Abstract
Because Bcl-2 family members inhibit the ability of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) to induce apoptosis, we investigated whether ABT-737, a small molecule Bcl-2 inhibitor, enhances TRAIL killing. We demonstrate that a combination of ABT-737 and TRAIL induced significant cell death in multiple cancer types, including renal, prostate, and lung cancers, although each agent individually had little activity in these tumor cells. All of these cell lines expressed the Mcl-1 protein that is known to block the activity of ABT-737 and TRAIL but did not block the synergy between these agents. However, Bax-deficient cell lines, including DU145 and HCT116 cells and those cell lines expressing low levels of TRAIL receptor, were resistant to apoptosis induced by these agents. To understand how ABT-737 functions to markedly increase TRAIL sensitivity, the levels of specific death-inducing signaling complex components were evaluated. Treatment with ABT-737 did not change the levels of c-FLIP, FADD, and caspase-8 but up-regulated the levels of the TRAIL receptor DR5. DR5 up-regulation induced by ABT-737 treatment occurred through a transcriptional mechanism, and mutagenesis studies demonstrated that the NF-κB site found in the DR5 promoter was essential for the ability of ABT-737 to increase the levels of this mRNA. Using luciferase reporter plasmids, ABT-737 was shown to stimulate NF-κB activity. Together, these results demonstrate that the ability of ABT-737 and TRAIL to induce apoptosis is mediated through activation of both the extrinsic and intrinsic pathways. Combinations of ABT-737 and TRAIL can be exploited therapeutically where antiapoptotic Bcl-2 family members drive tumor cell resistance to current anticancer therapies.
The recombinant TRAIL2 and agonist antibodies targeted against its receptor are capable of inducing the selective apoptotic death of human cancer cells while sparing normal human cells (1-4). TRAIL binds to two receptors, DR5 (TRAIL-R2) and DR4 (TRAIL-R1) (5), and when bound to the cell (6, 7) recruits intracellular FADD and caspase-8 to form a death-inducing signaling complex (DISC) (8). Activation of the DISC leads to the cleavage of caspase-8 and the BH3 protein BID that can function to stimulate the intrinsic mitochondrial pathway, which in turn releases cytochrome c, Smac/DIABLO (mitochondria-derived activator of caspase/direct inhibitor of apoptosis binding protein with low pI), and induces the cleavage of caspase-9 and subsequently caspase-3 (9, 10). Although TRAIL holds great promise as a potential chemotherapeutic agent, multiple tumor types grown in culture or implanted in animals are resistant to this agent (11, 12).
One common mechanism for the resistance to TRAIL-induced apoptosis is the overexpression of Bcl-2 or its family members that function to block the mitochondrial pathway. Bcl-2 protein overexpression protects prostate, breast, colon cancers, and melanoma from TRAIL-induced apoptosis (13-16). Other members of the Bcl-2 family, including Bfi-1/A1, Mcl-1, and Bcl-xL, can also function in a similar fashion (17-19). TRAIL treatment activates BH3-containing proteins, including PUMA, Bim, and Bak, that can be sequestered by Mcl-1, whereas TRAIL-activated Bax and Bid are bound to the Bcl-xL proteins (20). Lowering the level of Mcl-1 by the use of short hairpin RNAs or pharmacologic maneuvers, such as treatment with sorafenib, releases Bim from the Mcl-1 protein (21) and sensitizes tumor cells to TRAIL-induced apoptosis (22, 23).
The central role that Bcl-2 family members play in resistance to chemotherapy in multiple malignancies, including chronic lymphocytic leukemia, follicular lymphoma, and acute lymphocytic leukemia, has led to the development of a small molecule high affinity (nanomolar) inhibitor of this protein, ABT-737 (24, 25). ABT-737 binds to the hydrophobic groove in Bcl-2, Bcl-xL, and Bcl-w and prevents them from sequestering proapoptotic BH3-only proteins such as tBid, Bad, and Bim (25-27). ABT-737 binds with lower affinity to the Bcl-B, Mcl-1, and Bfi/A1 proteins. ABT-737 can induce concentration-dependent apoptosis when incubated with human leukemic cells (26) and is synergistic with paclitaxel in killing small cell lung cancer cells (28). In multiple myeloma, combining ABT-737 with the proteasome inhibitor, bortezomib, mephalan, or dexamethasone induces additive cytotoxic effects (29). Although some small cell lung carcinoma cell lines are moderately sensitive to this agent (30), the majority of cell lines of this tumor type and other solid tumors are resistant to ABT-737-induced apoptosis. An unbiased RNA interference-based screen demonstrated that Mcl-1, which does not bind ABT-737 with high affinity, is the protein causing resistance of small cell lung cancer to this compound (31). The importance of Mcl-1 to ABT-737 resistance has been demonstrated in multiple cell lines, including mouse lymphoma and human leukemia (26, 32). Knocking down the levels of this protein with RNA interference or by the addition of compounds, including the kinase inhibitor sorafenib (BAY 43-9006) and the cyclin-dependent kinase inhibitor roscovitine, sensitizes both solid and liquid tumor cells to death induced by this agent (27, 31). Additionally, the phosphorylation of Bcl-2 can also inhibit the activity of ABT-737 in leukemic samples and agents that block the mitogen-activated protein kinase pathway can reverse this resistance (26).
Given the role of the Bcl-2 family members in inhibiting TRAIL-induced apoptosis, we combined ABT-737 and TRAIL to examine their ability to synergize in multiple tumor types, including lung, prostate, and renal cancers. We find that these compounds are highly synergistic even in the face of elevated levels of Mcl-1. Solid tumor cell lines that are resistant to combination therapy prove to have low levels of either Bax protein or the TRAIL receptor DR5. In resistant cell lines, ABT-737 treatment up-regulates the levels of the TRAIL receptor, DR5, through a transcriptional mechanism based on activation of NF-κB. Therefore, our study demonstrates that ABT-737 through modulation of the intrinsic pathway can markedly enhance the apoptotic activity of the extrinsic pathway activated by TRAIL.
