Abstract
The replication terminator protein Tus of Escherichia coli promotes polar fork arrest at sequence-specific replication termini (Ter) by antagonizing DNA unwinding by the replicative helicase DnaB. Here, we report that Tus is also a polar antitranslocase. We have used this activity as a tool to uncouple helicase arrest at a Tus–Ter complex from DNA unwinding and have shown that helicase arrest occurred without the generation of a DNA fork or a bubble of unpaired bases at the Tus–Ter complex. A mutant form of Tus, which reduces DnaB–Tus interaction but not the binding affinity of Tus for Ter DNA, was also defective in arresting a sliding DnaB. A model of polar fork arrest that proposes melting of the Tus–Ter complex and flipping of a conserved C residue of Ter at the blocking but not the nonblocking face has been reported. The model suggests that enhanced stability of Tus–Ter interaction caused by DNA melting and capture of a flipped base by Tus generates polarity strictly by enhanced protein–DNA interaction. In contrast, the observations presented here show that polarity of helicase and fork arrest in vitro is generated by a mechanism that not only involves interaction between the terminator protein and the arrested enzyme but also of Tus with Ter DNA, without any melting and base flipping in the termination complex.
Keywords: protein–DNA interaction, protein–protein interaction, site-directed interstrand cross-linking
The replication of DNA in many prokaryotes and in certain regions of eukaryotic chromosomes is specifically terminated at specialized sequences called replication termini (Ter) (Fig. 1 A and B) that cause orientation-dependent fork arrest, and such arrest performs important physiological functions (1–3). In eukaryotes, sequence-specific replication termini are not present within every replication unit. Instead, the termini are found at specialized locations such as the nontranscribed spacers of rDNA (4) and at the mating type switch locus of Schizosaccharomyces pombe (5). We and others have shown by in vitro analyses that the replication termination proteins of Escherichia coli and Bacillus subtilis are polar contrahelicases, i.e., the proteins cause unidirectional arrest of the replicative helicase DnaB upon binding to the Ter sequences (6–10). The crystal structures of the replication terminator protein (RTP) of B. subtilis (11) and that of E. coli, called Tus (12), have been solved. Despite the fact that the proteins have very different structures, both proteins interact in vitro with their cognate binding sites to arrest DnaB helicase and RNA polymerase of E. coli in a polar mode (10, 13).
Fig. 1.
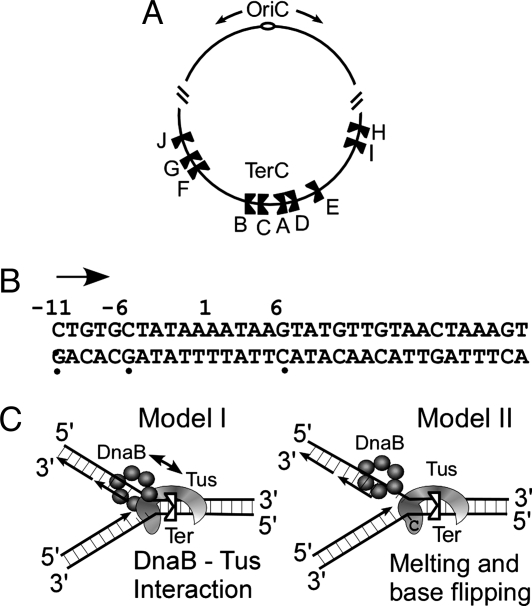
The replication termini of E. coli. (A) Diagram showing the relative locations and orientations of the known Ter sites of E. coli with respect to oriC. (B) The consensus nucleotide sequence of Ter, the C6, and its complementary G are shown. The locations for polar arrest of transcripts catalyzed by E. coli RNA polymerase are shown at −6 and −11. The arrow shows the direction of transcription (and replication). (C) Two models of polar fork arrest. The double-headed arrow in model I indicates Tus–DnaB interaction.
A satisfactory model of polar fork arrest should take into account the following biological observations. First, a Tus–Ter complex arrests only some helicases such as DnaB but not others such as PcrA, helicase I, and UvrD helicase (9, 14) in vitro. In fact, in vivo genetic experiments show that UvrD helicase removes Tus protein from Ter sites (15). Further evidence of helicase specificity is indicated by the observation that Tus either fails to arrest or arrests poorly the hexameric replicative helicase DnaC of B. subtilis in vivo (16). The Gram-positive DnaC protein is structurally related to DnaB of E. coli (17).
Second, the Tus–Ter complex also arrests RNA polymerase of E. coli in a polar mode at or near the coordinates −6 and −11, immediately upstream of the blocking face of Tus–Ter complex (13, 18) (see Fig. 1B). Finally, a variety of approaches show that Tus physically interacts with DnaB and there are mutants on the blocking face of Tus that reduce interactions in vitro that also are defective in arresting DnaB in vitro and the replication forks in vivo (19, 20).
Two models of fork arrest are shown in Fig. 1C. The first model postulates that Tus–DnaB protein–protein interaction and Tus–Ter binding both are necessary for polar fork arrest (19). The second one suggests that polarity is generated strictly by DNA–protein interaction caused by helicase-mediated remodeling of the Ter–Tus complex that not only involves DNA unwinding at the blocking end of Tus but also flipping of a C residue and its capture by Tus. Both of these steps have been reported to be essential for the generation of polarity. Forks approaching the nonblocking end are postulated to melt the DNA and dislodge Tus from the Ter site (21).
