Abstract
The aromatic polymer lignin protects plants from most forms of microbial attack. Despite the fact that a significant fraction of all lignocellulose degraded passes through arthropod guts, the fate of lignin in these systems is not known. Using tetramethylammonium hydroxide thermochemolysis, we show lignin degradation by two insect species, the Asian longhorned beetle (Anoplophora glabripennis) and the Pacific dampwood termite (Zootermopsis angusticollis). In both the beetle and termite, significant levels of propyl side-chain oxidation (depolymerization) and demethylation of ring methoxyl groups is detected; for the termite, ring hydroxylation is also observed. In addition, culture-independent fungal gut community analysis of A. glabripennis identified a single species of fungus in the Fusarium solani/Nectria haematococca species complex. This is a soft-rot fungus that may be contributing to wood degradation. These results transform our understanding of lignin degradation by wood-feeding insects.
Keywords: Asian longhorned beetle, Pacific dampwood termite, TMAH thermochemolysis, Anoplophora glabripennis, Zootermopsis angusticollis
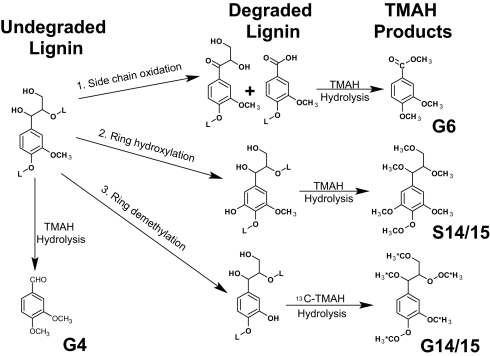
Lignin plays a central role in carbon cycling on Earth. Its heterogeneous structure imparts plants with structural rigidity and also serves to protect cellulose and hemicellulose from degradation (1). Most of what is known about lignin biodegradation is from pure culture studies with filamentous basidiomycete fungi, known as white-rot and brown-rot decay. Although both white-rot and brown-rot fungal degradation have been characterized, much more is known about the white-rot system (2, 3). White-rot fungi simultaneously degrade the three major components of the plant cell wall: lignin, cellulose, and hemicellulose. Analysis of white-rot–degraded wood shows that the reactions in lignin: (i) are oxidative, (ii) involve demethylation (or demethoxylation), (iii) include side-chain oxidation at Cα, and (iv) involve propyl side-chain cleavage between Cα and Cβ (Fig. 1) (4). In contrast to white-rot fungi, brown-rot fungi are able to circumvent the lignin barrier, removing the hemicellulose and cellulose with only minor modification to the lignin. Consequently, lignin remains a major component of the degraded plant cell wall (5). The remaining lignin is demethylated on aryl methoxy groups and contains a greater number of ring hydroxyl groups (6).
Fig. 1.
Lignin biodegradation reactions. Reactions include propyl side-chain oxidation/cleavage (reaction 1), ring hydroxylation (reaction 2), and demethylation (reaction 3). Reaction 1 is characteristic of white-rot fungus, whereas reactions 2 and 3 are characteristic of brown-rot fungus. Application of TMAH thermochemolysis to each of the degradative reactions is pictured following each reaction, showing chemical structures of compounds G4, G6, G14/15, and S14/15 that result from this analysis. Asterisks represent 13C-labeled carbons resulting from 13C-TMAH analysis.
Little is known about lignin degradation in complex ecosystems, such as insect guts, where a consortium of microbes may be involved in degradation rather than just a single species. Although cellulose degradation in insect guts is well documented (7, 8), the fate of lignin has not clearly been demonstrated (9, 10), and it is widely accepted that insect gut systems do not have the capacity to degrade lignin (10). Although the majority of previous reports suggest that many wood-feeding insects overcome the lignin barrier by feeding on predegraded wood (11) or through exosymbiotic relationships with wood-degrading fungi (12, 13), there are species of insects that feed on the inner wood of living, healthy trees. How these insects are able to circumvent the lignin barrier and gain access to the polymer carbohydrates has been a mystery. Herein, we show chemical changes in lignin upon passage of undegraded wood through two wood-feeding insect gut systems, one that naturally feeds on living healthy trees and a second that typically feeds on dead wood.
To determine the fate of lignin in these two systems, we analyzed the frass of both the Asian longhorned beetle (Anoplophora glabripennis) and the Pacific dampwood termite (Zootermopsis angusticollis). A. glabripennis is a wood-feeding cerambycid beetle that develops in the inner wood of a wide range of hardwood trees (14), feeding on living intact wood. Z. angusticollis is a lower termite that feeds on coniferous trees, attacking dead timber. Lignin structure of the frass was analyzed by unlabeled and 13C-tetramethylammonium hydroxide (TMAH) thermochemolysis, depolymerizing lignin into its aromatic subunits by breaking β-O-4 linkages and methylating all ring hydroxyls (15). The resulting product is then analyzed by GC/MS to determine how the lignin structure has been chemically modified. In undegraded wood, the predominant products from TMAH thermochemolysis are typically 3,4-dimethoxybenzldehyde (G4) from guaiacyl (G) lignin (Fig. 1) and 3,4,5-trimethoxybenzaldehyde (S4) from syringyl (S) lignin. Hardwood tree species contain both guaiacyl and syringyl lignin, but coniferous trees only contain guaiacyl lignin.