EXPERIMENTAL PROCEDURES
Cell Lines, Antibodies, and Reagents—Human cancer cell lines were grown in either Dulbecco's modified Eagle's medium or RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 100 units/ml penicillin, and 100 μg/ml streptomycin. Antibodies were obtained from the following sources: GAPDH, CHOP, β-actin, Bim, Bik, Puma and Mcl-1 from Santa Cruz Biotechnology (Santa Cruz, CA); Bcl-2, Bax, cytochrome c, poly(ADP-ribose) polymerase, Itch, TRAF2, and RIP from BD Biosciences; Bid and caspase-9 from Cell Signaling Technology (Danvers, MA); Bak (NT) from Upstate Biotechnology (Lake Placid, NY); caspase-3 from StressGen Bioreagents (Victoria, British Columbia, Canada); caspase-8 from MBL International (Nagoya, Japan); FLIP from Alexis; DR4 and DR5 from ProSci Inc. (Poway, CA); and FLAG antibody and FLAG-agarose beads from Sigma. HRP-conjugated goat anti-rabbit and goat anti-mouse antibodies were from GE Healthcare. Anti-mouse IgG2b-HRP was obtained from Southern Biotech (Birmingham, AL). The following reagents were obtained from the indicated sources: Z-VAD-fmkfromR&D Systems (Minneapolis, MN), human recombinant nontagged TRAIL from PeproTech (Rocky Hill, NJ), and FLAG-tagged TRAIL from Alexis were used. ABT-737 (A-779024.0) and its enantiomer (A-793844.0) were a gift of Abbott Laboratories. Both compounds were dissolved in dimethyl sulfoxide (DMSO; Sigma) at the concentration of 50 mm, and aliquots were stored at -80 °C.
Cytotoxicity Assays—Cells were seeded in 96-well plates or culture dishes and treated with recombinant TRAIL in the absence or presence of ABT-737. Cell viability was determined by an acid phosphatase assay (33, 34), and cellular apoptosis was quantitated under phase-contrast microscopy. Percentage of cell death was evaluated by trypan blue exclusion assay as described previously (36). The data shown reflect the percent activity when compared with vehicle-treated control cells.
FACS Analysis—Cell-surface DR5 expression was analyzed by flow cytometry (35). The procedure for direct antibody staining and subsequent flow cytometric analysis of this cell-surface protein was described previously (35). Phycoerythrin-conjugated mouse monoclonal anti-human DR5 (clone DJR2-4), anti-human DR4 (clone DJR1), and phycoerythrin-conjugated mouse IgG1 isotype control (MOPC21/P3) were purchased from eBioscience (San Diego, CA). Detection of Bax and Bak conformational change was carried out using cell pellets (1 × 106). Pellets were washed with phosphate-buffered saline and incubated for 40 min at 4 °C with either mouse IgG1 antibody as a negative control, a mouse monoclonal antibody against amino acids 1-52 of Bak (AM03, clone TC100; Oncogene Research Products), or a mouse monoclonal antibody against amino acids 12-24 of Bax (clone 6A7; BD Biosciences). After washing with phosphate-buffered saline, the binding of antibody was visualized with fluorescein isothiocyanate-conjugated anti-mouse IgG (1:200) (Sigma). 10,000 cells were analyzed using Cell Quest™ software (BD Biosciences).
Cytosolic Fractionation, DISC Immunoprecipitation, and Western Blotting—Cytosolic S100 fraction was prepared from cells according to a method described previously (36). TRAIL-induced DISC was immunoprecipitated and subjected to Western blot analysis according to a previously reported protocol (35). To examine protein expression, the cells were lysed in ice-cold lysis buffer (20 mmol/liter Tris-HCl (pH 7.4), 150 mmol/liter NaCl, 2 mmol/liter EDTA, 10% glycerol, 1% Triton X-100, 1% protease inhibitor mixture, and 1 mmol/liter phenylmethylsulfonyl fluoride). Equal amounts of proteins were subjected to SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were incubated with the antibodies as indicated overnight at 4 °C. The membranes were washed and incubated for 1 h at room temperature with the anti-mouse IgG2b-HRP, anti-mouse IgG-HRP, or anti-rabbit IgG-HRP. The blots were again washed and developed by enhanced chemiluminescence reagents (GE Healthcare).
Silencing of Gene Expression with Small Interfering RNA (siRNA) and Gene Transfection—Gene silencing was achieved by transfecting cells with siRNA duplexes using the Lipofectamine 2000 transfection reagent (Invitrogen) following the manufacturer's instructions and siRNAs duplexes targeting human DR5 (5′-AACTACCAGAAAGGTATACCT-3′), Mcl-1 (5′-AAAAGTATCACAGACGTTCTC-3′), Bcl-2 (5′-AACCGGGAGATAGTGATGAAG-3′), Bcl-xl (5′-TAGGGTGGCCCTTGCAGTTCA-3′), Bax (5′-AACATGGAGCTGCAGAGGATGA-3′) or siRNA duplexes targeting human Bak 5′-AAGCGAAGTCTTTGCCTTCTC-3′.
Scrambled sequence of nonsilencing control siRNA oligonucleotides, which does not match any human genome sequence, that target the sequence 5′-AATTCTCCGAACGTGTCACGT-3′ were purchased from Qiagen (Valencia, CA). Gene transfection of human FLAG-tagged Bax cDNA in pcDNA3 were described previously (37). The pRC/CMV-Bak vector was identical to one described previously (38).
Luciferase Activity Assay—Luciferase activities were measured with the dual-luciferase assay kits (Promega, Madison, WI). To examine the effects of ABT-737 on DR5 promoter activity, 6 × 105 cells were cotransfected with 4 μg of pGVB2-DR5 reporter plasmids (a gift of Dr. Toshyuki Sakai) (39) and as an internal control 0.01 μg of pEF-Renilla-luc using Lipofectamine 2000 reagent. Twenty hours later, cells were treated with ABT-737. Luciferase activities were determined by normalization of firefly luciferase to Renilla luciferase activity. The reporter constructs containing a 552-bp 5′-flanking region of the DR5 gene with a wild-type or mutated CHOP-binding site, NF-κB-binding site, or Elk-binding site were generously provided by Dr. H. G. Wang (University of South Florida College of Medicine, Tampa, FL) (40). The pNF-κB-luc (4 μg) plasmids and control vector plasmid were a gift of Drs. Kurtz and Nieminen (Medical University of South Carolina, Charleston, SC).
Quantitative Real Time-PCR—For quantitative real time-PCR, total RNA isolated from the cells using RNeasy kit (Qiagen, Valencia, CA) was reverse-transcribed using oligo(dT) and Superscript II RT (Invitrogen), and the resulting cDNA was used for PCR amplification using gene-specific primer pairs. PCR conditions for these reactions were as follows: 95 °C, 10 s; 58 °C, 30 s; 72 °C 10 s for 40 cycles. A Bio-Rad iQ5 multicolor PCR detection system and iQ5 optical system software analysis were used for quantifying gene expression (Bio-Rad, version 2.0). The expression level of DR5 was normalized to GAPDH. The primers used for real time PCR were as follows: DR5 forward ATCACCCAACAAGACCTAGC and reverse TTCTGAGATATGGTGTCCAGG; GAPDH forward CAGCCTCAAGATCATCAGCA and reverse GTCTTCTGGGTGGCAGTGAT.
