Abstract
Fatty acids are among the major building blocks of living cells, making lipid biosynthesis a potent target for compounds with antibiotic or antineoplastic properties. We present the crystal structure of the 2.6-MDa Saccharomyces cerevisiae fatty acid synthase (FAS) multienzyme in complex with the antibiotic cerulenin, representing, to our knowledge, the first structure of an inhibited fatty acid megasynthase. Cerulenin attacks the FAS ketoacyl synthase (KS) domain, forming a covalent bond to the active site cysteine C1305. The inhibitor binding causes two significant conformational changes of the enzyme. First, phenylalanine F1646, shielding the active site, flips and allows access to the nucleophilic cysteine. Second, methionine M1251, placed in the center of the acyl-binding tunnel, rotates and unlocks the inner part of the fatty acid binding cavity. The importance of the rotational movement of the gatekeeping M1251 side chain is reflected by the cerulenin resistance and the changed product spectrum reported for S. cerevisiae strains mutated in the adjacent glycine G1250. Platensimycin and thiolactomycin are two other potent inhibitors of KSs. However, in contrast to cerulenin, they show selectivity toward the prokaryotic FAS system. Because the flipped F1646 characterizes the catalytic state accessible for platensimycin and thiolactomycin binding, we superimposed structures of inhibited bacterial enzymes onto the S. cerevisiae FAS model. Although almost all side chains involved in inhibitor binding are conserved in the FAS multienzyme, a different conformation of the loop K1413–K1423 of the KS domain might explain the observed low antifungal properties of platensimycin and thiolactomycin.
Keywords: cerulenin, platensimycin, thiolactomycin, fatty acid synthesis, yeast
Three different schemes for de novo synthesis of fatty acids are found in nature. Eukaryotes and advanced prokaryotes generally use the type I fatty acid synthase system (FAS I), composed of complexes of large multifunctional enzymes. Bacteria, in contrast, use the dissociated FAS II system that consists of a set of separate enzymes, each catalyzing one of the reactions of the fatty acid synthase cycle (1). A third system exists in some parasites that use membrane-bound fatty acid elongases for the synthesis of aliphatic chains (2). Despite this considerable variation, the individual reaction steps of fatty acid biosynthesis are essentially conserved in all kingdoms of life. Four basic reactions constitute a single round of elongation. In the first step, an acceptor CoA or acyl carrier protein (ACP) associated acetyl unit is condensed with malonyl-ACP to form β-ketobutyryl-ACP, which is subsequently reduced by an NADPH-dependent ketoacyl-ACP reductase. The resulting β-hydroxyacyl-ACP is dehydrated to produce enoyl-ACP and finally reduced by an enoyl reductase (ER) to form the saturated acyl-ACP, which can be further elongated in a new cycle [see supporting information (SI) Fig. S1].
The importance of the fatty acid biosynthesis pathway makes the FAS systems attractive targets for the development of compounds with antibiotic or antitumor properties (3–5). This potential is further underlined by the potency of the classic drugs isoniazid and triclosan, both inhibiting the ER step of bacterial fatty acid biosynthesis (6, 7). Several inhibitors targeting the ketoacyl synthase (KS) step of the FAS cycle have also been described, including cerulenin (CER) (8), thiolactomycin (TLM) (9), and the recently discovered platensimycin (PLM) (10). The polyketide CER inhibits both FAS type I and II KS enzymes, by covalent modification of the active site cysteine and by occupying the long acyl-binding pocket (11, 12). TLM and PLM, in contrast, have been shown to be selective toward the FAS II system, blocking the malonyl and the CoA/ACP phosphopantetheine-binding sites (10, 13).
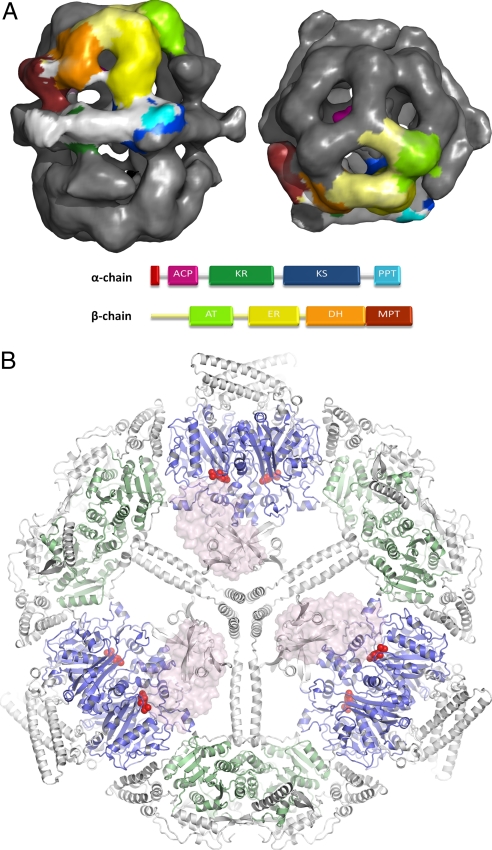
A wealth of knowledge about the structure, function, and inhibition of the distributed FAS type II system has been gathered over the last decade (14, 15). However, detailed structural studies of the large eukaryotic FAS type I multifunctional enzymes have only just been initiated. The architecture of the fatty acid synthase complexes of vertebrates and fungi are strikingly different. In mammals, the FAS type I domains form an X-shaped 540-kDa homodimer (16), whereas in fungi, the functional domains are organized in a 2.6-MDa barrel-shaped dodecameric complex (17). The fungal fatty acid synthase complex (FAS) is assembled from two polyfunctional proteins, α and β. In Saccharomyces cerevisiae, the 230-kDa β-chain harbors an acetyl transferase and a malonyl/palmitoyl transferase domain (AT/MPT), as well as a dehydratase and an ER domain. The 210-kDa α-chain contains a KS domain, a ketoacyl reductase (KR), and a phosphopantetheine transferase domain. In addition, the α-chain includes an N-terminal ACP, required for the transfer of intermediates between the different catalytic sites (1). In a recent major advance in our understanding of this system, Ban and colleagues presented an atomic model of the Thermomyces lanuginosus fatty acid synthase type I complex (18). The 3.1-Å electron density maps of the 230 × 260 Å multienzyme allowed tracing of all functional components of the FAS type I cycle except for the mobile ACP. In two following structures of the S. cerevisiae FAS (19, 20), the missing ACP domain was subsequently located, docked to the KS domain.
