Abstract
Human replication protein A (RPA) becomes phosphorylated on the RPA2 subunit by cyclin B-Cdc2 during mitosis, although the functional role of this modification is unclear. We find that this modification stimulates RPA2 to become hyperphosphorylated in response to mitotic DNA damage caused by bleomycin treatment. Cells in which endogenous RPA2 was replaced by a mutant subunit lacking both Cdc2 sites had a significant defect in mitotic release into a 2N G1 phase after exposure to bleomycin. An increased percentage of these mutant cells also was positive initially for cyclin B expression and BubR1 chromatin staining, indicative of an extended spindle assembly checkpoint. The mutant cells that experienced mitotic DNA damage also underwent apoptosis at higher levels than cells expressing the WT subunit. Even so, we did not find the mutation had any dramatic effects on the level of DNA repair in mitosis. Cells lacking ATM (a checkpoint factor and RPA2 kinase) also were severely defective in mitotic exit and were unable to support RPA hyperphosphorylation after mitotic DNA damage. Although checkpoint 1 effector kinase (Chk1) had a more complex role, inhibition of Chk1 activity with UCN-01 also reduced mitotic exit. Chk1 activation and mitotic RPA hyperphosphorylation were found to be independent events. Our results demonstrate that mitotic RPA hyperphosphorylation facilitates release of cells from a damaged mitosis into a 2N G1 phase, thereby increasing cell viability.
Keywords: checkpoint, mitosis, replication protein A, ATM, Chk1
DNA damage occurring during mitosis represents a daunting challenge to the integrity of the mammalian genome. Condensation of chromosomal DNA in preparation for cytokinesis constrains the activity of DNA repair reactions. Nevertheless, double-strand DNA breaks (DSBs) induced by exposure of prometaphase cells to ionizing radiation or topoisomerase II inhibitors cause a delay in mitotic exit (1, 2). The nature of this delay and its causes are only poorly understood.
An essential DNA replication factor that is involved intimately in the signaling and repair of DNA damage is replication protein A (RPA), a heterotrimeric single-stranded DNA-binding complex with subunits in human cells of 70 kDa (RPA1), 30 kDa (RPA2), and 14 kDa (RPA3) (3, 4). RPA acts during DNA replication stress conditions to recruit and activate the checkpoint kinase ATR (5, 6). The N terminus of the human RPA2 subunit also is a target of ATR and other members of the phosphatidylinositol 3-kinase-like kinase family (PIKKs; e.g., ATM and DNA-PK) and cyclin-Cdk complexes (Fig. 1A; e.g., refs. 7–14). RPA2 phosphorylation follows a preferred pathway so that initial modification by a kinase (e.g., cyclin A-Cdk2) stimulates the subsequent modification by another kinase (e.g., DNA-PK) (14). Recent cell-based studies have shown that human RPA activity in DNA replication and repair is regulated by changes in the pattern of RPA phosphorylation. For example, human cells in which endogenous RPA2 has been replaced with a RPA2 variant mutated at the two RPA2 cyclin-Cdk sites (S23A/S29A-RPA2) are defective in the repair of DSBs occurring during interphase (14).
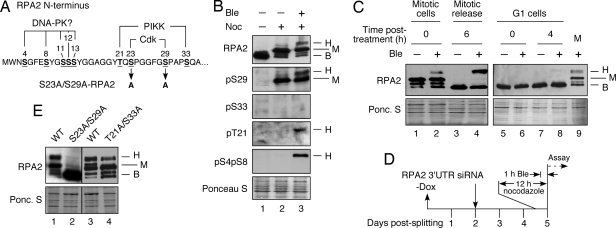
Fig. 1.
Mitotic phosphorylation of RPA2 cyclin-Cdk sites stimulates subsequent stress-dependent modification. (A) Schematic of RPA2 phosphorylation sites. (B) Lysates were isolated from asynchronous nonstressed cells (lane 1) or from nocodazole-arrested cells isolated by mitotic shake-off and then were mock treated (lane 2) or were treated with Ble (2 μg/ml) for 1 h (lane 3). (C) Nocodazole-arrested cells, either treated with Ble (2 μg/ml) for 1 h or not treated, were released into medium lacking both agents and were collected 6 h after release. To examine G1 cells, mitotic cells were released from the nocodazole block and 3 h after release were treated with Ble (2 μg/ml) for 1 h or were mock treated. Cells were collected immediately (0 h) or 4 h after the medium was replaced with medium lacking Ble. (D) Schematic indicating steps involved in cellular RPA2 replacement. −Dox = removal of doxycycline causing the induction of ectopic RPA2 expression. (E) Endogenous RPA2 was silenced in U2-OS cells replacing S23A/S29A-, T21A/S33A-, or WT-RPA2. After silencing (48 h), cells were treated with nocodazole for 12 h. The mitotic cells were collected by shake-off and were treated with Ble (2 μg/ml) for 1 h, followed by collection of cells. For all samples, lysates were prepared from the collected cells and analyzed by Western blotting using the indicated antibodies. The RPA2 species are indicated as follows: B, basal; H, hyperphosphorylated; M, mitotic.