Results
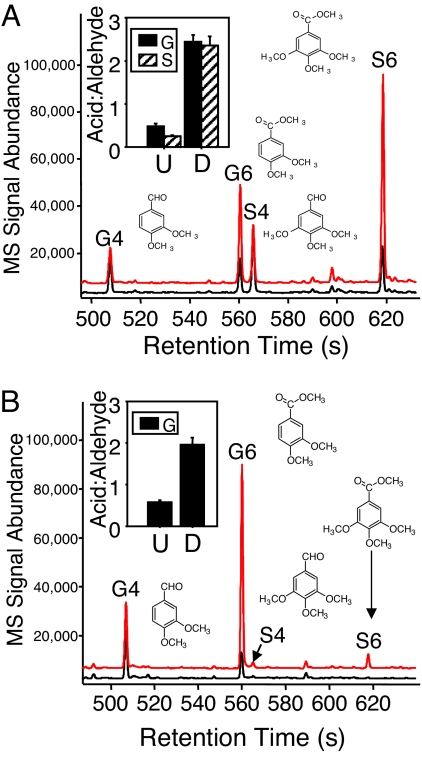
Studies with fungal lignin degradation show three predominant reactions: (i) propyl side-chain oxidation/cleavage (reaction 1, Fig. 1), (ii) ring hydroxylation (reaction 2, Fig. 1), and (iii) demethylation (reaction 3, Fig. 1) (15). All these processes can be detected by labeled or unlabeled TMAH analysis (Fig. 1). Side-chain oxidation is a predominant reaction with white-rot fungi resulting in Cα–Cβ cleavage/depolymerization of lignin (16). Previous studies demonstrate that oxidative alteration of the lignin propyl side chain results in the enhanced production of 3,4-dimethoxybenzoic acid (G6) and 3,4,5-trimethoxybenzoic acid (S6) (for hardwoods) (Fig. 2), and increases in G6/G4 and S6/S4 ratios indicate a higher degree of side-chain oxidation (16). Frass (feces) samples from A. glabripennis that fed on pin oak (Quercus palustris) (syringyl and guaiacyl lignin) and frass samples from Z. angusticollis that fed on ponderosa pine (Pinus ponderosa) (guaiacyl lignin only) were analyzed by TMAH thermochemolysis/GC/MS (Fig. 2 A and B). Both digested samples exhibited significant lignin side-chain oxidation (two-tailed Student's t test). For A. glabripennis-digested pin oak (n = 6), relative to undigested samples (n = 6), the G6/G4 ratio increased from 0.49 to 2.4 (P < 0.001) and the S6/S4 ratio increased from 0.26 to 2.4 (P < 0.0001) (Fig. 2a, inset). [Although all degraded samples showed an increase in the G4/G6 ratio, the ratio varied depending on the preparation of the wood. Because degradation of wood occurs on the outer surfaces, methods resulting in greater sampling of the inaccessible interior (fine grinding of sample) resulted in greater G4 abundance (lowered G6/G4 ratios).] For Z. angusticollis-digested ponderosa pine (n = 5), the G6/G4 ratio increased from 0.58 to 2.0 (P < 0.001) (Fig. 2b, inset). Interestingly, this oxidation and depolymerization of lignin occurred at similar levels to but much more rapidly (hours vs. weeks) than that observed with white-rot fungal degradation of loblolly pine (16). Thus, this indicates that the extent of lignin modification by these two insect systems is equivalent to that observed with wood-degrading fungi.
Fig. 2.
Side-chain oxidation and hydroxylation of wood after passage through insect guts. Chromatograms show increases in G6 and S6 in wood fed on by A. glabripennis (A) and Z. angusticollis (B) after passage through gut (red line = degraded [offset 5%], black line = undegraded). Inset graphs show significant changes in ratio, or side-chain oxidation of guaiacyl (solid) and syringyl (hatched) lignin between undegraded (U) and degraded (D) wood. Bars represent means ± 1 standard error. (B) In the Z. angusticollis chromatogram, hydroxylation of guaiacyl lignin can be seen through the production of S4 and S6 (from G4 and G6) in degraded samples, which were not present in the undegraded samples.