RESULTS
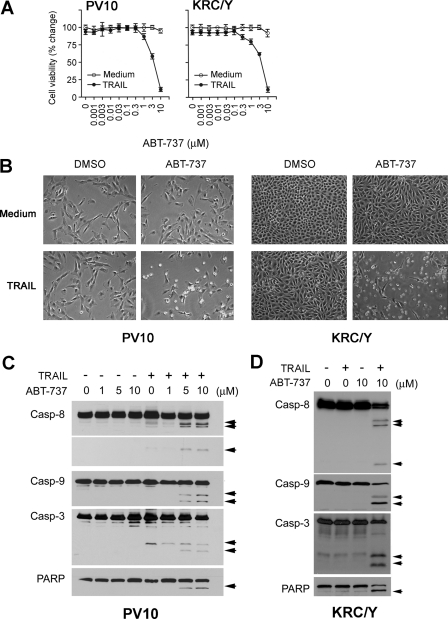
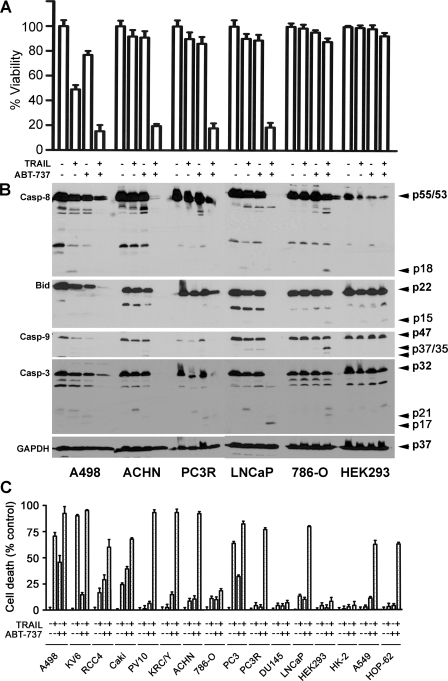
Synergy of TRAIL and ABT-737 to Induce Apoptosis—Adding TRAIL or ABT-737 alone to PV10 and KRC/Y renal cancer cells failed to induce cell death, but combinations of ABT-737 and TRAIL resulted in rapid apoptotic death beginning within 3 h (Fig. 1, A and B). A dose-response analysis showed increasing ABT-737-sensitized TRAIL-resistant renal carcinoma PV10 and KRC/Y cells to TRAIL (Fig. 1A). The cell death was easily visualized by phase-contrast microscopy (Fig. 1B). The dose-dependent induction of cleavage of caspase-8, -9, and -3 and PARP in both renal cancer cell lines PV10 and KRC/Y (Fig. 1, C and D) demonstrates that cell death was driven by both the intrinsic and extrinsic apoptotic pathways. The synergistic ability to induce cell death is not limited to these two renal cancer cell lines but is found in multiple kidney cancer cell lines, A498, ACHN, and prostate cancer cell lines PC3R and LNCaP (Fig. 2A) and is correlated with cleavage of caspase-8, -9, -3, and Bid (Fig. 2B). The slight differences in the extent of caspase cleavage between Fig. 1 and Fig. 2B are a result of the 3-versus 24-h incubation with TRAIL and ABT-737, as well as the increased overexposure of Fig. 2A to demonstrate all caspase cleavage products. The differences caused by different lengths of incubation are highlighted for a single cell line, A498 cells, in supplemental Fig. S1A and S1B. This loss of cell viability occurs with an increase in apoptosis in 12 different renal, prostate, and lung cancer cell lines (Fig. 2C). A number of cell lines were found to be resistant to combination treatment, including DU145 prostate cancer cells, 786-O renal cancer cells, human embryonic kidney 293 (HEK293) cells, and normal renal epithelial cell line HK-2 cells. All of the cell lines that were sensitive to the combination treatment demonstrated increased cleavage of caspase-8, Bid, caspase-9, and caspase-3, whereas resistant cell lines, including HK-2 and HEK293, did not demonstrate these changes.
FIGURE 1.
ABT-737 synergizes with TRAIL to induce apoptosis. A, PV10 cells and KRC/Y cells were treated with ABT-737 at the indicated doses in the absence or presence of TRAIL (100 ng/ml) for 24 h. Cell viability was determined by an acid phosphatase assay (see “Experimental Procedures”) (mean ± S.D., n = 4). B, phase-contrast microscopy of PV10 cells and KRC/Y cells after combined treatment with ABT-737 (10 μm) and TRAIL (100 ng/ml) for 3 h. C, Western blot of extracts of PV10 cells treated with ABT-737 at the indicated doses in the absence or presence of TRAIL (100 ng/ml) for 3 h. Arrows indicate caspase (Casp) or PARP cleavage products. D, Western blot analysis of KRC/Y cells treated with DMSO, 100 ng/ml TRAIL, 10 μm ABT-737, or the combination. Arrows indicate caspase and PARP cleavage products.
FIGURE 2.
ABT-737 enhances TRAIL-induced apoptosis in renal, prostate, and lung cancer cells but not in normal kidney cells. A, renal cancer (A498, ACHN, and 786-0), prostate cancer (PC3R and LNCaP), and human embryonic kidney cells 293 (HEK293) were treated with DMSO, 100 ng/ml TRAIL, 10 μm ABT-737, or a combination of the two agents for 24 h. Cell viability was determined by an acid phosphatase assay (mean ± S.D., n = 4). B, cleavage of caspase (Casp)-8, -9, and -3 and Bid was examined on the Western blots. Arrows denote procaspase-8 (p55 and p53), first cleavage fragments (p47 and p43), and the active p18 form of caspase-8; procaspase-9 (p47) processed to produce the active p37 and p35 forms; procaspase-3 (p32) processed to produce active p21 and p17 products; and full-length Bid (p22) and p15 truncated Bid. C, percentages of cell death in response to ABT-737, TRAIL, and ABT-737 plus TRAILs were assessed by the trypan blue exclusion assay. Each cell line was treated with either DMSO, 100 ng/ml TRAIL, 10 μm ABT-737, or the combination for 24 h (mean ± S.D., n = 4).
