Abstract
The von Hippel-Lindau (VHL) tumor suppressor gene regulates extracellular matrix deposition. In VHL negative renal cancer cells, VHL(-), the lack of fibronectin matrix assembly is thought to promote and maintain tumor angiogenesis allowing vessels to infiltrate tumors. Therefore, and considering the importance of this process in tumor growth, we aimed to study why VHL(-) renal cancer cells fail to form a proper extracellular matrix. Our results showed that VHL(-) cells were not defective in fibronectin production and that the fibronectin produced by these cells was equally functional in promoting cell adhesion and matrix assembly as that produced by VHL(+) cells. We have previously reported that VHL(-) cells fail to form β1 integrin fibrillar adhesions and have a diminished organization of actin stress fibers; therefore, we aimed to study if the small GTPase family is involved in this process. We found that activation of the RhoA GTPase was defective in VHL(-) cells, and this was possibly mediated by an increased activation of its inhibitor, p190RhoGAP. Additionally, the expression of constitutively active RhoA in VHL(-) cells resulted in formation of a fibronectin matrix. These results strongly suggest an important role for RhoA in some of the defects observed in renal cancer cells.
The von Hippel-Lindau disease is an autosomal dominant hereditary cancer syndrome caused by germ line mutations of the von Hippel-Lindau (VHL)4 tumor suppressor gene (1). According to the clinical manifestations, the disease has been classified into VHL type 1 and VHL type 2. VHL type 2 disease is further classified into three categories: type 2A, type 2B, and type 2C (1). VHL protein (pVHL) plays an important role in the oxygen-sensing pathway, and its best known function is to promote the ubiquitination and subsequent elimination by the proteasome of the hypoxia-inducible transcription factors HIF1α and HIF2α (2, 3). Loss of VHL leads to activation of the HIF pathway in normoxia (4), which in turn leads to excessive transcription of HIF-α target genes, including the angiogenic factor VEGF (5). Despite the latter providing an explanation for the high vascularization of VHL(-) tumors, it remains unclear how the loss of VHL leads to renal cancer.
A recent study has suggested that in fibrosarcoma tumor models, additional genetic changes other than dysregulation of HIF are necessary for the induction of tumorigenesis (6). Indeed, other pVHL functions independent of the regulation of HIF have been reported. These include regulation of cell motility and invasion (7-9), stability of microtubules (10-12), maintenance of an epithelial-like cell shape and monolayer organization (13-18), cell cycle and apoptosis (19-21), ciliogenesis (22, 23), and in addition, it has been recently shown that it serves as a protein kinase adaptor regulating the activity of NF-κB (24).
Adequate interaction of cells with the surrounding matrix regulates essential aspects of normal cell function, and breakdown of the basement membrane occurs in cancer progression and is often associated with solid tumors (25). With regard to RCC tumors, we and others have shown that RCCs lacking VHL fail to organize a normal extracellular fibronectin (FN) matrix (14, 26, 27). Therefore, the lack of a proper cell matrix might be involved in the aggressive behavior of VHL(-) tumors. In addition, highly angiogenic tumors are not only dependent on the pVHL-HIF 2α degradation pathway but are also a consequence of loss of a proper extracellular matrix assembly (28). Extracellular matrix assembly is a complex multistep process, which first requires binding of FN to cell surface integrins, mostly α5β1 (29). Additionally, other events such as actin stress fiber formation and cell contraction are required for the progressive ordination into detergent-insoluble fibrils (30, 31). These processes are mediated by the small Ras-type GTPase member RhoA (32, 33). In this respect, we have reported the inability of VHL(-) cells to form β1 fibrillar adhesions (14) and intracellular actin stress fibers (16), and other authors have shown that these cells also lack the proper assembly of actin and vinculin, which promotes actin stress fiber formation (8).
Although expression of VHL is necessary for proper extracellular matrix assembly, the mechanism by which pVHL mediates this process is not well understood. To approach this, we have evaluated the levels and functionality of the FN produced by VHL(+) and VHL(-) cells. We have also studied the role of the signaling pathway that controls formation of actin stress fibers and cell contraction in the regulation of FN matrix assembly in VHL(+) and VHL(-) cells. Our results demonstrated that FN expression levels in VHL(+) and (-) cells showed no correlation with the ability to assemble a FN matrix and that VHL(-) cells expressed functional FN that was properly assembled by VHL(+) cells. Additionally, we found that the lack of FN matrix in VHL(-) cells was partly due to sustained down-regulation of RhoA activity via p190RhoGAP-increased activation. These results provide an insight into novel mechanisms altered in VHL(-) renal cancer cells.
EXPERIMENTAL PROCEDURES
Cell Culture—786-O cells lacking VHL (RC3) and subclones stably producing wild-type VHL (WT8) (34), or different VHL mutants such as L188V (type 2C mutant) (35), RRR, and KRR (type 2C mutant) (36), Y112H (type 2A mutant), and C162F (type 2B) mutant. These mutants were provided by Dr. Michael Ohh (University of Toronto, Canada): parental RCC4 and RCC10 and subclones stably producing wild-type VHL (37) and parental UMRC cell lines and their stable VHL transfectants. Cell lines were all maintained as previously described (16).
Antibodies and Reagents—Polyclonal anti-FN antibody was from Sigma. Anti-RhoA was from Santa Cruz Biotechnology, anti-p190RhoGAP from Upstate, and anti-phosphotyrosine antibody PY20 was from Biomol. Anti-paxillin was from BD Biosciences, and anti-activated β1 integrin (HUTS21) was generously provided by Dr. Carlos Cabañas (CSIC). LPA and 2,3-butanedione-monoxime (BDM) were from Sigma, and the Rho kinase inhibitor Y-27632 was from Calbiochem.
Isolation of Deoxycholate-soluble and -insoluble Materials—Cells were cultured in 6-well plates with RPMI 1640 and GLUTAMAX-I (Invitrogen) supplemented with 5% FN-free fetal bovine serum. Deoxycholate-soluble and -insoluble material were obtained as described elsewhere (38). A detailed protocol is described under supplemental data.
Immunofluorescence Microscopy—Immunofluorescence staining was performed as previously described (14). A detailed protocol is described under supplemental data.
Quantitative Real Time PCR Analysis—Quantitative RNA analysis protocol and primer pairs for β-actin have been described elsewhere (39). FN primers used and a detailed protocol are described under supplemental data.