To accommodate all relevant biological information pertaining to fork arrest and to gain further insights into the replication termination mechanism, we investigated whether polar arrest of helicase translocation could be dissociated from actual DNA unwinding. The experimental strategy used to accomplish this objective takes into consideration the following known aspects of helicase structure, dynamics, and biochemistry. DnaB is a hexameric helicase (22). It is a toroid with a central channel (23, 24) that can accommodate either a single or both strands of the DNA double helix. Detailed published work has shown that when the hexameric DnaB ring is loaded onto a forked dsDNA, it causes unwinding of the duplex. However, a substrate with a 5′-single-stranded tail and no fork allows the enzyme to translocate on the DNA double helix by using the energy of ATP hydrolysis but presumably without melting the DNA duplex (25–27).
We spatially separated helicase arrest at the Tus-Ter complex from actual DNA unwinding at a downstream fork by using a triplex DNA substrate containing a 30-nt-long, 5′-single-stranded tail and a downstream annealed 45-mer oligo with a 30-nt-long, 3′-single-stranded tail that simulated a replication fork. Using this approach, we have discovered that the Tus-Ter complex promoted polar arrest of energy-dependent translocation of DnaB on the DNA duplex without causing net DNA unwinding or even without the formation of a transient denaturation bubble at the Ter site. The mechanistic implications of this and related observations are discussed.
Results
The chromosome of E. coli contains two sets of Ter sites present in opposite orientations. The sites act together to form a replication trap that forces the two replication forks to meet each other in a region called TerC (see Fig. 1A). The consensus Ter sequence is shown with the coordinates on the top (Fig. 1B). The C residue that is proposed to flip and its complementary G are shown at coordinate 6, and RNA polymerase arrest sites are shown at −6 and −11.
DnaB Sliding on a Triplex Substrate Did Not Cause Net Melting of Ter DNA.
Our experimental strategy to spatially uncouple helicase translocation from helicase-catalyzed DNA unwinding is shown in Fig. 2A. We investigated the effect of a range of concentrations of KCl on helicase activity in vitro to choose the concentrations that allowed DnaB to be active in vitro [supporting information (SI) Fig. S1]. On the basis of these experiments, we performed a helicase assay mostly in the standard helicase buffer and in some cases with the same buffer supplemented with 100 mM KCl. It should be noted that in the standard buffer without additional KCl we have reconstituted replication with purified proteins and have observed robust Tus-mediated replication termination in vitro (28).
Fig. 2.
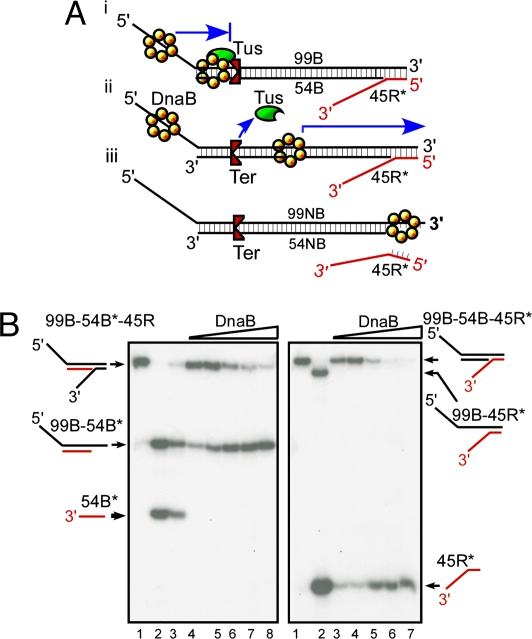
Experimental strategy for determining DnaB sliding on dsDNA and its arrest at a Tus–Ter complex. (A) Diagram showing the triplex substrates used for measurements of helicase sliding in the blocking orientation that arrests a sliding DnaB and is measured by the unwinding of the 45R reporter oligo (i) and the substrate with the nonblocking orientation of Ter (ii and iii). (B) Phosphorimagergrams showing the products generated by sliding of DnaB on 99B-54B*-45R triplex (Left) and on the 99B-54B-45R* (Right) (* indicates the location of the labeled 5′ end) triplexes in the absence of Tus. The labeled strands are shown in red. Three to four femtomoles of substrate DNA and 0–300 ng of DnaB were used in each reaction. (Left) Lanes 1–3, marker DNA; lanes 4–8, 0, 50, 100, 300, and 400 ng of DnaB, respectively. (Right) Lanes 1 and 2, marker DNA; lanes 3 and 4, DNA without DnaB; and lanes 5–7, 50, 100, and 300 ng of DnaB, respectively.