It has been found previously that RPA also undergoes constitutive modification of the two cyclin-Cdk sites during mitosis (15), suggesting a possible role of RPA phosphorylation in this cell-cycle phase. Examining mitotic RPA2 phosphorylation by cyclin B-Cdc2, we found this modification primes RPA2 to undergo additional phosphorylation events in response to genotoxic stress. The hyperphosphorylated RPA2 stimulates the release of damaged mitotic cells into G1 with normal 2N DNA content and increases cell viability.
Results
Mitotic Phosphorylation Primes RPA for Subsequent Modification.
Although mitotic RPA2 is phosphorylated by cyclin B-Cdk1 on S23 and S29 (14–16), it was unclear whether this RPA species also undergoes additional phosphorylation events in response to mitotic DNA damage. To test this, U2-OS cells were arrested in prometaphase with nocodazole, leading to formation of the mitotic RPA2 species (Fig. 1B, lane 2). Treatment of these mitotic cells with bleomycin (Ble) to cause DSBs (17) led to additional RPA2 modification at the S4/S8 and T21 residues (lane 3). S33 was not detectably modified, in contrast to observed formation of pS33 in S and G2 cells after Ble treatment (14). The additional phosphorylation events cause a significant percentage of the RPA2 pool to migrate at the hyperphosphorylated position. RPA2 therefore can become hyperphosphorylated in response to mitotic DNA damage.
We investigated whether the mitotic conditions sensitize RPA to become hyperphosphorylated in response to DNA damage. Nocodazole-arrested cells were treated with Ble for 1 h, and the RPA2 phosphorylation pattern then was assayed either immediately or 6 h after the medium was replaced with medium lacking nocodazole and Ble. As found previously, Ble treatment led to RPA2 hyperphosphorylation (Fig. 1C, lane 2). On release from the mitotic block, RPA2 from nondamaged cells reverted to a basal hypophosphorylated species (lane 3), but, even while losing the mitotic RPA2 form (lane 4), cells that were damaged in mitosis had a significant increase in the amount of hyperphosphorylated RPA2. This hyperphosphorylation was detected for at least 8 h after nocodazole release and disappeared thereafter [supporting information (SI) Fig. S1A]. In mitotic cells, most of the hyperphosphorylated RPA2 was negative for chromatin association (Fig. S1 B and C). After release from the nocodazole block (4 h), significant pS4pS8-RPA2 chromatin association was seen by immunofluorescence microscopy (Fig. S1C). RPA2 also was found in the insoluble cellular fraction by Western blot analysis (data not shown) but diminished thereafter. These data indicate that RPA has only limited effects on DNA repair in mitotic cells but plays a more significant role after mitotic release. In contrast to mitotic cells, Ble treatment of G1 cells caused no significant RPA2 hyperphosphorylation immediately after treatment (Fig. 1C, lane 6). Even at later times (4 h) after treatment, only a weak RPA2 hyperphosphorylation signal was seen (lane 8), and this signal was significantly less than that seen in mitotic cells treated with Ble (lane 9). These data indicate that mitotic cells are better able to induce RPA2 hyperphosphorylation in response to Ble treatment than are cells in G1.
We previously observed that phosphorylation of the two Cdk sites on RPA2 facilitated subsequent modification of S4, S8, and T21 by DNA-PK or a DNA-PK-dependent kinase in interphase cells (14). We therefore tested whether the formation of pS23/pS29 stimulated the RPA2 hyperphosphorylation seen after mitotic DNA damage. To achieve this, we used our RPA2 replacement strategy using U2-OS clones stably transfected with an inducible copy of control or mutant RPA2 (Fig. 1D) (14). Expression of ectopic RPA2 is coupled with knockdown of the endogenous RPA2 by the use of an siRNA specific to the endogenous message. As seen in our previous studies (14, 18), expression of the ectopic RPA2 is similar to that of the nonsilenced endogenous subunit (Fig. S2A). Tests of mitotic cells expressing an S23A/S29A-RPA2 mutant revealed that the mutant was almost completely defective in undergoing hyperphosphorylation in response to Ble treatment (Fig. 1E, lane 2). In contrast, a T21A/S33A-RPA2 variant mutated at both PIKK sites showed significant RPA2 hyperphosphorylation (lane 4). These data indicate that phosphorylation of the 2 Cdk sites stimulates additional phosphorylation events after mitotic DNA damage. Mutation of the two PIKK sites has a lesser effect, presumably because S33 is not targeted for modification in mitotic cells.
Mitotic RPA2 Phosphorylation Stimulates Release of Damaged Cells into a 2N G1 Phase.