Cell Adhesion Assay—FN purified from VHL(+) or (-) cells was absorbed at 5 μg/ml onto Maxisorb 96-well plates (Costar). The wells were aspirated and blocked with 1% BSA/PBS for 30 min. Cells (20 × 103/well) were resuspended in M199/0.1% BSA and incubated for 1 h at 37 °C in 5% CO2. After washing with PBS, cells were fixed with 1% glutaraldehyde/PBS and stained with 1% crystal violet/PBS. Cells were washed with distilled water, and the absorbance at 590 nm was determined on an ORION-2 plate reader. A negative control consisting of cells attached to BSA was subtracted from each sample.
RhoA Activity Assay—RhoA activity was measured using a GST-rhotekin-Rho binding domain (GST-C21), a gift of Dr. John Collard (The Netherlands Cancer Institute, Amsterdam, The Netherlands). Briefly, a total of 7 × 106 cells were lysed in radioimmune precipitation assay lysis buffer. After centrifugation (20,000 × g, 15 min, 4 °C), equal amounts of protein from the supernatant were incubated overnight at 4 °C with beads coated with GST-C21. Beads were washed, and bound protein was eluted by boiling in Laemmli buffer. Total and bound RhoA subjected to electrophoresis and transferred to nitrocellulose membranes were detected using an anti-RhoA mAb. For statistical purposes, gels from active and total RhoA normalized with respect to the loading control were subjected to densitometric analysis and analyzed using the Quantity-One™ program (Bio-Rad).
Analysis of p 190RhoGAP Phosphorylation—VHL(+) and (-) RCCs (6 × 106) were lysed in 500 μl of radioimmune precipitation assay buffer. Cell supernatants were incubated overnight at 4 °C with protein G-agarose beads (Pierce) coated with 4 μg of anti-p190RhoGAP mAb (Upstate) or with the IgG1 anti-CD45 T200 antibody (mock). Bound proteins were extracted by boiling in Laemmli buffer, subjected to electrophoresis, and transferred into nitrocellulose membranes. Phosphorylated proteins were detected using PY20 mAb followed by a horseradish peroxidase-labeled secondary antibody.
Retroviral Infection—Constructs of a constitutively active form (RhoA Q63L) or a dominant negative form (RhoA N19) of RhoA were kindly provided by Dr. Silvio Gutkind (Bethesda, MD). These constructions were subcloned into the BamH1/EcoR1 site of the retroviral vector pLZR IRES-GFP. 786-O cell infection was performed as described previously (16). siRNA Transfection—Control small interfering RNA (siRNA) or siRNA directed to human p190RhoGAP (5′GAACAGCGAUUUAAAGCAUTT-3′ and 3′AUGCUUUAAAUCGCUGUUCTT-5′) (Eurogentec) were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. Three days after transfection, cells were used for the experiment.
RESULTS
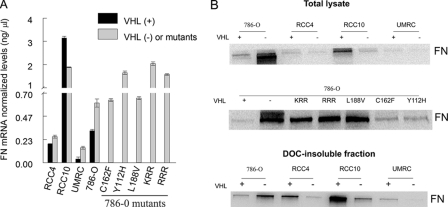
FN mRNA and Protein Expression Levels in VHL(+) and VHL(-) RCC Cells Reveal No Correlation with the Ability to Assemble an FN Matrix—The reduced FN matrix deposition previously observed by immunofluorescence and ELISA studies (14, 26, 40) could be partially explained by an aberrant production or a defective secretion of FN in VHL(-) RCC cells (11, 41). To determine whether this was a general phenomenon in RCCs, we first analyzed the FN mRNA and protein levels in several RCC cell lines and in cells with different pVHL mutations corresponding to types 2A, 2B, and 2C phenotype of the VHL disease. These included the mutants Y112H (type 2A), C162F (type 2B), L188V (type 2C), and a nonneddylateable version of pVHL termed RRR (type 2C), or the control termed KRR (36), all of them on the RCC 786-O background. Remarkably, FN mRNA levels measured by quantitative RT-PCR were significantly lower in 786-O and UMRC VHL(+) cells than in their corresponding VHL(-) counterparts (Fig. 1A). Similarly, FN mRNA levels in the various pVHL mutants were higher when compared with the corresponding levels in 786-O VHL(+) cells (Fig. 1A). In contrast, in RCC10 cells, FN mRNA expression was always higher in VHL(+) versus VHL(-) cells (Fig. 1A).
FIGURE 1.
Comparative study of FN mRNA and protein levels in several VHL(+) and VHL(-) RCC cells. A, FN gene transcription levels in 786-O, RCC4, UMRC, RCC10, and pVHL mutants were determined by quantitative real time PCR. FN mRNA levels were normalized to β-actin, and the number of copies were extrapolated from the corresponding standard curve expressed in ng/μl. Values represent the average from three different experiments (p ≤ 0.05). B, FN endogenous levels were determined by Western blots. Total cell lysates or deoxycholate-insoluble fractions were immunoblotted with an anti-FN polyclonal Ab. A representative experiment out of several performed with similar results is shown.
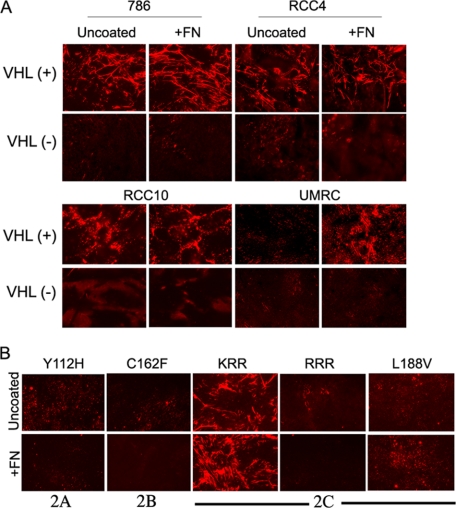
We also analyzed FN protein levels in cell lysates or deoxycholate-insoluble fractions derived from all these cell types. In agreement with the mRNA data, VHL(-) and mutants derived from 786-O parental cells consistently showed higher FN protein expression than 786-O VHL(+) cells (Fig. 1B). Because VHL(-) cells do not assemble FN, the FN encountered in the deoxycholate-insoluble fraction could be due to FN interacting with other cytoskeletal proteins. In contrast, RCC4, RCC10, and UMRC VHL(-) cells had lower FN protein levels than their VHL(+) counterparts, both in lysates and in deoxycholate-insoluble fractions (Fig. 1B). Therefore, protein levels did not always correspond with mRNA levels, and this might be partially explained by the fact that protein levels were measured in total cell lysates. Therefore, we discarded part of the FN contained in the conditioned medium. Interestingly, we purified FN from the conditioned medium of the different VHL(+) and (-) cell lines and found similar FN-secreted levels (data not shown). To further confirm the above conclusion, we analyzed the assembly of FN matrix in the absence or presence of exogenous FN. We have previously reported that addition of exogenous FN did not re-establish assembly in 786-O VHL(-) cells (14). Here we analyzed the assembly in three additional cell lines, RCC4, RCC10, and UMRC and in the VHL mutants described above. Our results showed that VHL(-) cells cultured on uncoated or FN-coated coverslips did not form visible FN fibrils, suggesting that FN availability is not exclusively responsible for their incapacity to form a matrix (Fig. 2A). Interestingly, VHL(+) but not VHL(-) UMRC cells, only formed a visible matrix when cultured on exogenous FN (Fig. 2A), indicating that, in this particular case, low secretion of FN (see Fig. 1, A and B) might be responsible for the lack of FN matrix in this VHL-expressing cell line. Additionally, none of the three different mutants, type 2A, 2B, or 2C, were able to assemble FN even when it was exogenously added (Fig. 2B). Altogether, these results indicated a heterogeneous expression of FN on the various VHL(+) and VHL(-) RCCs and most importantly, that lack of matrix assembly by VHL(-) cells could not always be attributed to low FN production in these cells.
