Abstract
We present a structurally dynamic model for nucleotide- and actin-induced closure of the actin-binding cleft of myosin, based on site-directed spin labeling and electron paramagnetic resonance (EPR) in Dictyostelium myosin II. The actin-binding cleft is a solvent-filled cavity that extends to the nucleotide-binding pocket and has been predicted to close upon strong actin binding. Single-cysteine labeling sites were engineered to probe mobility and accessibility within the cleft. Addition of ADP and vanadate, which traps the posthydrolysis biochemical state, influenced probe mobility and accessibility slightly, whereas actin binding caused more dramatic changes in accessibility, consistent with cleft closure. We engineered five pairs of cysteine labeling sites to straddle the cleft, each pair having one label on the upper 50-kDa domain and one on the lower 50-kDa domain. Distances between spin-labeled sites were determined from the resulting spin–spin interactions, as measured by continuous wave EPR for distances of 0.7–2 nm or pulsed EPR (double electron–electron resonance) for distances of 1.7–6 nm. Because of the high distance resolution of EPR, at least two distinct structural states of the cleft were resolved. Each of the biochemical states tested (prehydrolysis, posthydrolysis, and rigor), reflects a mixture of these structural states, indicating that the coupling between biochemical and structural states is not rigid. The resulting model is much more dynamic than previously envisioned, with both open and closed conformations of the cleft interconverting, even in the rigor actomyosin complex.
Keywords: double electron-electron resonance, dipolar interaction, actomyosin
Myosin motors use the energy derived from ATP hydrolysis to generate force for a diverse repertoire of biological tasks (1). Class II myosins produce muscle contraction and are involved in cytokinesis and cell motility. The present study focuses on Dictyostelium discoideum (Dicty) myosin II, which offers optimal facility for expression and purification of mutants, and is functionally similar to muscle myosin II (2, 3).
Background
Myosin ATPase Cycle.
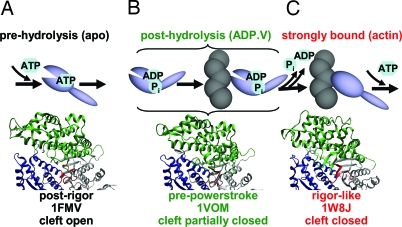
The myosin catalytic (motor) domain hydrolyzes ATP, interacts with actin, and produces force. During the ATPase cycle (Fig. 1), myosin alternates between states of weak and strong actin affinity, depending on the occupancy of the nucleotide-binding pocket. These biochemical transitions are coupled to protein structural transitions and force generation. ATP binding to myosin decreases actin affinity from nanomolar to millimolar, dissociating the actomyosin complex (Fig. 1A). Because the structure of this detached state is little affected by ATP (4), apo-myosin represents the prehydrolysis biochemical state in the present study. The posthydrolysis biochemical state [generated here by adding ADP and vanadate (ADP.V)] presumably exists both free and bound to actin (Fig. 1B) but is stable only in the actin-free state. Release of products, which is catalyzed by actin binding, accompanies the transition to a strongly bound actomyosin complex (Fig. 1C), which generates force. Fig. 1 illustrates a popular model in which biochemical states (Fig. 1 Upper) are tightly coupled to myosin structural states, defined by crystal structures (Fig. 1 Lower) (5–7). It is our goal to test this model by resolving these structural states in well defined biochemical states under noncrystalline conditions. The present study focuses on myosin's actin-binding cleft.
Fig. 1.
Actomyosin ATPase cycle (simplified and linearized) illustrating proposed changes in the actin-binding cleft, assuming tight coupling between biochemical states (listed at the top, with ligands used to produce these states in the present study in parentheses) and structural states defined by crystal structures (shown at bottom). 1FMV: Dicty myosin II, apo (5). 1VOM: Dicty myosin II, ADP.V (6). 1W8J: myosin V, apo (7). The illustration uses the following color scheme: green, U50; blue, L50; red, nucleotide pocket.
Actin-Binding Cleft.
A deep, solvent-filled cavity extends from the nucleotide-binding site to the actin-binding interface of myosin, dividing the interface into the upper (U50) and lower (L50) 50-kDa domains (Fig. 1). When first observed in the x-ray crystal structure of myosin subfragment-1 (S1) in the apo (no nucleotide) state [Protein Data Bank (PDB) ID code 2MYS], it was proposed that this open form of the cleft is not the “rigor-like” conformation that forms the strong actin-binding interface and catalyzes nucleotide release (4, 8). Thus, the 2MYS structure is usually considered to be in a “postrigor” conformation, in which the myosin head is approximately straight with the light-chain domain (lever arm) pointing “down” (away from the center of the sarcomere). The nucleotide-free and MgADP-bound Dicty myosin II crystal structures (5, 9) are similar to 2MYS; therefore, they are also considered postrigor structures that predominate in the prehydrolysis biochemical state (Fig. 1A). In the Dicty myosin II crystal structures obtained with posthydrolysis nucleotide analogs (e.g., ADP.V), the lever arm is in the “up” orientation, which is usually designated the “pre-powerstroke” structural state, in which the cleft is partially closed (Fig. 1B) (10, 11).
Actomyosin Complex.