We have recently purified the S. cerevisiae fatty acid synthase multienzyme and determined its structure in complex with the polyketide antibiotic CER by multiple isomorphous replacement with anomalous scattering (MIRAS) aided by cryo-EM reconstructions. The structure of this CER-inhibited eukaryotic FAS complex reveals both differences and similarities to the FAS type II systems, explains CER resistance for known fungal FAS mutants, and gives a rationale for fatty acid synthesis being rather nonselectively inhibited by CER but selectively inhibited by TLM and PLM.
Results and Discussion
Overall Structure.
The biosynthesis of the S. cerevisiae fatty acid synthase α- and β-chains is tightly balanced (21). Because expression of one or both subunits from extrachromosomal information might perturb this balance, we decided to His-tag the chromosomal FAS1 gene, coding for the β-chain. To facilitate analysis of the purified FAS complex, the six sets of active centers were synchronized by elongation of nonmature fatty acids through the addition of malonyl-CoA and NADPH and subsequent inhibition of the condensing step by CER (Fig. S2). The fatty acid synthase complex subjected to crystallization and EM studies was, thus, acetyl-depleted with a malonyl group occupying the central ACP thiol (S180) and CER covalently linked to the peripheral KS thiol (C1305). Initial MIRAS phases were obtained from Ta6Br12 and W18-cluster derivatized crystals and extended to ≈4.5 Å by density modification. The FAS–CER complex was built by using the atomic model of the S. cerevisiae FAS (19) and refined to a final resolution of 4 Å (Table S1; also see SI Materials and Methods for structural differences of S. cerevisiae FAS models).
As suggested from early EM micrographs (22, 23), the fungal FAS barrel consisted of two dome-shaped reaction chambers formed by six β-chains and an equatorial wheel-like structure constructed by six α-chains. The catalytic domains of the individual chains were embedded by a number of short and long expansion segments into the walls of the FAS particle (Fig. 1A). These intermediate segments formed a complex network that held the barrel together and oriented the catalytic centers toward the hollow reaction chambers. The mobile ACPs were diagonally tethered across the reaction chambers by two flexible linkers. Similar to what was seen in the two previous S. cerevisiae structures, we found the ACPs docked to the KS domains. The stalled acetyl-depleted state of our FAS preparation, thus, gives additional support to the notion that the KS/ACP interaction is intrinsically favored in yeast FAS (19, 20).
Fig. 1.
The fungal FAS multienzyme. (A) Cryo-EM reconstruction of S. cerevisiae FAS contoured at 1.7 σ. One single α- and one single β-chain are colored according to the attached domain representation. Expansion segments in between domains are colored white for the α-chain and pale yellow for the β-chain. (B) The central FAS α-wheel with six bound CER molecules is highlighted in red. Domains are colored according to A, and three docked ACP domains are represented by transparent surfaces.
FAS KS Domain.
As shown in Fig. 1B, the central α6-wheel consisted of three alternating KS and KR domains (also see Fig. S3). Similar to the FAS type II KSs, like the E. coli FabF/FabB enzymes (22, 23), the FAS KS domain formed a 80 × 60 × 50 Å thiolase-like dimer. To fit into the large multienzyme complex, the thiolase core fold was extended by three long insertions, increasing the buried dimer surface area of the KS by ≈400 Å2. The insertions connected the KS domain to the neighboring β-chain and form the three main spokes of the equatorial wheel, creating a docking site for the ACP.
KSs generally act via a Cys-His-His or a Cys-His-Asn triad to elongate an ACP- or CoA-associated acyl chain by adding C2 units through a ping-pong decarboxylating condensation mechanism. In the first step, the acyl group is attacked by an active site cysteine to form an acyl-enzyme intermediate. During the second step, malonyl-ACP binds to the active site where it is decarboxylated, creating a reactive carbanion. The carbanion is finally condensed with the bound acyl group, forming the C2-elongated β-ketoacyl-ACP product (24). Consistent with the multisubstrate mechanism, the KS active site was divided into three parts, one holding the catalytic residues and the acyl-binding pocket, a second responsible for malonyl binding, and a third part binding the CoA or ACP phosphopantetheine group that transfers the acyl and malonyl substrates.
