1 RNA in WT and mot1-42 cells harboring WT TATA sequence or the TGTAAA substitution. TGTA strains were transformed with plasmid vector or TBPm3 expression plasmid as shown. Error bars are standard deviations derived from three determinations.
To further address the role of TATA box identity and orientation in the Mot1 activation mechanism, the URA1 TATA was replaced with the sequence TGTAAA. This sequence is not stably bound by wild-type TBP but is recognized by the TBP allele TBPm3 (36). The G-C base pair substitution at the upstream end of the TBP-binding site and the locations of altered residues on the DNA binding surface of TBPm3 that confer binding enforce unique polarity on the TBPm3-DNA complex. As shown in Fig. 10B, substitution of the URA1 TATA with the TBPm3 site diminished URA1 expression in vivo, consistent with the inability of wild-type TBP to recognize the sequence. Similarly consistent was the stimulation of URA1 expression in cells expressing the TBPm3 allele. However, the results in mot1-42 cells show quite clearly that transcription directed by TBPm3 is still strongly Mot1-dependent. The results in Figs. 9 and 10 reveal that although relaxation of TBP binding polarity can partially bypass the requirement for Mot1, the Mot1 requirement is nonetheless relatively independent of the identity of the TATA box. The results indicate an important role for promoter sequence context and suggest that Mot1 activates URA1 transcription by more than one mechanism.
DISCUSSION
Mot1-mediated gene activation poses a conundrum. Multiple lines of genetic and molecular evidence indicate that the ATP hydrolysis-dependent displacement of TBP from DNA by Mot1 occurs in vivo (13, 34). Moreover, the effect of Mot1 on gene activation is direct in that it is present at Mot1-activated genes in vivo (13, 24, 25). One model for how Mot1 activates gene expression is that it displaces stable, kinetically trapped TBP-containing complexes from promoters to permit the assembly of functional complexes. This model is supported by the increased levels of TBP at Mot1-activated promoters in mot1 cells (26). TBP accumulation contrasts with the relative depletion of TFIIB, certain TAFs, and RNA pol II at these same promoters in mot1 cells (26). The model that Mot1 displaces inactive TBP complexes is also consistent with the observation that the global TBP population in cells is highly mobile, and the mobility is entirely Mot1-dependent.3 However, the dependence on Mot1 for establishment of functional PICs is also consistent with a co-activator function for Mot1. A co-activator function for Mot1 could involve a novel biochemical activity, as gene activation requires a functional Mot1 ATPase (13). Below we discuss a new, general model for Mot1 activation that accommodates these observations and the results presented here.
A Two-step Model for Mot1-mediated Gene Activation—Three types of biochemical results show that TBP alone binds to the URA1 promoter in the wrong orientation to support PIC formation. First, the hydroxyl radical footprints of TBP bound to the URA1 and AdMLP promoters are very similar to one another, provided that the DNA cleavage patterns are aligned such that the URA1 sequence is inverted compared with that of the AdMLP. Thus, URA1 utilizes the TATA box defined by the bottom strand sequence to bind TBP. Second, the TBP-FeBABE cleavage patterns on the two promoter sequences resemble one another, but again, only when the digestion patterns are compared when the promoter sequence of URA1 is inverted relative to the AdMLP. Third, the Mot1 binding and displacement of TBP from URA1 DNA required downstream DNA, rather than the upstream DNA required for removal of TBP from the AdMLP.
It is important to point out that the downstream DNA length requirement at URA1 is not a consequence of the cooperative assembly of Mot1 and TBP on the promoter, because the downstream length requirement was observed when Mot1 was added to preformed TBP-DNA complexes. The opposite orientation of the URA1 ternary complex is most simply rationalized by the biased binding orientation of TBP alone. No evidence indicates that Mot1 itself can discriminate among different flanking DNA sequences with which it interacts.
The results of the FeBABE cleavage studies, together with the additional studies summarized above, are consistent with TBP binding in unique, reciprocally biased orientations to the AdMLP and URA1 promoter sequences. A compelling aspect of the FeBABE reactivity pattern is its consistency with the TBPc-DNA structure (Fig. 7A) and its absence of symmetry with regard to the TATA box sequences. Our conclusion that TBP binding to the AdMLP is preferentially biased differs from that derived from similar studies using either 1,10-phenanthroline or fluorescence resonance energy transfer to probe the orientation of TATA box-bound TBP and TBPc, respectively, that demonstrated only a weak bias for TBP binding in the correct orientation (8, 48). There is no obvious reason for the differences observed between the studies. Among the possibilities are subtle differences in the experimental conditions, protein preparation, flanking DNA sequences, and the presence or absence of the N-terminal domain (TBP versus TBPc). Further investigation into the generality of the orientational bias of TBP-TATA box binding is warranted.
