Abstract
Transition from the vegetative phase to reproductive phase is a crucial process in the life cycle of higher plants. Although the molecular mechanisms of flowering regulation have been extensively characterized in a number of plant species, little is known regarding how the transition process initiates. Here, we show that the Rice Indeterminate 1 (RID1) gene acts as the master switch for the transition from the vegetative to reproductive phase. RID1 encodes a Cys-2/His-2-type zinc finger transcription factor that does not have an ortholog in Arabidopsis spp. A RID1 knockout (rid1), mutated by T-DNA insertion, never headed after growing for >500 days under a range of growth conditions and is thus referred to as a never-flowering phenotype. This mutation-suppressed expression of the genes is known to be involved in flowering regulation, especially in the Ehd1/Hd3a pathway and a series of RFT homologs. RID1 seems to be independent of the circadian clock. A model was proposed to place RID1 in the molecular pathways of flowering regulation in rice, for which there are two indispensable elements. In the first, RID1 is controlling the phase transition and initiation of floral induction. In the other, the Hd3a/RFL1/FTL complex acts as the immediate inducer of flowering. Loss of function in either element would cause never-flowering. Once the phase transition is induced with the activation of RID1, flowering signal is transduced and regulated through the various pathways and eventually integrated with FT-like proteins to induce flowering.
Keywords: never flowering, phase transition, rice indeterminate 1, flowering time
The transition of the shoot apical meristem (SAM) from vegetative to reproductive growth is a critical developmental switch in plants that is regulated by both environmental and endogenous factors (1, 2). Extensive molecular genetic analyses in Arabidopsis spp. have identified a set of flowering-time mutants that have been assigned to four major genetic pathways based on their functions. The autonomous and gibberellin (GA) pathways respond to endogenous factors, such as plant age or leaf number, and are largely independent of environmental signals. The photoperiod and vernalization pathways respond to environmental factors (3, 4).
Among the environmental factors, photoperiod is one of the most important regulators of flowering. Long-day (LD) or short-day (SD) photoperiod-sensitive plants can be induced to flower with the appropriate day length. Comparison of flowering pathways in Arabidopsis, a LD plant, and rice, a SD plant, has revealed that major genes involved in photoperiod flowering are highly conversed between these two species. For instance, OsGI, Hd1, and Hd3a in rice, like their orthologs of Arabidopsis, GI, CO, and FT, respectively, play important roles in flowering (5–7). The Arabidopisis homolog of Hd6, a QTL that controls photoperiod sensitivity in rice and encodes a subunit of protein kinase CK2, is also involved in the photoperiod pathway by regulating the circadian rhythm in Arabidopsis (8, 9). RCN1 and RCN2, which delay flowering in rice when overexpressed, have conserved functions similar to that of their Arabidopsis homolog TFL1 (10). However, distinct effects of some regulators were also observed. For instance, Hd1 activates the expression of Hd3a and promotes flowering under SD conditions, which is similar to CO under inductive LD conditions in Arabidopsis. Under noninductive LDs, Hd1 suppresses the expression of Hd3a and delays flowering in rice, whereas CO has no effect on flowering in Arabidopsis under noninductive SDs (11–13). Furthermore, some genes are unique to the flowering pathway in rice or Arabidopsis. It was shown that Ehd1, a B-type response regulator, is specific to floral induction in rice (6), whereas the MADS box gene, FLC, which plays a central role in integration of the autonomous and vernalization pathways in Arabidopsis (14), has no ortholog in rice (6, 15). It was recently reported that OsMADS51 and Ghd7 involved in flowering-time regulation in rice lacks homologs in Arabidopsis (16, 17).
Although a large number of flowering-time mutants have been identified in both Arabidopsis spp. and rice (15, 18), all of the individual mutants can only affect flowering time quantitatively by either accelerating or delaying flowering. An interesting question is whether there exists an upstream master regulator that controls the phase transition such that inactivating the master regulator would block the main flowering pathways for flowering transition and, thus, would cause a never-flowering phenotype.
The transition to flowering in maize is affected by indeterminate1 (id1). Unlike Arabidopsis spp. and rice, maize is determinate, producing a set number of leaves before flowering. The id1 mutant has prolonged vegetative growth and retains vegetative features in the inflorescence (19). ID1 encodes a Cys-2/His-2 zinc-finger protein (19), one of the largest classes of transcription factor families in eukaryotes, playing roles in diverse biological processes (20, 21). Expression of ID1 is limited to developing leaves, indicating a noncell autonomous effect on the vegetative-to-reproductive state of the apical meristem (19, 22, 23). The ID1 protein defines a new family of zinc-finger proteins named the ID-domain (IDD) gene family, which is found in both monocots and dicots. Biochemical analysis showed that IDD proteins can bind to an 11-bp DNA sequence with a consensus motif TTTGTCG/CT/CT/aT/aT (24). However, apart from ID1, the biological function of the IDD genes and the potential role of these genes in flowering time have not been reported to date.
In this study, we identified a rice mutant (rid1) caused by a T-DNA insertion at the RID1 locus that encoded an IDD protein. Plants homozygous for the mutant allele produced a never-flowering phenotype. Expression of the genes known to be involved in rice-flowering pathways was largely suppressed in the rid1 mutant. The results suggest that RID1 functions as a master switch for flowering induction in rice.