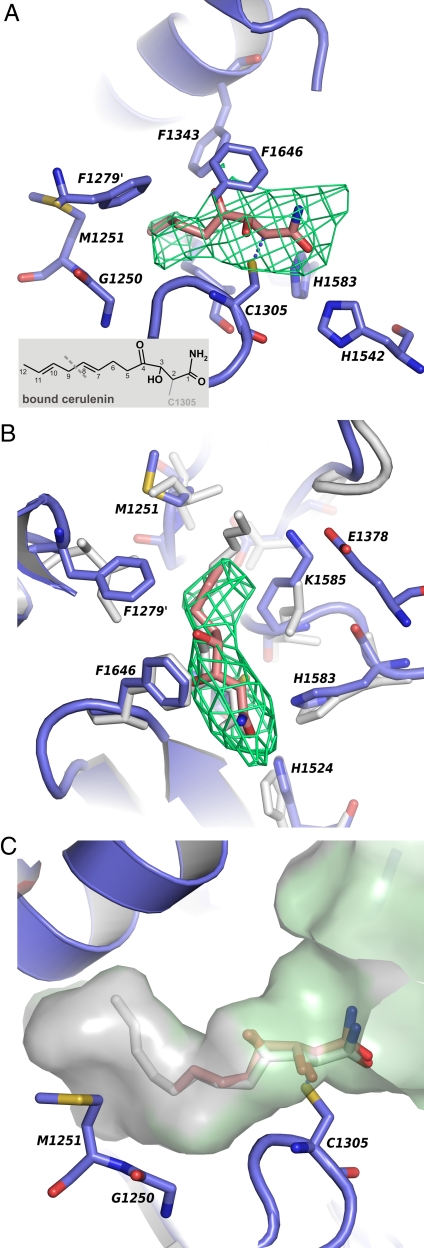
In the large fungal fatty acid complex, the KS dimers were located with one active site tunnel directed toward the upper and the other directed toward the lower FAS reaction chamber (Fig. 1B). The central part of each KS tunnel was occupied by the side chains of C1305, H1583, and H1542, forming a conserved KS catalytic triad. As in the FAS II enzymes, the active cysteine was positioned at the N-terminal end of a long α-helix, allowing it to exploit the helix dipole moment to increase its nucleophilicity (22). The two histidines responsible for the decarboxylation step (25, 26) are similar to the FAS II KSs positioned at two loops, ≈3 and 5 Å apart from the nucleophilic cysteine (Fig. 2A).
Fig. 2.
The FAS KS acyl-binding pocket with bound CER. (A) Active site of FAS KS. The prime indicates a residue from an adjacent KS molecule. An averaged Fo − Fc omit map (green) reveals the CER molecule (red) covalently bound to the FAS KS C1305. The map is contoured at 5 σ, within 3 Å from the CER. A chemical sketch of CER is shown (Inset) with a dotted line indicating the truncated structure used for refinement. (B) Superposition of the FabF–CER complex (white) (PDB ID code 1B3N) onto the FAS KS active site (blue). (C) Surface representation of the residue conservation between FAS KS and FabF with FAS CER in red, E. coli FabF CER in white, conserved side-chains in green, and nonconserved residues in gray.
In contrast to the well conserved catalytic site, the makeup of the curved acyl-binding pocket of the yeast FAS KS differed significantly from the FAS type II FabF/B enzymes. As shown in the superposition of FAS KS and FabF in Fig. 2B, the side chains of F1279 and K1585 made the entrance of the FAS KS acyl-binding tunnel significantly narrower than the Ile and Leu counterparts of the bacterial enzyme. K1585 was stretched along the side of the cavity to form a salt bridge to E1378, thereby increasing the hydrophobic character of the cavity. The nearby F1343 represented one of the few amino acids that were conserved also in the two bacterial enzymes and was responsible for the sharp kink of the acyl-binding tunnel (see Fig. 2 A and C). Amino acids V1254, E1342, and G1339 in the inner part of the acyl-binding tunnel shared some remote similarity to the bacterial FabF/FabB structures. However, the end of the pocket created by the FAS KS dimer partner had a completely unique lining, tailored to fit the wide range of saturated acyl intermediates of the S. cerevisiae fatty acid synthase type I system.
CER Binding.
The natural product CER, derived from the filamentous fungus Cephalosporium caerulens (8, 27), effectively targets Cys-His-His-like KS enzymes and forms a covalent modification of the catalytic cysteine (11, 12, 28). CER shows a broad spectrum of antimicrobial activity (29, 30), exhibits antitumor activity (31, 32), and reduces food intake and body weight in mice (33).
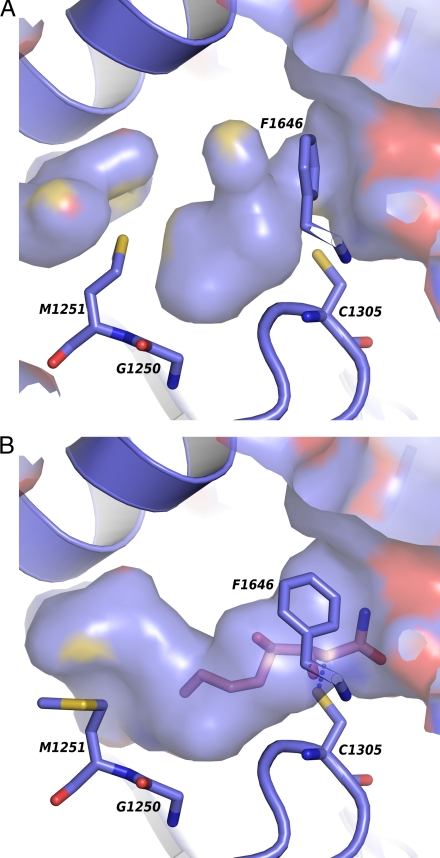
Inspection of the deep hydrophobic substrate-binding pocket of the FAS KS domain revealed strong Fo − Fc difference density located in the vicinity of C1305. A superposition of two bacterial type II KS-CER complex structures (28, 34) onto the FAS condensing enzyme showed that the amide half of the inhibitor colocalizes with the 4 to 5 σ Fo − Fc density peaks in the different KS active sites of the FAS particle. As was shown for the CER-inhibited FabF/FabB enzymes, the C2 atom of CER formed a covalent bond with the SH group of the catalytic cysteine, whereas the CER amide was located close to the two active site histidine residues, H1542 and H1583. The hydroxyl group at C3 was positioned to interact with the backbone nitrogen of F1646. A similar main chain interaction has been suggested to stabilize the oxyanion formed during the acetyl transfer step of the condensation reaction (28). As compared with the native FAS structure (19), the active site of the KS–CER complex is in an open conformation (10, 34), with the phenyl group of F1646 flipped (Fig. 3 A and B). This conformation provides free access to the nucleophilic cysteine and enables CER to mimic the acyl-enzyme intermediate of the condensation reaction (34).