FIGURE 2.
Analysis of FN matrix organization in RCCs and pVHL mutants in the presence of exogenous FN. A, 786-O, RCC4, RCC10, and UMRC RCC cells and their counterparts expressing VHL were grown to confluence on uncoated or FN (10 μg/ml)-coated coverslips. After 3-4 days, cells were lysed, and the FN matrix was analyzed by indirect immunofluorescence using a polyclonal anti-FN antibody. B, type 2A (Y112H), 2B (C162F), and 2C (L188V, RRR and control KRR) pVHL mutants were grown under the same conditions as in A, and the FN matrix was analyzed after 4 days of cell culture. The images shown are representative of at least three experiments.
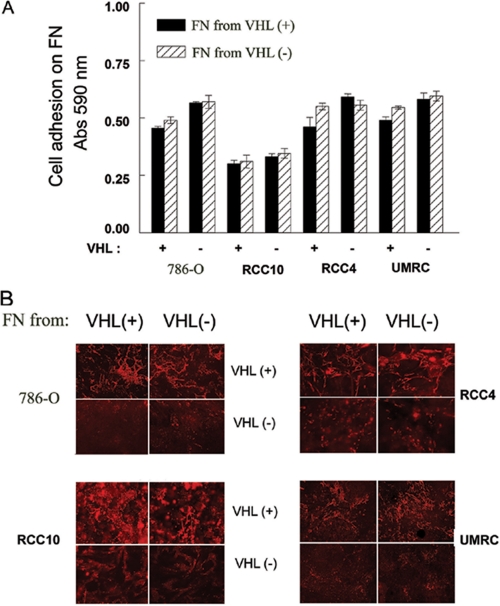
Fibronectin Produced by VHL(-) Cells Is Functional and Normally Assembled into Fibrils—If FN production and secretion do not always explain the failure to assemble FN by VHL(-) cells, the possibility exists that FN produced by cells lacking VHL is malfolded or malprocessed and hence not functional for its assembly. We therefore studied whether the FN produced by VHL(-) cells was able to bind to its receptor, the α5β1 integrin, and to form fibrils. To this end, we first performed cell adhesion experiments using FN purified from VHL(+) or VHL(-) cell-conditioned medium. As shown in Fig. 3A, all cell lines showed similar levels of adhesion to FN from either source, and this was independent of their VHL status. These results indicated that FN synthesized by VHL(-) cells is able to mediate cell adhesion as efficiently as that synthesized by VHL(+) cells. Moreover, they also indicated that the lack of FN assembly by VHL(-) cells is not due to a decreased α5β1 integrin activity, further extending our previous observations (14) as well as those of others (13), showing that expression and activation of α5β1 on VHL(-) cells are normal.
FIGURE 3.
Functional evaluation of FN purified from VHL(+) and VHL(-) RCC cells. A, cell adhesion on FN purified from VHL(+) and VHL(-) cell-conditioned medium using gelatin-agarose affinity matrices. Cells (2 × 104/well) were incubated on 96-well FN (5 μg/ml)-coated plates for 1 h at 37 °C in 5% CO2, fixed, and stained as described under “Experimental Procedures.” Attached cells were then quantitated at 590 nm. A negative control BSA background was used. The average from three different experiments is represented. B, FN assembly was analyzed by indirect immunofluorescence staining. 786-O, RCC4, RCC10, and UMRC (2 × 105 cells) were plated on uncoated or FN (10 μg/ml)-coated coverslips, using FN purified from VHL(+) or VHL(-) cells. After 3-4 days of culture at 37 °C and 5% CO2, FN matrix formation was analyzed with a conventional immunofluorescence microscope. A representative experiment of at least four performed is shown.
We also compared the ability of FN purified from the conditioned medium of VHL(+) and VHL(-) cells to assemble into fibrils. Our results confirmed that VHL(-) RCCs did not assemble a FN matrix, even in the presence of exogenous FN from VHL(+) cells (Fig. 3B). In contrast, VHL(+) RCCs consistently formed a FN matrix when cultured either on FN purified from their own conditioned medium or from that of VHL(-) cells (Fig. 3B). To discern between assembly of endogenous and exogenous FN, we used UMRC VHL(+) cells, because they barely produce FN, and hence they do not assemble unless we provide them with exogenous FN (see Fig. 2A). As shown in Fig. 3B, UMRC VHL(+) cells showed a similar assembly pattern when cultured on FN from VHL(+) or VHL(-) cells. Therefore, these results indicated that FN produced by VHL(-) cells does not interfere with normal FN function in VHL(+) cells. Additionally, they clearly established that FN secreted by VHL(-) cells is fully functional and able to be assembled by VHL(+) cells.