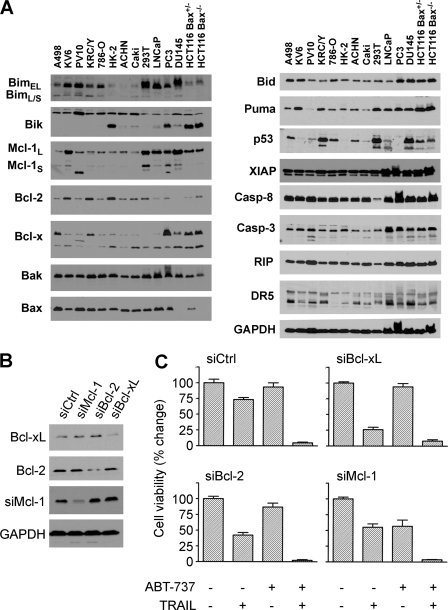
Regulation of Tumor Cell Sensitivity to Combined or Single Agent Treatment—To explore to the reason for differences in drug sensitivity between these cell lines, we examined the expression of components of the apoptotic cascade by Western blotting, including DISC proteins and Bcl-2 family members (Fig. 3A). Both the renal cancer cell line 786-O and the normal renal epithelial cells HK-2 display significantly lower levels of the TRAIL receptor DR5 expression than other cell lines. Because the other TRAIL receptor DR4 is not expressed in these cell lines or the majority of cell lines examined, an inability to bind TRAIL to the DR5 receptor could make these cells resistant to the combination therapy. Western blot analysis demonstrated that DU145 and a subset of colon cancer HCT116 cells (Fig. 3A), which did not contain Bax protein, were resistant to combination therapy. In comparison, the parental HCT116 cells that contained Bax were sensitive to the combination therapy (supplemental Fig. S2A and S2B). Bax has been shown to be essential for TRAIL killing (41, 42), and the addition of ABT-737 does not overcome this blockade. ABT-737 occupies a hydrophobic pocket at one end of the BH3 binding groove that interferes with Bcl-2 family protein-protein interactions (25). However, because of the difference in groove structure, Mcl-1 does not bind ABT-737 and thus functions to inhibit the activity of this agent. The Western blots shown in Fig. 3A demonstrate that all cell lines express the Bcl-2 family member Mcl-1. To examine whether Mcl-1 functions similarly in the renal cancer cell line PV10, specific siRNA duplexes targeting Bcl-2, Bcl-xL, and Mcl-1 were transfected into ABT-737-resistant cells (Fig. 3B). siRNA-mediated inhibition of each of these three Bcl-2 family members increased TRAIL killing, supporting the notion that TRAIL-induced apoptosis can be blocked by Bcl-2 family proteins (Fig. 3C). However, only knockdown of Mcl-1 protein rendered PV10 cells susceptible to ABT-737 when administered as a single agent (Fig. 3C) supporting the hypothesis that these renal and prostate cell lines are in part resistant to ABT-737 based on Mcl-1 expression. Chemical inhibitors of caspase-8 (Z-IETD) and caspase-9 (Z-LEHD) individually block the apoptotic activity of combination therapy with ABT-737 and TRAIL (supplemental Fig. S2C and S2D), suggesting that both the intrinsic and extrinsic pathways are needed for cell death induced by these agents.
FIGURE 3.
BH3 proteins play a role in controlling resistance to ABT-737 and TRAIL. A, expression patterns determined by Western blot analysis of Bcl-2 family and apoptosis pathway proteins in 14 cell lines. B, PV10 cells were treated individually with siRNAs to either Bcl-2, Bcl-xl, or Mcl-1, and extracts of these cells were examined by Western blotting for the levels of these proteins. C, PV10 cells transfected with either control, Bcl-2, Bcl-xl, or Mcl-1, and siRNA duplexes were then treated with DMSO, 100 ng/ml TRAIL, ABT-737 10 μm, or the combination for 24 h. Cell viability was measured by the acid phosphatase assay (mean ± S.D., n = 4).
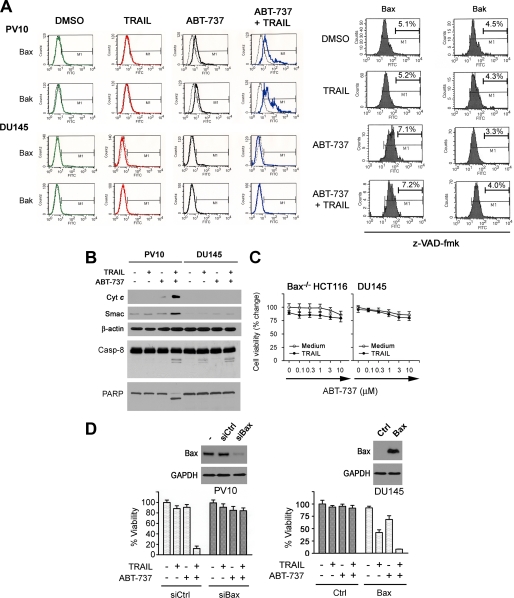
ABT-737 treatment of multiple cell lines did not change the level of Bax and Bak proteins (data not shown). Activation of the mitochondrial pathway occurs through induction of a conformational change in Bax or Bak, resulting in the exposure of the NH2 terminus of each molecule (43-45). Flow cytometric analysis with an antibody against the activated form of these proteins revealed that treatment with ABT-737 induced a conformational change in both the Bax and Bak proteins in PV10 renal carcinoma cells (Fig. 4A). In contrast, DU145 cells treated with ABT-737 did not demonstrate a change in either protein (Fig. 4A). The inactive enantiomer of ABT-737 (25) did not show cytotoxicity and was not synergistic with TRAIL (supplemental Fig. S3) Bax and Bak were not activated by treatment of tumor cells with this compound (data not shown) (25). As demonstrated in Fig. 4A, conformational changes in both Bax and Bak were significantly increased with combined ABT-737 and TRAIL treatment, and this increase was inhibited by the pan-caspase inhibitor Z-VAD-fmk (Fig. 4A). In contrast, treatment with this caspase inhibitor had no effect on the small changes induced by ABT-737 alone (Fig. 4A). These results suggest that caspase activation was essential for the enhanced activation of Bax and Bak seen in combination treatment. To determine whether the changes in Bax and Bak induced by ABT-737 and/or TRAIL were associated with activation of the mitochondrial apoptotic pathway, we measured the release of the mitochondrial enzyme cytochrome c and the protein Smac/DIABLO into the cytosol of PV10 and DU145 cells after exposure to ABT-737, TRAIL, and the combination (Fig. 4B). In PV10 cells, ABT-737 treatment alone did not release these proteins nor induce PARP cleavage, a marker of apoptosis, suggesting that although there were low level of Bax activation, changes in Bax alone were not sufficient to activate apoptosis. In contrast, combination therapy when administered to PV10 cells induced markers of activation of the mitochondrial pathway, as well as apoptosis.