There is no crystal structure for any actin–myosin complex. Fitting the 2MYS crystal structure to cryo-EM maps of S1-decorated actin filaments is improved by movement of the U50 in the direction of a more closed cleft (12). When the crystal structure of myosin V (PDB ID code 1W8J) (Fig. 1C) revealed a fully closed cleft, it was proposed to be the long sought-after “rigor-like” structural state (13). Myosin V is a processive nonmuscle motor that binds actin in a diffusion-limited, temperature-independent manner, suggesting little or no structural rearrangement, and the myosin V structure fits well into cryo-EM maps of myosin II-decorated actin filaments (14). However, the extent to which actin-bound cleft conformations are similar in myosin II and myosin V is not known. In the present study, the myosin V crystal structure (Fig. 1C Lower) is tested as a model for the structure that occurs upon strong actin binding (Fig. 1C Upper). Several other myosin crystal structures have closed cleft conformations that differ dramatically (15, 16). Spectroscopic probes are needed to obtain data under solution conditions that are not accessible to crystallography.
Site-Directed Spin Labeling and Computational Simulation.
Site-directed spin labeling and electron paramagnetic resonance (SDSL-EPR) have recently benefited from advances in understanding spin label dynamics, accessibility, and distance in relation to protein structure (17, 18). Twentieth-century EPR methods, based on continuous wave (CW) spectroscopy, were sensitive only to distances in the range of 0.7–2.5 nm (19), but now the pulsed electron double resonance technique called double electron–electron resonance (DEER) is sensitive in the range of 1.7–6 nm (20). Computational simulations of protein dynamics, explicitly including the bound spin label (21), are effective in increasing the reliability of EPR data analysis, especially in the case of DEER (22). To examine cleft closure in myosin, we have engineered cysteine spin labeling sites in a Dicty myosin II construct lacking other reactive Cys. We measured probe mobility, accessibility, and spin–spin distance as a function of biochemical state. To determine whether our experimental data are consistent with structural models based on crystal structures (Fig. 1), we performed computational simulations of experimental observables, explicitly including the spin label.
Results
Functional Properties of Spin-Labeled S1dC Mutants.
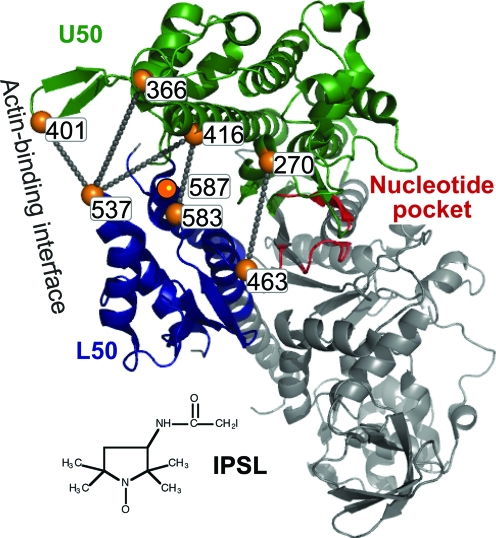
Fig. 2 illustrates spin labeling sites (Cys mutations) in this study. ATPase activity and actin binding were measured for all 10 singly and doubly spin-labeled Dicty myosin II S1 (S1dC) mutants [supporting information (SI) Table S1]. All had actin-activated ATPase activities comparable with that of the unlabeled S1dC control, and all bound actin in rigor (low ionic strength, no ATP), as measured by cosedimentation (23). In most cases, all sites were fully labeled, so the measured activities are those of labeled protein (Table S1).
Fig. 2.
Sites of Cys mutants used for labeling with IPSL, illustrated in the 1FMV crystal structure of Dicty S1dC. Lines indicate pairs used for distance measurements.
Spin Label Mobility and Accessibility.
Single-Cys mutants were labeled within the cleft at positions 416, 583, and 587 (Fig. 2). The spin label 3-(2-iodoacetamido)-PROXL (IPSL) was chosen, rather than the more standard methanethiosulfonate spin label (MTSL), which proved less reliable because of its reversible reactivity.
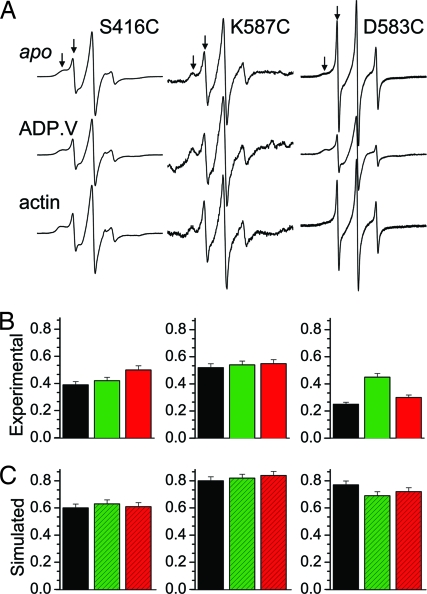
At every site tested, IPSL produced spectra containing two or more resolved components (Fig. 3A, arrows), suggesting multiple conformations of the protein or spin label side chain. The order parameter S, a measure of restriction on the nanosecond rotational motion of IPSL, was calculated as the weighted average obtained from a two-component fit by using standard EPR dynamics software (Fig. 3B and SI Text). The experimental S is directly compared with S (Fig. 3C) calculated from the MD simulation's orientational trajectory (data not shown), which showed less evidence for conformational heterogeneity than did the experimental data. The S values obtained from MD simulation were consistently greater than experimental values, suggesting that the MD simulation, based on the crystal structure, does not sample the low-order conformation evident in experimental spectra. A change in populations of the two components with ADP.V binding at D583C, which was not observed in MD simulations, suggests the presence of at least two cleft conformations. For S416C and K587C, there was little or no dependence on biochemical state (Fig. 3B), which was consistent with MD simulations (Fig. 3C).