Fig. 3.
Conformational changes. (A) The blocked hydrophobic acyl cavity of the native FAS KS (19). The surface is colored according to atom type. (B) The side chains of F1646 and M1251 flip upon CER binding, opening the active site, and extending the long curved fatty acid binding pocket.
Because the FAS KS elongates saturated fatty acids, the unsaturated acyl chain of CER might not be expected to have an optimal fit in the curved KS binding tunnel. However, the 1,4-nonadienoic CER was shown to be the most effective inhibitor of S. cerevisiae FAS of 11 saturated and unsaturated CER analogs (35). In the FAS–CER complex CER C5–C8 stretches into the long KS acyl pocket, interacting with the side chains of F1279, K1585, and F1343 (Fig. 3 A and B). CER C7 is positioned in the vicinity of F1279, a residue that is conserved in almost all fungal FAS KSs. This side chain may form π-π stacking interactions with the CER C7–C8 double bond, which in part might explain the observed FAS preference for the unsaturated CER.
The last part of the CER acyl chain, including the second diene moiety, could not be seen in electron density maps. However, a B-factor sharpened 2Fo − Fc omit map of the active site revealed that the side-chain of M1251, positioned in the center of the fatty acid binding pocket, was flipped, as compared to the native structure, indicating the influence of C9–C12 of CER (Figs. 3 A and B and S4). A similar change of rotamer was observed in the M1251 counterpart I108 of the FabF–CER complex (34). This suggests that the acyl chain conformation of CER bound to FAS and to FabF might be related, directed away from the KS dimer interface instead of toward it, as seen in the FabB–CER complex. The FabF I108 has been proposed to be responsible for the low CER sensitivity compared to FabB, which has a glycine residue at this position (28, 34). Because the IC50 value of FAS CER inhibition (35) is in the range of FabB, the FAS KS has to be able to compensate for the rearrangement of the methionine, for example, by stacking interactions to the CER C7–C8 double bond and a better fit of the rigid CER tail to the end of the acyl-binding pocket.
Implications on FAS KS Product Specificity.
The influence of methionine M1251 on KS acyl binding can be deduced from a systematic study on CER inhibition of S. cerevisiae FAS with respect to the length of the CER hydrophobic tail (35). Morisaki et al. found that inhibition efficiencies of CER analogs truncated at C8, C9, and C10 decrease constantly (IC50 of 60, 170, and 300 μM), whereas it is significantly increased for C11–C16 analogs (IC50 between 30 and 4.5 μM), indicating the rotational barrier of M1251 and its compensation at longer chain lengths. The importance of this position is further underlined by S. cerevisiae strains that are mutated in the residue preceding the methionine, G1250. Replacing this glycine with a serine makes yeast 20–80 times more resistant to CER than the wild-type strain (36). The resistance might be attributable to new steric restraints, distorting the main-chain conformation of the G1250-M1251-G1252 turn and, thereby, affecting the necessary movement of M1251 (Fig. 3 A and B).
The human mitochondrial KS HsmtKAS is responsible for the production of the lipoic acid precursor octanoyl-ACP. Interestingly, HsmtKAS features a methionine side chain partly blocking the acyl-binding pocket (37), similar to the FAS KS M1251. This methionine was suggested to be involved in determining the bimodal product specificity of the enzyme. Testing acyl-ACP of various chain lengths revealed markedly weak binding for octanoyl- and tetradecanoyl-ACP, which is attributed to the energy barrier associated with the required rearrangement of the methionine (38).
Intriguingly, the S. cerevisiae FAS G1250S mutant strain was also reported to produce high amounts of decanoic acid and ethyl caproate, the methyl ester of octanoic acid (39). Because the C8 atom of CER is positioned at about the C7 of a Cys-1305 bound acyl intermediate, one might speculate that, in line with binding of the truncated CER analogs, octanoyl-ACP would have decreased affinity to the FAS KS cavity, giving octanoic acid as a side product. However, such a simplistic picture does not find support in the observed narrow C16–C18 product specificity of the S. cerevisiae FAS (40). It must be pointed out that the product spectrum of type I fatty acid synthesis does not solely reflect the enzymatic characteristics of the KS domain but is a result of the whole type I framework. As reported for the thioesterase domain of human FAS I, it can be assumed that the functionally similar MPT domain in the fungal FAS complex is involved in chain length determination, only releasing fatty acid chains of a certain length (20, 41, 42).
Docking of TLM and PLM.
The highly conserved interactions between the hydrophilic part of the CER and the S. cerevisiae FAS perfectly agree with the conservation of the fine-tuned KS elongating mechanism across eukaryotic and prokaryotic FAS systems. Because CER has been shown not to discriminate between FAS type I and FAS type II KSs, the differences of the acyl-binding pockets are obviously limited in their ability to impose binding selectivity. TLM and the recently discovered PLM also anchor at the catalytic residues. However, instead of binding to the hydrophobic acyl pocket, TLM and PLM bind to the malonate portion of the active site (10, 13). Despite the conservation of the malonyl and the phosphopantetheine binding sites (Fig. 2C), both TLM and PLM showed an apparent higher selectivity toward the FAS type II KSs. TLM exhibits an IC50 exceeding 1 mM for the S. cerevisiae FAS complex (43), whereas PLM does not show any inhibitory effect on Candida albicans cells (10).