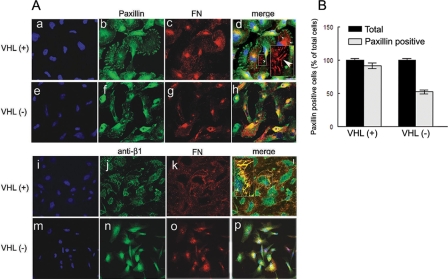
VHL(-) Cells Are Unable to Trigger the FN Stretching Necessary for Its Assembly—We have previously shown that VHL(-) cells fail to assemble proper β1 fibrillar adhesions (FAs), a cell matrix adhesion structure essential for FN assembly (14). Therefore, the lack of proper formation of actin stress fibers and cell contraction could explain the defective assembly of FN by VHL(-) cells, because these events are required for the exposure of cryptic sites in FN and its progressive ordination into detergent-insoluble fibrils (33). To further analyze this process, we repeated our previous studies but now in the presence of exogenous FN to determine if VHL(-) cells were able to initialize the first steps of FN stretching required for its assembly into fibrils. After 4 h in culture, we analyzed by confocal microscopy the formation of focal contacts (FCs) by staining with paxillin, a major component of these structures, and with a FN antibody to see how FN was being stretched from these structures. Our results showed that VHL(+) cells arranged proper FCs, and the initial steps of fibril formation with short fibrils appearing at the edge of the FCs were very evident (Fig. 4A, panels c and d, white arrow-heads). In contrast, VHL(-) cells, although forming proper FCs, were not as efficient as VHL(+) cells, and the number of cells forming proper FCs were lower compared with VHL(+) cells (Fig. 4, A, panels f-h and B). These results strongly support our findings in that the impaired or defective formation of focal contacts may affect the proper formation of fibrillar adhesions required for FN assembly into fibrils. After 24 h, these structures became more mature and organized elongated FAs, which colocalized with FN fibrils in VHL(+) cells (Fig. 4, panels j-l, see magnified area). On the contrary, and even in the presence of exogenous FN, VHL(-) cells failed to form FA structures as well as arranging FN into fibrils, (Fig. 4, panels n-p). These results and our previous observation of a diminished organization of actin stress fibers in VHL(-) cells (16) support the hypothesis that a decreased FN stretching could contribute to the lack of FN assembly in these cells.
FIGURE 4.
FN stretching and cell matrix contacts in VHL(+) versus VHL(-). A, 786-O VHL(+) and VHL(-) cells were plated on coverslips previously coated with FN (20 μg/ml) and cultured for either 4 h or 24 h. Cells were fixed and permeabilized before DAPI staining (panels a, e, i, and m) and double immunostaining with antibodies against Paxillin (panels b, f, d, and h), and FN (panels c, g, d, and h) or activated β1 integrins (panels j, n, l, and p) and FN (panels k, o, l, and p). Samples were then analyzed with a confocal microscope. Horizontal sections are presented (at least six different fields were evaluated). A representative experiment is shown. Bar, 25 μm. Magnified areas at zoom 3×. B, positive cells for paxillin staining were counted, at least 10 cells per field (at least three fields) in three independent experiments. Results are represented as percentage of total cells.
Activation of the Small GTPase RhoA Is Defective in VHL(-) Cells—It is well established that the GTP-binding protein RhoA mediates the formation of actin stress fibers and regulates cellular functions such as cell adhesion and contractility (42). Furthermore, inhibitors of RhoA-mediated cellular contractility prevent FN assembly into fibrils (32), and a constitutively active form of RhoA restores the assembly of FN into fibrils in cells lacking active β1 integrins (43). Accordingly, we hypothesized that RhoA activation and/or function could be defective in VHL(-) cells.
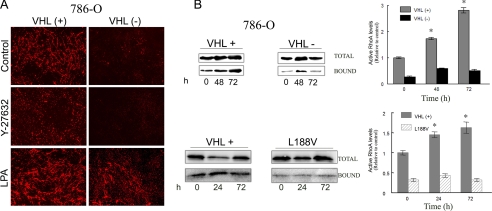
To determine whether RhoA was directly involved in FN matrix assembly in RCC, we first performed immunofluorescence experiments to analyze the assembly of FN by 786-O VHL(+) and VHL(-) RCCs in the presence of inhibitors or activators of the RhoA signaling pathway. In the presence of Y-27632 or 2-3-BDM, inhibitors of RhoA downstream effectors, we found that the assembly of FN was completely abolished in VHL(+) cells (Fig. 5A and data not shown). Interestingly, stimulation of RhoA by LPA resulted in increased FN fibril formation in VHL(+) cell cultures and induction of a few FN fibrils by VHL(-) cells (Fig. 5A). We next performed RhoA activity assays using the C21-GST fusion protein. We evaluated RhoA activity in a time course from 0 to 72 h in culture. As shown in Fig. 5B, the RCC cell line 786-O stably expressing wild-type pVHL showed a basal RhoA activity, which progressively increased over time. In contrast, 786-O VHL(-) cells and the VHL mutant L188V, had a very low basal RhoA activity that did not increase over time (Fig. 5B). Similar results were obtained for the RCC4 cell line, which displayed higher levels of active RhoA in VHL(+) compared with VHL(-) cells (supplemental Fig. S1). These results suggested that the RhoA signaling pathway was playing a role in the functional alterations observed on these VHL(-) cells. Furthermore, the results with the type 2C mutants, which retained the ability to regulate HIF as the VHL wild type, indicated that RhoA activity in these cells seems to be independent of HIF.
FIGURE 5.
RhoA activation state in VHL(+) and (-) RCCs. A, 786-O RCCs were cultured for 3-4 days in the absence or presence of the RhoA signaling inhibitor, Y27632 (5 μm) or the activator LPA (6 μm). FN matrix formation was analyzed by immunofluorescence experiments performed as described under “Experimental Procedures.” B, 786-O VHL(+), VHL(-) cells and the mutant L188V (7 × 106) were incubated for 3 h in serum-free medium, and this was considered as time 0 for basal levels of RhoA activity. Cells were then maintained in culture in complete medium for an additional period of 24, 48, and 72 h. After each time point, cells were lysed, and the lysates were incubated overnight with C21-GST. Bound and total RhoA were analyzed by Western blot using an anti-RhoA mAb; VHL(+) (gray bars), VHL(-) (black bars), and L188V (striped bars) bound versus total are represented; a representative experiment is shown, and the average of three different experiments of VHL(+) and VHL(-) is represented; *, p ≤ 0.05.