Results
Identification of a Never-Flowering Mutant.
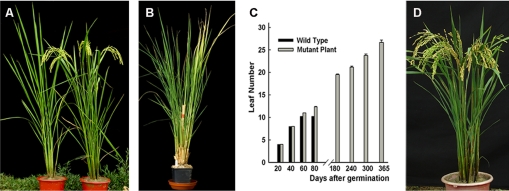
We generated an enhancer trap mutant library of rice for functional analysis of the rice genome (25, 26), from which progenies of 2,000 independent T0 plants (primary transformants) were screened for heading time in 2004 at Wuhan, China. Progenies from eight families showed flowering-time segregation compared with the WT plants, and some plants from one family failed to head in the rice growing season (Fig. 1A).
Fig. 1.
Phenotype and complementation of the rid1 mutant. (A) The phenotypes of rid1 mutant (Left) and WT (Right) plants at maturity stage. (B) The rid1 plant at 370 d after germination. (C) Numbers of leaves in the main culm of WT and rid1 plants (average of six plants for both WT and rid1) during a whole year. (D) A phenotype-rescued rid1 plant by genetic complementation.
The latter mutant plants were maintained by ratooning and then planted on Hainan Island (South China Sea) in mid-November of 2004, where they were exposed to natural SDs, inducers of flowering in rice. The mutant plants still had vegetative growth and did not head during this season until early May, 2005. Two artificial day-length treatments were also applied: neutral day-length (12 h light/12 h dark) and SD (10 h light/14 h dark), under which WT plants headed at ≈60 d and 55 d after germination, respectively. However, the mutant did not head after growing for 370 d (Fig. 1B) and instead continued to produce leaves with more than 30 leaves produced on the main culms of mutant plants (Fig. 1C). We designated this never-flowering mutant as rid1.
RID1 Encodes a Cys-2/His-2 Zinc-Finger Transcription Factor.
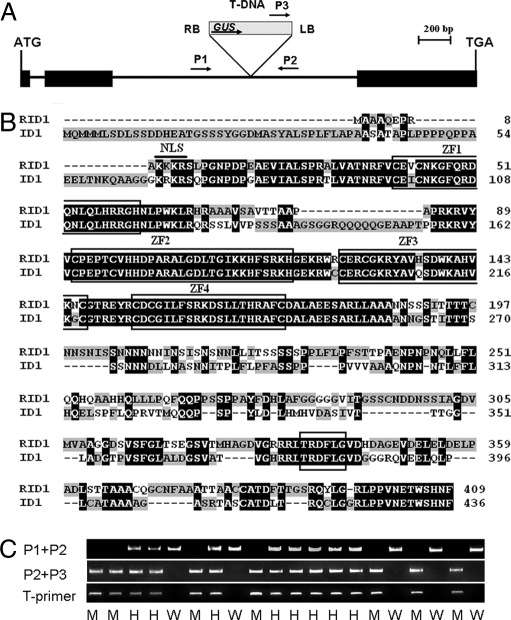
Using the thermal asymmetric interlaced PCR (TAIL-PCR) method (27), we isolated genomic fragment flanking of the T-DNA insertion site from the rid1 mutant plant. BLAST search of the flanking sequence against the TIGR Rice Genome Annotation database (http://rice.plantbiology.msu.edu/index.shtml) retrieved a sequence that was annotated as a Cys-2/His-2 zinc finger gene (LOC_Os10g28330), which we named RID1. RID1 consisted of three exons, and the T-DNA was inserted into the second intron (Fig. 2A). The RID1-encoded protein showed 58% amino-acid-sequence identity with the maize ID1 (Fig. 2B). DNA blot hybridization showed that there is only one copy of T-DNA insertion in this mutant line (data not shown).
Fig. 2.
Molecular features of RID1. (A) Structure of RID1 and T-DNA insertion site. Three exons (filled boxes) and two introns (lines between the filled boxes) are shown. T-DNA was inserted into the second intron. Arrows indicate the primers used for analyzing the insertion site. LB and RB represent the left and right borders of T-DNA. (B) Alignment of amino acid sequences of RID1 and ID1 proteins. Clustalx software was used for the alignment. The identical amino acids are shown with white text on a black background. The “TRDFLG” motif and four zinc-finger motifs are boxed. A thick bar at the N-terminal region shows the putative nuclear localization signal motif. (C) PCR genotyping RID1 segregants with primers as indicated in A (Table S2): M, homozygous for T-DNA insertion; W, WT; H, hemizygous. Primers P1 and P2 flank the T-DNA insertion and amplify a product from homozygous or WT allele. PCR-positive plants with P2 and P3 indicate T-DNA insertion in the examined site. Presence of a product with P2 and P3 and not with P1 and P2 indicates a plant homozygous for the insertion. PCR-positive plants amplified by T-primers indicate T-DNA insertion.