Fig. 3.
Spin label mobilities. (A) CW EPR spectra of single-Cys mutants in three biochemical states. (B) Order parameter of IPSL from the spectra in A, in biochemical states apo (prehydrolysis, black), ADP.V (posthydrolysis, green), and actin-bound (red). (C) Order parameter from MD trajectories based on crystal structures 1FMV (postrigor, black), 1VOM (pre-powerstroke, green), and 1W8J (rigor-like, red).
Solvent accessibility was measured by EPR power saturation (Fig. 4A). There was little or no effect of ADP.V (green bars vs. black bars), but actin (red bars vs. black bars) increased accessibility markedly. At first glance, this result seemed opposite to what would be expected for actin-induced cleft closure, because simulated solvent accessibility to the native side chain decreased for the rigor-like myosin V (Fig. 4C, red bars). However, when the spin-labeled Cys side chain was included explicitly (Fig. 4B, red bars), the simulations were in good agreement with experiment; accessibility increased for myosin V (Fig. 4, red bars) relative to postrigor (Fig. 4, black bars). A closer look at MD trajectories provided an explanation: The larger spin-labeled side chain was excluded from the cleft, increasing solvent exposure. However, the magnitude of change predicted by the myosin V crystal structure (Fig. 4B, red bars vs. black bars) is greater than that observed experimentally upon actin binding (Fig. 4A), which suggests that it is a mixture of conformations including those of lower accessibility or that the cleft in the actin-bound biochemical state of Dicty myosin is not as fully closed, as in the myosin V crystal structure.
Fig. 4.
Solvent accessibility of spin-labeled cleft sites (see Fig. 2). (A) Experimental. (B) Simulated, including Cys-attached nitroxide spin label. (C) Simulated for unlabeled native residue. The color codes for biochemical (A) and structural (B and C) states are as in Fig. 3.
Spin–Spin Distance Measurements.
Fig. 2 illustrates the double-Cys mutants used for distance measurements. Two pairs [F270C(U50):V463C(L50) and S416C(U50):D583C(L50)] located in the “inner cleft” adjacent to the nucleotide-binding site were selected to detect cleft closure directly. Three pairs of sites in the “outer cleft” near the actin-binding interface were selected to detect cleft closure indirectly through sensitivity to the counterclockwise rotation of U50 relative to L50. These three pairs all include N537C of L50, which is located on a fully exposed loop that also contains the hydrophobic triplet shown to be critical for strong actin-binding (24). The corresponding sites in U50 are S416C (in HO helix), G366C (in loop 4), and G401C (in cardiomyopathy loop). G401C has been used previously to probe the actomyosin interface (23). S416C and N537C were used previously as labeling sites for both FRET and DEER distance measurements (25, 26).
Spin–Spin Distance Measurements in the Inner Cleft.
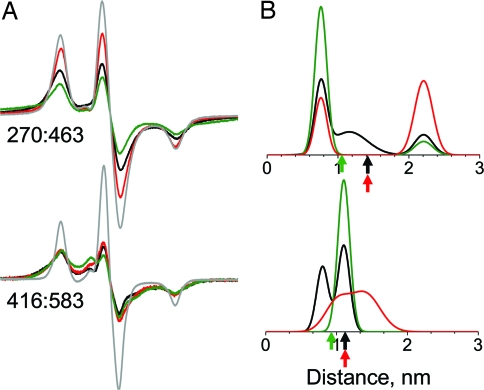
For these two probe pairs, only CW EPR was used, because the predominant distances were found to be on the order of 2 nm or less, as predicted by crystal structures, resulting in considerable spectral broadening (Fig. 5A). For 270:463, analysis in terms of Gaussian distance distributions (Fig. S1 and Table S2) resolves three populations, centered at distances of 0.8, 1.2, and 2.2 nm; all three are present in the apo state, and two of them are present in each of the other biochemical states (Fig. 5B). Similarly, at 416:583, populations are centered at 0.8, 1, and 1.4 nm, with two populations present in both apo and actin-bound biochemical states (Fig. 5B). In contrast, MD simulations based on the dynamics of probes attached to crystal structures predict more homogenous distributions (Fig. S2), suggesting that the experimentally observed heterogeneity is not because of multiple probe conformations but because of multiple cleft conformations present in each biochemical state. Attempts to acquire DEER data from these samples gave very rapid decays that were not reliably analyzable, as expected for distances of <2 nm.
Fig. 5.