Ring hydroxylation of guaiacyl units results in formation of syringyl units in lignin; an increase in syringyl compounds is indicative of ring hydroxylation. Hydroxylation can occur on intact lignin or side-chain–oxidized lignin. The diastereomeric pair of the enantiomers 1-(3,4-dimethoxy)-1,2,3-trimethoxypropane [peaks G14 and G15 from G lignin or 1-(3,4,5-trimethoxyphenyl)-1,2,3-trimethoxypropane (S14/15) from S lignin] represents the intact lignin polymer (these compounds contain the fully methylated glycerol side chains). In Z. angusticollis frass samples fed on pine, which contains only guaiacyl lignin, detection of syringyl units in the degraded samples that were not present in undegraded samples demonstrates ring hydroxylation. Ion counts attributed to hydroxylated products S4 and S6 (0.04% and 0.11%, respectively, n = 4) and intact polymer products S14 and S15 (0.03% and 0.01%, n = 4) were observed in the degraded sample (Fig. 2b). Because the most abundant hydroxylated product is S6, this suggests that ring hydroxylation occurred after depolymerization in the Z. angusticollis gut. Ring hydroxylation was observed previously with brown-rot fungal lignin degradation (17). In A. glabripennis, there was a significant decrease in the ratio of total syringyl lignin-to-guaiacyl lignin in the degraded wood (S/G = 2.53) compared with control wood (S/G = 2.07; P = 0.011, n = 4, two-tailed Student's t test). This decrease is not necessarily attributable to a lack of ring hydroxylation but, instead, could be attributable to preferential degradation of syringyl lignin over guaiacyl lignin. Syringyl lignin is known to be more easily degraded and depolymerized; thus, preferential removal of syringyl compounds would be expected, making hydroxylation difficult to detect (18).
To determine whether demethylation of ring methoxyl groups had occurred and also to confirm that syringyl units found in the digested pine were derived from ring hydroxylation, we used 13C-TMAH thermochemolysis. Unlabeled TMAH cannot be used to assay for demethylation because it methylates all ring hydroxyls and they become indistinguishable from the original methoxyl groups (Fig. 1, reaction 3). With 13C-TMAH, the molecular weight of the demethylated product exhibits an increase in one mass unit for each site of demethylation (Fig. 1, reaction 3). Freshly collected A. glabripennis frass showed significant amounts of demethylation (n = 4) and compounds G4, S4, and G6 were isotopically enriched, whereas undegraded wood (n = 4) showed no enrichment (Table 1). Inspection of 1-(3,4-dimethoxyphenyl)-1,2,3-trimethoxypropane (G14/15) and S14/15, which are proxies for the intact lignin polymer, showed little or no demethylation, consistent with demethylation occurring after side-chain oxidation. In Z. angusticollis frass samples (n = 4), compared with undegraded wood (n = 4), 13C-TMAH analysis provided no evidence of demethylation of G4. For G6, 13C-TMAH analysis actually showed a decrease in the relative amount of the methylated versus demethylated product (Table 1). This is consistent with consumption of any original demethylated units (15). In contrast, the guaiacyl units from intact lignin (G14/15) of degraded wood were more extensively demethylated than fresh wood (Table 1), indicating that demethylation reactions occurred on the intact lignin polymer. The syringyl units generated from hydroxylation of guaiacyl units exhibited a high percentage of demethylation (Table 1). Note that in guaiacyl lignin, there is only one site for ring demethylation, whereas in syringyl lignin, there is an additional aromatic methoxyl group; thus, syringyl-derived structures have one or two sites for demethylation.
Table 1.
Demethylation of lignin by A. glabripennis and Z. angusticollis
| Product | No. OH |
A. glabripennis |
Z. angusticollis |
||
|---|---|---|---|---|---|
| Undegraded (% yield on sample) | Degraded (% yield on sample) | Undegraded (% yield on sample) | Degraded (% yield on sample) | ||
| G4 | 1 OH | 4.57 ± 1.42 | 8.28 ± 1.02* | 5.52 ± 0.45 | 3.77 ± 0.15* |
| G6 | 1 OH | 25.1 ± 2.1 | 35.6 ± 3.1* | 23.5 ± 1.1 | 9.14 ± 0.43† |
| G14 | 1 OH | 0.57 ± 0.41 | 1.34 ± 0.52 | 0.56 ± 0.16 | 1.58 ± 0.15‡ |
| G15 | 1 OH | 1.21 ± 0.54 | 1.06 ± 0.51 | 0.70 ± 0.30 | 1.51 ± 0.29§ |
| S4 | 1 OH | 2.57 ± 0.19 | 2.56 ± 0.20 | NP | 29.4 |
| S4 | 2 OH | 0.52 ± 0.02 | 0.19 ± 0.09* | NP | 3.38 |
| S16 | 1 OH | 6.70 ± 0.40 | 6.42 ± 0.07 | NP | 25.6 |
| S16 | 2 OH | 56.9 ± 2.2 | 70.2 ± 9.2 | NP | 5.35 |
Percentage of compounds with phenolic hydroxyl (1- or 2-OH) groups resulting from demethylation of ring methoxyl groups. In guaiacyl lignin, there is only one site for ring demethylation, whereas in syringyl lignin, there is an additional aromatic methoxyl group; thus, syringyl-derived structures have one or two sites for demethylation. Data shown are the mean ± standard error. NP, not present in sample.
*P < 0.05, significant changes between degraded and undegraded samples (two-tailed Student's t test).
†P < 0.001.
‡P < 0.01.
§Nearly significant, P = 0.0508.