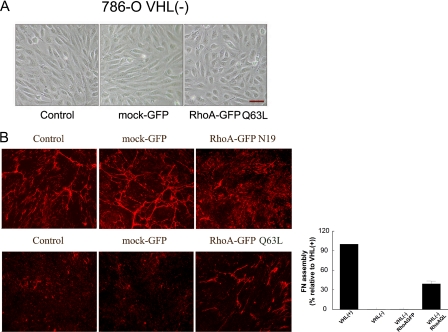
Constitutive Activation of RhoA Stimulates FN Assembly by VHL(-) Cells—To obtain further evidence for the involvement of RhoA in the lack of FN assembly in VHL(-) cells, we introduced a constitutively active (GFP RhoA-Q63L) or a dominant negative (GFP RhoA-N19) recombinant RhoA form in VHL(-) and VHL(+) cells, respectively. The expression of RhoA was increased at least three times (supplemental Fig. S2). Three days after infection, we analyzed changes on cell morphology and observed that infection of VHL(+) cells with GFP RhoA-N19 did not induce phenotypic alterations (data not shown). In contrast, infection of VHL(-) cells with GFP RhoA-Q63L, induced an epithelial cytoarchitecture that resembled that of VHL(+) cells. Cell dimensions changed from 70 ± 5-μm length and 20-μm width to 35 ± 10-μm length and 20-μm width after infection with RhoA-Q63L, whereas infection with the empty vector did not have any effect (Fig. 6A). We next analyzed whether infection with these mutants induced alterations in the assembly of FN. Immunofluorescence experiments with VHL(+)/GFP RhoA-N19 cells revealed a decrease of FN fibrils when compared with the control VHL(+) cells or with cells infected with the empty vector (Fig. 6B, top). The GFP RhoA-Q63L mutant did not increase the constitutive FN matrix assembly observed on VHL(+) (data not shown). In contrast, infection of VHL(-) cells with GFP RhoA-Q63L stimulated the assembly of FN into fibrils, with a recovery of at least 39% with respect to that observed in VHL(+) cells. However, infection with the empty vector had no effect (Fig. 6B, bottom). Altogether, these results further supported the conclusion that the inability of VHL(-) cells to properly assemble a FN matrix is due, at least in part, to an impaired activation of RhoA.
FIGURE 6.
Effect of expressing a recombinant dominant negative or constitutively active RhoA in VHL(+) and (-) cells. A, 786-O cells were infected with a retroviral vector expressing GFP alone or with a constitutively active (RhoA-Q63L) mutant. One week after infection, 786-O cells were grown to confluence and changes in cell morphology were analyzed with a phase contrast microscope. The scale bar represents 50 μm. B, FN matrix formation by 786-O VHL(+) and VHL(-) cells infected with a retroviral vector expressing GFP alone or with a dominant negative (RhoA N19) or a constitutively active (RhoA Q63L) mutants, respectively, was analyzed by indirect immunofluorescence. Cells (2 × 105 cells) were grown for 3-4 days at 37 °C and 5% CO2, and the FN matrix was detected with a polyclonal anti-FN antibody. Quantification of FN assembly recovery in VHL(-) cells with RhoA Q63L is also represented. Error bars represent the standard deviation of at least three independent experiments.
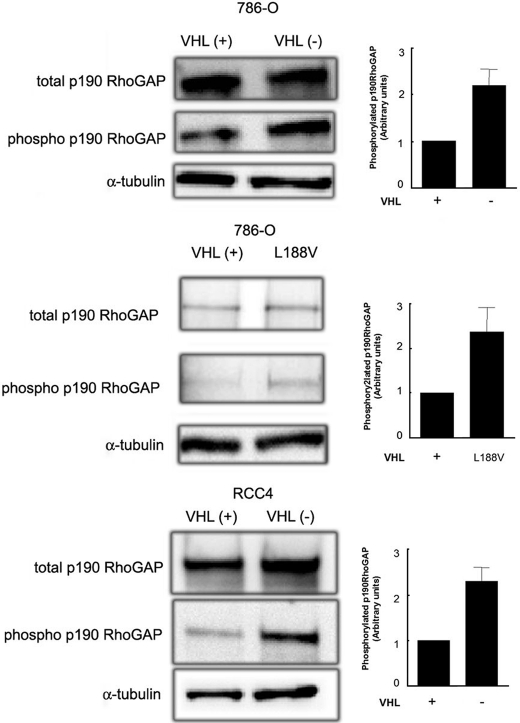
Down-regulation of RhoA Activity in VHL(-) RCC Cells Correlates with Increased p190RhoGAP Phosphorylation—Down-regulation of RhoA activity has been correlated with activation of GTPase-activating proteins such as p190RhoGAP (44). Therefore, we asked whether the decreased RhoA activity encountered in VHL(-) cells was due to an increased activation of p190RhoGAP. To this end, p190RhoGAP was immunoprecipitated from cell lysates, and tyrosine phosphorylation levels were analyzed. Fig. 7 shows that phosphorylated p190RhoGAP was increased in VHL(-) cells and in the L188V mutant (2.3 ± 0.3- and 2.4 ± 0.5-fold, respectively) derived from 786-O cell lines, and 2.35 ± 0.25-fold in VHL(-) cells derived from RCC4 cell lines, with respect to their counterparts stably expressing wild-type pVHL VHL(+) (Fig. 7, top, middle, and bottom panels). These results indicated that the enhanced phosphorylation of p190RhoGAP in VHL(-) cells may explain the decreased RhoA activity in these cells.
FIGURE 7.
Analysis of p190RhoGAP phosphorylation state in VHL(+) and VHL(-) RCCs. 786-O, VHL(+) and (-) cells (top panel), the mutant L188V (medium panel), and RCC4 VHL(+) and (-) cells (bottom panel) were grown for 72 h in complete medium, then serum-starved for 3 h and p190RhoGAP was immunoprecipitated from cell lysates (7 × 106 cells). Total p190RhoGAP constitutive expression was analyzed by Western blot using a specific anti-p190RhoGAP mAb, and phosphorylated p190RhoGAP was measured with the anti-phosphotyrosine mAb PY20. Phospho-p190RhoGAP versus total p190RhoGAP is represented. A representative experiment of several performed is shown.
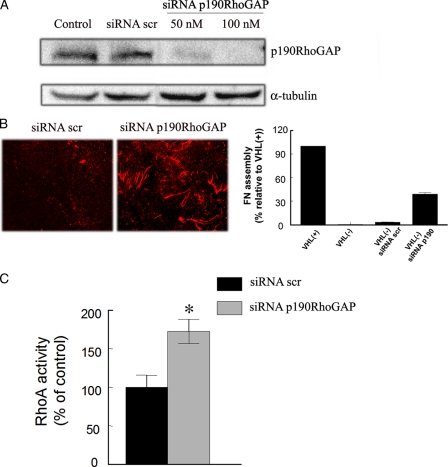
Gene Silencing p190RhoGAP in VHL(-) Cells Partially Restores the Assembly of FN—Because increased activation of p190RhoGAP correlated with decreased RhoA activity in VHL(-) cells, we next addressed the importance of this protein in the assembly of FN. To this aim, we silenced p190RhoGAP expression in VHL(-) cells, and then tested their capability to form FN fibrils. We achieved an 85-95% interference detected by immunoblotting (Fig. 8A). Silencing p190RhoGAP partially restored the ability of VHL(-) cells to form FN fibrils, with a 39% of recovery (Fig. 8B), and the resulting pattern was similar to that observed for (Q63L)RhoA-transfected VHL(-) cells. We also tested whether this effect was due to increased RhoA activity. We found that VHL(-) cells transfected with siRNA to p190RhoGAP showed increased RhoA activity (1.7 ± 0.15-fold) compared with cells transfected with scramble siRNA (Fig. 8C).