Although previous work showed that DnaB can load on a 5′-tailed substrate in the absence of a fork and slide on dsDNA (25), we determined that such sliding does not occur on a substrate consisting of M13 single-stranded circular DNA that formed partial duplexes with linear ssDNA of length from 15 bps to >1,000 bps (14). Instead, we observed that DnaB unwinds the partial duplexes. Therefore, before proceeding further with the experiment, we sought to ascertain whether ATP hydrolysis-dependent DnaB sliding would occur under our experimental conditions. Our strategy was to incubate a triplex substrate containing a 30-nt-long, 5′-single-stranded tail with DnaB and ATP. The 5′ tail was expected to promote the loading of the enzyme, and because of the absence of a fork, the hexameric toroid of DnaB was expected to accommodate both strands of the DNA in its central channel and slide over the duplex. In the absence of Tus, the helicase was expected to slide past the Ter site until it encountered the 30-nt-long 3′ tail located downstream, closer to the 3′ end (Fig. 2A). The 3′-tailed structure simulates a fork that should cause DnaB to enter into the unwinding mode and melt the 45-mer reporter strand.
To experimentally verify these predictions, we constructed two triplexes, one formed by annealing the oligonucleotides (called oligos here after) 54B and 45R to the 99B top strand. This annealing generated 99B-54B*-45R with the label at the 5′ end of the 54-mer (54B*) (the oligos are listed in (Table S1) and the second one, identical in sequence and construction, but having the label at the 5′-end of 45R (99B-54B-45R*). The labeled strands of the triplexes are shown in red in Fig. 2B, and the location of the label is indicated by * in the text. DnaB was loaded separately onto the two triplexes. Helicase movement was monitored by measuring the unwinding of the downstream reporter 45-mer, after resolution of the reaction products by nondenaturing 12% PAGE. A representative image from a phosphorimager (phosphorimagergram) showed that helicase progression on the triplex 99B-54B*-45R (Fig. 2B Left) did not melt and release the 54-mer* (54B*) from the triplex. On the other hand, the 3′-tailed 45-mer (45R) located downstream of Ter was unwound by DnaB, generating the 99B-54B* partial duplex (Fig. 2B Left).
In the complementary experiment we used the 99B-54B-45R* triplex and found that incubation with DnaB and ATP caused release of the 45R* without any detectable formation of the 99B-45R* partial duplex. The results are consistent with sliding of the DnaB on the double-stranded substrate, past the Ter site until the helicase reached the downstream fork and unwound the reporter, resulting in the release of 45R* (Fig. 2B Right). In summary, these results confirmed that DnaB was able to slide over the duplex region without causing DNA unwinding, and the sliding was successfully monitored by the melting of the downstream reporter.
Tus Is a Polar Antitranslocase.
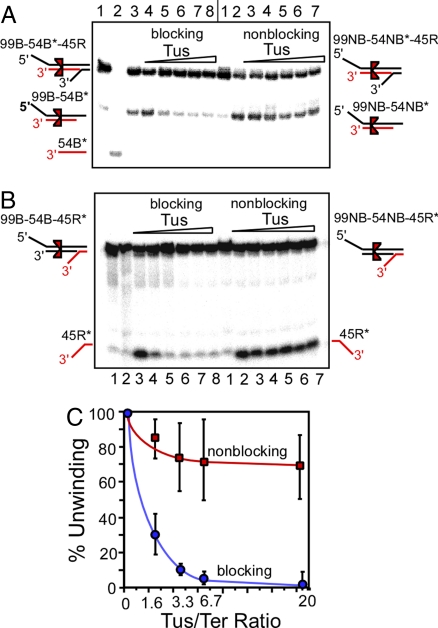
Having determined that helicase sliding was taking place on the DNA substrates at the appropriate salt concentrations, we proceeded to investigate whether polar arrest of a sliding helicase could be effected by assembling a Tus–Ter complex separately on the triplexes 99B-54B*-45R, 99B-54B-45R* that contained the Ter site in the blocking orientation and two others, namely 99NB-54NB*-45R and 99NB-54NB-45R*, that contained the Ter site in the opposite, nonblocking orientation with respect to the 5′ tailed end of either 99B or 99NB (Fig. 2A). The triplexes were preincubated with increasing amounts of Tus at the molar ratios of Tus to Ter DNA of 0.5–20, then ATP and sufficient DnaB (to melt at least 50% of the reporter 45-mer from the triplexes) were added and the reaction was allowed to proceed for 15 min at 37°C after which the products were resolved by 12% nondenaturing PAGE. The phosphorimagergrams of the gels showed that the Tus—Ter complex impeded helicase sliding on the duplex DNA in those triplexes that had the Ter in the blocking but not in those with the site in the nonblocking orientation. Once again, helicase sliding caused no detectable net melting of the 54-mer/99-mer duplex region either in the blocking or the nonblocking triplexes (Fig. 3 A and B).
Fig. 3.