FIGURE 4.
Bax is required for apoptosis induced by ABT-737 and TRAIL. A, activation of Bax or Bak was determined by FACS analysis using monoclonal N-Bax or N-Bak antibodies. PV10 or DU145 cells were treated with DMSO, 100 ng/ml TRAIL, 10 μm ABT-737, or the combination of TRAIL and ABT-737 for 24 h followed by FACS analysis. Isotype mouse IgG1 antibody was used as a negative control. PV10 cells were treated with 20 μm Z-VAD-fmk prior to the addition of ABT-737, TRAIL, or the combination as described in the first part of this panel. B, S100 cytosol fractions from PV10 and DU145 cells were isolated and examined by Western blot analysis for the release of cytochrome c (Cyt c) and Smac, levels of cleaved caspase-8 (Casp-8) and PARP, and β-actin. C, Bax-deficient (Bax-/-) HCT116 cells and DU145 cells were treated with TRAIL in combination of various doses of ABT-737 for 24 h. Compared with nontreated control, cell viability was assessed by an acid phosphatase assay (mean ± S.D., n = 4). D, left panel, PV10 cells were transfected with either siRNA control (siCtrl) or siRNA Bax (siBax) for 36 h and then treated with DMSO, ABT-737 (10 μm), TRAIL (100 ng/ml), or the combination of ABT-737 and TRAIL for 24 h (mean ± S.D., n = 4). Changes in cell viability were determined by acid phosphatase assay. The level of Bax was examined in extracts of these cells by Western blot. Right panel, DU145 cells were transiently transfected with pcDNA3-Bax for 36 h and then treated with DMSO, ABT-737 (10 μm), TRAIL (100 ng/ml), or the combination of ABT-737 and TRAIL for 24 h (mean ± S.D., n = 4). Changes in cell viability were determined by the acid phosphatase assay and the level of Bax examined by Western blot.
As demonstrated in Fig. 2C, DU145 cells proved resistant to this therapy (Fig. 4C). To further confirm the importance of Bax in mediating sensitization, PV10 cells were transfected with siRNA duplexes targeting Bax. The knockdown of this protein rendered the PV-10 cells resistant to the combination treatment (Fig. 4D, left panel). In comparison, enforced expression of Bax in DU145 cells increased cell sensitivity to TRAIL, as well as ABT-737 (Fig. 4D, right panel), and restored synergy between ABT-737 and TRAIL. To further investigate the role of Bak in this sensitization process, PV10 cells were transfected with siRNA directed at Bak. Results demonstrate that the knockdown of Bak protein was not sufficient to inhibit apoptosis induced by treatment with TRAIL and ABT-737 (supplemental Fig. S4A and S4B). However, Bax negative cell lines, DU145 and HCT116 Bax-/- cells, became sensitive to this therapy when Bak is overexpressed (supplemental Fig. S4C and S4D). It is possible that Bak is binding Mcl-1 in these cells and allowing the induction of cell death.
Together, these results confirm that Bax is a critical protein for the ABT-737-mediated sensitization to TRAIL-induced apoptosis. The importance of Bax in these solid tumor cell lines may arise, in part, because Mcl-1 binds and inhibits the proapoptotic activity of Bak. Overexpression of Bak may saturate Mcl-1 binding and allow this protein to enhance TRAIL-mediated apoptosis. However, it does not appear to be sufficient to regulate cell death in PV10 cells.
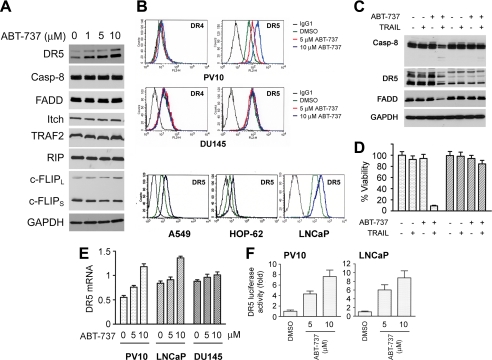
ABT-737-mediated Up-regulation of TRAIL Receptor DR5—Our observation in Fig. 2 that ABT-737 treatment of renal and prostate cell lines increases TRAIL-mediated cleavage of caspase-8 and Bid suggested that the DISC assembly could be modulated by ABT-737 treatment. Western blot analysis of PV10 cells did not show any significant changes in the levels of caspase-8, FADD, c-FLIP, RIP, Itch, and TRAF2 (Fig. 5A) after ABT-737 treatment. In contrast, incubation with ABT-737 in a dose-dependent fashion up-regulated DR5 protein expression (Fig. 5A). FACS analysis using a phycoerythrin-conjugated anti-DR4 or DR5 antibody confirmed cell surface increases in DR5 but not DR4 expression after ABT-737 treatment of the PV10 renal cancer, LNCaP prostate cancer, A549 and HOP-62 lung cancer cell lines (Fig. 5B) but not in DU145 prostate cancer cells. Treatment of ABT-737 also increased DR5 protein in Caki cells that are sensitized by ABT-737 but not in 786-O cells that are resistant to this drug (supplemental Fig. S5A). In contrast to ABT-737, the inactive enantiomer of ABT-737 even at high concentrations did not affect the levels of DR4 or DR5 (supplemental Fig. S5B). Lowering the levels of DR5 protein by siRNA transfection abolished ABT-737-mediated sensitization to TRAIL (Fig. 5, C and D), demonstrating the importance of this receptor to the enhancement of ABT-737 killing, although enforced expression of DR5 in 786-O cells increased the cell sensitivity to TRAIL, supporting role of DR5 (supplemental Fig. S5C and S5D).
FIGURE 5.