PLM was shown to preferably bind to the acyl-loaded intermediate state of E. coli FabF (10). The acyl enzyme intermediate is characterized by the flipped phenylalanine adjacent to the catalytic cysteine, turning the active site into an open conformation. Similar to the FabF–PLM complex, TLM was observed to bind to the open conformation of the E. coli FabB active site, forming an edge–face interaction with the flipped aromatic ring of F392 (28). Because the FAS-CER complex mimics the acyl-enzyme intermediate and, as such, the conformation accessible to PLM and TLM binding, we compared the two homologous bacterial inhibitor structures with the KS domain of our S. cerevisiae fatty acid synthase type I complex.
A superposition of the TLM and the PLM complexes onto the FAS KS domain revealed that almost all amino acid side chains involved in protein–inhibitor interactions were conserved in the three enzymes. Except for a single hydrogen bond of the PLM complex (FabF T270), all hydrogen bond acceptors and donors were formally retained in the two modeled KS inhibitor complexes. However, one side chain substitution, FabF/FabB G310/G305 to FAS N1549, in the vicinity of the C10 methyl group of the TLM complex, might imply a suboptimal fit of the TLM thiolactone ring to the FAS KS.
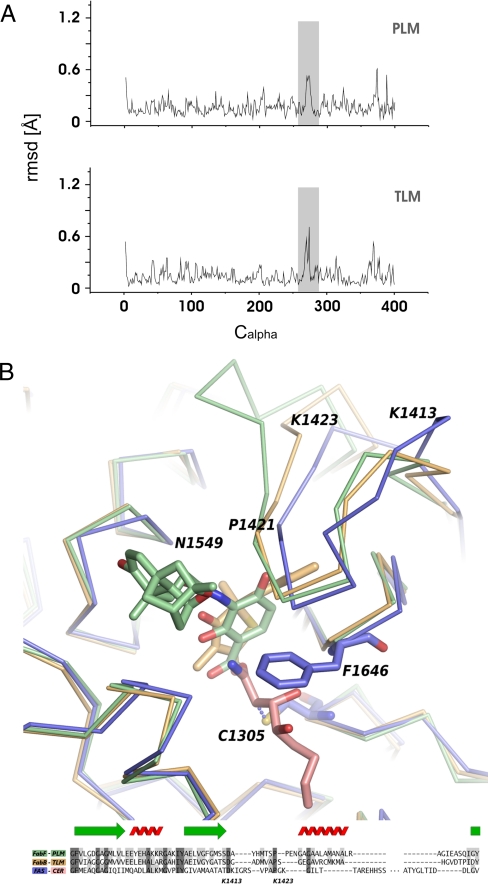
Besides the flipping of F400/F392 and minor shifts of the active site residues to improve the hydrogen bonding geometry, binding of PLM and TLM involves a change in the main chain conformation of the two KS enzymes. As seen in Fig. 4A, the rmsd values between structures of the noninhibited and inhibited bacterial enzymes are significantly raised in loops A266–A277 (FabF) and G266–E275 (FabB), indicating the induced fit of the PLM and the TLM molecules. This main chain region, corresponding to K1413–K1423 of the FAS KS, also accounts for some of the largest differences in the three superimposed proteins (Fig. 4B). The beginning of the FAS KS loop is extended as compared to the FabF and FabB structures, creating a bulge that interacts with the neighboring α-chain, whereas the end of the loop is shorter than in both of the bacterial enzymes. Although the side chain of the loop that forms the most extensive interactions with the two inhibitor molecules (P272/P272/P1421) is conserved in all three structures, these main chain differences could well represent a rationale for the observed fungal resistance toward both PLM and TLM.
Fig. 4.
TLM and PLM. (A) Induced fit binding of TLM and PLM. Root mean squared deviations between FabF with and without PLM (PDB codes 2GFX and 2GFV) and FabB with and without TLM (PDB codes 1FJ8 and 1FJ4). (B) Superposition of the FabF–PLM (green) and FabB–TLM (gold) complexes onto the FAS–CER structure (blue/red). A structure-based sequence alignment around the FAS KS loop, K1413–K1423, is included for reference.
Affinity Versus Inhibition Efficiency.
The binding affinity of a drug molecule to its target protein is a key characteristic for an inhibitor. However, inhibition efficiency is also defined by other parameters, as, for example, the effective concentration in the cell interior. Indeed, for some organisms resistance toward the inhibitors TLM and CER was revealed to be caused by efflux pumps, which keep the inhibitor concentration below a critical level (44–46). In the case of FAS KS inhibition, the properties of the KS condensation mechanism and the compartmentalization of the type I fungal fatty acid synthase system might also affect inhibitor efficiency. KS inhibitors can generally be classified by their mode of action. “Ping” inhibitors like CER inhibit the initial transfer of the acyl moiety onto the catalytic cysteine, whereas “pong” inhibitors block the binding of the malonyl-ACP (Fig. S1). Accordingly, inhibition of the S. cerevisiae FAS by the CER analog iodoacetamide was reported to be impaired by preincubation with acetyl-CoA (47). TLM was in contrast shown to compete with malonyl-ACP for binding to the malonyl binding site (13). PLM attacked both the malonyl- and the phosphopantetheine-binding sites and, thus, is likely to compete not only with acyl-ACP and malonyl-ACP but also with unloaded ACP.