As the footprint extension on the AdMLP resulted in near complete protection of upstream DNA, and as Mot1 was able to displace essentially all of the DNA-bound TBP with only an upstream DNA extension (31), the analyses of Mot1 function further support the conclusion that TBP is capable of binding to the AdMLP in essentially one orientation. The alternative possibility is that TBP binds in both orientations but that somehow Mot1 nonetheless binds in only one orientation with respect to the DNA sequence. This latter possibility is difficult to envision stereochemically, and it is also inconsistent with results showing that Mot1 contacts specific residues on the top surface of TBP, which are required for ternary complex formation on both the AdMLP and URA1 templates (29) (Fig. 3 and data not shown).
Does reverse binding of TBP to the URA1 promoter occur in vivo? There is no method to measure TBP binding orientation in vivo, but it is likely that TBP binds with this orientational bias in vivo as well. Prior chromatin mapping analyses demonstrated that the URA1 TATA box is accessible in vivo (26, 50). Thus, the binding behavior of TBP to naked DNA in vitro appears to be a reasonable model for understanding its binding behavior in vivo. One argument for how reverse binding of TBP may be minimized in vivo is that TBP may bind to the URA1 promoter as a constituent of the TFIID complex, rather than as free TBP. Consistent with this suggestion, URA1 transcription is TFIID-dependent (26). Association of TAFs with other promoter sequences may help properly orient TBP (5). On the other hand, a substantial proportion of TBP is not stably associated with TAFs in vivo.3 Therefore, whereas functional PICs may assemble from a complex nucleated by TFIID rather than TBP, the high proportion of free TBP in cells implies that binding of TAF-free TBP to the URA1 promoter is almost certain to occur in vivo. This being the case, because the polarity of the PIC is determined by the binding orientation of TBP, we propose that one critical step in URA1 activation by Mot1 involves the clearing of incorrectly oriented TBP from the promoter. It is noteworthy that the results presented here show that the TBP-URA1 DNA complex is extraordinarily stable in vitro, which may render the URA1 promoter especially sensitive to Mot1 in vivo. Presumably, TBP that interacts with the TATA box in the correct orientation is then stabilized by interactions with other PIC components, and thus becomes refractory to Mot1-mediated dissociation. Support for this model comes from the partial suppression of the Mot1 requirement for URA1 transcription by TBP A100P, an allele with relaxed binding orientation specificity that can direct transcription from a reversed TATA element in vivo (35). Regulation of transcription by a reverse TATA element does not appear to be a general feature of Mot1-activated promoters; however, we suggest that transcription may well be affected by spurious binding of TBP to inappropriate sites within core promoters susceptible to Mot1 action.
Although the results presented here strongly suggest that Mot1 functions in part by facilitating redistribution of TBP binding orientation, this cannot explain entirely how Mot1 activates URA1. Substitution of the URA1 TATA box with either of two other TATA sequences failed to diminish the requirement for Mot1, even in the case in which transcription was largely dependent on TBPm3 binding to TGTAAA, which fixes the orientation of TBP binding. Caveats to the TATA substitution experiments are worth pointing out, however. First, although replacement with the AdMLP TATA did not bypass the requirement for Mot1 in vivo, it is unclear if replacement with the AdMLP TATA in the context of the URA1 chromatin environment would be sufficient to strongly bias TBP binding polarity. For example, in some cases, the effects of flanking sequences are even more important for determining TBP binding affinity than the sequence of the TATA box itself (51). Because the “rules” that determine these effects are not well understood, the extent to which the AdMLP TATA element perturbs TBP binding orientation in vivo is unknown. Although TBPm3 only interacts with the TGTAAA sequence in the proper orientation, the cells also possess wild-type TBP that may compete or interfere with TBPm3 binding via interaction with neighboring or overlapping sites. Thus, the Mot1 requirement for URA1 transcription driven by TBPm3 may reflect a role for Mot1 in regulating wild-type TBP binding else-where on the URA1 promoter rather than, or in addition to, regulation of TBPm3-containing PICs.