DR5 protein levels are up-regulated by ABT-737 treatment. A, effects of ABT-737 treatment of PV10 cells on the protein levels of key DISC proteins. Cells were treated with ABT-737 at the indicated dose for 24 h followed by Western blotting of cell extracts with specific antibodies. B, PV10, DU145, A549, HOP-62, and LNCaP cells were treated with DMSO or ABT-737 (5 or 10 μm) for 24 h, and the expression of cell surface DR4 and DR5 was determined by FACS analysis. Isotype-matched IgG1 monoclonal antibody was used as a negative control. C, PV10 cells were transfected with siRNA duplexes targeting DR5 for 36 h and then were treated with DMSO, 100 ng/ml TRAIL, 10 μm ABT-737, or the combination, and extracts were subjected to Western blotting with multiple antibodies. D, cells treated as in C and then were assessed for cell viability by an acid phosphatase assay (mean ± S.D., n = 4). E, RNA was extracted from PV10 cells treated with DMSO (0) or ABT-737 (5 or 10 μm) for 24 h. The level of DR5 mRNA was then determined by quantitative real time-PCR. F, PV10 cells were transfected with the vector PGVB2-DR5 (-1188)-luc encoding the upstream region of the DR5 promoter cloned in front of a luciferase reporter and then treated with DMSO or ABT-737 for 24 h (mean ± S.D., n = 3). DR5 luciferase activity was determined by dual luciferase assay (see “Experimental Procedures”).
To determine how ABT-737 regulates DR5 levels, we carried out quantitative real time PCR on RNA samples from tumor cells treated with varying doses of ABT-737. Shown in Fig. 5E is our observation that both PV10 and LNCaP cells demonstrated a dose-dependent increase in the level of DR5 mRNA after ABT-737 treatment, and in contrast the unresponsive DU145 cells showed little change in mRNA levels. Treatment with ABT-737 did not cause any change in the half-life of the DR5 mRNA (data not shown), but treatment with this agent was capable of inducing the luciferase activity of a reporter plasmid containing 1188 bp of the upstream portion of DR5 promoter, pGVB2-DR5 (-1188) (Fig. 5F) (39). Together these results suggest that ABT-737 treatment of tumor cells regulates the transcription of the DR5 gene.
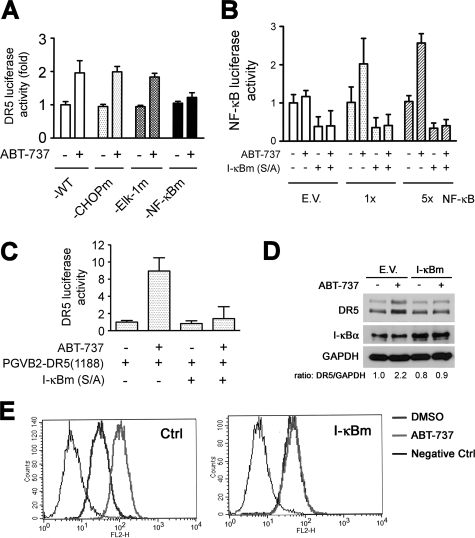
ABT-737 Activation of NF-κB to Stimulate DR5 Transcription—When PV10 cells were transfected with the reporter constructs with different lengths of the 5′-flanking region of the DR5 gene, ABT-737 failed to increase the luciferase activity of pGVB2-DR5(-605) and pGVB2-DR5(-115) while significantly increasing the luciferase activity of pGVB2-DR5(-605) and pGL3-DR5(-1188) (supplemental Fig. S6A). To examine the specific sequences regulated in the DR5 promoter by ABT-737, we focused on the NF-κB, CHOP, and Elk-1-binding sites within this region (40). Previous results have suggested that DR5 gene transcription is regulated by the transcription factors that bind to NF-κB and CHOP sites (40, 46). We compared the effects of ABT-737 on the transactivation of reporter constructs carrying all wild-type-binding sites or singly mutated sites. ABT-737 treatment increased the luciferase activity of the wild type and those promoter constructs carrying the mutated CHOP- or Elk-binding sites (Fig. 6A) but did not increase the luciferase activity of the transfected construct carrying the mutated NF-κB-binding site. Using reporter plasmids that contain either a single copy or five copies of the NF-κB-binding site and empty vector as a control, we demonstrated that ABT-737 treatment is capable of inducing up to a 3-fold increase in the activity of this promoter (Fig. 6B), suggesting that this compound can activate the NF-κB pathway. Transfection into PV10 cells of a dominant-negative mutant I-κBα (I-κBm S32/36A) abrogated the ability of ABT-737 to induce DR5 luciferase activity (Fig. 6C). In contrast, siRNA directed at the CHOP transcription factor did not change ABT-737-mediated DR5 promoter activity but did block the ability of tunicamycin to stimulate this promoter (supplemental Fig. S6B). We next examined whether modulation of NF-κB activity inhibited the ability of ABT-737 to increase the levels of DR5 protein expression. PV10 cells were transfected with dominant-negative IκB or empty vector as a control, and then the cells were treated overnight with ABT-737. The addition of ABT-737 increased DR5 expression levels, and this increase in protein as well as cell-surface DR5 expression was blocked by the dominant-negative IκB expression (Fig. 6, D and E). These results further support the notion that NF-κB is directly involved in DR5 protein up-regulation in ABT-737-treated cells.
FIGURE 6.
NF-κB activation by ABT-737 is required for DR5 up-regulation. A, PV10 cells were transfected with wild-type pGL3-DR5(-552)-luc (WT), pGL3-DR5(-552)-CHOPm, pGL3-DR5(-552)-NF-κBm, or pGL3-DR5(-552)-Elk-1m-luc. Luciferase activity was normalized to Renilla-luciferase activity (mean ± S.D., n = 3). B, PV10 cells were transfected with luciferase plasmids containing no insert empty vector (E.V.) (control), 1 (1×), or 5 (5×) copies of an NF-κB reporter plasmid pEF-Renilla-luc (RLuc), and firefly luciferase was normalized to RLuc. After 36 h, these cells were treated with DMSO or ABT-737 (10 μm) for 24 h prior to assay. C, PV10 cells were cotransfected with I-κBm (S/A) and PGVB2-DR5(-1188)-luc, and after 36 h were treated with DMSO or ABT-737 (10 μm) for an additional 24 h. Luciferase assays were carried out as described under “Experimental Procedures.” D, PV10 cells were transfected with empty vector (E.V.) control or pcDNA3-I-κBm, and after 36 h were treated with DMSO or ABT-737 (10 μm) for an additional 24 h. The expression of DR5, I-κBα, and GAPDH was determined by Western blotting. These Western blots were scanned, and the ratio of DR5 to GAPDH was determined by densitometry. E, again, cell surface DR5 expression was determined by FACS analysis. Isotype-matched IgG1 monoclonal antibody was used as a negative control.