In the fungal FAS, the domains of the fatty acid synthase cycle are kept in a small compartment that enhances their concentrations. Although ACP is reported to be one of the most abundant proteins in E. coli, with a cellular concentration of ≈100 μM (48), the ACP concentrations in the fungal fatty acid type I system exceed this level by at least an order of magnitude, given the 70–100 Å radius of the FAS reaction chambers. In addition, the restricted movement of the ACP by the flexible linkers, as well as the observed intrinsic affinity between the ACP and the KS domain, further increase the effective concentration of the ACP. Thus, a FAS KS inhibitor must compete with very high concentrations of ACP and ACP-associated intermediates. Because the FAS type I system is regulated by the concentration of malonyl-CoA via acetyl-CoA carboxylase (49), a down-regulated FAS complex will have acylated KS domains. This implies that ping and pong FAS inhibitors will not only face different types of substrate competition, the competition rate will also differ from a FAS in resting state compared with a FAS working at a high rate.
Concluding Remark.
In this report, we present an independent structure solution of the S. cerevisiae fatty acid synthase multienzyme and provide a first structural study of an inhibited FAS type I system. The inhibitor complex structure explains some of the biochemical properties of CER inhibition, gives insight into the FAS chain length determination, and paves a way for modeling and structure based drug design of other eukaryotic fatty acid synthase type I systems.
Materials and Methods
Cloning and Purification.
To facilitate the purification of the S. cerevisiae FAS, we modified the FAS1 gene to code for a C-terminally His8-tagged β-chain. Before crystallization, fatty acids of the purified FAS were elongated and the FAS enzymatic activity inhibited by CER (Fig. S2). For a detailed description of cloning, purification, and synchronization of the S. cerevisiae FAS, see SI Materials and Methods.
Crystallization, Data Collection, and Processing.
Diffracting crystals of the CER inhibited FAS grew in 0.1 M Hepes (pH 7), 1 M ammonium sulfate and PEG of various chain lengths at 4°C. X-ray crystallographic data of native and Ta6Br12- and W18-soaked crystals were collected at beamlines X10SA (Swiss Light Source, Villigen, Switzerland) and ID14 EH1–4, ID23 EH1–2, and ID29 (European Synchrotron Radiation Facility, Grenoble, France) to resolutions of 4, 6, and 7 Å, respectively (Table S1). A number of heavy atom cluster sites were located by means of direct and difference Fourier methods and refined as spherically averaged superatoms. Initial MIRAS phases were improved by using a combination of solvent-flipping, solvent-flattening, averaging, and statistical density modification to a resolution of 4.5 Å (Fig. S5). The complete structure was built by using the S. cerevisiae fatty acid synthase [PDB ID code 2UV8 (19)] and subjected to NCS-restrained domain-wise, rigid-body refinement with grouped B-factors. For a detailed description of phasing and refinement, see SI Materials and Methods with the data collection and refinement statistics in Table S1. For a brief comparison of the S. cerevisiae fatty acid synthase structures, see also Structural Differences of S. cerevisiae FAS Models in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Ronnald Vollrath for purification and crystallization of FAS and Kornelius Zeth for initial data collection. We are grateful to Ehmke Pohl and Clemens Schulze-Briese (Swiss Light Source), as well as Joanne McCarthy and Didier Nurizzo (European Synchrotron Radiation Facility), for beamline support. We also thank Deryck Mills for help with electron microscopy, Rasmus Schroeder for advice on image processing, Remco Wouts for computer support, and Werner Kühlbrandt for discussions.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors of the CER-inhibited FAS structure have been deposited in the protein data bank, www.pdb.org (PDB ID code 2VKZ).
This article contains supporting information online at www.pnas.org/cgi/content/full/0805827105/DCSupplemental.
References
- 1.Schweizer E, Hofmann J. Microbial type I fatty acid synthases (FAS): Major players in a network of cellular FAS systems. Microbiol Mol Biol Rev. 2004;68:501–517. doi: 10.1128/MMBR.68.3.501-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee SH, Stephens JL, Paul KS, Englund PT. Fatty acid synthesis by elongases in trypanosomes. Cell. 2006;126:691–699. doi: 10.1016/j.cell.2006.06.045. [DOI] [PubMed] [Google Scholar]
- 3.Campbell JW, Cronan JE., Jr Bacterial fatty acid biosynthesis: Targets for antibacterial drug discovery. Annu Rev Microbiol. 2001;55:305–332. doi: 10.1146/annurev.micro.55.1.305. [DOI] [PubMed] [Google Scholar]
- 4.Heath RJ, White SW, Rock CO. Inhibitors of fatty acid synthesis as antimicrobial chemotherapeutics. Appl Microbiol Biotechnol. 2002;58:695–703. doi: 10.1007/s00253-001-0918-z. [DOI] [PubMed] [Google Scholar]