To test whether this T-DNA insertion was responsible for the mutant phenotype, 20 T1 plants were assayed for cosegregation between the flowering phenotype and T-DNA insertion by PCR, using primers P1, P2, and P3 to amplify the T-DNA insertion site (Fig. 2C). All of the seven plants homozygous for T-DNA insertion showed the never-flowering phenotype, whereas the remaining 13 plants, either homozygous for WT or hemizygous, headed at the same time as the WT. The cosegregation result strongly suggested that the rid1 mutant phenotype was caused by the T-DNA insertion.
To further verify that the never-flowering phenotype is because of the loss of function of RID1, a 5.7-kb KpnI-BamHI fragment harboring the entire RID1 coding region and a 1.9-kb 5′-upstream sequence were introduced into a rid1 mutant background. Of 143 plants regenerated, 117 headed with varying heading dates (Fig. 1D), whereas no heading was observed among the 14 plants transformed with the empty vector (the negative control). DNA blot hybridization analysis of 12 T0 RID1-transgenic plants chosen at random revealed that six plants contained one copy of the transgene, and the remaining plants carried two or more copies. All of the T1 progenies produced by self-pollination of the six single-copy transformants showed segregation of heading date under both SDs and LDs. As expected, all of the transgene-positive segregants headed, whereas all of the negative plants never headed (data not shown). We therefore concluded that loss of function of RID1 is the cause of the never-flowering phenotype.
Expression Pattern of RID1 and Subcellular Localization of Its Protein.
To examine the temporal and spatial expression pattern of RID1, we performed RT-PCR analysis [with the primers in supporting information (SI) Table S1] with total RNA prepared from blades and sheaths of mature leaves and immature leaves, roots, shoot apices, and floral meristems from Zhonghua 11 WT plants grown under neutral-day-length conditions. The analysis revealed a low but detectable level of RID1 transcript in immature leaves, but none in the root, mature leaves, SAM, or other tissues examined (Fig. S1). This expression pattern was similar to the previous result in maize (19, 22).
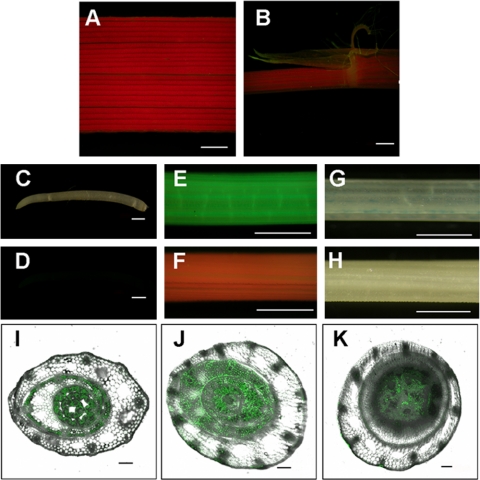
To further examine the expression pattern of RID1, the N-terminal sequence was fused in frame with a GFP, which was then fused with a 1.9-kb RID1 upstream fragment (amplified with RG-F primers in Table S2) (RID1::RID1:GFP). This construct was introduced into rice by Agrobacterium-mediated transformation. GFP signal was clearly detected in the outer epidermal cell of immature leaves and in the region immediately beneath the meristem where internodes are visible (Fig. 3E and I–K) but not in other tissues.
Fig. 3.
Expression of RID1 detected by GFP and GUS reporters. GFP assay of RID1::RID1:GFP transgenic plants in mature leaf blade (A), mature leaf ligule (B), root under light background (C) and dark background (D), and immature leaf (E), respectively, with the immature leaf of a WT plant as the control (F). GUS assay in immature leaf of T-DNA insertion rid1 plant (G) and WT plant (H), respectively. (A–H, Scale bar, 20mm.) Confocal composite images of GFP and transmission channels show transverse sections: ≈2 cm above the SAM (I), across the top of the SAM (J), and the stem below the SAM (K) of RID1::RID1:GFP transgenic plant (I–K, Scale bar, 100 μm.)
The construct used for generating the mutant library carried a GUS reporter (25). It is anticipated that insertion of the T-DNA in certain manners would result in expression of the GUS gene in the same pattern as the target gene. In the rid1 mutant, the T-DNA was inserted in such a way that GUS was in the same orientation as RID1 (Fig. 2A). We therefore assayed the pattern of GUS expression in this line. Indeed, GUS expression in the rid1 mutant was only detected in immature leaves, similar to that of GFP (Fig. 3G).
We also assayed the subcellular localization of the RID1 protein. There is a putative nuclear localization signal motif near the N terminus of RID1 (22). Therefore, a construct was made to include the 112 amino acids at the N terminus of RID1, which was fused to the GFP protein and driven by the maize Ubiquitin promoter (Ubi::ΔRID1:GFP). We also made a construct for the control that contained GFP alone under the control of the Ubiquitin promoter. In the onion epidermal cells bombarded with the Ubi::ΔRID1:GFP construct, the GFP signal was observed in the nuclei (Fig. S2 A–C). But in the cells bombarded with the Ubi::GFP construct, the GFP signal was in the cytosol and plasma membrane (Fig. S2 D and E). This result confirmed the nuclear localization of the RID1 protein and is consistent with the presence of a predicted nuclear localization signal in the N terminus of ID1-like proteins (22).
RID1 Regulates Genes in Flowering Pathways but Not in the Circadian Clock.