We examined the effects of mitotic DNA damage on cell-cycle progression in the parental U2-OS cell line. After mitotic Ble treatment, cells were released into medium lacking both Ble and nocodazole for various periods of time, and the DNA content profile was determined by FACS (Fig. S2B). Although ≈80% of nondamaged cells entered a 2N G1 phase at 6 h after release, this percentage was reduced to 32% after mitotic damage. The percentage of cells that became positive for bromodeoxyuridine incorporation 20 h after release also decreased after mitotic damage (i.e., ≈15% of damaged cells compared with ≈25% of nondamaged cells; Table S1), although the average levels of bromodeoxyuridine incorporation in these positive cells were similar (data not shown). Testing the mitotic marker mitotic protein monoclonal 2 (MPM2), we observed that ≈5% of the damaged cells remained in mitosis at 3 h, but this percentage returned to control levels at 6 h (Table S2). Collectively, these data indicate that mitotic DNA damage causes a delay in mitotic exit. At later times, a significant percentage of cells underwent a failed cytokinesis and left mitosis with a 4N DNA content, a phenomenon seen by others (19). Our FACS analysis did not reveal significant levels of cells with increased ploidy at these later times (e.g., 8N; data not shown), indicating that the postmitotic 4N cells do not subsequently enter S phase.
We examined the functional consequences of mitotic RPA2 phosphorylation on the mitotic response to DNA damage. Mitotic U2-OS cells in which endogenous RPA was replaced with WT- or S23A/S29A-RPA2 were treated with Ble and released into medium lacking both Ble and nocodazole. The DNA content then was determined by FACS. Six hours after release, ≈57% of Ble-treated cells replaced with WT-RPA2 retained a 4N DNA content when damaged in mitosis, compared with 22% of WT-RPA2 cells not treated with Ble (Fig. 2A). Such data are consistent with reports by others that mitotic damage slows mitotic exit (20). After DNA damage, a greater percentage (77%) of cells replaced with S23A/S29A-RPA2 than with WT-RPA2 cells retained a 4N DNA content. Even 20 h after release, the Ble-treated mutant cells showed a similar higher fraction of 4N cells (72% vs. 46%, respectively; Fig. S3A). Thus, cells expressing an RPA2 phosphorylation mutant show a defective release into a 2N G1 phase after mitotic DNA damage.
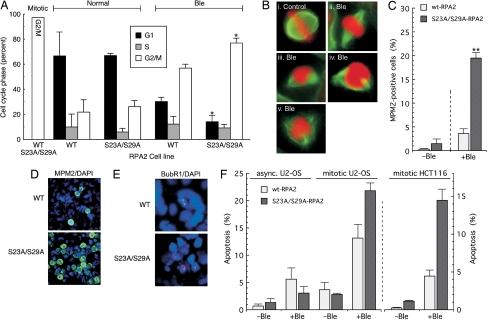
Fig. 2.
Loss of mitotic RPA2 phosphorylation inhibits mitotic exit and stimulates cellular apoptosis. (A) Endogenous RPA2 was silenced in U2-OS cells ectopically expressing WT- or S23A/S29A-RPA2. After silencing (48 h), cells were treated with nocodazole for 12 h; then mitotic cells were collected by mechanical shake-off. Cells were mock treated or treated with Ble (2 μg/ml Ble for 1 h), followed by release into medium lacking nocodazole and Ble. Cells were collected 7 h after release and were analyzed by FACS for DNA content. (B) After mitotic release (2 h), cells expressing S23A/S29A- or WT-RPA2 were stained with α-tubulin (to visualize spindles) and 7-amino-actinomycin D (to stain DNA). i shows untreated cells undergoing metaphase. Panels ii through v show Ble-treated cells displaying abnormal spindle assembly and globular DNA appearance. (C and D) Cells were silenced for endogenous RPA2 and ectopic RPA2 was induced. After silencing (48 h), cells were treated with nocodazole for 12 h and were mock treated or treated with Ble (2 μg/ml for 1 h). Then the medium was replaced with medium lacking nocodazole and Ble for 3 h. Cells then were stained and imaged for MPM2 (green) and DAPI (blue) and were quantitated. (E) Mitotic cells expressing ectopic RPA2 and silenced for endogenous RPA2 were mock treated or treated with Ble (2 μg/ml for 1 h) and, 3 h after release from nocodazole and Ble, were stained for BubR1 (red) and DAPI (blue). (F) Asynchronous U2-OS clones were mock treated or treated with Ble (30 μg/ml Ble for 2.5 h); then the medium was replaced with fresh medium. Cells then were analyzed for apoptosis by a TUNEL assay 20 h after release. Mitotically arrested cells were treated with 10 μg/ml Ble for 1 h and then were similarly treated. An identical treatment was used for HCT116 clones, except that cells were scored 8 h after release.
Cells examined after release from a Ble-treated mitosis invariably had unusual mitotic figures (Fig. 2B). Rather than a symmetrical pattern of mitotic spindles and an orderly alignment of chromosomes along the equatorial plate, as seen in nondamaged cells (Fig. 2Bi), Ble-treated cells often had asymmetric spindles (Fig. 2Biii–v). Such cells also showed a broad orientation of chromosomes on the metaphase plate (Fig. 2Bii) or had a globular characteristic to the DNA without observable chromosomal structure (Fig. 2B ii–v; Fig. S3B). Even so, a high percentage of cells with this globular DNA appearance stained positive for the mitotic marker MPM2 (Fig. S3C). The abnormal mitotic figures were seen in Ble-treated cells regardless of the RPA2 variant expressed. Although a larger percentage of cells replaced with S23A/S29A-RPA2 showed spindle defects (data not shown), this presumably reflects the greater percentage of mutant cells that are stalled in mitosis after DNA damage.