Spin–spin distance measurements in the inner cleft, with color codes referring to biochemical and structural states as defined in Figs. 1 and 3. (A) CW EPR spectra. A noninteracting control (single-Cys mutant) is shown in gray. Scan width, 200 G. (B) Distance distributions obtained by fitting CW EPR spectra, assuming a sum of multiple Gaussians. Arrows indicate distances between Cβ atoms in crystal structures.
Spin–Spin Distance Measurements in the Outer Cleft.
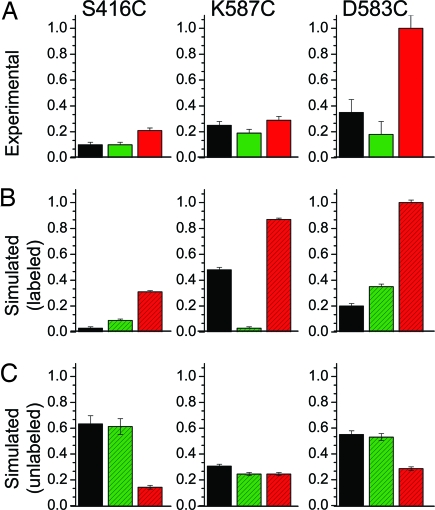
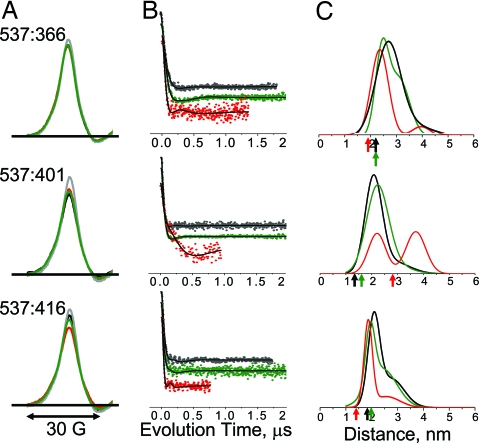
At pairs of sites near the actin-binding interface, both CW EPR spectra and DEER decays were obtained (Fig. 6), because distances were predicted to be in the 2- to 3-nm range, where both methods should have some sensitivity and are complementary. CW spectra (Fig. 6A) showed only slight attenuation in amplitude, with insignificant broadening in spectral wings, indicating that there were no distances substantially below 1.5 nm, a conclusion that could not be reached by DEER alone because it has negligible sensitivity in this range. Both experiments gave consistent results; i.e., for each sample, CW EPR (Fig. 6A) and DEER (Fig. 6B) showed the same qualitative correlation of biochemical state with distance. However, the sensitivity of DEER for r > 2 nm was clearly superior, justifying quantitative analysis in terms of distance distributions (Fig. 6C and Table S3). DEER was sensitive to changes in biochemical state at these sites, indicating distance changes in the 2- to 5-nm range (Fig. 6B). Generally, ADP.V produced subtle distance changes, whereas actin had more dramatic effects. For each of the three probe pairs, the effects of actin were in the direction predicted by crystal structures based on Cβ atom positions, although the experimentally measured distances were consistently greater (arrows in Fig. 6C and Table S3). For two of the pairs (537:366 and 537:416), ADP.V decreased slightly both the mean distance and width, and actin induced more pronounced effects in the same direction, consistent with cleft closure and ordering. The other pair (537:401) exhibited the opposite behavior: ADP.V increased slightly the distance and width, whereas actin substantially increased the distance (Fig. 6C; robustness of fit documented in Fig. S3). Inspection of the crystal structures reveals that these increased distances for this pair are, in fact, predicted because of the scissors-like movement of the cleft. Thus, the DEER results for all three pairs are consistent with slight cleft closure because of ADP.V and more substantial closure because of actin. The distance distributions indicate conformational heterogeneity; each biochemical state shows evidence for at least two conformations, and this is especially clear on actin addition. In particular, for one pair (537:401) there is a clear split into two equally populated conformations in which the mean distance differs by ≈1.6 nm. This separation is too large to be explained by probe flexibility.
Fig. 6.
Spin–spin distance measurements in the outer cleft, with color codes referring to biochemical and structural states as defined in Fig. 3. (A) CW EPR spectra (low field portion). A noninteracting control (labeled single-Cys mutant) is shown in gray. (B) Background-corrected DEER decays, normalized to constant modulation depth and offset vertically for clarity. (C) Distance distributions from DEER fits, assuming a sum of two Gaussians. Arrows indicate Cβ–Cβ distances in crystal structures.