To further investigate the role of RID1 in the transition to flowering, we performed RT-PCR analysis of Hd1, Ehd1, and Hd3a, all of which play important roles in rice flowering (5–7). It was shown that although expression of Hd1 was detectable in mature as well as immature leaves in both WT and rid1 plants, expression of Ehd1 and Hd3a was detected only in the mature leaves of WT plants (Fig. S1).
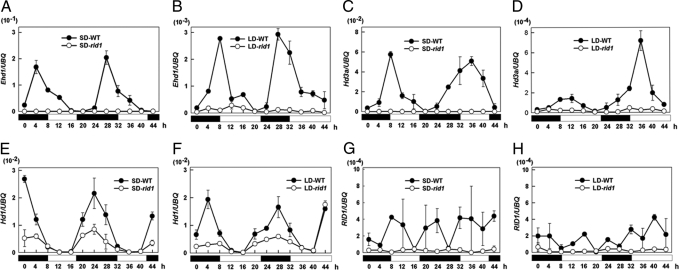
We performed quantitative RT-PCR analysis (with the primers in Table S3) to assay the diurnal expression patterns of these genes together with RID1 in the rid1 mutant and WT plants under both SDs and LDs (Fig. 4). Transcript levels of Ehd1 and Hd3a were largely reduced in the rid1 mutants (Fig. 4 A–D) under both SDs and LDs, suggesting that Ehd1 and Hd3a are regulated by RID1. The transcript level of Hd1 in the rid1 mutant was lower than in WT, especially during the dark period, under both SDs and LDs (Fig. 4 E and F), suggesting that expression of Hd1 is partly regulated by RID1. The RID1 transcript level was very low in the WT plant and did not seem to show diurnal or circadian patterns under either SDs or LDs (Fig. 4 G and H). These results suggested that RID1 transcription may not be controlled by circadian clocks.
Fig. 4.
Diurnal expression patterns of RID1, Ehd1, Hd1, and Hd3a in WT (filled circles) and rid1 (open circles) plants under SDs (A, C, E, and G) and LDs (B, D, F, and H) by quantitative RT-PCR analysis. The expression levels are relative to the UBQ mRNA. The average values (means ± SEM) are based on two separate RNA extractions each with at least three technical repeats. The open and filled bars at the bottom represent the light and dark periods, respectively.
It was recently reported that Ghd7, encoding a CCT domain protein, has a key role in photoperiod flowering by regulating the putative Ehd1–Hd3a pathway under LDs (17). We compared the expression patterns of Ghd7 in the rid1 mutant and WT plants under both SDs and LDs. The transcript level of Ghd7 in the rid1 mutant was lower than in WT, especially during the light period under both SDs and LDs (Fig. S3), suggesting that expression of Ghd7 is partly regulated by RID1.
It has been shown that the expression of FT-like (FTL) genes around dawn can be attributed to Hd1 and that Ehd1 may cause the expression of FTL genes during daytime (6, 28). There are 13 FTL genes in the rice genome (29), six of which (FTL1/FTL, FTL3/RFT1, FTL4, FTL5, FTL6, and FTL11), in addition to Hd3a (FTL2), showed expression 35 d after germination under SDs (30). We thus compared the expression of these six FTL genes in WT and rid1 plants. Like Hd3a, expression of RFT1 (FTL3) was dramatically reduced in the rid1 mutant compared to WT under both LDs and SDs. FTL1 and FTL4 were slightly reduced under both LDs and SDs (Fig. S4 A–F). Expression of FTL5 was down-regulated in rid1 only under LDs, whereas FTL6 was down-regulated only under SDs (Fig. S4 H and I). In contrast, the transcript level of FTL11 in the rid1 mutant was slightly higher than that in WT under both SDs and LDs (Fig. S4 K and L). Thus, all of the six FTL genes are regulated, at least partly, by RID1.
It has also been reported that overexpression of RCN1 and RCN2, the rice homologs of TERMINAL FLOWER 1/CENTRORADIALIS, delays the transition to reproductive phase in rice (10). We investigated the expression of these two genes. Their transcript levels were not obviously affected in the rid1 mutant (Fig. S5), indicating that RCN1 and RCN2 are likely independent of RID1 in regulating rice flowering.
We also investigated the effect of the rid1 mutation on the circadian clock by examining the diurnal expression of Cab1R, a well characterized clock-controlled gene (31). The results showed that expression of Cab1R is not affected in the rid1 mutant under either LDs or SDs (Fig. S6), suggesting that the circadian clock may not be affected by the rid1 mutation.
Taken together, the mutation in RID1 affected expression of the genes known to be involved in flowering. RID1 seems to be independent of the circadian clock, as it neither showed a circadian rhythm in expression pattern nor affected the circadian clock.
Possible Targets of the RID1 Protein.
Kozaki et al. (24) found 16 oligonucleotides, with an 11-bp consensus motif TTTGTCG/CT/CT/aT/aT, that showed binding activity with the IDD protein in vitro. To identify possible targets of the RID1 protein, we searched the rice genomic sequences for this motif and found positive hits in the putative promoter regions of 761 predicted genes. Among the genes surveyed in this work, only one positive hit, 5′-TTTGTCGTAAT-3′, was found in the promoter region of Hd1. Because Hd1 mRNA was detected in all of the examined tissues, including the SAM and immature leaves (Fig. S1), it may have a partly overlapping expression domain with RID1.