It is possible that the loss of mitotic RPA phosphorylation inhibits release of cells from the spindle assembly checkpoint under conditions of mitotic DNA damage. Such a mechanism leads to the prediction that mutant cells would have increased staining of the mitotic marker MPM2 and the spindle assembly checkpoint factor BubR1. To test, we found that, in the absence of Ble treatment, only a small percentage of cells showed MPM2 staining 3 h after release (Fig. 2C). Although the percentage of MPM2-positive cells expressing WT-RPA2 increased to 3.7% after Ble treatment, nearly one-fifth (19.5%) of cells replaced with S23A/S29A-RPA2 were MPM2 positive (Fig. 2 C and D). At later times, the percentage of MPM2-positive cells decreased (data not shown) even though the 4N fraction remained elevated (see Fig. S3A), again indicating that cells eventually decay from the mitotic block and enter a tetraploid state. Nearly all MPM2-positive cells also were positive for cyclin B staining (Fig. S4A Upper). However, we did observe cells that appeared to have entered interphase with a G2-like state and showed a flat nuclear morphology (e.g., see ref. 19). These cells were positive for cyclin B and negative for MPM2 staining (Fig. S4A Lower). When the RPA2 variants were tested in HCT116 cells, the percentage of of cyclin B-positive cells expressing S23A/S29A-RPA2 was larger than the percentage expressing WT-RPA2 (Fig. S4 B and C), indicating that the effects of the RPA2 mutant also occur in a different cell line.
Examining BubR1 in Ble-treated cells, we saw that the fraction of cells showing BubR1 chromatin staining was significantly higher in cells expressing the mutant RPA2 than in those expressing WT-RPA2 (Fig. 2E). In the absence of Ble treatment, the number of cells expressing either WT- or S23A/S29A-RPA2 and showing BubR1 staining was insignificant (data not shown). In sum, these data suggest that the cells expressing mutant RPA2 are defective in release from a DNA damage-dependent spindle assembly checkpoint, causing these cells to be maintained in mitosis.
The effect of RPA phosphorylation on cellular apoptosis after mitotic DNA damage was examined. Mitotic U2-OS cells replaced with WT- or S23A/S29A-RPA2 were treated with Ble and then released from the nocodazole block in the absence of Ble. In the absence of Ble treatment, similar levels of TUNEL-positive cells were observed, regardless of the RPA2 variant expressed (Fig. 2F). In contrast, although mitotic Ble treatment caused an ≈3.5-fold increase in WT-RPA2 cells undergoing apoptosis, cells replaced with the S23A/S29A-RPA2 mutant showed a nearly 8-fold increase in apoptosis figures. A similar difference was observed when HCT116 clones were tested (Fig. 2F). Apoptosis in the mitotic mutant cells treated with Ble also was ≈8-fold higher than in treated asynchronous mutant cells, even though the mitotic cells were exposed to 3-fold less drug (Fig. 2F). Along with being defective in G1 entry, cells expressing the S23A/S29A-RPA2 mutant had increased levels of apoptosis after mitotic DNA damage.
Mitotic RPA Phosphorylation Has Minor Effects on DNA Repair.
It has been observed that interphase RPA phosphorylation stimulates chromosomal DNA repair (14). We therefore investigated whether the increased apoptosis and reduced entry to a 2N G1 phase in cells expressing the mutant RPA2 was a result of defective association of DNA repair proteins with DNA. Immediately after Ble treatment in mitotic cells, we noted a selective increase in the amount of insoluble RPA2 (i.e., chromatin-bound RPA; ref. 21) in cells replaced with S23A/S29A-RPA2 (Fig. 3A). (Note also the lack of detectable mitotic RPA2 phosphorylation, again attesting to the efficiency of RPA2 replacement.) Under these conditions, however, the RPA2 mutation did not have a significant effect on the amount of chromatin-bound Ku80 (a component of the non-homologous end-joining pathway) (Fig. 3A), and no Rad51 binding was detected for either RPA2 variant regardless of Ble treatment (data not shown). As an indicator of the amounts of DNA damage and DNA repair, we assayed γH2AX levels. At the initial time point (0 h), we observed only a slight (1.3-fold) increase in γH2AX levels in Ble-treated cells replaced with S23A/S29A-RPA2 compared with WT-RPA2 (Fig. 3A).
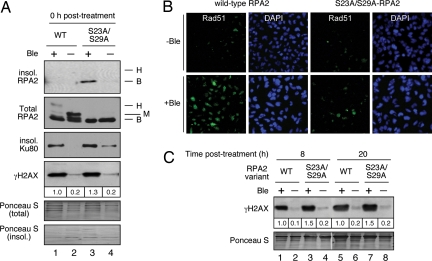
Fig. 3.