DISCUSSION
We find that resistance to TRAIL treatment of renal, prostate, and lung cancer cells in culture can be overcome by the addition of ABT-737. These two agents are highly synergistic in inducing apoptosis within hours. This remarkable killing is dependent on the activity of TRAIL, because knocking down the receptor with RNA interference completely inhibited the apoptosis induced by this combination treatment, and three cell lines with low levels of the DR5 receptor, 786-O renal cancer cells, HEK293 cells, and the normal kidney cell line HK-2 were resistant to this combination therapy. However, Caki cells that also had a low level of DR5 demonstrated an increased level of DR5 protein after ABT-737 treatment and were sensitive to the combination. Other resistant cell lines included two Bax negative cell lines DU145 and HCT116 Bax-/- cells. Bax has been noted previously to be essential for TRAIL-mediated cell death (41, 42), as suggested by the observation that colon cancer and leukemic T cells deficient in this protein are TRAIL-resistant (42, 47). DU145 cells become sensitive to the combination therapy when Bax is reexpressed. Similarly, it has been shown that chemotherapeutic agents, including etoposide and bortezomib, that elevate the levels of Bak or increase DR5 protein levels can also overcome TRAIL resistance (36, 41, 48).
Recently, similar results have been obtained in pancreatic cancer cells in which ABT-737 and TRAIL were found to be highly synergistic (49). These experiments described the ability of these two compounds to work synergistically because ABT-737 is able to displace the Bim protein from its binding partners. However, these researchers also demonstrate that this combination induced increased caspase-8 cleavage in the two pancreatic cancer cell lines studied. This result supports the possibility that in pancreatic cancer cells like renal, prostate, and lung cancer cells that DR5 induction by ABT-737 enhances TRAIL-induced caspase-8 cleavage. We further showed that the combination of TRAIL and ABT-737 kills TRAIL-resistant cells despite varied levels of Mcl-1. Decreasing the level of Mcl-1 sensitizes PV10 renal carcinoma cells to both TRAIL and ABT-737 consistent with the ability of this protein to inhibit cell death induced by both agents. These inhibitory findings with Mcl-1 are consistent with what has been demonstrated by several other investigators (22-24, 26, 32), including results obtained with pancreatic cancer cells (48). Decreasing Bcl-2 and Bcl-xL by siRNA treatment sensitizes the resistant cell line to TRAIL killing alone but not ABT-737. It has been suggested that the ability of ABT-737 to induce apoptosis is dependent on the levels of Bcl-2 and the amount of Bim bound to Bcl-2 (50), whereas others have suggested that low levels of Mcl-1 and high Bcl-2 predict ABT-737 sensitivity (27). Although these levels might be predicative of ABT-737 sensitivity alone, combination therapy was equally active in conditions of low and high Bim, Mcl-1, and Bcl-2, suggesting that another mechanism was at work.
The addition of ABT-737 to all tumor cells examined enhanced the function of the TRAIL-DISC and led to increased cleavage of caspase-8 upon the addition of TRAIL. This result is consistent with the induction by ABT-737 of the DR5 receptor protein in PV10 renal cancer, LNCaP prostate cancer, and two lung cancer cell lines, A549 and HOP-62. Interestingly, we find that ABT-737 transcriptionally activates the DR5 promoter and stimulates an increase in DR5 mRNA. DR5 transcription has been shown to be regulated by stress stimuli that through the unfolded protein response increase the levels of the transcription factor CHOP, which in turn binds to the DR5 promoter (40). ABT-737 treatment of tumor cells did not induce an increase in CHOP protein nor did an siRNA directed at CHOP decrease the ability of ABT-737 to stimulate an increase in DR5 mRNA (data not shown). Instead, we find that mutation of the NF-κB site in the DR5 promoter (51) decreased the ability of ABT-737 to activate the transcription of this gene. Using reporter constructs that contain multiple NF-κB-binding sites, we further demonstrate that ABT-737 is able to activate NF-κB. One possibility to explain these results might be that ABT-737 induces the production of reactive oxygen species (ROS). ROS have been shown to up-regulate the expression of DR5, whereas pretreatment with N-acetylcysteine causes a significant inhibition of ROS-induced up-regulation of DR5 gene and protein expression (52, 53). Although some recent reports (52, 54) have suggested that ROS increased DR5 is dependent on CHOP expression, we did not find any changes in the levels of this protein.
Activation of NF-κB has complex effects on the TRAIL-induced apoptotic cascade. Blocking NF-κB activation in cell lines with constitutive activation, for example multiple myeloma, pancreatic, and renal cancer (55-57), enhances the ability of TRAIL to induce apoptotic cell death. One explanation for these results might be that NF-κB activation increases the expression of Bcl-xL protein, and this protein is known to inhibit TRAIL-induced mitochondrial activation (58). Inhibition of NF-κB activity by preventing the degradation of IκB has also been shown to down-regulate the level of c-FLIP, a DISC-inhibitory protein, thus sensitizing to TRAIL (59). However, this combination therapy may be successful because ABT-737 may function to inhibit the activity of Bcl-2 family proteins induced by NF-κB activation and to increase TRAIL receptor DR5 overcoming endogenous levels of c-FLIP.
Thus, we find that combinations of ABT-737 and TRAIL can be exploited therapeutically in a diverse set of solid tumors where antiapoptotic Bcl-2 family members drive tumor cell resistance to proapoptotic therapy. The ability of ABT-737 to regulate the transcription of specific genes in cell lines where it does not activate cell death suggests a novel mechanism where it could enhance the effects of a diverse set of chemotherapeutic agents.
Supplementary Material
Acknowledgments
We thank Abbott Laboratories for providing ABT-737 and its enantiomer; Dr. H.-G. Wang (H. Lee Moffitt Cancer Center and Research Institute, University of South Florida College of Medicine) for kindly providing pGL3-DR5 firefly luciferase reporter plasmids; and Dr. B. Vogelstein (Howard Hughes Medical Institute, The Johns Hopkins University School of Medicine) for supplying the HCT116 cell lines. We also thank Drs. R. Gemmill and H. Drabkin for help in providing the renal cancer cell lines and Drs. D. Kurtz and A. Nieminen for the NF-κB firefly luciferase reporter plasmids (Medical University of South Carolina).
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA104710 (to A. S. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S6.
Footnotes
The abbreviations used are: TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; DISC, death-inducing signaling complex; DR5, death receptor 5; ROS, reactive oxygen species; siRNA, small interfering RNA; FACS, fluorescence-activated cell sorter; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HRP, horseradish peroxidase; Z, benzyloxycarbonyl; fmk, fluoromethyl ketone; HEK293, human embryonic kidney 293; PARP, poly(ADP-ribose) polymerase.