To examine whether Hd1 may be directly regulated by RID1, we tested the binding activity of the IDD1 domain in RID1 with this motif by using a one-hybrid yeast assay. The cotransformants could not grow on the SD/Leu−/Trp−/His− medium with 30 mM 3-AT, whereas the transformed positive and negative controls showed the expected results in three independent repeats (data not shown). Detailed comparison of this motif with the published data (24) showed that the sequence of this motif is not exactly the same as those of any of the 16 motifs identified previously. These results indicated that the IDD domain in RID1 could not bind to this putative motif, suggesting that none of these genes is a direct target of the IDD protein.
Discussion
RID1 Acts as a Master Switch of Phase Transition.
Genetic analyses in Arabidopsis spp. have established multiple pathways that independently regulate flowering time, including autonomous, photoperiod, vernalization, and hormone signaling. Although a large number of flowering-regulation mutants have been identified, all of the mutants only showed quantitative effects by either accelerating or delaying flowering under certain environmental conditions, and none of the mutants alone could cause a never-flowering phenotype. Moreover, even the co-2/fca-1/ga1-3 triple mutant, having simultaneous defects in photoperiod, autonomous, and GA-dependent flowering pathways and never flowering under both LDs and SDs, flowered after a 7-week vernalization treatment (32). Thus, none of the mutants identified in Arabidopsis corresponded to a gene that can singly act as the master switch for the phase transition to produce a never-flowering phenotype equivalent to the rid1 mutant in rice identified in this study.
In rice, photoperiod is the most important environmental cue for signaling flowering, as vernalization is not required for flowering (6). Two independent photoperiod pathways, one involving Hd1 and the other involving Ehd1 that control heading date by regulating Hd3a (and/or RFT1) have been identified. Our analysis clearly demonstrated that the main flowering pathways previously identified in rice were blocked in the rid1 mutant, such that expression of Ehd1 and Hd3a/RFT1 was completely repressed and expression of Hd1 partly reduced. Although there may exist additional flowering pathway(s) in rice that have not been identified presently, the never-flowering phenotype of this mutant indicated that RID1 acts as the master switch of phase transition and that loss of function of this regulator would result in failure of flowering.
RID1 Regulates a Distinct Flowering Pathway.
Previous studies in maize showed that plants homozygous for the loss-of-function id1 mutation remained in a prolonged vegetative phase and eventually flowered (19, 33). However, the inflorescences in the flowered plants exhibited a vegetative characteristic referred to as floral reversion. This observation indicates that the ID1 gene plays important roles both in controlling phase transition and in maintaining the flowering state. The phenotypic difference between the maize id1 and rice rid1 mutants may reflect functional redundancy of the ID-related genes in the two species, such that this gene may be partly redundant in maize (19, 34), but not in rice.
Phylogenetic analysis indicated that none of the 16 IDD genes identified in the Arabidopsis genome belonged to the ID1/OsID/SbID group (22). There has been no evidence of an ID-related gene in Arabidopsis that contributes to flowering transition (22), despite the extensive progresses made in understanding the regulatory mechanisms of flowering (35). In addition, Ehd1, which regulates floral induction, is found only in rice; no ortholog of Ehd1 is evident in Arabidopsis (6). This evidence suggests that the RID1/Ehd1 represents a distinct floral induction pathway in rice but not in Arabidopsis spp. Moreover, an ID-related gene was also reported in sorghum (22), another SD plant. However, because Ehd1-regulated flowering has only been reported in rice, it remains to be determined whether the RID1/Ehd1 pathway represents a distinction between SD and LD plants or is conserved in the grasses but not in cress species.
A Model to Place RID1 in the Pathways.
Expression analyses showed that transcript levels of Ehd1, Hd3a, and RFT1 were reduced to an undetectable level in the rid1 mutant under both SDs and LDs, suggesting that Ehd1, Hd3a, and RFT1 are downstream of RID1. The expression of Hd1 during the night and Ghd7 during the day was also partly affected by RID1 under both SDs and LDs, indicating that Hd1 and Ghd7 are also downstream of RID1. It should be noted that the partly reduced expression of the Hd1 and Ghd7 in the rid1 mutant seems to suggest that these genes may have other function(s) in addition to photoperiod flowering. In fact, Ghd7 was shown to have major pleiotropic effects on heading date, plant height, and grain number (17).
Apart from Hd3a and RFT1, mRNA levels of five additional FTL genes examined in this study were also affected by rid1, although to lesser degrees. Recent studies showed that RFT1 can complement the flowering function of Hd3a in rice. Double suppression of Hd3a and RFT1 by RNAi resulted in never-flowering, whereas RFT1-RNAi plants flowered normally and Hd3a-RNAi plants flowered ≈30 days later than the WT (36). There has been evidence that other FTL genes may also function in flowering regulation in rice, as overexpression of FTL1 may promote flowering (28). Our observation that expression of the FTL-genes was affected in the rid1 mutant indicated that these genes are also likely downstream of RID1.