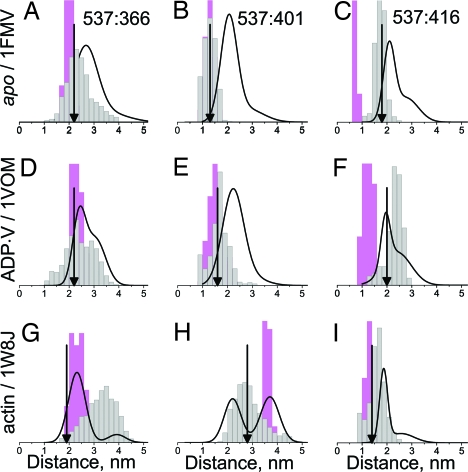
Just as MD simulations were needed to interpret measurements of mobility, accessibility, and CW EPR distance (Figs. 3 and 4 and Fig. S2), they serve to clarify the interpretation of DEER distance measurements (Fig. 7) (22). When backbone carbon atoms were restrained, the distance distributions were narrower than the experimental ones, but relaxation of restraints gave broader distributions, with widths closer to the observed ones. Thus, the protein backbone and side chains sample a wider range of conformational space in solution than in the crystal. The best agreement between experiment and simulation is observed for the ADP.V biochemical state (Fig. 7 Middle); whereas in the apo biochemical state (Fig. 7 Top) experimental distances are consistently greater than simulated. In most cases the experimental data indicate heterogeneity (at least two Gaussian components, as shown in Table S3) more clearly than the simulations predict. In particular, in the actin-bound state (Fig. 7 Bottom), a bimodal distribution is detected experimentally for all three pairs. For two of these cases (Fig. 7 G and H), this bimodal distribution matches remarkably well the sum of the two simulations: restrained and unrestrained, suggesting that both of these structural states (presumably corresponding to open and closed cleft) are populated in solution.
Fig. 7.
Comparison of DEER distance distributions (black curves) (see Fig. 6) with those derived from MD simulations based on crystal structures (see Fig. 1). Pink indicates restrained Cα atoms. Gray indicates unrestrained Cα atoms. Arrows indicate Cβ–Cβ distances in crystal structures.
Discussion
Nucleotide- and Actin-Induced Cleft Closure.
Whereas probe mobility is relatively insensitive to biochemical state (Fig. 3) and accessibility changes only slightly with nucleotide, accessibility increases substantially upon actin binding at all three singly labeled sites (Fig. 4A). This effect of actin seems surprising at first, in light of the expectation of cleft closure, but it is predicted by MD simulations (Fig. 4B) assuming that the actin-bound state is similar to the myosin V “rigor-like” crystal structure, which has a closed cleft (Fig. 1). In the inner cleft, both ADP.V and actin binding affect distance measurements in the CW EPR range (Fig. 5). Each of the three biochemical states gives rise to a multicomponent distance distribution, suggesting a mixture of structural states. Based on MD results (Fig. S2), these structural states probably correspond to those represented by crystal structures (Fig. 1) postrigor (cleft open), pre-powerstroke (partially closed cleft), and rigor-like (cleft closed). All three structural states are populated in the apo biochemical state, ADP.V is mainly in the open or partially closed state, and actin stabilizes two states, one of which corresponds to complete cleft closure.
In the outer cleft (Figs. 6 and 7), ADP.V has only a slight effect on distances, in agreement with simulations (Fig. 7 Middle), so the outer cleft pairs resolve only two cleft structural states: open and closed. Actin binding has a pronounced effect in the outer cleft (Fig. 7 Bottom), in agreement with the myosin V (rigor-like) structural state, in which the outer cleft is primarily closed (Fig. 1). However, the actin-bound biochemical state in solution contains both open and closed cleft conformations, and all three biochemical states have components that correspond to the closed cleft conformation (Table S3), indicating that actin's presence is not necessary to form the closed cleft.
SDSL-EPR Methodology and Computational Simulation.
This study underscores several principles about the power of site-directed spin labeling, especially when combined with MD simulations. Virtually every experimental measurement capable of resolving conformational states revealed the occupation of two or more conformations (Figs. 3, 5, and 6), whereas the MD simulation based on a spin label attached to a single crystal structure usually did not, suggesting that each biochemical state in solution is populated by more than one structural state. Mobility at cleft sites was relatively insensitive to changes in biochemical state (Fig. 3), whereas solvent accessibility proved more sensitive (Fig. 4). The agreement of experimental and simulated solvent accessibility was much better when the spin label was included in the simulation (Fig. 4), as observed previously for spin-labeled T4 lysozyme (27). Experimental distance measurements are in better agreement with computational simulations not only when the spin-label is included in place of the native side chain (22) but also when the backbone atoms are unrestrained (Fig. 7). Figs. 5 and 6 confirm that CW EPR and DEER are quite complementary techniques for determining distance distributions (28). For all cases, CW EPR and DEER data were consistent, but CW EPR was superior in sensitivity and resolution for distances of <2 nm (Fig. 5), and DEER was superior for distances of >2 nm (Fig. 6).
Comparison of Rigor-Like Structural Models.
The variety of published crystal structures proposed to be rigor-like suggests that the extent of cleft closure is not conserved among myosins (15). For example, the cleft structure of a proposed rigor-like nucleotide-free Dicty myosin II construct (PDB ID code 1Q5G) (16), is significantly different from that of the myosin V structure (PDB ID code 1W8J) (Fig. 1) (29). Our observed changes in distance induced by actin binding, relative to the apo state, are strikingly more consistent with 1W8J than with 1Q5G (Table S4).
Relationship to Previous Spectroscopy.