FIGURE 8.
Effect of gene-silencing p190RhoGAP on VHL(-) cells assembly of FN. A, silencing of p190RhoGAP was achieved by transfecting 786 VHL(-) cells with a specific siRNA (at the indicated concentrations). The level of silencing was verified by immunoblotting with an anti-p190RhoGAP antibody 3 days after cell transfection. B, FN matrix formation by 786 VHL(-) cells transfected with scramble (scr) RNA or with siRNA to p190RhoGAP (100 nm) was analyzed by indirect immunofluorescence using a polyclonal anti-FN antibody, and quantification of FN assembly recovery is represented. C, RhoA activity in 786 VHL(-) cells transfected with scramble RNA or with siRNA to p190RhoGAP was determined by a RhoGELISA assay.
DISCUSSION
In VHL(-) RCC cells, the lack of FN assembly has been shown to promote and maintain tumor angiogenesis allowing vessels to infiltrate tumors (27). Considering that regulation of FN assembly by pVHL seems to be essential for its tumor suppressor function (27), the aim of our studies was to understand why VHL(-) RCC cells fail to form a proper extracellular matrix. An altered integrin expression/function or insufficient FN synthesis could account for this defect. However, we previously demonstrated (14) that integrin expression or function did not significantly differ in VHL(+) or VHL(-) cells, and in this work we confirm that FN production did not correlate with its assembly. With respect to this, some RCC tumors accumulated FN in the tumor cell cytoplasm, and little or none in the stroma (41), suggesting a defective transport and secretion of FN that could account for the lack of FN assembly by these cells. However, our previous results using cultured cells showed no evidence of intracellular accumulation of FN (14). Additionally, recent studies have demonstrated that VHL(-) cells maintain the ability to secrete FN (27). These findings support our present results showing that the levels of FN produced by VHL(-) cells are apparently independent of its ability to assemble into a matrix.
Thus, other possibilities have to be considered regarding the regulation of FN assembly by pVHL. First, it has been proposed that VHL interaction with FN might be necessary for FN matrix assembly (26, 40, 45). A direct interaction between pVHL and collagen IV was recently reported (46, 47), indicating that the role of pVHL in the remodeling of extracellular matrix components is not exclusive for FN. Kurban et al. (47) also demonstrated that the pVHL/collagen interaction seems to be necessary for collagen assembly into fibrils. However, it remains unclear how pVHL mediates this process, although these authors demonstrate that the effect of pVHL on the regulation of collagen assembly is independent of its ligase activity. With respect to FN, and given the role of pVHL in proteasomal-mediated degradation, pVHL could be involved in the removal of abnormally processed FN molecules; thus, regulating FN at the post-transcriptional level. Although this is still a possibility Kurban et al. also showed that the pVHL-FN interaction is indirect; hence it could be in fact mediated through collagen IV. Therefore, although a direct or indirect pVHL-FN interaction might be important, it may not always be sufficient to support FN assembly.
A second possibility is that pVHL indirectly regulates FN assembly by regulating other extracellular proteins that degrade this matrix. In this respect, it has been proposed that extracellular FN in VHL(-) cells is degraded by the increased activation of metalloproteinases such as MMP-2 (27). This possibility agrees with the fact that increased expression of MMPs is a consequence of pVHL loss of function (7, 48). Finally, and considering our present results in which VHL(-) cells were unable to assemble exogenously added FN produced by VHL(+), we can suggest that additional functional defects associated with the lack of VHL and different from FN itself might also be taking part in this process. Therefore, in this report, we analyzed the possibility that intracellular events such as cytoskeleton reorganization and cell contractility might be regulating the assembly of FN in VHL(+) cells. In this regard, VHL(-) cells do not form proper structures related to cytoskeleton organization and cell contractility (8, 14, 16). These facts support our present observations showing that VHL(-) cells plated on exogenously added FN were unable to make the initial FN stretching necessary for its assembly. This process might be partly caused by a deficient activation of the small GTPase RhoA in these cells. Because RhoA is an important regulator of cell cytoskeleton organization (42), the altered activity of this GTPase in VHL(-) cells may also account for the fibroblastoid phenotype observed in VHL(-) cells. This phenotype, also caused by the lack of proper cell-cell adhesion structures (16), may also contribute to the lack of matrix assembly in these cells. In this respect, we clearly show that VHL(-) cells have a deficient RhoA activity, probably due to increased activity of the RhoA inhibitor p190RhoGAP. This was also supported by our results in which we partially restored the ability to form FN fibrils in VHL(-) cells, in which we silenced the p190RhoGAP gene. Besides these results, no relationship between pVHL and Rho-GTPases has been established to date, and thus it is still unclear how pVHL regulates this process. One possibility is that pVHL regulates Rho-GTPases indirectly, through a second VHL-interacting protein. For example, pVHL interacts with several atypical PKCs (49, 50) and may regulate its cellular activity in certain cases (Okuda and co-workers, Ref. 8). Atypical PKCs have been involved in the regulation of the GTPase RhoA activity (51-53). Moreover, PKC-δ was shown to interact with p190RhoGAP in endothelial cells (54). It is therefore possible that pVHL interaction with atypical PKCs regulates the activity of Rho-GTPases by modulating the phosphorylation state of RhoA inhibitors (GDI, p190RhoGAP) or activators (GEF). This hypothesis would be in agreement with our present results showing an increased p190Rho-GAP phosphorylation in VHL(-) cells. Another remaining question would be how decreased RhoA activity in VHL(-) cells may account for their phenotypic changes and contribute to tumorigenesis. It is well known that Rho GTPases have been involved in the pathology of various human diseases including cancer (55), and interfering RhoA activation via p190RhoGAP leads to melanoma cell migration (56). Therefore, it is reasonable to think that decreased Rho GTPase activity may be also involved in the pathogenesis of renal carcinomas.
In summary, our present study demonstrates that FN production and function are not deficient in VHL(-) cells but rather that altered intracellular signals regulating cell contractility contributes to the inability of these cells to assemble an FN matrix. Furthermore, our results with type 2C mutants indicated that these mechanisms are independent of HIF. Thus, we provide a mechanism that explains some of the defects in extracellular matrix assembly observed in VHL(-) cells. The precise role of pVHL in this mechanism and whether this is a general mechanism in other cell lines remains to be established.