It was also shown that rice plants carrying the nonfunctional alleles at both Hd1 and Ehd1 loci still flowered under both SDs and LDs (6), suggesting the existence of alternative flowering pathway(s). However, the never-flowering phenotype indicates that all of the essential flowering pathways were ineffective in the rid1 mutant and thus are downstream of RID1.
Based on the above discussion, a model was proposed to place RID1 in the molecular pathways of flowering regulation in rice (Fig. 5). There are two indispensable elements in the system. The first is the RID1 gene controlling the phase transition and initiation of floral induction. The other is the Hd3a/RFL1/FTL complex acting as the immediate inducer of flowering. Completely abolishing the function of either element would cause never-flowering. Once the phase transition is induced with the activation of RID1, flowering signaling is transduced and regulated through the various pathways, including ones yet to be identified. Signals from these pathways are eventually integrated with proteins of FTL genes such as Hd3a and RFT1 and thus induce flowering.
Fig. 5.
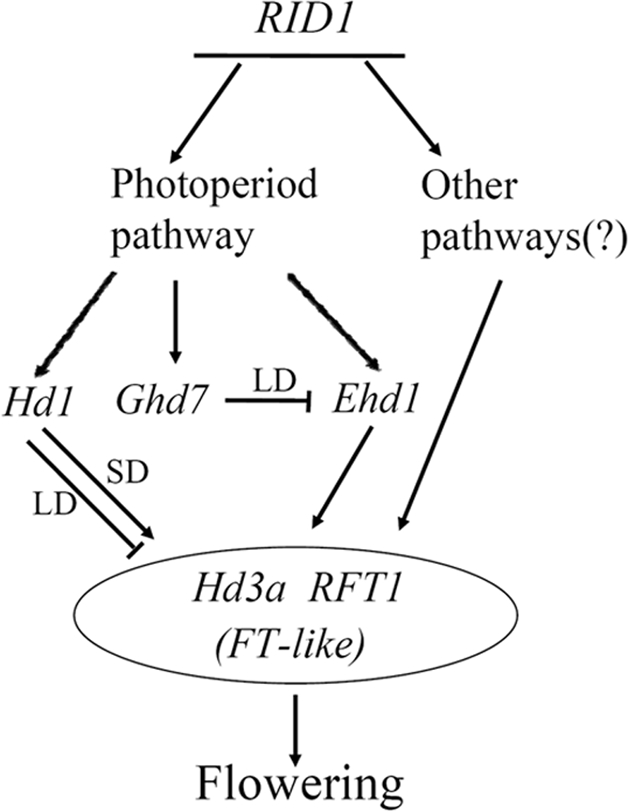
A proposed model for the initiation and integration of the flowering pathways in rice.
The challenge for future studies is to elucidate how RID1 regulates the phase transition. RID1 is expressed in immature leaves, whereas its downstream components, Hd3a/RFT1 and Ehd1, are expressed in mature leaves, and the phase transition takes place in SAM. Thus, the product of RID1 has to travel to mature leaves to regulate the expression of the other components, whose products will in turn travel to SAM to regulate floral initiation (37, 38). Moreover, because Hd3a/RFT1 and Ehd1 may not be the direct targets of RID1, as indicated by our data, activation of these downstream components may require cofactor(s) that are bound to the promoters of these genes and are activated by RID1. Furthermore, very little is known about when and how RID1 is activated in this process, which may be another focal point for understanding the mechanism of floral induction in future studies.
Materials and Methods
Plant Materials and Growth Conditions.
Screening of heading time mutants was carried out by planting the mutant materials under natural LDs in the experimental field of Huazhong Agriculture University at Wuhan, China, in the summer of 2004. The rid1 plants were propagated vegetatively by ratooning them in the winter on Hainan Island (South China Sea) under natural SDs. We also planted the rid1 and WT plants under artificial SDs (14 h light/10 h dark, 28°C), LDs (10 h light/14 h dark, 28°C), and neutral day-length (12 h light/12 h dark, 28°C). Heading date was recorded as the number of days from germination to the day of panicle emergence from the leaf sheath.
Genotyping of Mutant Plants.
Genotyping of the rid1-segregating population was performed by PCR using the following primers: P1, P2, P3, and T (Table S2). The PCR was conducted with an initial step of 94°C incubation for 2 min; a second step of 30 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1.5 min; and a final extension of 1 kb, 660 bp, and 611 bp, respectively.
Vector Construction and Plant Transformation.
A 5.7-kb genomic DNA fragment containing the entire RID1 coding region and the 1920-bp upstream and 955-bp downstream sequences was isolated by digestion of the Clemson BAC clone OSJNBa0018A14 (kindly provided by R. Wing, University of Arizona) and inserted into the binary vector pCAMBIA2301. An empty pCAMBIA2301 vector was used as a control. The transformation recipient was callus culture that was induced from seeds homozygous for rid1. We obtained 55 rid1 callus cultures from 188 T2 seeds that were harvested from RID1/rid1 heterozygous T1 pants.