DNA damage and repair factors following mitotic DNA damage. (A) Mitotic cells expressing WT- or S23A/S29A-RPA2 were mock treated or treated with Ble (2 μg/ml for 1 h) in the presence of nocodazole, followed by extraction of the soluble fraction with CSK buffer containing 0.5% Triton X-100. The insoluble and total fractions then were subjected to Western blot analysis using indicated antibodies. (B) Mitotic cells, as described in A, were released from nocodazole and Ble treatment and were fixed 8 h after release. Cells were then analyzed for Rad51 chromatin association by immunofluorescence microscopy. (C) Mitotic cells as described in A were released into fresh media lacking nocodazole and Ble for either 8 or 20 h and then were isolated. Samples were analyzed for γH2AX levels by Western blot.
Eight hours after Ble treatment, the most significant effect was an increase in the level of Rad51 chromatin binding in cells expressing WT-RPA2 (Fig. 3B). Cells replaced with the mutant RPA2 had comparatively weaker Rad51 chromatin staining that was only slightly higher than seen in nondamaged cells. This increased Rad51 staining in cells replaced with WT-RPA2 occurs in interphase, because at this time point, nearly all cells have exited mitosis (Table S2 and data not shown). The modest increase in γH2AX levels in the mutant cells was maintained through the two later time points (Fig. 3C). Combined, our data show only small differences in γH2AX levels and Ku80 chromatin binding in damaged mitotic cells expressing either WT- or S23A/S29A-RPA2. Significant effects of the RPA2 mutation on DNA repair are seen only after cells re-enter interphase. We therefore argue that increased G1 entry of WT-RPA2 cells after mitotic DNA damage is unlikely to result solely from greater DNA repair.
Role of Checkpoint Kinases in the Response to Mitotic DNA Damage.
ATM is a key sensor of DSBs (22) and therefore probably is a factor mediating the response to mitotic DNA damage. We examined the response of mitotic cells lacking the ATM kinase (AT cells) to Ble treatment. Similar to the effect seen in U2-OS cells, entry into G1 was lower for Ble-treated mitotic control cells (i.e., cells expressing ATM) than for untreated cells (4N/2N = 2.5 and 0.4, respectively) (Fig. 4A). Surprisingly, compared with untreated WT cells, G1 entry was reduced significantly for mitotic AT cells that were not exposed to Ble (4N/2N = 2.3). We take these results to indicate that the compromised G2/M checkpoint (23) allows AT cells with endogenous DNA damage to enter mitosis, which then causes a partial mitotic arrest. After Ble treatment, only a small fraction of AT cells entered G1 (5%; 4N/2N = 15). When the influence of ATM on mitotic RPA2 hyperphosphorylation in response to DNA damage was examined, AT cells were seen to have no apparent RPA2 migrating at the hyperphosphorylated position (Fig. 4B). This result is consistent with the defective RPA2 phosphorylation in interphase AT cells seen by others (8). Note that the partial reduction of mitotic phosphorylation (e.g., lane 4) probably results from a minor loss of phosphate during extract preparation that we occasionally observe. Overall, AT cells have a reduced ability to exit mitosis. The action of ATM can be explained, at least in part, by its ability to support RPA2 hyperphosphorylation in response to mitotic DNA damage.
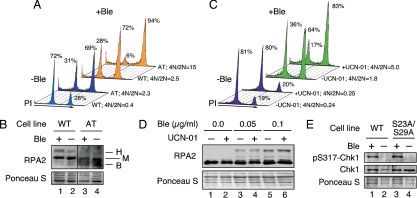
Fig. 4.
Chk1 and ATM enhance release into G1 on release from genotoxic stress during mitosis. (A) Mitotic WT (GM637) or AT cells (GM5849) were mock treated or treated with Ble (0.05 μg/ml for 1 h). Cells were released into medium lacking nocodazole and Ble and, 3 h after release, were analyzed for DNA content by FACS. (B) Treatment of cells was performed as described in A. Immediately after treatment, samples were collected and analyzed by Western blot for RPA2. (C) Nocodazole-arrested U2-OS cells were mock treated or treated with Ble (0.05 μg/ml for 1 h) in the presence or absence of 300 nM UCN-01. Cells then were released into medium lacking Ble but containing UCN-01 for 3 h and were analyzed by FACS for DNA content. (D) Samples prepared as described in C were treated with either 0.05 or 0.1 μg/ml Ble and were analyzed for RPA2 (and RPA2 hyperphosphorylation) by Western blot. (E) Cells replaced for WT- or S23A/S29A-RPA2 were mock treated or treated with Ble (10 μg/ml for 1 h). Lysates were prepared and analyzed for total Chk1 and pS317-Chk1 by Western blot. Similar results were observed using 2 μg/ml Ble (data not shown). B, basal (nonphosphorylated); H, hyperphosphorylated; M, mitotic.
Previous studies by others have found differing roles for checkpoint 1 effector kinase (Chk1) in the regulation of mitotic progression. Down-regulation of mammalian Chk1 by siRNA caused an increased release into G1 following mitotic DNA damage, indicating that Chk1 normally prevents cytokinesis in a damaged mitosis (24). In contrast, functional null mutations in the Drosophila grp gene (i.e., the Chk1 homolog) caused defects both in chromosome congression at metaphase and segregation at anaphase, leading to a failure in cytokinesis (25), suggesting that Chk1 stimulates mitotic exit. Generally similar results were found in Chk1-deficient chicken and human cells, tying Chk1 to the spindle assembly checkpoint (26). We therefore examined whether Chk1 also plays a role in the mitotic arrest seen after Ble treatment. Down-modulation of Chk1 by siRNA caused a small but reproducible increase in the 2N G1 population (Fig. S5 A and B), consistent with the results of Huang, et al. (24).