Solvent accessibility in the smooth muscle myosin cleft was probed with an engineered tryptophan at F425W (S416 in Dicty myosin II) (30). Fluorescence quenching showed that F425W was more solvent-exposed in weak-binding than in strong-binding states, and an increase in fluorescence with actin suggested a shift in the direction of cleft closure. In the present study, we found no change in spin label solvent accessibility for two of three labeled cleft sites with binding of ADP.V (Fig. 4) or ADP (data not shown), but we did observe a large increase in accessibility with actin-binding (Fig. 4A), a result consistent with simulated actin-induced cleft closure (Fig. 4B). We conclude that accessibility data are most reliable when several sites are probed and models are tested with explicit MD simulations of probe accessibility. Evidence for actin-induced cleft movement has been observed for spin (31) and fluorescent (26) probes attached to sites 537 and 416. We observed an actin-induced distance decrease between these sites (Fig. 6 Bottom) and showed that this decrease is predicted from the crystal structures by simulations (Fig. 7 and Table S3). By making multiple measurements across the cleft, including mobility and accessibility as well as distance distributions, we have obtained a more complete model of cleft closure, including clear evidence for structural heterogeneity within single biochemical states. In particular, these results add to the spectroscopic evidence that the strong actin–myosin interface is surprisingly dynamic (23).
Coupling Between Biochemical and Structural States.
Unlike crystallography, spectroscopy can resolve multiple structural states (conformations) in solution for a single biochemical state. EPR of spin-labeled nucleotides has shown that in both prehydrolysis and posthydrolysis biochemical states, the nucleotide pocket (switch 1) is in a closed structural state, whereas strong actin binding induces an equilibrium between open and closed states (32). The present study resolved at least two distinct cleft conformations, probably corresponding to open (postrigor) and closed (rigor-like) structures (Fig. 6). In the case where these two conformations were most clearly resolved (Fig. 6, 537:401), the apo and ADP.V states are mainly in the open cleft conformation, whereas the actin-bound state is equally populated by both open and closed conformations. This result is strikingly parallel to that detected by nucleotide spin labels, suggesting that the switch I conformation (near the nucleotide pocket) is coupled to the cleft conformation near the actin-binding interface. In the inner cleft, three conformations are resolved (Fig. 5), suggesting that the inner cleft is more directly coupled to switch II, which differs in all three structural states (7). Similarly, in both rabbit (33) and Dicty (34), a spin label in the force-generating region of myosin resolves three distinct structural states that change their populations with biochemical state. Continued analysis of the complex structural and biochemical coupling among the subdomains of myosin and actin, in space and time, is needed to understand the mechanism of actomyosin function. High-resolution EPR techniques will continue to play a leading role in this effort.
Methods
Protein Preparations and Spin Labeling.
Cysteine mutations for spin labeling were introduced into a Dicty myosin II gene truncated at residue 762, containing only a single (unreactive) Cys at position 655 (SI Text). These S1dC derivatives were expressed and purified (23). F-actin was prepared from rabbit skeletal muscle (35). Unless otherwise indicated, experiments were carried out at 4°C in EPR buffer (50 mM KCl/3 mM MgCl2/10 mM MOPS, pH 7.5). Spin labeling was carried out overnight at 100 μM myosin and 800 μM IPSL or 4-maleimido-TEMPO (MSL). Unreacted label was removed by exchange into EPR buffer, and functional properties were measured (SI Text) and are tabulated in Table S1.
CW EPR: Mobility and Accessibility.
EPR samples contained 100 μM myosin S1dC. The posthydrolysis state was formed by addition of 5 mM ADP and 5 mM Na3VO4. The actomyosin state was formed by mixing 100 μM S1dC with 200 μM F-actin. For accessibility measurements, duplicate samples were prepared, one containing 5 mM NiEDDA as the paramagnetic relaxation agent, and accessibility was measured by EPR power saturation (27, 36–38). Order parameters were measured from EPR spectra by simulation and fitting with the program NLSL (39).
Spin–Spin Distance Measurements.
Samples were the same as in other EPR experiments, except that the buffer also contained 10% (vol/vol) glycerol, volume was 100 μl, and samples were flash-frozen in liquid nitrogen. EPR acquisition parameters are described in SI Text. Spin–spin distances were determined by fitting the experimental EPR data by simulations assuming that the distance distribution is a sum of Gaussians (SI Text). After 4–12 h of acquisition, 100-μl samples at 100 μM concentration yielded background-corrected DEER signals with sufficient signal-to-noise ratio for reliable analysis, and analysis was performed by using the software DEERAnalysis2006.1 (40, 41) and DEFit (25, 42).
Computational Simulations Based on Crystal Structures.
Crystal structures (Fig. 1) were modified to include missing loops and residues by using InsightII, and native residues were mutated to spin-labeled Cys by using Visual Molecular Dynamics (VMD) (43). Parameters for IPSL and MSL were based on CHARMM19 united-atom force fields (22). Metropolis Monte Carlo minimization (22, 44) was used to determine starting points for MD simulations. MD simulations were carried out essentially as described previously (21, 25) (see also SI Text). The simulated spin label order parameter was calculated from the MD trajectory (21). Solvent-accessible surface area (SASA) was calculated in VMD for both the unlabeled (native) residue and IPSL (SI Text). Distances between spin label nitroxide oxygen atoms were measured in VMD, binned (1-Å width), and normalized by area.
Supplementary Material
Acknowledgments.