Phosphorimagergrams showing the polar arrest of a sliding DnaB without base pair melting by a Tus–Ter complex in the blocking but not in the non-blocking orientation. (A) Representative phosphorimagergrams of products generated by sliding DnaB at a fixed concentration (300 ng) on the 99B-54B*-45R triplex (4 fmole), in the presence of an increasing range of concentration of Tus (molar ratio 0–20) and the same on the 99NB-54NB*-45R triplex. The products are identified by the diagrams on the margins. Blocking Tus; lanes 1–3, marker DNA; lanes 4–8, substrate plus 300 ng DnaB plus Tus at molar ratios of Tus/DNA of 0, 1.6, 3.3, 6.7 and 20 molar. Non blocking Tus; lane 1, marker DNA; lanes 2 and 3, DNA plus 100 and 300 ng of DnaB, respectively; lanes 4–7, DNA plus DnaB (300 ng) plus Tus at molar ratios of, 1.6, 3.3, 6.7, and 20, respectively. B, lanes, 1 and 2, marker DNA; lanes 3–8, DNA substrate plus DnaB (300 ng), plus Tus at molar ratios of 0, 1.6, 3.3, 6.7, 6.7 and 20 respectively.
Three independent sets of experiments were carried out with each set of triplexes with the label on 54B* (or 54NB*) (Fig. 3A) or on the 45R* (Fig. 3B). The data from the experiments using 99B-54B-45R* and 99NB-54NB-45R* are shown with the error bars (SDs) (Fig. 3C). We also analyzed similar data (data not presented) using the triplexes having the label on 54B* and 54NB*, and we obtained results identical to those shown in Fig. 3C. These results lead us to conclude that sliding of DnaB on dsDNA was arrested in a polar mode by the Tus–Ter complex and that there was no net melting of DNA duplex on which the helicase sliding had occurred. In the nonblocking orientation of Ter, and in the absence of DNA unwinding, it would appear that a sliding DnaB must displace the Tus from the Tus–Ter complex without melting the Ter sequence.
We then asked the question as to whether helicase sliding on the dsDNA would cause formation of a bubble of unpaired bases at Tus–Ter by attempting to trap the denatured region, if any, with either 10% HCHO or 2 mM KMnO4 but were unable to detect any DNA melting (see Fig. S2 and SI Text). We wanted to ascertain further whether DNA melting at the blocking end of Ter was necessary for arresting the sliding helicase by performing the following definitive experiment.
DnaB Could Translocate Over a Duplex Region Containing Interstrand Cross-Links That Immediately Preceded C6.
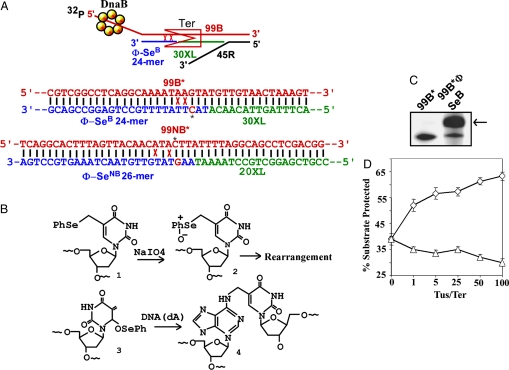
A definitive approach to testing whether DNA melting is needed in front of the blocking end of Tus–Ter for polar DnaB arrest is by preventing any DNA strand separation adjacent to GC6 by inducing site-directed, covalent, interstrand cross-links at two predetermined sites immediately preceding C6. Using such a substrate one could ask the question as to whether DnaB would slide unimpeded over the interstrand cross-links and be arrested by the Tus–Ter complex in a polar mode. To perform this experiment, we synthesized a 24-mer oligo containing phenyl-selenide-dTTP (24Φ-SeB) (29–31) at the positions corresponding to the T residues at coordinates 4 and 5 of the bottom strand of Ter. This oligo was used to construct the duplex 99B-24Φ-SeB (Fig. 4A and Table S1) by inducing site-directed interstrand cross-linking by oxidation with 10 mM Na-periodate for 4 h at room temperature at ≈22°C. The chemical reaction pathway is shown in Fig. 4B. The yield of the cross-linked product that migrated in the gel just above the 99B* marker varied from ≈40% to >95% (Fig. 4C). Further details are provided in SI Text.
Fig. 4.
Interstrand cross-linking of residues in front of Ter did not abolish arrest of helicase translocation. (A) (Top) Schematic representation of the cross-linked triplex with Ter in the blocking orientation showing the locations of the various oligos and the cross-links (red x). (Middle) Sequence of the triplex with Ter in the blocking orientation about the region of the cross-links; the phenyl selenide substituted oligo is shown in blue except for the GC6 pair that is shown in red with an asterisk; a part of the 30XL sequence is shown in green. (Bottom) Sequence of the control triplex with Ter in the nonblocking orientation; the 26-mer Φ-SeNB oligo sequence is shown in blue except for GC6 that is shown in red with an asterisk; the oligo 20-XL is shown in green; the red X shows the location of the cross-link. (B) The reaction pathway for interstrand T-to-A cross-linking caused by oxidation of an oligonucleotide containing two phenyl-selenide-derivatized T residues. C, autoradiogram of a preparative gel showing the separation of residual noncross-linked 99*-mer from the cross-linked 99*-mer with 24Φ-SeB (arrow). (D) Pooled data from four independent sets of experiments with standard error bars showing the protection of the substrate from melting at the reporter strand by Tus in the blocking and nonblocking orientations of Ter.