To prepare GFP fusions, a fragment containing the RID1 promoter and coding region was amplified by PCR using the BAC clone OSJNBa0018A14 as the template with the primer pair RG-F (Table S2). The PCR product was cloned into a pENTR/D-TOPO vector by using a pENTR Directional TOPO Cloning Kit (Invitrogen). An LR recombination reaction between the attL and attR site was performed in between the entry clone and the destination vector, pGWB4 (39), which contains the GFP gene, to generate the construct RID1::RID1:GFP. The resulting construct was introduced into Agrobacterium tumefaciens EHA105 and then into the Japanese rice Zhonghua 11 by Agrobacterium-mediated transformation as previously described (25).
DNA Blot Hybridization.
The probes for Southern blot hybridization were the PCR products amplified with T-primers (Table S2). The experimental procedures for Southern analysis followed methods described previously (40).
GFP and GUS Imaging.
GFP and GUS images of transgenic rice plants were viewed under a stereo light microscope (LEICA MZFL III), and the photos were taken by using a digital camera (Nikon E5400). For preparing the transverse sections, the shoot apex of RID1::RID1:GFP transgenic plants was fixed into a hole of a potato block. Transverse sections were then made by hand-sectioning. The sections were analyzed by using a laser-scanning confocal microscope (LEICA TCS SP2). Fluorescence was excited with a 488-nm argon laser and emission images were collected in the 510–540 nm range.
Real Time Quantitative RT-PCR.
For diurnal expression analysis, plants were grown for 28 d in the greenhouse under natural-day-length conditions. For SD samples, the plants were transferred to a growth chamber set for SDs (10 h light/14 h dark, 28°C), and entrained for 3 d. Then, penultimate and immature leaves were harvested every 4 h during a 48 h period from WT and rid1 plants, respectively. For LD samples, plants were transferred to a growth chamber set for LDs (14 h light/10 h dark, 28°C) and entrained for 3 d. Samples were collected in the same way as SDs.
The TRIzol reagent (Invitrogen) was used according the manufacturer's instructions to extract total RNAs from various tissues or organs of rid1 and the WT plants. Relative quantification of gene expression was performed on an ABI PRISM 7500 PCR instrument (Applied Biosystems). A rice Ubiquitin gene was used as the endogenous control. The real time PCR primers for the RID1, Ehd1, Hd3a, Hd1, Cab1R, RCN1, RCN2, and FTL genes are listed in Table S3. Real-time PCR was performed in an optical 96-well plate that included SYBR Premix EX Taq and 0.5 μl of Rox Reference Dye II (Takara), 1 μl of the cDNA sample, and 0.2 μM each gene-specific primer, in a final volume of 25 μl. The reactions followed the following temperature profile: 95°C for 10 seconds, 50 cycles of 95°C for 5 seconds, and 60°C for 40 seconds. Disassociation curve analysis was performed as follows: 95°C for 15 seconds, 60°C for 20 seconds, and 95°C for 15 seconds. Data were collected by using the ABI PRISM 7500 sequence detection system following the instruction manual.
Yeast One-Hybrid Assay.
Yeast one-hybrid assay was carried out by using a Clontech system. Triple tandem repeats of putative motif 5′-TTTGTCGTAAT-3′ from the Hd1 promoter was inserted into pHIS2. Putative DNA-binding domain of zinc finger Z2 and Z3 in IDD domain was amplified by using the primer pair RID1-Rec2 (Table S2) and fused to the GAL4 activation domain in the vector pGADT7-Rec2. The two constructs were sequence-verified and cotransformed to the yeast strain Y187 for determination of the DNA-protein interactions.
Supplementary Material
Acknowledgments.
This work was supported by grants from the National Natural Science Foundation of China, the National Special Key Project of China on Functional Genomics of Major Plants and Animals, and a Royal Society United Kingdom-China Fellowship.
Footnotes
Conflict of interest statement: A patent was filed based on this work.
Data deposition: The RID1 sequences reported in this paper have been deposited in the GenBank database [accession nos. FJ009578 (cDNA) and FJ009579 (genomic)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0806019105/DCSupplemental.