We thank Piotr Fajer and Marco Bonora for expert advice on DEER experiments and analysis; Sarah Blakely, Erin M. Hoffman, Benjamin Matzke, and Eunice Song for excellent assistance with protein preparations; Kurt Torgersen for EPR spectroscopy; and Octavian Cornea for help with manuscript preparation. This work was supported by National Institutes of Health Grants AR32961, AG26160, and RR22362 (to D.D.T.) and by the Minnesota Supercomputing Institute. J.C.K. and B.S. were supported by National Institutes of Health Training Grants GM08700 and AR07612, respectively. D.J.K. was supported by National Science Foundation Training Grant CHE-0452204.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802286105/DCSupplemental.
References
- 1.Baker JP, Titus MA. Myosins: Matching functions with motors. Curr Opin Cell Biol. 1998;10:80–86. doi: 10.1016/s0955-0674(98)80089-6. [DOI] [PubMed] [Google Scholar]
- 2.Sasaki N, Ohkura R, Sutoh K. Dictyostelium myosin II as a model to study the actin-myosin interactions during force generation. J Muscle Res Cell Motil. 2002;23:697–702. doi: 10.1023/a:1024415409406. [DOI] [PubMed] [Google Scholar]
- 3.Uyeda TQ, Spudich JA. A functional recombinant myosin II lacking a regulatory light chain-binding site. Science. 1993;262:1867–1870. doi: 10.1126/science.8266074. [DOI] [PubMed] [Google Scholar]
- 4.Rayment I, et al. Three-dimensional structure of myosin subfragment-1: A molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 5.Bauer CB, Holden HM, Thoden JB, Smith R, Rayment I. X-ray structures of the apo and MgATP-bound states of Dictyostelium discoideum myosin motor domain. J Biol Chem. 2000;275:38494–38499. doi: 10.1074/jbc.M005585200. [DOI] [PubMed] [Google Scholar]
- 6.Smith CA, Rayment I. X-ray structure of the magnesium(II). ADP.vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 Å resolution. Biochemistry. 1996;35:5404–5417. doi: 10.1021/bi952633+. [DOI] [PubMed] [Google Scholar]
- 7.Coureux PD, Sweeney HL, Houdusse A. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J. 2004;23:4527–4537. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rayment I, et al. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- 9.Gulick AM, Bauer CB, Thoden JB, Rayment I. X-ray structures of the MgADP, MgATPgammaS, and MgAMPPNP complexes of the Dictyostelium discoideum myosin motor domain. Biochemistry. 1997;36:11619–11628. doi: 10.1021/bi9712596. [DOI] [PubMed] [Google Scholar]
- 10.Fisher AJ, et al. Structural studies of myosin:nucleotide complexes: A revised model for the molecular basis of muscle contraction. Biophys J. 1995;68:19S–26S. discussion (1995) 68:27S–28S. [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher AJ, et al. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP. BeFx and MgADP.AlF4. Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 12.Holmes KC, Angert I, Kull FJ, Jahn W, Schröder RR. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 2003;425:423–427. doi: 10.1038/nature02005. [DOI] [PubMed] [Google Scholar]
- 13.Coureux PD, et al. A structural state of the myosin V motor without bound nucleotide. Nature. 2003;425:419–423. doi: 10.1038/nature01927. [DOI] [PubMed] [Google Scholar]
- 14.Holmes KC, Schröder RR, Sweeney HL, Houdusse A. The structure of the rigor complex and its implications for the power stroke. Philos Trans R Soc London B. 2004;359:1819–1828. doi: 10.1098/rstb.2004.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y, et al. Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor. Structure. 2007;15:553–564. doi: 10.1016/j.str.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 16.Reubold TF, Eschenburg S, Becker A, Kull FJ, Manstein DJ. A structural model for actin-induced nucleotide release in myosin. Nat Struct Biol. 2003;10:826–830. doi: 10.1038/nsb987. [DOI] [PubMed] [Google Scholar]
- 17.Hubbell WL, Altenbach C, Hubbell CM, Khorana HG. Rhodopsin structure, dynamics, and activation: A perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Adv Protein Chem. 2003;63:243–290. doi: 10.1016/s0065-3233(03)63010-x. [DOI] [PubMed] [Google Scholar]
- 18.Fanucci GE, Cafiso DS. Recent advances and applications of site-directed spin labeling. Curr Opin Struct Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Rabenstein MD, Shin YK. Determination of the distance between two spin labels attached to a macromolecule. Proc Natl Acad Sci USA. 1995;92:8239–8243. doi: 10.1073/pnas.92.18.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeschke G, Bender A, Paulsen H, Zimmermann H, Godt A. Sensitivity enhancement in pulse EPR distance measurements. J Magn Reson. 2004;169:1–12. doi: 10.1016/j.jmr.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 21.LaConte LE, Voelz V, Nelson W, Enz M, Thomas DD. Molecular dynamics simulation of site-directed spin labeling: Experimental validation in muscle fibers. Biophys J. 2002;83:1854–1866. doi: 10.1016/S0006-3495(02)73950-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sale K, Song L, Liu YS, Perozo E, Fajer P. Explicit treatment of spin labels in modeling of distance constraints from dipolar EPR and DEER. J Am Chem Soc. 2005;127:9334–9335. doi: 10.1021/ja051652w. [DOI] [PubMed] [Google Scholar]