- 5.Wright HT, Reynolds KA. Antibacterial targets in fatty acid biosynthesis. Curr Opin Microbiol. 2007;10:447–453. doi: 10.1016/j.mib.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee A, et al. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 7.McMurry LM, Oethinger M, Levy SB. Triclosan targets lipid synthesis. Nature. 1998;394:531–532. doi: 10.1038/28970. [DOI] [PubMed] [Google Scholar]
- 8.Hata T, Matsumae A, Nomura S, Kim T, Ryan K. Study of new antifungal antibiotic. Japan J Bact. 1960;15:1075–1076. [Google Scholar]
- 9.Oishi H, et al. Thiolactomycin, a new antibiotic I Taxonomy of the producing organism, fermentation and biological properties. J Antibiot. 1982;35:391–395. doi: 10.7164/antibiotics.35.391. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, et al. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature. 2006;441:358–361. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- 11.D'Agnolo G, Rosenfeld IS, Awaya J, Omura S, Vagelos PR. Inhibition of fatty acid synthesis by the antibiotic cerulenin. Specific inactivation of beta-ketoacylacyl carrier protein synthetase. Biochim Biophys Acta. 1973;326:155–166. doi: 10.1016/0005-2760(73)90241-5. [DOI] [PubMed] [Google Scholar]
- 12.Kauppinen S, Siggaard-Andersen M, von Wettstein-Knowles P. beta-ketoacyl-ACP synthase I of Escherichia coli: Nucleotide sequence of the fabB gene and identification of the cerulenin binding residue. Carlsberg Res Commun. 1988;53:357–370. doi: 10.1007/BF02983311. [DOI] [PubMed] [Google Scholar]
- 13.Nishida I, Kawaguchi A, Yamada M. Effect of thiolactomycin on the individual enzymes of the fatty acid synthase system in Escherichia coli. J Biochem. 1986;99:1447–1454. doi: 10.1093/oxfordjournals.jbchem.a135614. [DOI] [PubMed] [Google Scholar]
- 14.White SW, Zheng J, Zhang Y-M, Rock CO. The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem. 2005;74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y-M, White SW, Rock CO. Inhibiting bacterial fatty acid synthesis. J Biol Chem. 2006;281:17541–17544. doi: 10.1074/jbc.R600004200. [DOI] [PubMed] [Google Scholar]
- 16.Maier T, Jenni S, Ban N. Architecture of mammalian fatty acid synthase at 4.5 Å resolution. Science. 2006;311:1258–1262. doi: 10.1126/science.1123248. [DOI] [PubMed] [Google Scholar]
- 17.Jenni S, Leibundgut M, Maier T, Ban N. Architecture of a fungal fatty acid synthase at 5 Å resolution. Science. 2006;311:1263–1267. doi: 10.1126/science.1123251. [DOI] [PubMed] [Google Scholar]
- 18.Jenni S, et al. Structure of fungal fatty acid synthase and implications for iterative substrate shuttling. Science. 2007;316:254–261. doi: 10.1126/science.1138248. [DOI] [PubMed] [Google Scholar]
- 19.Leibundgut M, Jenni S, Frick C, Ban N. Structural basis for substrate delivery by acyl carrier protein in the yeast fatty acid synthase. Science. 2007;316:288–290. doi: 10.1126/science.1138249. [DOI] [PubMed] [Google Scholar]
- 20.Lomakin IB, Xiong Y, Steitz TA. The crystal structure of yeast fatty acid synthase, a cellular machine with eight active sites working together. Cell. 2007;129:319–332. doi: 10.1016/j.cell.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 21.Wenz P, Schwank S, Hoja U, Schueller H-J. A downstream regulatory element located within the coding sequence mediates autoregulated expression of the yeast fatty acid synthase gene FAS2 by the FAS1 gene product. Nucleic Acids Res. 2001;29:4625–4632. doi: 10.1093/nar/29.22.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang W, et al. Crystal structure of beta-ketoacyl-acyl carrier protein synthase II from E. coli reveals the molecular architecture of condensing enzymes. EMBO J. 1998;17:1183–1191. doi: 10.1093/emboj/17.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsen JG, et al. The x-ray crystal structure of beta-ketoacyl [acyl carrier protein] synthase I. FEBS Lett. 1999;460:46–52. doi: 10.1016/s0014-5793(99)01303-4. [DOI] [PubMed] [Google Scholar]
- 24.Heath RJ, Rock CO. The claisen condensation in biology. Nat Prod Rep. 2002;19:581–596. doi: 10.1039/b110221b. [DOI] [PubMed] [Google Scholar]
- 25.von Wettstein-Knowles P, Olsen JG, McGuire KA, Henriksen A. Fatty acid synthesis. Role of active site histidines and lysine in Cys-His-His-type beta-ketoacyl-acyl carrier protein synthases. FEBS J. 2006;273:695–710. doi: 10.1111/j.1742-4658.2005.05101.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y-M, Hurlbert J, White SW, Rock CO. Roles of the active site water, histidine 303, and phenylalanine 396 in the catalytic mechanism of the elongation condensing enzyme of Streptococcus pneumoniae. J Biol Chem. 2006;281:17390–17399. doi: 10.1074/jbc.M513199200. [DOI] [PubMed] [Google Scholar]
- 27.Bills GF, Platas G, Gams W. Conspecificity of the cerulenin and helvolic acid producing ‘Cephalosporium caerulens', and the hypocrealean fungus Sarocladium oryzae. Mycol Res. 2004;108:1291–1300. doi: 10.1017/s0953756204001297. [DOI] [PubMed] [Google Scholar]
- 28.Price AC, et al. Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism. J Biol Chem. 2001;276:6551–6559. doi: 10.1074/jbc.M007101200. [DOI] [PubMed] [Google Scholar]
- 29.Nomura S, Horiuchi T, Hata T, Omura S. Inhibition of sterol and fatty acid biosyntheses by cerulenin in cell-free systems of yeast. J Antibiot. 1972;25:365–368. doi: 10.7164/antibiotics.25.365. [DOI] [PubMed] [Google Scholar]