Surprisingly, use of the Chk1 inhibitor 7-hydroxystaurosporine (UCN-01) shows a more complex role for Chk1 in the response to mitotic DNA damage. Mitotic U2-OS cells were treated with Ble in the presence or absence of UCN-01. In the absence of Ble, UCN-01 had no apparent effect on mitotic release into G1 (Fig. 4C). Surprisingly, although Ble treatment led to a loss of cells entering G1 (4N/2N = 1.8), incubation of cells with UCN-01 led to a further reduction in G1 entry (4N/2N = 5.0). These data indicate that Chk1 activity is important for cells to exit mitosis into a 2N G1 phase after mitotic DNA damage.
Because the inhibition of Chk1 by UCN-01 and the mutation of RPA2 cyclin-Cdk sites yield similar effects in response to mitotic DNA damage, we examined whether the two pathways are directly linked. At either of the two Ble concentrations tested, no effect of UCN-01 on RPA phosphorylation was seen (Fig. 4D). Similar levels of activated Chk1 (i.e., Chk1 phosphorylated on S317) were observed in cells replaced with either WT- or S23A/S29A-RPA2 following Ble treatment (Fig. 4E). These data indicate that the effects of RPA2 hyperphosphorylation and Chk1 activation on mitotic release are separable and involve distinct pathways.
Discussion
DNA damage in a mitotic cell reduces the ability of cells to exit mitosis properly and to enter G1 with a 2N DNA content. Recent study of this process indicates an interplay between the DNA damage and spindle assembly checkpoints, with damage causing a fraction of mitotic cells to exit mitosis into a G2-like phase with 4N DNA content and cyclin B expression (19). Our study has shown that mitotic RPA hyperphosphorylation has a role both in stimulating cellular exit from a damaged mitosis into a normal 2N state G1 phase and in decreasing cell death from DNA damage-dependent apoptosis. G1 entry after mitotic DNA damage also is stimulated by ATM and by Chk1 activity, the latter acting in a pathway distinct from that involving RPA hyperphosphorylation.
Expression of the S23A/S29A-RPA2 mutant had robust effects on mitotic exit in response to mitotic DNA damage. We propose that mitotic phosphorylation at S23 and S29 primes RPA2 to undergo additional phosphorylation events in response to such damage. We and others previously demonstrated that RPA2 modification at particular sites stimulates the subsequent modification of other sites on the same RPA2 molecule (8, 9, 14). Hyperphosphorylated RPA2 therefore would be the active entity in regulating mitotic exit in response to DNA damage.
Although RPA phosphorylation stimulates DNA repair in interphase cells (14), the loss of mitotic phosphorylation results in only a slight increase in γH2AX levels. That RPA lacks a significant role in mitotic DNA repair is suggested also by the low levels of chromatin-associated RPA in mitotic cells following DNA damage. Our data suggest instead that the mitotic RPA2 hyperphosphorylation provides a signal that stimulates release of cells from a mitotic arrest caused by DNA damage. Upon the generation of this signal, the spindle assembly checkpoint can be relieved to allow mitotic exit into a 2N G1 phase and RPA-mediated DNA repair. In the absence of the hyperphosphorylation signal, the spindle assembly checkpoint is maintained, causing a larger fraction of cells to be arrested in mitosis. These cells eventually can decay into a 4N state but, because we do not observe further increases in ploidy, do not proceed subsequently into S phase. Because of the abnormal tetraploid state or because of reduced DNA repair after mitotic exit, cells expressing the mutant S23A/S29A-RPA2 mutant are prone to cell death through apoptosis.
Mikhailov, et al. observed that DNA damage caused defects in kinetochore attachment that activated the spindle assembly checkpoint, although in an ATM-independent manner (20). Because we observe that cells expressing WT-RPA2 have a reduced number of cells with BubR1 associated with chromosomal structures, it is possible that RPA phosphorylation either facilitates kinetochore attachment under damage conditions or, instead, reduces the sensitivity of spindle assembly checkpoint factors for kinetochore alterations.
The strong inhibitory effects of ATM mutation and Chk1 inactivation on mitotic exit to G1 further demonstrate a role for the DNA damage checkpoint machinery during mitosis. As suggested by others (24, 25) we found both positive and negative roles for the Chk1 kinase in response to mitotic DNA damage. That is, a reduction of Chk1 protein levels by siRNA increases exit from a damaged mitosis into a 2N G1 phase. In contrast, use of a Chk1 inhibitor (UCN-01) resulted in a severe reduction in the fraction of cells able to exit from a damaged mitosis. The simplest interpretation of these data is that the presence of the Chk1 protein, but not its activity, is necessary to inhibit mitotic exit in response to DNA damage (i.e., because UCN-01 reduces mitotic release). However, to facilitate mitotic exit, Chk1 activity is required. Recent work by Zachos et al. (26) led the authors to predict that Chk1 acts directly at kinetochores to sense tension, potentially explaining a need for the Chk1 protein. Once the spindle assembly checkpoint is activated, Chk1 activity may be required to down-regulate this mitotic checkpoint, allowing repair of DNA lesions in G1.