Limitations of the in vivo experiments notwithstanding, the simplest interpretation is that Mot1 performs at least two different functions in activation of URA1 transcription. The first activity is to displace inappropriately bound TBP from the promoter, which would otherwise interfere with productive PIC formation. The second function has not been delineated biochemically, but because it cannot be bypassed by replacement of the TATA box with other TATAs with different properties, we suggest that Mot1 also functions as a co-activator in some specific step in PIC assembly subsequent to the recruitment of appropriately oriented TBP to the promoter. Although there is no experimental evidence to date to speculate on what such an activity may be, the URA1 promoter appears to provide an excellent model system for tackling this problem biochemically.
Diversity in Mot1 Biochemical Activity on Different Promoter DNAs—Previous results established a model of how Mot1 recognizes TBP-DNA and uses docking of its ATPase on upstream DNA to propel dissociation of TBP from DNA (31). The model was derived from multiple lines of biochemical investigation and was consistent with general features of DNA translocases. However, virtually all of the biochemical studies were performed using model DNA templates that are not physiologic targets of Mot1 action in vivo. Given the complexity of the transcriptional regulatory activity of Mot1 in vivo, a key question was whether the biochemical studies on such DNA templates accurately reflect Mot1 behavior at natural target promoters. The results here show that basic features of the Mot1 mechanism are similar at a natural target promoter, arguing for a consistency in TBP-DNA recognition and ATP-dependent catalytic activity. For instance, binding and displacement of TBP bound to URA1 DNA require flanking DNA, just like at the AdMLP template. The requirements for the same residues on TBP and a similar length in DNA extension are consistent with a similar mode of recognition, and the requirement for a downstream DNA extension rather than upstream can be explained entirely by the opposite orientation of TBP.
Interestingly, the Mot1-TBP-URA1 DNA complex is distinguishable from ternary complexes formed on the AdMLP in possessing very high stability. Two populations of ternary complex were detectable on the AdMLP in the absence of ATP; about half the complexes were much less stable than the TBP-DNA complex alone, whereas about half the ternary complexes were much more stable than TBP-DNA alone (31). The uniformly high stability of Mot1-TBP-URA1 DNA ternary complexes (Fig. 8B) shows that ternary complex stability is influenced by DNA sequence. The high stability of the URA1 ternary complex may be somehow related to the proposed co-activator function of Mot1 described above. Moreover, the downstream DNA length requirement for Mot1 on the URA1 promoter (≤15 bp) is slightly less than the upstream DNA length requirement on the AdMLP (17 bp), suggesting that Mot1 may have a somewhat different conformation when associated with the two DNAs. Biochemical results also show that although the TBP-URA1 DNA complex is much more stable than the TBP-AdMLP DNA complex, ATP-dependent dissociation of TBP by Mot1 occurs with essentially the same rate on both templates. Thus, the Mot1 catalytic rate is determined by some mechanistic step that is unrelated to the stability of the TBP-DNA interaction or the DNA sequence. One possibility is that the rate is determined by a sub-step in the ATPase cycle (e.g. ATP binding, hydrolysis, or ADP/Pi release) or a conformational change in the ATPase.
Supplementary Material
Acknowledgments
We are grateful to Kevin Struhl, Karen Arndt, and Michael Resnick for providing plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grants GM55763 (to D. T. A.) and GM39929 (to M. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Fig. 1.
Footnotes
The abbreviations used are: TBP, TATA-binding protein; WT, wild type; PIC, preinitiation complex; pol, polymerase; 5-FOA, 5-fluoroorotic acid; AdMLP, adenovirus major late promoter; TAF, TATA-binding protein-associated factor.
Sprouse, R. O., Karpova, T. S., Mueller, F., Dasgupta, A., McNally, J. G., and Auble, D. T. (2008) Proc. Natl. Acad. Sci. U. S. A., in press.