- 30.Omura S. The antibiotic cerulenin, a novel tool for biochemistry as an inhibitor of fatty acid synthesis. Bacteriol Rev. 1976;40:681–697. doi: 10.1128/br.40.3.681-697.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhajda FP, et al. Fatty acid synthesis: A potential selective target for antineoplastic therapy. Proc Natl Acad Sci USA. 1994;91:6379–6383. doi: 10.1073/pnas.91.14.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhajda FP, et al. Synthesis and antitumor activity of an inhibitor of fatty acid synthase. Proc Natl Acad Sci USA. 2000;97:3450–3454. doi: 10.1073/pnas.050582897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loftus TM, et al. Reduced food intake and body weight in mice treated with fatty acid synthase inhibitors. Science. 2000;288:2379–2381. doi: 10.1126/science.288.5475.2379. [DOI] [PubMed] [Google Scholar]
- 34.Moche M, Schneider G, Edwards P, Dehesh K, Lindqvist Y. Structure of the complex between the antibiotic cerulenin and its target, beta-ketoacyl-acyl carrier protein synthase. J Biol Chem. 1999;274:6031–6034. doi: 10.1074/jbc.274.10.6031. [DOI] [PubMed] [Google Scholar]
- 35.Morisaki N, et al. Effect of side-chain structure on inhibition of yeast fatty-acid synthase by cerulenin analogs. Eur J Biochem. 1993;211:111–115. doi: 10.1111/j.1432-1033.1993.tb19876.x. [DOI] [PubMed] [Google Scholar]
- 36.Inokoshi J, et al. Cerulenin-resistant mutants of Saccharomyces cerevisiae with an altered fatty acid synthase gene. Mol Gen Genet. 1994;244:90–96. doi: 10.1007/BF00280191. [DOI] [PubMed] [Google Scholar]
- 37.Christensen CE, Kragelund BB, von Wettstein-Knowles P, Henriksen A. Structure of the human beta-ketoacyl [ACP] synthase from the mitochondrial type II fatty acid synthase. Protein Sci. 2007;16:261–272. doi: 10.1110/ps.062473707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Joshi AK, Hofmann J, Schweizer E, Smith S. Cloning, expression, and characterization of the human mitochondrial beta-ketoacyl synthase: Complementation of the yeast CEM1 knock-out strain. J Biol Chem. 2005;280:12422–12429. doi: 10.1074/jbc.M413686200. [DOI] [PubMed] [Google Scholar]
- 39.Aritomi K, et al. Self-cloning yeast strains containing novel FAS2 mutations produce a higher amount of ethyl caproate in Japanese sake. Biosci Biotechnol Biochem. 2004;68:206–214. doi: 10.1271/bbb.68.206. [DOI] [PubMed] [Google Scholar]
- 40.Sumper M, Oesterhelt D, Riepertinger C, Lynen F. Synthesis of various carboxylic acids by the multienzyme complex of fatty acid synthesis from yeast, and clarification of their structure. Eur J Biochem. 1969;10:377–387. doi: 10.1111/j.1432-1033.1969.tb00701.x. [DOI] [PubMed] [Google Scholar]
- 41.Pirson W, Schuhmann L, Lynen F. Specificity of yeast fatty acid synthetase with respect to the priming substrate. Decanoyl-CoA and derivatives as primers of fatty acid synthesis in vitro. Eur J Biochem. 1973;36:16–24. doi: 10.1111/j.1432-1033.1973.tb02879.x. [DOI] [PubMed] [Google Scholar]
- 42.Chakravarty B, Gu Z, Chirala SS, Wakil SJ, Quiocho FA. Human fatty acid synthase: Structure and substrate selectivity of the thioesterase domain. Proc Natl Acad Sci USA. 2004;101:15567–15572. doi: 10.1073/pnas.0406901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayashi T, Yamamoto O, Sasaki H, Kawaguchi A, Okazaki H. Mechanism of action of the antibiotic thiolactomycin: Inhibition of fatty acid synthesis of Escherichia coli. Biochem Biophys Res Commun. 1983;115:1108–1113. doi: 10.1016/s0006-291x(83)80050-3. [DOI] [PubMed] [Google Scholar]
- 44.Calabrese D, Bille J, Sanglard D. A novel multidrug efflux transporter gene of the major facilitator superfamily from Candida albicans (FLU1) conferring resistance to fluconazole. Microbiology. 2000;146:2743–2754. doi: 10.1099/00221287-146-11-2743. [DOI] [PubMed] [Google Scholar]
- 45.Furukawa H, Tsay JT, Jackowski S, Takamura Y, Rock CO. Thiolactomycin resistance in Escherichia coli is associated with the multidrug resistance efflux pump encoded by emrAB. J Bacteriol. 1993;175:3723–3729. doi: 10.1128/jb.175.12.3723-3729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schweizer HP. Intrinsic resistance to inhibitors of fatty acid biosynthesis in Pseudomonas aeruginosa is due to efflux: Application of a novel technique for generation of unmarked chromosomal mutations for the study of efflux systems. Antimicrob Agents Chemother. 1998;42:394–398. doi: 10.1128/aac.42.2.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oesterhelt D, Bauer H, Kresze GB, Steber L, Lynen F. Reaction of yeast fatty acid synthetase with iodoacetamide. 1. Kinetics of inactivation and extent of carboxamidomethylation. Eur J Biochem. 1977;79:173–180. doi: 10.1111/j.1432-1033.1977.tb11795.x. [DOI] [PubMed] [Google Scholar]
- 48.Tang L, Weissborn AC, Kennedy EP. Domains of Escherichia coli acyl carrier protein important for membrane-derived-oligosaccharide biosynthesis. J Bacteriol. 1997;179:3697–3705. doi: 10.1128/jb.179.11.3697-3705.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wakil SJ, Stoops JK, Joshi VC. Fatty acid synthesis and its regulation. Annu Rev Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.