Although we have shown that lignin is degraded by both insect gut systems, the source of the enzymes/reagents responsible for these reactions is not known. Because fungi are the predominant degraders of lignin, we also investigated the potential role of fungi in these processes. In A. glabripennis, we analyzed total gut DNA of A. glabripennis larvae fed in Q. palustris for the presence of fungi using PCR amplification of the internal transcribed spacer (ITS) region with fungus-specific primers. Cloning and sequencing of 48 clones yielded only one strain of fungus, with all clones having an identical DNA sequence. Basic local alignment search tool analysis identified this sequence as an ascomycete fungus, falling into the same Fusarium solani/Nectria haematococca species complex as the Saperda vestita-related fungus. We also identified this fungal isolate from guts of A. glabripennis larvae infesting different tree species in New York and China (data not shown). F. solani/N. haematococca is a species complex known to have lignin-degrading capabilities (19), and although not closely related to basidiomycete wood-degrading fungi (Fig. 3), ascomycetes are known to degrade lignin and are responsible for wood decay referred to as “soft rot,” a process not well understood (20). Recent work by Shary et al. (20) using model lignin compounds demonstrated the ability of ascomycetes to catalyze propyl side-chain oxidation, similar to reactions shown in the A. glabripennis and Z. angusticollis systems. To substantiate participation of fungi in the A. glabripennis gut digestive system further, we performed metatranscriptomic analysis of the A. glabripennis gut microbial community. Both eukaryotic and prokaryotic transcripts were obtained, including fungal transcripts. These data are consistent with these fungi being metabolically active and playing a role in lignocellulose degradation in the beetle gut community.
Fig. 3.
Soft-rot fungal species associated with the gut of A. glabripennis. Fungal community analysis of A. glabripennis gut contents yielded only a single strain of fungus based on ITS sequence, falling into the same species complex as a previously identified cerambycid related fungus. Phylogenetic analysis using the ITS region and performing a neighbor-joining analysis demonstrates that these fungi fall into the Ascomycota along with other known “soft-rot” degrading fungi (37), with both brown-rot and white-rot fungi being phylogenetically distinct. GenBank accession number for each sequence is listed next to the species name on the tree.
Discussion
In this study, we have shown definitive evidence for lignin degradation in the guts of two unrelated xylophagous species, A. glabripennis and Z. angusticollis. This work demonstrates lignin degradation in wood-feeding beetles; previous evidence of lignin degradation by termites was summarized by Breznak and Brune (7) as “scarce… and somewhat ambiguous.” These authors state that the ambiguity comes from the nature of the lignin preparation used for these studies and the “various conventions when referring to a substance as lignin.” Much of the “lignin” used in prior studies came from infusion of 14C-lignin precursors into growing plants with the assumption that the majority of the label is incorporated into polymeric lignin. The scepticism that termites degrade lignin was reinforced by the recent metagenome sequencing of the higher termite species Nasutitermes corniger hindgut community. No genes encoding known lignin-degrading enzymes were found (21). However, the section of the hindgut investigated, although microbe-rich, is also anaerobic, excluding possible aerobic microbes that may be able to degrade lignin. Also, previous investigation of lignin modification by the lower termite Cryptotermes brevis noted only minor changes in the lignin molecule, with no evidence for modifications to side chains (22). In contrast to our work here, where the entire sample was analyzed, these earlier experiments characterized only the isolated (Björkman) lignin fraction from insect feces, limiting detection of monomeric units formed from side-chain oxidation.
Our results using TMAH thermochemolysis show that two different insect gut systems cause significant alteration of the lignin polymer. This analysis detects chemical modification of the entire lignin fraction of the sample; thus, changes detected can be attributed to the entire lignocellulose sample. All reactions known to occur in lignin biodegradation—side-chain oxidation, ring hydroxylation, and demethylation—occur in these insects. Side-chain oxidation, a reaction that leads to lignin depolymerization, is the predominant reaction observed with A. glabripennis, a process that can be catalyzed by white-rot fungi (3) and by ascomycetes (20). Many other species of cerambycids feed on decaying wood that is infested with wood-rot fungi. These “lower” cerambycids are not able to feed on fungus-free wood (23). Although A. glabripennis does not feed on wood infested with fungi, our data and those of others suggest that this insect has evolved a fungal symbiosis in the gut to allow digestion of living woody tissue. Interestingly, a fungal ascomycete, Nectria haematococca, was found in the gut of a closely related cerambycid species (Fig. 3), but the role of this fungus was not investigated (24).
For Z. angusticollis, our data show side-chain oxidation along with demethylation. Most notably, we detected ring hydroxylation as evidenced by formation of syringyl products from a wood that only contains guaiacyl lignin. All three reactions observed in the Z. angusticollis gut are more consistent with brown-rot fungal decay, but neither brown-, white-, nor soft-rot fungi has been shown previously to be associated with termite guts. The participation of fungi in Z. angusticollis wood degradation is a topic of ongoing research in our laboratories. Aside from fungi, the presence of aromatic degrading bacteria (actinomycetes) in termite guts has been well documented (25). Although these bacteria could possibly play a role in circumventing the lignin barrier (25, 26), the biochemical abilities of these bacteria are not as well defined as fungal systems. Researchers have yet to demonstrate extensive depolymerization of lignin by bacteria, and we suggest that bacteria are unlikely to be involved in side-chain oxidation. The presence of actinomycetes related to those harbored by termites was reported previously in the gut of A. glabripennis, and these bacteria may have similar aromatic degrading abilities (27).