References
- 1.Ranish, J. A., and Hahn, S. (1996) Curr. Opin. Genet. Dev. 6 151-158 [DOI] [PubMed] [Google Scholar]
- 2.Reese, J. C. (2003) Curr. Opin. Genet. Dev. 13 114-118 [DOI] [PubMed] [Google Scholar]
- 3.Hahn, S. (2004) Nat. Struct. Mol. Biol. 11 394-403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee, T. I., and Young, R. A. (1998) Genes Dev. 12 1398-1408 [DOI] [PubMed] [Google Scholar]
- 5.Matangkasombut, O., Auty, R., and Buratowski, S. (2004) Adv. Protein Chem. 67 67-92 [DOI] [PubMed] [Google Scholar]
- 6.Kim, J. L., Nikolov, D. B., and Burley, S. K. (1993) Nature 365 520-527 [DOI] [PubMed] [Google Scholar]
- 7.Kim, Y., Geiger, J. H., Hahn, S., and Sigler, P. B. (1993) Nature 365 512-520 [DOI] [PubMed] [Google Scholar]
- 8.Cox, J. M., Hayward, M. M., Sanchez, J. F., Gegnas, L. D., van der Zee, S., Dennis, J. H., Sigler, P. B., and Schepartz, A. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 13475-13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pugh, B. F. (2000) Gene (Amst.) 255 1-14 [DOI] [PubMed] [Google Scholar]
- 10.Li, B., Carey, M., and Workman, J. L. (2007) Cell 128 707-719 [DOI] [PubMed] [Google Scholar]
- 11.Chitikila, C., Huisinga, K. L., Irvin, J. D., Basehoar, A. D., and Pugh, B. F. (2002) Mol. Cell 10 871-882 [DOI] [PubMed] [Google Scholar]
- 12.Pereira, L. A., Klejman, M. P., and Timmers, H. T. M. (2003) Gene (Amst.) 315 1-13 [DOI] [PubMed] [Google Scholar]
- 13.Dasgupta, A., Darst, R. P., Martin, K. J., Afshari, C. A., and Auble, D. T. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 2666-2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geisberg, J. V., Holstege, F. C., Young, R. A., and Struhl, K. (2001) Mol. Cell Biol. 21 2736-2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prelich, G. (1997) Mol. Cell Biol. 17 2057-2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meisterernst, M., and Roeder, R. G. (1991) Cell 67 557-567 [DOI] [PubMed] [Google Scholar]
- 17.Willy, P. J., Kobayashi, R., and Kadonaga, J. T. (2000) Science 290 982-985 [DOI] [PubMed] [Google Scholar]
- 18.Schluesche, P., Stelzer, G., Piaia, E., Lamb, D. C., and Meisterernst, M. (2007) Nat. Struct. Mol. Biol. 14 1196-1201 [DOI] [PubMed] [Google Scholar]
- 19.Eisen, J. A., Sweder, K. S., and Hanawalt, P. C. (1995) Nucleic Acids Res. 23 2715-2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis, J. L., Kunisawa, R., and Thorner, J. (1992) Mol. Cell Biol. 12 1879-1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Auble, D. T., Hansen, K. E., Mueller, C. G. F., Lane, W. S., Thorner, J., and Hahn, S. (1994) Genes Dev. 8 1920-1934 [DOI] [PubMed] [Google Scholar]
- 22.Patikoglou, G. A., Kim, J. L., Sun, L., Yang, S.-H., Kodadek, T., and Burley, S. K. (1999) Genes Dev. 13 3217-3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoopes, B. C., LeBlanc, J. F., and Hawley, D. K. (1992) J. Biol. Chem. 267 11539-11547 [PubMed] [Google Scholar]
- 24.Andrau, J.-C., Van Oevelen, C. J. C., Van Teeffelen, H. A. A. M., Weil, P. A., Holstege, F. C. P., and Timmers, H. T. M. (2002) EMBO J. 21 5173-5183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geisberg, J. V., Moqtaderi, Z., Kuras, L., and Struhl, K. (2002) Mol. Cell. Biol. 22 8122-8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dasgupta, A., Juedes, S. A., Sprouse, R. O., and Auble, D. T. (2005) EMBO J. 24 1717-1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darst, R. P., Wang, D., and Auble, D. T. (2001) EMBO J. 20 2028-2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pereira, L. A., van der Knaap, J. A., van den Boom, V., van den Heuvel, F. A., and Timmers, H. T. (2001) Mol. Cell Biol. 21 7523-7534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darst, R. P., Dasgupta, A., Zhu, C., Hsu, J.-Y., Vroom, A., Muldrow, T. A., and Auble, D. T. (2003) J. Biol. Chem. 278 13216-13226 [DOI] [PubMed] [Google Scholar]