Materials and Methods
Cell Culture.
U2-OS and HCT116 cell lines were cultured in McCoy's medium containing 10% FBS. RPA2 mutagenesis, replacement, and silencing were performed as described by Anantha et al. (14). To silence Chk1, the siGenome SMARTpool (Dharmacon) specific to human Chk1 was used. The AllStars Negative Control siRNA (Qiagen) was used for the negative control. Human fibroblast cell lines positive (GM637) or null (GM5849) for ATM (Coriel Institute) were cultured in McCoy's medium containing 15% FBS. For synchronization experiments, cells were arrested in prometaphase by treatment with 100 ng/ml nocodazole (Sigma) for 12 h, and the mitotic cells were isolated by shake-off. Cells were mock treated or treated with Ble (Calbiochem) in the continued presence of nocodazole. Cells then were washed in PBS and released into medium devoid of nocodazole and Ble.
Immunoblotting and Antibodies.
The insoluble (i.e., chromatin-bound) fraction was obtained by washing cells with cytoskeletal (CSK) buffer (14), followed by treatment of cells with CSK buffer containing 0.5% (vol/vol) Triton X-100 for 5 min on ice. Cells then were centrifuged at 400 × g in a benchtop centrifuge at 4°C for 5 min to separate the soluble and insoluble fractions. The insoluble pellet was washed once with CSK buffer devoid of Triton X-100. The antibodies used in this study were against γH2AX (Upstate Biotechnology), Ku (p80) (Ab-7; NeoMarkers), Chk1 (Santa Cruz Biotechnology), general RPA2 (NeoMarkers), pT21 (Abcam), and pChk1, pS4/pS8-, pS33-, and pS29-RPA2 antibodies (Bethyl Laboratories). The pS29-RPA2 antibody was custom-synthesized as described in Anantha et al. (14). Western blot analysis was performed as indicated in Anantha et al. (14).
Immunofluorescence Microscopy.
For visualization of BubR1, cells were fixed with methanol and acetone (1:1) for 10 min on ice and then were stained with an anti-BubR1 antibody (BD Transduction Laboratories). For detection of chromatin-bound RPA, cells first were extracted with CSK buffer with 0.5% (vol/vol) Triton X-100 for 5 min on ice, then washed with CSK buffer lacking Triton X-100, and fixed with 4% (wt/vol) paraformaldehyde for 20 min. For detection of Rad51, paraformaldehyde-fixed cells were permeabilized by incubation in 10 mM Tris·HCl, pH 7.4, 150 mM NaCl, 1% (wt/vol) BSA, and 0.2% (vol/vol) Triton X-100 (TNB-T) for 30 min and then blocked in TNB (TNB-T without Triton X-100) for 30 min at room temperature. Cells then were stained with the anti-Rad51 antibody (Ab-1, Oncogene Research) in TNB. For detection of cyclin B and MPM2, cells first were fixed with 4% (wt/vol) paraformaldehyde followed by extraction with 0.5% (vol/vol) Triton X-100 in PBS. Cells then were stained with antibodies against cyclin B (a gift from M. Pagano, New York University School of Medicine, New York), MPM2 (Upstate Biotechnology), or α-tubulin (Sigma). Images were captured by epifluorescence microscopy, and the fraction of cyclin B or MPM2-positive cells was quantitated.
Flow Cytometry.
FACS analysis for DNA content was performed as described by Anantha et al. (14). FACS was performed on a Becton Dickinson flow cytometer using the Cell Quest software. Cell cycle analysis was performed using the Mod-Fit software.
Quantification of Apoptosis.
Apoptotic cells were measured by a TUNEL assay, using the In Situ Cell Death Detection Kit (Roche Diagnostics) following the manufacturer's instructions.
Supplementary Material
Acknowledgments.
We thank Drs. Michele Pagano, Nick Cowan, and Susan Smith (all from New York University School of Medicine) for reagents; Peter Lopez and Gelo de la Cruz for assistance with acquisition and interpretation of FACS data; Catherine Potenski for aid with epifluorescence microscopy; and Drs. Susan Smith, Jane Skok, Angus Wilson, and Anjana Saxena for helpful comments on the study. This work was supported by National Institutes of Health Grant AI29963, the New York University Cancer Institute, the Rita J. and Stanley Kaplan Comprehensive Cancer Center, and National Cancer Institute Grant P30CA16087.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803001105/DCSupplemental.