Although fungal degradation of woody biomass has been viewed as the paradigm for lignocellulose degradation, our work here shows that lignin degradation takes place in an ecosystem in which it was previously thought not to occur. These two insect systems displayed similarities to the pathways described for white-, brown-, and soft-rot fungi, but they are instead occurring in insect-microbial systems rather than by the action of a single fungal species. Known lignin-degrading pathways are aerobic processes. Despite the belief that termites ferment cellulose anaerobically, aerobic regions of termite guts, in addition to those of other insects, are known to exist in which large populations of aerobic microbes are housed, which can outnumber anaerobic microbial populations in the gut in some cases (28–30). Although we cannot directly measure a rate of lignin degradation, or identify specific genes or organisms responsible for this degradation at this time, the striking modifications to lignin detected in wood after passage through these insect guts is quite astonishing in regard to the short period that wood is held in the insect digestive system (only several hours), compared with the weeks for which similar degradation has been reported for wood-rot fungal systems (15, 16). Being controlled systems, it is likely that these insect guts have evolved optimal conditions for lignocellulose degradation that cannot be obtained in free-living systems. Discovery that these gut microbial communities can degrade lignin provides considerable enthusiasm for bioprospecting these systems, through metagenomic or proteomic approaches, for new lignin-degrading enzymes and systems. Further understanding of these systems will contribute to our understanding of the ecological roles that these insects play in carbon cycling in natural systems.
Materials and Methods
Sample Collection and Preparation.
A. glabripennis and Z. angusticollis were obtained from colonies maintained at the Department of Entomology, Pennsylvania State University. A. glabripennis adults were allowed to oviposit into four potted pin oak (Q. palustris) trees in a quarantine greenhouse, and larvae were allowed to develop in trees for 6 weeks. At this point, trees were cut down, larvae were removed, and fresh frass samples were collected from each insect, which were immediately frozen in liquid nitrogen to halt wood degradation. Wood samples adjacent to but not in contact with larval galleries were also collected as control samples. In total, 10 frass samples and accompanying controls were collected. For termite frass collection, five pairs of Z. angusticollis workers were removed from the colony and placed into cups containing 1 × 1 × 0.4 cm of ponderosa pine (P. ponderosa) blocks and allowed to feed for 4 days. During this time, frass was collected from each cup every 12 h and frozen. Samples were then pooled over the 4 days to provide enough material for analysis. Five control wood blocks were placed in additional cups, and a small sample was collected from them during each collection point and pooled. Frass samples and control wood tissue were lyophilized overnight. For unlabeled TMAH performed off-line, frass samples were not ground before analysis, but for online 13C-TMAH, samples were ground to a fine powder utilizing a freezer mill before being used for analysis. The percentage (%) of organic carbon content was determined for each sample using a Carlo Erba 1108 elemental analyzer (Carlo Erba) to normalize quantitative TMAH thermochemolysis analysis results.
Standard TMAH Thermochemolysis Analysis.
Tetramethylammonium hydroxide thermochemolysis was performed as modified by Frazier et al. (31). Approximately 0.5 mg of each sample and 150 μl of TMAH (25 wt % TMAH in methanol; Fisher Scientific) were placed in borosilicate glass tubes and mixed by vortexing for 30 sec. The samples were dried under nitrogen and vacuum-sealed on a manifold before being baked at 250°C for 30 min. The sealed tubes were allowed to cool before cutting the glass and adding an internal standard, linolenic acid methyl ester (Sigma Aldrich) at 54.2 ng/μl in ethyl acetate. Samples were extracted from tubes by washing the sides of the tubes with ethyl acetate (Fisher Scientific). All washings (approximately 1 ml per sample) were filtered through glass wool, combined, and concentrated to 200 μl under a stream of nitrogen. The samples were analyzed by capillary GC-MS on a Hewlett Packard 6890 series GC system connected to a Pegasus III time-of-flight mass spectrometer (LECO Corporation). The column used for the GC separation was a 15-m × 0.25-mm (i.d.) fused silica capillary column with a film thickness of 0.25 μm (5% phenylpolysiloxane − 95% methylpolysiloxane, Restek Rtx-5). The samples (1 μl) were injected onto a split/splitless injector operating in splitless mode at a temperature of 280°C. The column, using helium as the carrier gas, was run with a constant flow (1.0 ml/min), and the oven temperature was held initially at 50°C for 2 min, then ramped from 50°C to 300°C at 15°C per minute, and then held at 300°C for 6.33 min, giving a total run time of 25 min. The ionization mode on the mass spectrometer was electron ionization at 70 eV, and the ion source temperature was 200°C, whereas the transfer line temperature was 280°C. Mass spectra were collected at a rate of 20 spectra per second after the 120-sec solvent delay. Masses were acquired between m/z 45 and 500, and data acquisition and analysis were performed by use of the LECO Pegasus III software (version 3.22). Chromatograms obtained from GC/TOF-MS were integrated using masses 166, 196, and 226, which are specific for G4, G6, S4, and S6. This was done to remove small peaks that coelute with G6 from the total ion count. After integration, the peak area of each of these four compounds was determined, and G6:G4 and S6:S4 were calculated (32).