Supplementary Material
This work was supported in part by SAF2006-04013 from the Ministerio de Educación y Ciencia (MEC) (to M. J. C.), SAF2007-60592 and RECAVA C03/01 (to M. O. L.), and PI060400 from the Ministerio de Sanidad y Consumo and the Fundacion de Investigacion Medica Mutua Madrileña (to A. G. -P.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental methods and Figs. S1 and S2.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: VHL, von Hippel-Lindau; FN, fibronectin; BSA, bovine serum albumin; PBS, phosphate-buffered saline; GST, glutathione S-transferase; GFP, green fluorescent protein; PKC, protein kinase C; Ab, antibody; siRNA, small interfering RNA; RCC, renal cancer cells; HIF, hypoxia-inducible transcription factors; FA, fibrillar adhesion; FC, focal contacts.
References
- 1.Kaelin, W. G., Jr. (2002) Nat. Rev. Cancer 2 673-682 [DOI] [PubMed] [Google Scholar]
- 2.Ivan, M., Kondo, K., Yang, H., Kim, W., Valiando, J., Ohh, M., Salic, A., Asara, J. M., Lane, W. S., and Kaelin, W. G., Jr. (2001) Science 292 464-468 [DOI] [PubMed] [Google Scholar]
- 3.Jaakkola, P., Mole, D. R., Tian, Y. M., Wilson, M. I., Gielbert, J., Gaskell, S. J., Kriegsheim, A., Hebestreit, H. F., Mukherji, M., Schofield, C. J., Maxwell, P. H., Pugh, C. W., and Ratcliffe, P. J. (2001) Science 292 468-472 [DOI] [PubMed] [Google Scholar]
- 4.Maxwell, P. H., Wiesener, M. S., Chang, G. W., Clifford, S. C., Vaux, E. C., Cockman, M. E., Wykoff, C. C., Pugh, C. W., Maher, E. R., and Ratcliffe, P. J. (1999) Nature 399 271-275 [DOI] [PubMed] [Google Scholar]
- 5.Maher, E. R., and Kaelin, W. G., Jr. (1997) Medicine (Balt.) 76 381-391 [DOI] [PubMed] [Google Scholar]
- 6.Mack, F. A., Rathmell, W. K., Arsham, A. M., Gnarra, J., Keith, B., and Simon, M. C. (2003) Cancer Cell 3 75-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koochekpour, S., Jeffers, M., Wang, P. H., Gong, C., Taylor, G. A., Roessler, L. M., Stearman, R., Vasselli, J. R., Stetler-Stevenson, W. G., Kaelin, W. G., Jr., Linehan, W. M., Klausner, R. D., Gnarra, J. R., and Van de Woude, G. F. (1999) Mol. Cell Biol. 19 5902-5912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamada, M., Suzuki, K., Kato, Y., Okuda, H., and Shuin, T. (2001) Cancer Res. 61 4184-4189 [PubMed] [Google Scholar]
- 9.Hsu, T., Adereth, Y., Kose, N., and Dammai, V. (2006) J. Biol. Chem. 281 12069-12080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hergovich, A., Lisztwan, J., Barry, R., Ballschmieter, P., and Krek, W. (2003) Nat. Cell Biol. 5 64-70 [DOI] [PubMed] [Google Scholar]
- 11.Bluyssen, H. A., Lolkema, M. P., van Beest, M., Boone, M., Snijckers, C. M., Los, M., Gebbink, M. F., Braam, B., Holstege, F. C., Giles, R. H., and Voest, E. E. (2004) FEBS Lett. 556 137-142 [DOI] [PubMed] [Google Scholar]
- 12.Lolkema, M. P., Mans, D. A., Snijckers, C. M., van Noort, M., van Beest, M., Voest, E. E., and Giles, R. H. (2007) FEBS Lett. 581 4571-4576 [DOI] [PubMed] [Google Scholar]
- 13.Davidowitz, E. J., Schoenfeld, A. R., and Burk, R. D. (2001) Mol. Cell Biol. 21 865-874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esteban-Barragan, M. A., Avila, P., Alvarez-Tejado, M., Gutierrez, M. D., Garcia-Pardo, A., Sanchez-Madrid, F., and Landazuri, M. O. (2002) Cancer Res. 62 2929-2936 [PubMed] [Google Scholar]
- 15.Lewis, M. D., and Roberts, B. J. (2003) Oncogene 22 3992-3997 [DOI] [PubMed] [Google Scholar]
- 16.Calzada, M. J., Esteban, M. A., Feijoo-Cuaresma, M., Castellanos, M. C., Naranjo-Suarez, S., Temes, E., Mendez, F., Yanez-Mo, M., Ohh, M., and Landazuri, M. O. (2006) Cancer Res. 66 1553-1560 [DOI] [PubMed] [Google Scholar]
- 17.Esteban, M. A., Tran, M. G., Harten, S. K., Hill, P., Castellanos, M. C., Chandra, A., Raval, R., O'Brien, T. S., and Maxwell, P. H. (2006) Cancer Res. 66 3567-3575 [DOI] [PubMed] [Google Scholar]
- 18.Krishnamachary, B., Zagzag, D., Nagasawa, H., Rainey, K., Okuyama, H., Baek, J. H., and Semenza, G. L. (2006) Cancer Res. 66 2725-2731 [DOI] [PubMed] [Google Scholar]
- 19.Kim, M., Yan, Y., Lee, K., Sgagias, M., and Cowan, K. H. (2004) Biochem. Biophys. Res. Commun. 320 945-950 [DOI] [PubMed] [Google Scholar]
- 20.Bindra, R. S., Vasselli, J. R., Stearman, R., Linehan, W. M., and Klausner, R. D. (2002) Cancer Res. 62 3014-3019 [PubMed] [Google Scholar]
- 21.Roe, J. S., Kim, H., Lee, S. M., Kim, S. T., Cho, E. J., and Youn, H. D. (2006) Mol. Cell 22 395-405 [DOI] [PubMed] [Google Scholar]
- 22.Esteban, M. A., Harten, S. K., Tran, M. G., and Maxwell, P. H. (2006) J. Am. Soc. Nephrol. 17 1801-1806 [DOI] [PubMed] [Google Scholar]
- 23.Lolkema, M. P., Mans, D. A., Ulfman, L. H., Volpi, S., Voest, E. E., and Giles, R. H. (2007) Eur. J. Hum. Genet. 3 3. [DOI] [PubMed] [Google Scholar]