References
- 1.Sanchez Y, et al. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science. 1999;286:1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- 2.Skoufias DA, Lacroix FB, Andreassen PR, Wilson L, Margolis RL. Inhibition of DNA decatenation, but not DNA damage, arrests cells at metaphase. Mol Cell. 2004;15:977–990. doi: 10.1016/j.molcel.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 3.Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): The eukaryotic SSB. Crit Rev Biochem Mol Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 4.Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst) 2004;3:1015–1024. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 5.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 6.Namiki Y, Zou L. ATRIP associates with replication protein A-coated ssDNA through multiple interactions. Proc Natl Acad Sci USA. 2006;103:580–585. doi: 10.1073/pnas.0510223103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutta A, Stillman B. cdc2 Family kinases phosphorylate a human cell DNA replication factor, RPA, and activate DNA replication. EMBO J. 1992;11:2189–2199. doi: 10.1002/j.1460-2075.1992.tb05278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu VF, Weaver DT. The ionizing radiation-induced replication protein A phosphorylation response differs between ataxia telangiectasia and normal human cells. Mol Cell Biol. 1993;13:7222–7231. doi: 10.1128/mcb.13.12.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan ZQ, Amin AA, Gibbs E, Niu H, Hurwitz J. Phosphorylation of the p34 subunit of human single-stranded-DNA-binding protein in cyclin A-activated G1 extracts is catalyzed by cdk-cyclin A complex and DNA-dependent protein kinase. Proc Natl Acad Sci USA. 1994;91:8343–8347. doi: 10.1073/pnas.91.18.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Block WD, Yu Y, Lees-Miller SP. Phosphatidyl inositol 3-kinase-like serine/threonine protein kinases (PIKKs) are required for DNA damage-induced phosphorylation of the 32 kDa subunit of replication protein A at threonine 21. Nucleic Acids Res. 2004;32:997–1005. doi: 10.1093/nar/gkh265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu JS, Kuo SR, Melendy T. DNA damage-induced RPA focalization is independent of gamma-H2AX and RPA hyper-phosphorylation. J Cell Biochem. 2006;99:1452–1462. doi: 10.1002/jcb.21066. [DOI] [PubMed] [Google Scholar]
- 12.Olson E, Nievera CJ, Klimovich V, Fanning E, Wu X. RPA2 is a direct downstream target for ATR to regulate the S-phase checkpoint. J Biol Chem. 2006;281:39517–39533. doi: 10.1074/jbc.M605121200. [DOI] [PubMed] [Google Scholar]
- 13.Manthey KC, et al. NBS1 mediates ATR-dependent RPA hyperphosphorylation following replication-fork stall and collapse. J Cell Sci. 2007;120:4221–4229. doi: 10.1242/jcs.004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anantha RW, Vassin VM, Borowiec JA. Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J Biol Chem. 2007;282:35910–35923. doi: 10.1074/jbc.M704645200. [DOI] [PubMed] [Google Scholar]
- 15.Oakley GG, et al. RPA phosphorylation in mitosis alters DNA binding and protein–protein interactions. Biochemistry. 2003;42:3255–3264. doi: 10.1021/bi026377u. [DOI] [PubMed] [Google Scholar]
- 16.Fang F, Newport JW. Distinct roles of cdk2 and cdc2 in RP-A phosphorylation during the cell cycle. J Cell Sci. 1993;106:983–994. doi: 10.1242/jcs.106.3.983. [DOI] [PubMed] [Google Scholar]
- 17.Povirk LF, Wubter W, Kohnlein W, Hutchinson F. DNA double-strand breaks and alkali-labile bonds produced by bleomycin. Nucleic Acids Res. 1977;4:3573–3580. doi: 10.1093/nar/4.10.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vassin VM, Wold MS, Borowiec JA. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol Cell Biol. 2004;24:1930–1943. doi: 10.1128/MCB.24.5.1930-1943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chow JP, et al. DNA damage during the spindle-assembly checkpoint degrades CDC25A, inhibits cyclin-CDC2 complexes, and reverses cells to interphase. Mol Biol Cell. 2003;14:3989–4002. doi: 10.1091/mbc.E03-03-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mikhailov A, Cole RW, Rieder CL. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr Biol. 2002;12:1797–1806. doi: 10.1016/s0960-9822(02)01226-5. [DOI] [PubMed] [Google Scholar]
- 21.Dimitrova DS, Gilbert DM. Stability and nuclear distribution of mammalian replication protein A heterotrimeric complex. Exp Cell Res. 2000;254:321–327. doi: 10.1006/excr.1999.4770. [DOI] [PubMed] [Google Scholar]
- 22.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 23.Shiloh Y. ATM and related protein kinases: Safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 24.Huang X, Tran T, Zhang L, Hatcher R, Zhang P. DNA damage-induced mitotic catastrophe is mediated by the Chk1-dependent mitotic exit DNA damage checkpoint. Proc Natl Acad Sci USA. 2005;102:1065–1070. doi: 10.1073/pnas.0409130102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sibon OC, Kelkar A, Lemstra W, Theurkauf WE. DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat Cell Biol. 2000;2:90–95. doi: 10.1038/35000041. [DOI] [PubMed] [Google Scholar]
- 26.Zachos G, et al. Chk1 is required for spindle checkpoint function. Dev Cell. 2007;12:247–260. doi: 10.1016/j.devcel.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.