13C-TMAH Thermochemolysis Analysis.
13C-labeled TMAH was synthesized as previously described (33). 13C-TMAH thermochemolysis was performed in-line using a Shimadzu Pyr-4a pyrolysis unit, which also doubles as a stand-alone injection port [modified from Filley et al. (34)]. For each treatment, three replicate samples were analyzed from unique insect pools along with three corresponding controls. A total of 150 μg of each sample was weighed into a 3 × 3-mm platinum bucket that contained eicosane internal standard. Next, 3 μl of a 25-wt % solution of 13C-TMAH in water was added to the bucket and placed in the sample holder on the top of the Pyr-4a pyrolysis unit. The TMAH was allowed to soak into the sample for 5 min, and the bucket was then dropped into the pyrolysis unit maintained isothermal at 350°C. The injector base of the Pyr-4a was maintained at 320°C with a split flow of 25/1. Chromatography and mass spectral analysis were performed on a Shimadzu GC17A interfaced to a Shimadzu QP5050A quadrupole mass spectrometer collecting in mass range 40–550 amu. Chromatographic separation was performed on a fused silica column [Restek RTx-5 (Restek), 30 m, 0.25 mm i.d., 0.25-μm film thickness]. The oven was temperature programmed from 60°C (1 min stationary) to 300°C at 7°C per minute and maintained at that temperature for 15 min. Chromatograms obtained from this analysis were analyzed [as described previously (16, 34)] for S/G to measure hydroxylation and % 13C-labeled methyl groups on G4, G6, G14, G15, S4, and S6 to determine the extent of demethylation of each of these compounds in each sample.
Fungal Community Analysis of the A. glabripennis Gut.
Adult A. glabripennis were allowed to oviposit onto Q. palustris trees, and larvae were allowed to grow and develop for 90 days as described previously. After 90 days, each tree was dissected and living larvae were collected for gut community analysis. Ten living larvae were used for gut dissections, which were performed in a laminar flow hood to maintain sterility using sterile dissection tools. Larvae removed from trees were immediately chilled and dissected within 1 h of removal from trees. Larvae were surface sterilized in 70% ethanol for 1 min and rinsed in sterile water before dissection. Whole guts were dissected by cutting the cuticle open laterally, ligating the gut at the anterior midgut and posterior hindgut, and carefully transferring the entire gut into a sterile microcentrifuge tube. Ten guts were pooled into a single microcentrifuge tube for DNA extraction for each tree species to reduce individual variation within trees. Total DNA was extracted using the FastDNA SPIN for Soil Kit (MP Biomedicals) using the FastPrep Instrument for tissue homogenization following the manufacturer's protocol. This kit was used because of the complexity of the A. glabripennis gut contents (containing wood, bacteria, and fungi) to ensure complete DNA extraction from all organisms. A control DNA extraction was also performed by using the sterile water rinsate to confirm that no contaminating DNA was extracted. DNA concentration was determined by absorbance at 260 nm and stored at −20°C until use. The fungal ITS region was amplified using PCR from total gut DNA extractions. Fungal ITS primers ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) were used to amplify ≈540 bases of the ITS region (35). Polymerase chain reactions were performed in 25-μl volumes with the following components: 5 μl of 5× GoTaq green reaction buffer, 0.5 μl of GoTaq DNA polymerase (1.25 U, Promega), 2 μl of 10 μM forward primer (ITS1), 2 μl of 10 μM reverse primer (ITS4), and 20 ng of template DNA. Polymerase chain reaction conditions were 95°C denaturation for 3 min, 25 cycles of 95°C for 30 sec, 55°C for 1 min, and 72°C for 1.5 min, with a final extension at 72°C for 5 min. Control DNA extraction samples also underwent PCR to ensure that there was no contaminating DNA during extraction. Polymerase chain reaction amplification revealed a single band from ITS amplification, but that single band may contain more than one fungal sequence. To resolve this, the PCR product was ligated into pCR 2.1 TOPO vector (Invitrogen) following the manufacturer's protocol. The vector was then transformed into chemically competent Escherichia coli cells (TOP10, Invitrogen Corp.) by heat-shock, and clone libraries were created. Insert DNA from clones was amplified from the M13 priming site of the vector using direct PCR. Forty-eight 25-μl PCRs were set up with the following components for each reaction: 5 μl of 5× GoTaq green reaction buffer, 0.5 μl of GoTaq DNA polymerase (1.25 U; Promega), 2 μl of 10 μM forward primer (M13 Universal), and 2 μl of 10 μM reverse primer (M13Rev). Individual colonies were picked from the clone library using a sterile pipette tip and immersed into the PCR mix to allow the bacteria cells to be added to the PCR. The PCR program had an initial 95°C denaturation for 10 min to rupture bacteria cells, followed by 30 cycles of 95°C for 30 sec, 55°C for 1 min, and 72°C for 1.5 min, with a final extension at 72°C for 5 min. Four microliters of the PCR product was cleaned up for sequencing by addition of 0.8 μl of ExoSAP-IT (USB Corporation) and incubating the sample at 37°C for 15 min, followed by 80°C for 30 min. Two microliters of this reaction was then used to sequence from the forward direction, from the M13 universal priming site, and 2 μl was used for the reverse from the M13 reverse site. Sequencing using the BigDye Terminator (Applied Biosystems) method was performed at the Penn State Nucleic Acid Facility. Internal transcribed spacer sequences were constructed over the regions amplified based on consensus of the forward and reverse sequences for each clone analyzed. Alignment of forward and reverse sequences was performed using MEGA 4.0.1 (36), and the vector sequence was removed and a consensus sequence was created. All sequences from the clone library were identical; thus, a representative sequence (deposited into GenBank, accession no. EU862818) was then aligned with known white-, brown-, and soft-rot fungi (37). The ITS sequences obtained from the National Center for Biotechnology Information database, with a fungal ITS sequence from a closely related beetle gut (24) and each sequence's closest BLAST match. Sequences were aligned by ClustalX (Conway Institute, UCD Dublin) in MEGA, and from this alignment, a neighbor-joining tree was created and clustered with a bootstrap test (500 replicates) (38, 39).