References
- 1.Baurle I, Dean C. The timing of developmental transitions in plants. Cell. 2006;125:655–664. doi: 10.1016/j.cell.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Corbesier L, Coupland G. The quest for florigen: A review of recent progress. J Exp Bot. 2006;57:3395–3403. doi: 10.1093/jxb/erl095. [DOI] [PubMed] [Google Scholar]
- 3.Mouradov A, Cremer F, Coupland G. Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell. 2002;14:S111–S130. doi: 10.1105/tpc.001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parcy F. Flowering: A time for integration. Int J Dev Biol. 2005;49:585–593. doi: 10.1387/ijdb.041930fp. [DOI] [PubMed] [Google Scholar]
- 5.Yano M, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12:2473–2483. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doi K, et al. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls. FT-like gene expression independently of Hd1. Genes Dev. 2004;18:926–936. doi: 10.1101/gad.1189604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kojima S, et al. Hd3a, a rice ortholog of the Arabidopsis FT Gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002;43:1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi Y, Shomura A, Sasaki T, Yano M. Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subunit of protein kinase CK2. Proc Natl Acad Sci USA. 2001;98:7922–7927. doi: 10.1073/pnas.111136798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugano S, Andronis C, Ong MS, Green RM, Tobin EM. The protein kinase CK2 is involved in regulation of circadian rhythms in. Arabidopsis. Proc Natl Acad Sci USA. 1999;96:12362–12366. doi: 10.1073/pnas.96.22.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakagawa M, Shimamoto K, Kyozuka J. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 2002;29:743–750. doi: 10.1046/j.1365-313x.2002.01255.x. [DOI] [PubMed] [Google Scholar]
- 11.Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature. 2003;422:719–722. doi: 10.1038/nature01549. [DOI] [PubMed] [Google Scholar]
- 12.Putterill J, Robson F, Lee K, Simon R, Coupland G. The. CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- 13.Suárez-López P, et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 14.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izawa T, Takahashi Y, Yano M. Comparative biology comes into bloom: Genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr Opin Plant Biol. 2003;6:113–120. doi: 10.1016/s1369-5266(03)00014-1. [DOI] [PubMed] [Google Scholar]
- 16.Kim SL, Lee S, Kim HJ, Nam HG, An G. OsMADS51 is a short-day flowering promoter that functions upstream of. Ehd1, OsMADS14, and Hd3a. Plant Physiol. 2007;145:1484–1494. doi: 10.1104/pp.107.103291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue WY, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 18.Izawa T. Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J Exp Bot. 2007;58:3091–3097. doi: 10.1093/jxb/erm159. [DOI] [PubMed] [Google Scholar]
- 19.Colasanti J, Yuan Z, Sundaresan V. The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell. 1998;93:593–603. doi: 10.1016/s0092-8674(00)81188-5. [DOI] [PubMed] [Google Scholar]
- 20.Englbrecht CC, Schoof H, Böhm S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome BMC. Genomics. 2004;5:39. doi: 10.1186/1471-2164-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agarwal P, et al. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol Biol. 2007;65:467–485. doi: 10.1007/s11103-007-9199-y. [DOI] [PubMed] [Google Scholar]
- 22.Colasanti J, et al. The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics. 2006;7:158. doi: 10.1186/1471-2164-7-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong A, Colasanti J. Maize floral regulator protein INDETERMINATE1 is localized to developing leaves and is not altered by light or the sink/source transition. J Exp Bot. 2007;58:403–414. doi: 10.1093/jxb/erl206. [DOI] [PubMed] [Google Scholar]
- 24.Kozaki A, Hake S, Colasanti J. The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res. 2004;32:1710–1720. doi: 10.1093/nar/gkh337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu C, et al. Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 2003;35:418–427. doi: 10.1046/j.1365-313x.2003.01808.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang JW, et al. RMD: A rice mutant database for functional analysis of the rice genome. Nucl Acids Res. 2006;34:D745–D748. doi: 10.1093/nar/gkj016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, et al. Non-random distribution of T-DNA insertions at various levels of the genome hierarchy as revealed by analyzing 13804 T-DNA flanking sequences from an enhancer-trap mutant library. Plant J. 2007;49:947–959. doi: 10.1111/j.1365-313X.2006.03001.x. [DOI] [PubMed] [Google Scholar]
- 28.Izawa T, et al. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 2002;16:2006–2020. doi: 10.1101/gad.999202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chardon F, Damerval C. Phylogenomic analysis of the PEBP gene family in cereals. J Mol Evol. 2005;61:579–790. doi: 10.1007/s00239-004-0179-4. [DOI] [PubMed] [Google Scholar]
- 30.Ishikawa R, et al. Suppression of the floral activator. Hd3a is the principal cause of the night break effect in rice. Plant Cell. 2005;17:3326–3336. doi: 10.1105/tpc.105.037028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugiyama N, Izawa T, Oikawa T, Shimamoto K. Light regulation of circadian clock-controlled gene expression in rice. Plant J. 2001;26:607–615. doi: 10.1046/j.1365-313x.2001.01063.x. [DOI] [PubMed] [Google Scholar]
- 32.Reeves PH, Coupland G. Analysis of flowering time control in Arabidopsis by comparison of double and triple mutants. Plant Physiol. 2001;126:1085–1091. doi: 10.1104/pp.126.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tooke F, Ordidge M, Chiurugwi T, Battey N. Mechanisms and function of flower and inflorescence reversion. J Exp Bot. 2005;56:2587–2599. doi: 10.1093/jxb/eri254. [DOI] [PubMed] [Google Scholar]
- 34.Muszynski MG, et al. Delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 2006;142:1523–1536. doi: 10.1104/pp.106.088815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corbesier L, Coupland G. Photoperiodic flowering of. Arabidopsis: Integrating genetic and physiological approaches to characterization of the floral stimulus. Plant Cell Environ. 2005;28:54–66. [Google Scholar]
- 36.Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. Hd3a and RFT1 are essential for flowering in rice. Development. 2008;135:767–774. doi: 10.1242/dev.008631. [DOI] [PubMed] [Google Scholar]
- 37.Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- 38.Corbesier L, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 39.Nakagawa T, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 40.Liu KD, et al. A genome-wide analysis of wide compatibility in rice and the precise location of the S5 locus in the molecular map. Theor Appl Genet. 1997;95:809–814. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.