The DNA triplex 99B*-24-Φ-SeB-30XL-45R* with Ter in the blocking orientation was constructed by annealing the purified 99B*-24-Φ-SeB duplex with molar excess of unlabeled 30XL and labeled 45R* (Fig. 4A Top and Middle). We also constructed the control substrate with Ter in the nonblocking orientation by annealing and cross-linking the 26-mer Φ-SeNB with the labeled 99NB* upper strand followed by hybridization of the partial duplex with a molar excess of the 20-XL and 45R (see Fig. 4A Bottom).
The measurements of the percentage of the input triplex in which the labeled reporter oligo was protected from helicase activity in both orientations of the Ter, as a function of increasing molar ratio of Tus/Ter are shown with the standard error bars in Fig. 4D. The data were collected from four independent experiments for each orientation of Ter and showed that interaction of Tus with Ter in the blocking orientation impeded helicase progression despite the presence of two interstrand cross-links immediately preceding the C6. The helicase progression in the control substrate with Ter in the reverse orientation was much less responsive to increasing Tus concentration. The locations of the cross-links in the blocking orientation (Fig. 4A Top and Middle) were such that a denaturation bubble such as that required by model II could not have formed in such a triplex.
We also asked the question as to whether there would be a more robust arrest of DNA sliding in a substrate that was provided with a 7-bp bubble of unpaired bases including GC6 in comparison with a substrate that lacked such an unpaired bubble and found that such an enhancement did not occur (Fig. S3 and SI Text).
The Tus Mutant Form E49K Was Defective in Arresting Helicase Sliding.
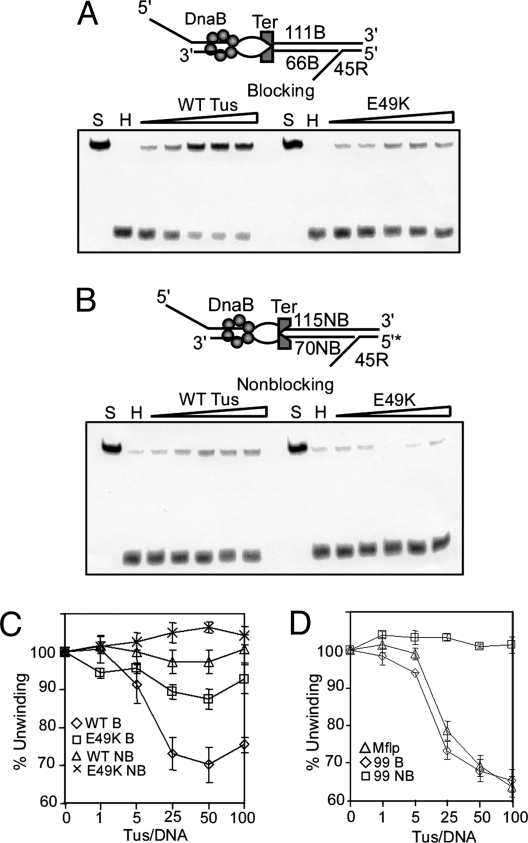
It is generally believed that helicase sliding and DNA unwinding are caused by the same ATP hydrolysis-dependent locomotion of DnaB on both strands or on only one strand of the DNA duplex, respectively (25, 27). Consistent with this postulate, one would expect that mutations in Tus, which are defective in arresting helicase-catalyzed DNA unwinding, should also be similarly defective in arresting a sliding helicase. To verify this prediction, we measured the magnitude of arrest of a sliding DnaB by the mutant form E49K in comparison with the WT Tus in a triplex substrate with a bubble of five unpaired bases that did not include GC6 (Fig. 5).
Fig. 5.
Effects of a mutation in Tus and another in Ter on the arrest of helicase sliding. (A) Representative phosphorimagergrams of gels showing arrest of sliding DnaB by the WT and the E49K mutant form of Tus in the blocking orientation of Ter. S, substrate, H substrate plus DnaB. The wedge indicates addition of an increasing range of molar ratios of Tus to DNA of 1 to 100. (B) The same as A except that the substrate contained the Ter site in the nonblocking orientation. (C) Pooled data from three independent sets of experiments shown with standard error bars. (D) Effect of the transversion GC6 to AT6 in Ter on arrest of DnaB sliding. The triplex was constructed by annealing Mflp1-Mflp2–45R* oligos (Table S1). The data are pooled from eight independent sets of experiments and plotted with standard error bars.
Preliminary experiments showed that this triplex was somewhat better melted at a given concentration of DnaB at the reporter oligo in comparison with the standard triplex without a bubble (data not shown). We prepared 111B-66B-45R* with Ter in the blocking and the corresponding triplex 115NB-70NB-45R* (Table S1) with the Ter site in the nonblocking orientation and performed three sets of helicase sliding/arrest measurements. Representative phosphorimagergrams are shown in Fig. 5 A and B, and the data from three independent sets of experiments with standard error bars are shown in Fig. 5C. The results described above showed that the E49K mutant form, which was previously known to be partially defective in interaction with DnaB and in arresting helicase-catalyzed unwinding, without any detectable loss of binding to Ter DNA (19), was also partially defective in arresting a sliding helicase. These data appear to be consistent with the notion that sliding and unwinding are just two manifestations of the same helicase translocation mechanism. Moreover, the results provided further support to the conclusion that the loss of DnaB–Tus interaction led to a decrease in the ability of Tus to impede helicase translocation. On the basis of the experiments presented above, we conclude that DNA melting at the blocking end of the Tus–Ter complex was not essential for causing polar arrest of DnaB translocation. The data suggest further that DnaB–Tus interaction appears to be an important and necessary component of the polar fork arrest mechanism. We have obtained identical results by using the DNA substrates 99B-54B-45R* and 99NB-54NB-45R* that lacked a bubble of unpaired bases at Ter (data not shown).