Acknowledgments.
We thank D. Gamblin for assistance with sample analysis, I. Ramos and C. Delphia for assistance with insects and frass collection, Penn State Nucleic Acid Facility for DNA sequencing, and G. W. Felton and R. Jabbour for their comments on the manuscript. This project was funded by the Alphawood Foundation and the Pennsylvania State University College of Agricultural Sciences Seed Grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The A. glabripennis gut fungal ITS nucleotide sequence has been deposited in the GenBank database (accession no. EU862818).
References
- 1.Jeffries T. Biodegradation of lignin-carbohydrate complexes. Biodegradation. 1990;1:163–176. [Google Scholar]
- 2.Goodell B. In: Goodell B, Nicholas DD, Schultz TP, editors. Wood Deterioration and Preservation: Advances in Our Changing World; ACS Symposium Series; Washington, DC: Am Chem Soc; 2003. pp. 97–118. [Google Scholar]
- 3.Kirk TK, Farrell RL. Enzymatic combustion—The microbial-degradation of lignin. Annu Rev Microbiol. 1987;41:465–505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- 4.Chen CL, Chang H, Kirk TK. Carboxylic-acids produced through oxidative cleavage of aromatic rings during degradation of lignin in spruce wood by phanerochaete-chrysosporium. J Wood Chem Technol. 1983;3:35–57. [Google Scholar]
- 5.Green F, Highley TL. Mechanism of brown-rot decay: Paradigm or paradox. Int Biodeter Biodegr. 1997;39:113–124. [Google Scholar]
- 6.Kirk TK, Highley TL. Quantitative changes in structural components of conifer woods during decay by white-rot and brown-rot fungi. Phytopathology. 1973;63:1338–1342. [Google Scholar]
- 7.Breznak JA, Brune A. Role of Microorganisms in the digestion of lignocellulose by termites. Annu Rev Entomol. 1994;39:453–487. [Google Scholar]
- 8.Martin M. Cellulose digestion in insects. Comp Biochem Physiol. 1983;75A:313–324. [Google Scholar]
- 9.Brune A. Microbiology: Woodworker's digest. Nature. 2007;450:487–488. doi: 10.1038/450487a. [DOI] [PubMed] [Google Scholar]
- 10.Ohkuma M. Termite symbiotic systems: Efficient bio-recycling of lignocellulose. Appl Microbiol Biot. 2003;61:1–9. doi: 10.1007/s00253-002-1189-z. [DOI] [PubMed] [Google Scholar]
- 11.Kukor J, Cowan D, Martin M. The role of ingested fungal enzymes in cellulose digestion in the larvae of cerambycid beetles. Physiol Zool. 1988;61:364–371. [Google Scholar]
- 12.Johjima T, Taprab Y, Noparatnaraporn N, Kudo T, Ohkuma M. Large-scale identification of transcripts expressed in a symbiotic fungus (Termitomyces) during plant biomass degradation. Appl Microbiol Biot. 2006;73:195–203. doi: 10.1007/s00253-006-0570-8. [DOI] [PubMed] [Google Scholar]
- 13.Taprab Y, et al. Symbiotic fungi produce laccases potentially involved in phenol degradation in fungus combs of fungus-growing termites in Thailand. Appl Environ Microbiol. 2005;71:7696–7704. doi: 10.1128/AEM.71.12.7696-7704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morewood WD, Hoover K, Neiner PR, Sellmer JC. Complete development of Anoplophora glabripennis (Coleoptera: Cerambycidae) in northern red oak trees. Can Entomol. 2005;137:376–379. [Google Scholar]
- 15.Filley TR. In: Goodell B, Nicholas DD, Schultz TP, editors. Wood Deterioration and Preservation: Advances in Our Changing World; ACS Symposium Series; Washington, DC: Am Chem Soc; 2003. pp. 119–139. [Google Scholar]
- 16.Filley TR, Hatcher PG, Shortle WC, Praseuth RT. The application of 13C-labeled tetramethylammonium hydroxide (13C-TMAH) thermochemolysis to the study of fungal degradation of wood. Org Geochem. 2000;31:181–198. [Google Scholar]
- 17.Kirk TK, Adler E. Methoxyl deficient structural elements in lignin of sweetgum decayed by a brown-rot fungus. Acta Chem Scand. 1970;24:3379–3390. [Google Scholar]
- 18.Chiang VL, Funaoka M. The difference between guaiacyl and guaiacyl-syringyl lignins in their responses to Kraft delignification. Holzforschung. 1990;44:309–313. [Google Scholar]
- 19.Lozovaya VV, et al. Lignin degradation by Fusarium solani f. sp glycines. Plant Dis. 2006;90:77–82. doi: 10.1094/PD-90-0077. [DOI] [PubMed] [Google Scholar]