- 30.Durr, H., Korner, C., Muller, M., Hickmann, V., and Hopfner, K.-P. (2005) Cell 121 363-373 [DOI] [PubMed] [Google Scholar]
- 31.Sprouse, R. O., Brenowitz, M., and Auble, D. T. (2006) EMBO J. 25 1492-1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller, F., Demeny, M. A., and Tora, L. (2007) J. Biol. Chem. 282 14685-14689 [DOI] [PubMed] [Google Scholar]
- 33.Sikorski, R. S., and Hieter, P. (1989) Genetics 122 19-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Auble, D. T., Wang, D., Post, K. W., and Hahn, S. (1997) Mol. Cell Biol. 17 4842-4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spencer, J. V., and Arndt, K. M. (2002) Mol. Cell Biol. 22 8744-8755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strubin, M., and Struhl, K. (1992) Cell 68 721-730 [DOI] [PubMed] [Google Scholar]
- 37.Schmitt, M. E., Brown, T. A., and Trumpower, B. L. (1990) Nucleic Acids Res. 18 3091-3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Storici, F., Lewis, L. K., and Resnick, M. A. (2001) Nature Biotech. 19 773-776 [DOI] [PubMed] [Google Scholar]
- 39.Gupta, S., Cheng, H., Mollah, A. K. M. M., Jamison, E., Morris, S., Chance, M. R., Khrapunov, S., and Brenowitz, M. (2007) Biochem. 46 9886-9898 [DOI] [PubMed] [Google Scholar]
- 40.Li, F., and Mullins, J. I. (2002) Methods Mol. Biol. 182 19-27 [DOI] [PubMed] [Google Scholar]
- 41.Hsieh, M., and Brenowitz, M. (1996) Methods Enzymol. 274 478-493 [DOI] [PubMed] [Google Scholar]
- 42.Fenton, H. J. H. (1894) J. Chem. Soc. 65 899-910 [Google Scholar]
- 43.Losson, R., Fuchs, R. P. P., and Lacroute, F. (1985) J. Mol. Biol. 185 65-81 [DOI] [PubMed] [Google Scholar]
- 44.Basehoar, A. D., Zanton, S. J., and Pugh, B. F. (2004) Cell 116 699-709 [DOI] [PubMed] [Google Scholar]
- 45.Brenowitz, M., Senear, D. F., Jamison, E., and Dalma-Weiszhausz, D. D. (1993) in Footprinting Techniques for Studying Nucleic Acid-Protein Complexes (Revzin, A., ed) pp. 1-43, Academic Press, New York
- 46.Buratowski, S., Hahn, S., Guarente, L., and Sharp, P. A. (1989) Cell 56 549-561 [DOI] [PubMed] [Google Scholar]
- 47.Pastor, N., Weinstein, H., Jamison, E., and Brenowitz, M. (2000) J. Mol. Biol. 304 55-68 [DOI] [PubMed] [Google Scholar]
- 48.Liu, Y., and Schepartz, A. (2001) Biochem. 40 6257-6266 [DOI] [PubMed] [Google Scholar]
- 49.Das, R., Laederach, A., Perlman, S. M., Herschlag, D., and Altman, R. B. (2005) RNA (N. Y.) 11 344-354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Albert, I., Mavrich, T. N., Tomsho, L. P., Zanton, S. J., Schuster, S. C., and Pugh, B. F. (2007) Nature 446 572-576 [DOI] [PubMed] [Google Scholar]
- 51.Faiger, H., Ivanchenko, M., Cohen, I., and Haran, T. E. (2006) Nucleic Acids Res. 34 104-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roy, A., Exinger, F., and Losson, R. (1990) Mol. Cell Biol. 10 5257-5270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang, S. D., Marmorstein, R., Harrison, S. C., and Ptashne, M. (1996) Mol. Cell Biol. 16 3773-3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ranish, J. A., Lane, W. S., and Hahn, S. (1992) Science 255 1127-1129 [DOI] [PubMed] [Google Scholar]
- 55.Takamoto, K., Chance, M. R., and Brenowitz, M. (2004) Nucleic Acids Res. 32 E119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.