A Ter mutant with a GC-to-TA transversion at position 6 did not reduce Tus-mediated arrest of helicase movement in vitro. We reasoned that if C6 flipping were not necessary for arresting a sliding DnaB in vitro a transversion of CG6 to AT6 should not have had a noticeable impact on the magnitude of the process. To test this hypothesis, we constructed a triplex with the mutated Ter site out of the 99-mer called Mflp1, a 54-mer called Mflp2 and the reporter 45R* (Table S1). We compared the relative abilities of the Mflp triplex, the normal triplex 99B-54B-45R*, and the control nonblocking 99NB-54NB-45R* to arrest sliding DnaB in the presence of WT Tus. Eight independent sets of experiments were carried out, and the data with standard errors were plotted as a function of Tus/Ter molar ratios (Fig. 5D). No significant difference in the polarity of fork arrest was observed between the WT and the mutant form of Ter. This result is consistent with our in vivo data (32) but appears to be at variance with in vivo data that revealed retention of up to ≈33% of polar fork arrest activity in this mutant (33). We do not know the reason for the different in vivo observations.
Discussion
The data presented here support the conclusion that polar arrest of helicase translocation in vitro does not depend on DNA melting and flipping of GC6. In vitro experiments with the E49K mutant form of Tus supports the conclusion that DnaB–Tus contacts are involved in helicase arrest. The mutation causes loss of binding with DnaB without causing any loss of binding affinity for Ter DNA (19, 20). These findings do not support model II but are consistent with a variety of in vivo and in vitro data pertaining to replication termination.
We have previously reported that a termination complex arrests transcription catalyzed by RNA polymerase of E. coli in vitro at the −6 and −11 positions of Ter (18) (Fig. 1B). Such arrest has also been observed in vivo (34). The data are consistent with the possibility that transcriptional arrest occurs when the leading edge of the enzyme either contacts or approaches closely the Tus–Ter complex. Because the open complex formation is known to occur deep within the body of RNA polymerase (35), it follows that RNA polymerase arrest should occur before the enzyme causes base-pair melting within the Ter sequence. Furthermore, if the transcribing RNA polymerase were allowed to proceed further and melt the Ter site, up to and including GC6, the leading edge of the enzyme would have displaced Tus from Ter, thereby breaching through the terminus and nullifying its arrest. These results are more easily explained on the basis of model I shown in Fig. 1C. Does RNA polymerase arrest involve Tus–RNA polymerase interaction? Although this question has not been experimentally addressed with Tus, we have investigated this question with regard to RTP, the terminator protein of B. subtilis, and have found that a single point mutant that reduced RTP–DnaB interaction and failed to arrest the helicase was also defective in arresting RNA polymerase, suggesting possibly a common or overlapping interaction domain (36).
As described earlier, the Tus–Ter complex does not arrest all helicases in vitro (14) and in vivo, and the termination complex either does not arrest or arrests very poorly the hexameric helicase DnaC of B. subtilis (16). This relative, albeit limited, specificity of arrest is more easily explained on the basis of a model that does not require a mechanism exclusively dependent on strong and stable binding of Tus to Ter. The RTP of B. subtilis also arrests forks in a polar mode, and experiments using a fusion protein of RTP with a fragment of GFP suggests a mechanism of fork arrest that is more consistent with interaction of RTP with a component of the replisome, possibly the DnaC helicase rather than just RTP-Ter binding (37).
X-ray crystallography of a branched Ter DNA bound to Tus, in the absence of other replisomal components, has revealed a C6 flip (21). Although both our in vitro results reported here and our in vivo data (32) showed that a C6 to A6 in transversion in Ter did not manifest any loss of arrest of a sliding or unwinding DnaB, another group (33) has reported that the same mutation can cause a 3-fold reduction in fork arrest. This incremental reduction in fork arrest perhaps can be reconciled with other observations as discussed below.
We propose that the GC6 base pair and its interaction with Tus might function as a fail-safe mechanism in situations where the DnaB helicase might unwind past the C6 and the T7 residues of Tus, resulting in potential loss of vital Tus–Ter contacts and abolition of fork arrest. In such a situation, the interaction of C6 with Tus might compensate for the loss of protein–protein contacts caused by the extra unwinding and restore fork arrest by a mechanism that not only involves Tus–Ter interaction but also Tus–DnaB contact. We want to state clearly that generation of polarity solely by enhanced DNA–protein interaction resulting from C6–Tus interaction does not appear to be a viable model. In this context, a sliding helicase that does not unwind DNA would not appear to need such an additional backup mechanism and that might explain why mutations in GC6 had no detectable effect on arresting helicase translocation in vitro.
Experimental Procedures
Oligonucleotides and Triplex Construction.