- 20.Shary S, Ralph SA, Hammel KE. New insights into the ligninolytic capability of a wood decay ascomycete. Appl Environ Microbiol. 2007;73:6691–6694. doi: 10.1128/AEM.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warnecke F, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 22.Katsumata KS, Jin ZF, Hori K, Iiyama K. Structural changes in lignin of tropical woods during digestion by termite, Cryptotermes brevis. J Wood Sci. 2007;53:419–426. [Google Scholar]
- 23.Kukor JJ, Martin MM. Cellulose digestion in monochamus-marmorator Kby (Coleoptera, Cerambycidae)—Role of acquired fungal enzymes. J Chem Ecol. 1986;12:1057–1070. doi: 10.1007/BF01638996. [DOI] [PubMed] [Google Scholar]
- 24.Delalibera I, Handelsman J, Raffa KF. Contrasts in cellulolytic activities of gut microorganisms between the wood borer, Saperda vestita (Coleoptera: Cerambycidae), and the bark beetles, Ips pini and Dendroctonus frontalis (Coleoptera: Curculionidae) Environ Entomol. 2005;34:541–547. [Google Scholar]
- 25.Watanabe Y, Shinzato N, Fukatsu T. Isolation of actinomycetes from termites' guts. Biosci Biotechnol Biochem. 2003;67:1797–1801. doi: 10.1271/bbb.67.1797. [DOI] [PubMed] [Google Scholar]
- 26.Harazono K, et al. Isolation and characterization of aromatics-degrading microorganisms from the gut of the lower termite coptotermes formosanus. Biosci Biotechnol Biochem. 2003;67:889–892. doi: 10.1271/bbb.67.889. [DOI] [PubMed] [Google Scholar]
- 27.Schloss PD, Delalibera I, Handelsman J, Raffa KF. Bacteria associated with the guts of two wood-boring beetles: Anoplophora glabripennis and Saperda vestita (Cerambycidae) Environ Entomol. 2006;35:625–629. [Google Scholar]
- 28.Brune A, Emerson D, Breznak JA. The termite gut microflora as an oxygen sink—Microelectrode determination of oxygen and pH gradients in guts of lower and higher termites. Appl Environ Microbiol. 1995;61:2681–2687. doi: 10.1128/aem.61.7.2681-2687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson KS, Barbehenn RV. Oxygen levels in the gut lumens of herbivorous insects. J Insect Physiol. 2000;46:897–903. doi: 10.1016/s0022-1910(99)00196-1. [DOI] [PubMed] [Google Scholar]
- 30.Tholen A. The gut microflora of Reticulitermes flavipes, its relation to oxygen, and evidence for oxygen-dependent acetogenesis by the most abundant Enterococcus sp. FEMS Microbiol Ecol. 1997;24:137–149. [Google Scholar]
- 31.Frazier SW, et al. Characterization of organic matter from natural waters using tetramethylammonium hydroxide thermochemolysis GC-MS. J Anal Appl Pyrol. 2003;70:99–128. [Google Scholar]
- 32.Hatcher PG, Nanny MA, Minard RD, Dible SD, Carson DM. Comparison of two thermochemolytic methods for the analysis of lignin in decomposing gymnosperm wood: The CuO oxidation method and the method of thermochemolysis with tetramethylammonium hydroxide (TMAH) Org Geochem. 1995;23:881–888. [Google Scholar]
- 33.Filley TR, Minard RD, Hatcher PG. Tetramethylammonium hydroxide (TMAH) thermochemolysis: Proposed mechanisms based upon the application of C-13-labeled TMAH to a synthetic model lignin dimer. Org Geochem. 1999;30:607–621. [Google Scholar]
- 34.Filley TR, Nierop KGJ, Wang Y. The contribution of polyhydroxyl aromatic compounds to tetramethylammonium hydroxide lignin-based proxies. Org Geochem. 2006;37:711–727. [Google Scholar]
- 35.White TJ, Bruns T, Lee S, Taylor J. In: PCR Protocols: A Guide to Methods and Applications. Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. San Diego: Academic; 1990. pp. 315–322. [Google Scholar]
- 36.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 37.Worrall JJ, Anagnost SE, Zabel RA. Comparison of wood decay among diverse lignicolous fungi. Mycologia. 1997;89:199–219. [Google Scholar]
- 38.Saitou N, Nei M. The neighbor-joining method—A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 39.Felsenstein J. Confidence-limits on phylogenies—An approach using the bootstrap. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]