- 23.Korman VL, Anderson SE, Prochniewicz E, Titus MA, Thomas DD. Structural dynamics of the actin-myosin interface by site-directed spectroscopy. J Mol Biol. 2006;356:1107–1117. doi: 10.1016/j.jmb.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 24.Onishi H, Mikhailenko SV, Morales MF. Toward understanding actin activation of myosin ATPase: The role of myosin surface loops. Proc Natl Acad Sci USA. 2006;103:6136–6141. doi: 10.1073/pnas.0601595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fajer PG, et al. Myosin cleft closure by double electron-electron resonance and dipolar EPR. J Phys Condens Matter. 2007;19:285208. [Google Scholar]
- 26.Conibear PB, Bagshaw CR, Fajer PG, Kovacs M, Malnasi-Csizmadia A. Myosin cleft movement and its coupling to actomyosin dissociation. Nat Struct Biol. 2003;10:831–835. doi: 10.1038/nsb986. [DOI] [PubMed] [Google Scholar]
- 27.Altenbach C, Froncisz W, Hemker R, McHaourab H, Hubbell WL. Accessibility of nitroxide side chains: Absolute Heisenberg exchange rates from power saturation EPR. Biophys J. 2005;89:2103–2112. doi: 10.1529/biophysj.105.059063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banham JE, et al. Distance measurements in the borderline region of applicability of CW EPR and DEER: A model study on a homologous series of spin-labelled peptides. J Magn Reson. 2008;191:202–218. doi: 10.1016/j.jmr.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 29.Kull FJ, Endow SA. A new structural state of myosin. Trends Biochem Sci. 2004;29:103–106. doi: 10.1016/j.tibs.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Yengo CM, De La Cruz EM, Chrin LR, Gaffney DP, II, Berger CL. Actin-induced closure of the actin-binding cleft of smooth muscle myosin. J Biol Chem. 2002;277:24114–24119. doi: 10.1074/jbc.M111253200. [DOI] [PubMed] [Google Scholar]
- 31.Fajer MI, Li H, Yang W, Fajer PG. Mapping electron paramagnetic resonance spin label conformations by the simulated scaling method. J Am Chem Soc. 2007;129:13840–13846. doi: 10.1021/ja071404v. [DOI] [PubMed] [Google Scholar]
- 32.Naber N, Purcell TJ, Pate E, Cooke R. Dynamics of the nucleotide pocket of myosin measured by spin-labeled nucleotides. Biophys J. 2007;92:172–184. doi: 10.1529/biophysj.106.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nesmelov YE, Agafonov RV, Burr A, Weber RT, Thomas DD. Structure and dynamics of the force generating domain of myosin probed by site-directed spin labeling and multifrequency electron paramagnetic resonance. Biophys J. 2008;95:247–256. doi: 10.1529/biophysj.107.124305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agafonov RV, Titus MA, Nesmelov YE, Thomas DD. Muscle and nonmuscle myosins probed by a spin label at equivalent sites in the force-generating domain. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0801342105. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prochniewicz E, Zhang Q, Janmey PA, Thomas DD. Cooperativity in F-actin: binding of gelsolin at the barbed end affects structure and dynamics of the whole filament. J Mol Biol. 1996;260:756–766. doi: 10.1006/jmbi.1996.0435. [DOI] [PubMed] [Google Scholar]
- 36.Altenbach C, Marti T, Khorana HG, Hubbell WL. Transmembrane protein structure: spin labeling of bacteriorhodopsin mutants. Science. 1990;248:1088–1092. doi: 10.1126/science.2160734. [DOI] [PubMed] [Google Scholar]
- 37.Kirby TL, Karim CB, Thomas DD. Electron paramagnetic resonance reveals a large-scale conformational change in the cytoplasmic domain of phospholamban upon binding to the sarcoplasmic reticulum Ca-ATPase. Biochemistry. 2004;43:5842–5852. doi: 10.1021/bi035749b. [DOI] [PubMed] [Google Scholar]
- 38.Surek JT, Thomas DD. A paramagnetic molecular voltmeter. J Magn Reson. 2008;190:7–25. doi: 10.1016/j.jmr.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Budil DE, Lee S, Saxena S, Freed JH. Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg-Marquardt algorithm. J Magn Reson A. 1996;120:155–189. [Google Scholar]
- 40.Jeschke G, Koch A, Jonas U, Godt A. Direct conversion of EPR dipolar time evolution data to distance distributions. J Magn Reson. 2002;155:72–82. doi: 10.1006/jmre.2001.2498. [DOI] [PubMed] [Google Scholar]
- 41.Jeschke G. Distance measurements in the nanometer range by pulse EPR. Chemphyschem. 2002;3:927–932. doi: 10.1002/1439-7641(20021115)3:11<927::AID-CPHC927>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 42.Fajer PG, Brown L, Song L. Practical Pulsed Dipolar EPR (DEER) Berlin: Springer; 2006. [Google Scholar]
- 43.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 44.Sale K, Sar C, Sharp KA, Hideg K, Fajer PG. Structural determination of spin label immobilization and orientation: A Monte Carlo minimization approach. J Magn Reson. 2002;156:104–112. doi: 10.1006/jmre.2002.2529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.