Phenyl-selenide oligos were prepared as described (29, 30, 38). For all of the oligos used in this work and the details of the substrate construction see Table S1 and SI Text.
Enzymes.
DnaB and Tus were purified as described (6).
Helicase Sliding and S1 Nuclease Analysis.
Helicase sliding measurements were carried out as described (6). S1 nuclease analyses are described in detail in SI Text.
Supplementary Material
Acknowledgments.
We thank Drs. Stanley Cohen, Paul Modrich, Jerry Hurwitz, Richard Novick, and Dhruba. Chattoraj for useful comments on our manuscript. This work was supported by grants from the National Institute of General Medical Sciences and National Institute of Allergy and Infectious Diseases (to D.B.) and National Institute of General Medical Sciences Grant GM-054996 (to M.M.G.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805898105/DCSupplemental.
References
- 1.Bastia D, Mohanty BK. In: DNA Replication in Eukaryotic Cells. DePamphilis M, editor. Plainview, NY: Cold Spring Harbor Lab Press; 1996. pp. 177–215. [Google Scholar]
- 2.Bastia D, Mohanty B. K. In: DNA Replication and Human Disease. DePamphilis M, editor. Plainview, NY: Cold Spring Harbor Lab Press; 2006. pp. 155–174. [Google Scholar]
- 3.Rothstein R, Michel B, Gangloff S. Replication fork pausing and recombination or “gimme a break”. Genes Dev. 2000;14:1–10. [PubMed] [Google Scholar]
- 4.Brewer BJ, Fangman WL. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 5.Dalgaard JZ, Klar AJ. Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature. 1999;400:181–184. doi: 10.1038/22139. [DOI] [PubMed] [Google Scholar]
- 6.Khatri GS, MacAllister T, Sista PR, Bastia D. The replication terminator protein of E. coli is a DNA sequence-specific contra-helicase. Cell. 1989;59:667–674. doi: 10.1016/0092-8674(89)90012-3. [DOI] [PubMed] [Google Scholar]
- 7.Lee EH, Kornberg A, Hidaka M, Kobayashi T, Horiuchi T. Escherichia coli replication termination protein impedes the action of helicases. Proc Natl Acad Sci USA. 1989;86:9104–9108. doi: 10.1073/pnas.86.23.9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill TM, Marians KJ. Escherichia coli Tus protein acts to arrest the progression of DNA replication forks in vitro. Proc Natl Acad Sci USA. 1990;87:2481–2485. doi: 10.1073/pnas.87.7.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaul S, et al. The replication terminator protein of the gram-positive bacterium Bacillus subtilis functions as a polar contrahelicase in gram-negative Escherichia coli. Proc Natl Acad Sci USA. 1994;91:11143–11147. doi: 10.1073/pnas.91.23.11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahoo T, Mohanty BK, Patel I, Bastia D. Termination of DNA replication in vitro: Requirement for stereospecific interaction between two dimers of the replication terminator protein of Bacillus subtilis and with the terminator site to elicit polar contrahelicase and fork impedance. EMBO J. 1995;14:619–628. doi: 10.1002/j.1460-2075.1995.tb07038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bussiere DE, Bastia D, White SW. Crystal structure of the replication terminator protein from B. subtilis at 2.6 A. Cell. 1995;80:651–660. doi: 10.1016/0092-8674(95)90519-7. [DOI] [PubMed] [Google Scholar]
- 12.Kamada K, Horiuchi T, Ohsumi K, Shimamoto N, Morikawa K. Structure of a replication-terminator protein complexed with DNA. Nature. 1996;383:598–603. doi: 10.1038/383598a0. [DOI] [PubMed] [Google Scholar]
- 13.Mohanty BK, Sahoo T, Bastia D. The relationship between sequence-specific termination of DNA replication and transcription. EMBO J. 1996;15:2530–2539. [PMC free article] [PubMed] [Google Scholar]
- 14.Sahoo T, Mohanty BK, Lobert M, Manna AC, Bastia D. The contrahelicase activities of the replication terminator proteins of Escherichia coli and Bacillus subtilis are helicase-specific and impede both helicase translocation and authentic DNA unwinding. J Biol Chem. 1995;270:29138–29144. doi: 10.1074/jbc.270.49.29138. [DOI] [PubMed] [Google Scholar]
- 15.Bidnenko V, Lestini R, Michel B. The Escherichia coli UvrD helicase is essential for Tus removal during recombination-dependent replication restart from Ter sites. Mol Microbiol. 2006;62:382–396. doi: 10.1111/j.1365-2958.2006.05382.x. [DOI] [PubMed] [Google Scholar]
- 16.Andersen PA, Griffiths AA, Duggin IG, Wake RG. Functional specificity of the replication fork-arrest complexes of Bacillus subtilis and Escherichia coli: Significant specificity for Tus-Ter functioning in E. coli. Mol Microbiol. 2000;36:1327–1335. doi: 10.1046/j.1365-2958.2000.01945.x. [DOI] [PubMed] [Google Scholar]
- 17.Velten M, et al. A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol Cell. 2003;11:1009–1020. doi: 10.1016/s1097-2765(03)00130-8. [DOI] [PubMed] [Google Scholar]