- 24.Yang, H., Minamishima, Y. A., Yan, Q., Schlisio, S., Ebert, B. L., Zhang, X., Zhang, L., Kim, W. Y., Olumi, A. F., and Kaelin, W. G., Jr. (2007) Mol. Cell 28 15-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanahan, D., and Weinberg, R. A. (2000) Cell 100 57-70 [DOI] [PubMed] [Google Scholar]
- 26.Ohh, M., Yauch, R. L., Lonergan, K. M., Whaley, J. M., Stemmer-Rachamimov, A. O., Louis, D. N., Gavin, B. J., Kley, N., Kaelin, W. G., Jr., and Iliopoulos, O. (1998) Mol. Cell 1 959-968 [DOI] [PubMed] [Google Scholar]
- 27.Kurban, G., Hudon, V., Duplan, E., Ohh, M., and Pause, A. (2006) Cancer Res. 66 1313-1319 [DOI] [PubMed] [Google Scholar]
- 28.Tang, N., Mack, F., Haase, V. H., Simon, M. C., and Johnson, R. S. (2006) Mol. Cell Biol. 26 2519-2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hynes, R. O. (1992) Cell 69 11-25 [DOI] [PubMed] [Google Scholar]
- 30.Wierzbicka-Patynowski, I., and Schwarzbauer, J. E. (2003) J. Cell Sci. 116 3269-3276 [DOI] [PubMed] [Google Scholar]
- 31.Pankov, R., Cukierman, E., Katz, B. Z., Matsumoto, K., Lin, D. C., Lin, S., Hahn, C., and Yamada, K. M. (2000) J. Cell Biol. 148 1075-1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong, C., Chrzanowska-Wodnicka, M., Brown, J., Shaub, A., Belkin, A. M., and Burridge, K. (1998) J. Cell Biol. 141 539-551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, Q., Magnusson, M. K., and Mosher, D. F. (1997) Mol. Biol. Cell 8 1415-1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iliopoulos, O., Kibel, A., Gray, S., and Kaelin, W. G., Jr. (1995) Nat. Med. 1 822-826 [DOI] [PubMed] [Google Scholar]
- 35.Ohh, M., Takagi, Y., Aso, T., Stebbins, C. E., Pavletich, N. P., Zbar, B., Conaway, R. C., Conaway, J. W., and Kaelin, W. G., Jr. (1999) J. Clin. Investig. 104 1583-1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stickle, N. H., Chung, J., Klco, J. M., Hill, R. P., Kaelin, W. G., Jr., and Ohh, M. (2004) Mol. Cell Biol. 24 3251-3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krieg, M., Haas, R., Brauch, H., Acker, T., Flamme, I., and Plate, K. H. (2000) Oncogene 19 5435-5443 [DOI] [PubMed] [Google Scholar]
- 38.Sechler, J. L., Takada, Y., and Schwarzbauer, J. E. (1996) J. Cell Biol. 134 573-583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naranjo-Suarez, S., Castellanos, M. C., Alvarez-Tejado, M., Vara, A., Landazuri, M. O., and del Peso, L. (2003) J. Biol. Chem. 278 31895-31901 [DOI] [PubMed] [Google Scholar]
- 40.Clifford, S. C., Cockman, M. E., Smallwood, A. C., Mole, D. R., Woodward, E. R., Maxwell, P. H., Ratcliffe, P. J., and Maher, E. R. (2001) Hum. Mol. Genet. 10 1029-1038 [DOI] [PubMed] [Google Scholar]
- 41.He, Z., Liu, S., Guo, M., Mao, J., and Hughson, M. D. (2004) Cancer Genet. Cytogenet. 152 89-94 [DOI] [PubMed] [Google Scholar]
- 42.Ridley, A. J. (2001) Trends Cell Biol. 11 471-477 [DOI] [PubMed] [Google Scholar]
- 43.Cali, G., Mazzarella, C., Chiacchio, M., Negri, R., Retta, S. F., Zannini, M., Gentile, F., Tarone, G., Nitsch, L., and Garbi, C. (1999) J. Cell Sci. 112 957-965 [DOI] [PubMed] [Google Scholar]
- 44.Arthur, W. T., Petch, L. A., and Burridge, K. (2000) Curr. Biol. 10 719-722 [DOI] [PubMed] [Google Scholar]
- 45.Hoffman, M. A., Ohh, M., Yang, H., Klco, J. M., Ivan, M., and Kaelin, W. G., Jr. (2001) Hum. Mol. Genet. 10 1019-1027 [DOI] [PubMed] [Google Scholar]
- 46.Grosfeld, A., Stolze, I. P., Cockman, M. E., Pugh, C. W., Edelmann, M., Kessler, B., Bullock, A. N., Ratcliffe, P. J., and Masson, N. (2007) J. Biol. Chem. 282 13264-13269 [DOI] [PubMed] [Google Scholar]
- 47.Kurban, G., Duplan, E., Ramlal, N., Hudon, V., Sado, Y., Ninomiya, Y., and Pause, A. (2007) Oncogene 13 13. [DOI] [PubMed] [Google Scholar]
- 48.Jiang, Y., Zhang, W., Kondo, K., Klco, J. M., St Martin, T. B., Dufault, M. R., Madden, S. L., Kaelin, W. G., Jr., and Nacht, M. (2003) Mol. Cancer Res. 1 453-462 [PubMed] [Google Scholar]
- 49.Iturrioz, X., Durgan, J., Calleja, V., Larijani, B., Okuda, H., Whelan, R., and Parker, P. J. (2006) Biochem. J. 397 109-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pal, S., Claffey, K. P., Dvorak, H. F., and Mukhopadhyay, D. (1997) J. Biol. Chem. 272 27509-27512 [DOI] [PubMed] [Google Scholar]
- 51.Holinstat, M., Mehta, D., Kozasa, T., Minshall, R. D., and Malik, A. B. (2003) J. Biol. Chem. 278 28793-28798 [DOI] [PubMed] [Google Scholar]
- 52.Mehta, D., Rahman, A., and Malik, A. B. (2001) J. Biol. Chem. 276 22614-22620 [DOI] [PubMed] [Google Scholar]
- 53.Pang, H., and Bitar, K. N. (2005) Am. J. Physiol. Cell Physiol. 289 C982-C993 [DOI] [PubMed] [Google Scholar]
- 54.Harrington, E. O., Shannon, C. J., Morin, N., Rowlett, H., Murphy, C., and Lu, Q. (2005) Exp. Cell Res. 308 407-421 [DOI] [PubMed] [Google Scholar]
- 55.Li, X., and Lim, B. (2003) Oncol Res. 13 323-331 [DOI] [PubMed] [Google Scholar]
- 56.Moyano, J. V., Maqueda, A., Casanova, B., and Garcia-Pardo, A. (2003) Mol. Biol. Cell 14 3699-3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.