- 18.Mohanty BK, Sahoo T, Bastia D. Mechanistic studies on the impact of transcription on sequence-specific termination of DNA replication and vice versa. J Biol Chem. 1998;273:3051–3059. doi: 10.1074/jbc.273.5.3051. [DOI] [PubMed] [Google Scholar]
- 19.Mulugu S, et al. Mechanism of termination of DNA replication of Escherichia coli involves helicase-contrahelicase interaction. Proc Natl Acad Sci USA. 2001;98:9569–9574. doi: 10.1073/pnas.171065898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson TA, Nilles AF, Valjavec-Gratian M, Hill TM. Site-directed mutagenesis and phylogenetic comparisons of the Escherichia coli Tus protein: DNA-protein interactions alone can not account for Tus activity. Mol Genet Genomics. 2001;265:941–953. doi: 10.1007/s004380100501. [DOI] [PubMed] [Google Scholar]
- 21.Mulcair MD, et al. A molecular mousetrap determines polarity of termination of DNA replication in E. coli. Cell. 2006;125:1309–1319. doi: 10.1016/j.cell.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 22.LeBowitz JH, McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J Biol Chem. 1986;261:4738–4748. [PubMed] [Google Scholar]
- 23.San Martin MC, Stamford NP, Dammerova N, Dixon NE, Carazo JM. A structural model for the Escherichia coli DnaB helicase based on electron microscopy data. J Struct Biol. 1995;114:167–176. doi: 10.1006/jsbi.1995.1016. [DOI] [PubMed] [Google Scholar]
- 24.Yu X, Jezewska MJ, Bujalowski W, Egelman EH. The hexameric E. coli DnaB helicase can exist in different quaternary states. J Mol Biol. 1996;259:7–14. doi: 10.1006/jmbi.1996.0297. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan DL. The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J Mol Biol. 2000;301:285–299. doi: 10.1006/jmbi.2000.3965. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan DL, O'Donnell M. Twin DNA pumps of a hexameric helicase provide power to simultaneously melt two duplexes. Mol Cell. 2004;15:453–465. doi: 10.1016/j.molcel.2004.06.039. [DOI] [PubMed] [Google Scholar]
- 27.Kaplan DL, O'Donnell M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol Cell. 2002;10:647–657. doi: 10.1016/s1097-2765(02)00642-1. [DOI] [PubMed] [Google Scholar]
- 28.Abhyankar MM, Zzaman S, Bastia D. Reconstitution of R6K DNA replication in vitro using 22 purified proteins. J Biol Chem. 2003;278:45476–45484. doi: 10.1074/jbc.M308516200. [DOI] [PubMed] [Google Scholar]
- 29.Hong IS, Greenberg MM. DNA interstrand cross-link formation initiated by reaction between singlet oxygen and a modified nucleotide. J Am Chem Soc. 2005;127:10510–10511. doi: 10.1021/ja053493m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong IS, Ding H, Greenberg MM. Oxygen independent DNA interstrand cross-link formation by a nucleotide radical. J Am Chem Soc. 2006;128:485–491. doi: 10.1021/ja0563657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong IS, Ding H, Greenberg MM. Radiosensitization by a modified nucleotide that produces DNA interstrand cross-links under hypoxic conditions. J Am Chem Soc. 2006;128:2230–2231. doi: 10.1021/ja058290c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sista PR, Hutchinson CA, 3rd, Bastia D. DNA–protein interaction at the replication termini of plasmid R6K. Genes Dev. 1991;5:74–82. doi: 10.1101/gad.5.1.74. [DOI] [PubMed] [Google Scholar]
- 33.Coskun-Ari FF, Hill TM. Sequence-specific interactions in the Tus-Ter complex and the effect of base pair substitutions on arrest of DNA replication in Escherichia coli. J Biol Chem. 1997;272:26448–26456. doi: 10.1074/jbc.272.42.26448. [DOI] [PubMed] [Google Scholar]
- 34.Roecklein B, Pelletier A, Kuempel P. The tus gene of Escherichia coli: Autoregulation, analysis of flanking sequences, and identification of a complementary system in Salmonella typhimurium. Res Microbiol. 1991;142:169–175. doi: 10.1016/0923-2508(91)90026-7. [DOI] [PubMed] [Google Scholar]
- 35.Murakami KS, Darst SA. Bacterial RNA polymerases: The whole story. Curr Opin Struct Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 36.Gautam A, Mulugu S, Alexander K, Bastia D. A single domain of the replication termination protein of Bacillus subtilis is involved in arresting both DnaB helicase and RNA polymerase. J Biol Chem. 2001;276:23471–23479. doi: 10.1074/jbc.M009537200. [DOI] [PubMed] [Google Scholar]
- 37.Duggin IG. DNA replication fork arrest by the Bacillus subtilis RTP-DNA complex involves a mechanism that is independent of the affinity of RTP-DNA binding. J Mol Biol. 2006;361:1–6. doi: 10.1016/j.jmb.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 38.Hong IS, Greenberg MM. Efficient DNA interstrand cross-link formation from a nucleotide radical. J Am Chem Soc. 2005;127:3692–3693. doi: 10.1021/ja042434q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.