References
- 1.Ashkenazi, A., Pai, R. C., Fong, S., Leung, S., Lawrence, D. A., Marsters, S. A., Blackie, C., Chang, L., McMurtrey, A. E., Hebert, A., DeForge, L., Koumenis, I. L., Lewis, D., Harris, L., Bussiere, J., Koeppen, H., Shahrokh, Z., and Schwall, R. H. (1999) J. Clin. Invest. 104 155-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walczak, H., Miller, R. E., Ariail, K., Gliniak, B., Griffith, T. S., Kubin, M., Chin, W., Jones, J., Woodward, A., Le, T., Smith, C., Smolak, P., Goodwin, R. G., Rauch, C. T., Schuh, J. C., and Lynch, D. H. (1999) Nat. Med. 5 157-163 [DOI] [PubMed] [Google Scholar]
- 3.Hao, C., Song, J. H., Hsi, B., Lewis, J., Song, D. K., Petruk, K. C., Tyrrell, D. L., and Kneteman, N. M. (2004) Cancer Res. 64 8502-8506 [DOI] [PubMed] [Google Scholar]
- 4.Ichikawa, K., Liu, W., Zhao, L., Wang, Z., Liu, D., Ohtsuka, T., Zhang, H., Mountz, J. D., Koopman, W. J., Kimberly, R. P., and Zhou, T. (2001) Nat. Med. 7 954-960 [DOI] [PubMed] [Google Scholar]
- 5.Walczak, H., and Krammer, P. H. (2000) Exp. Cell Res. 256 58-66 [DOI] [PubMed] [Google Scholar]
- 6.Pan, G., Ni, J., Wei, Y. F., Yu, G., Gentz, R., and Dixit, V. M. (1997) Science 277 815-818 [DOI] [PubMed] [Google Scholar]
- 7.Sheridan, J. P., Marsters, S. A., Pitti, R. M., Gurney, A., Skubatch, M., Baldwin, D., Ramakrishnan, L., Gray, C. L., Baker, K., Wood, W. I., Goddard, A. D., Godowski, P., and Ashkenazi, A. (1997) Science 277 818-821 [DOI] [PubMed] [Google Scholar]
- 8.Kischkel, F. C., Lawrence, D. A., Chuntharapai, A., Schow, P., Kim, K. J., and Ashkenazi, A. (2000) Immunity 12 611-620 [DOI] [PubMed] [Google Scholar]
- 9.Igney, F. H., and Krammer, P. H. (2002) Nat. Rev. Cancer 2 277-288 [DOI] [PubMed] [Google Scholar]
- 10.Li, H., Zhu, H., Xu, C. J., and Yuan, J. (1998) Cell 94 491-501 [DOI] [PubMed] [Google Scholar]
- 11.Jin, H., Yang, R., Fong, S., Totpal, K., Lawrence, D., Zheng, Z., Ross, J., Koeppen, H., Schwall, R., and Ashkenazi, A. (2004) Cancer Res. 64 4900-4905 [DOI] [PubMed] [Google Scholar]
- 12.Nicholson, D. W. (2000) Nature 407 810-816 [DOI] [PubMed] [Google Scholar]
- 13.Fulda, S., Meyer, E., and Debatin, K. M. (2002) Oncogene 21 2283-2294 [DOI] [PubMed] [Google Scholar]
- 14.Munshi, A., Pappas, G., Honda, T., McDonnell, T. J., Younes, A., Li, Y., and Meyn, R. E. (2001) Oncogene 20 3757-3765 [DOI] [PubMed] [Google Scholar]
- 15.Sinicrope, F. A., Penington, R. C., and Tang, X. M. (2004) Clin. Cancer Res. 10 8284-8292 [DOI] [PubMed] [Google Scholar]
- 16.Thomas, W. D., Zhang, X. D., Franco, A. V., Nguyen, T., and Hersey, P. (2000) J. Immunol. 165 5612-5620 [DOI] [PubMed] [Google Scholar]
- 17.Bai, J., Sui, J., Demirjian, A., Vollmer, C. M., Jr., Marasco, W., and Callery, M. P. (2005) Cancer Res. 65 2344-2352 [DOI] [PubMed] [Google Scholar]
- 18.Taniai, M., Grambihler, A., Higuchi, H., Werneburg, N., Bronk, S. F., Farrugia, D. J., Kaufmann, S. H., and Gores, G. J. (2004) Cancer Res. 64 3517-3524 [DOI] [PubMed] [Google Scholar]
- 19.Werner, A. B., de Vries, E., Tait, S. W., Bontjer, I., and Borst, J. (2002) J. Biol. Chem. 277 22781-22788 [DOI] [PubMed] [Google Scholar]
- 20.Willis, S. N., Chen, L., Dewson, G., Wei, A., Naik, E., Fletcher, J. I., Adams, J. M., and Huang, D. C. (2005) Genes Dev. 19 1294-1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang, W., Konopleva, M., Ruvolo, V. R., McQueen, T., Evans, R. L., Bornmann, W. G., McCubrey, J., Cortes, J., and Andreeff, M. (2008) Leukemia (Basingstoke), 22 808-818 [DOI] [PubMed] [Google Scholar]
- 22.Ricci, M. S., Kim, S. H., Ogi, K., Plastaras, J. P., Ling, J., Wang, W., Jin, Z., Liu, Y. Y., Dicker, D. T., Chiao, P. J., Flaherty, K. T., Smith, C. D., and El-Deiry, W. S. (2007) Cancer Cell 12 66-80 [DOI] [PubMed] [Google Scholar]
- 23.Meng, X. W., Lee, S. H., Dai, H., Loegering, D., Yu, C., Flatten, K., Schneider, P., Dai, N. T., Kumar, S. K., Smith, B. D., Karp, J. E., Adjei, A. A., and Kaufmann, S. H. (2007) J. Biol. Chem. 282 29831-29846 [DOI] [PubMed] [Google Scholar]
- 24.Certo, M., Del Gaizo Moore, V., Nishino, M., Wei, G., Korsmeyer, S., Armstrong, S. A., and Letai, A. (2006) Cancer Cell 9 351-365 [DOI] [PubMed] [Google Scholar]
- 25.Oltersdorf, T., Elmore, S. W., Shoemaker, A. R., Armstrong, R. C., Augeri, D. J., Belli, B. A., Bruncko, M., Deckwerth, T. L., Dinges, J., Hajduk, P. J., Joseph, M. K., Kitada, S., Korsmeyer, S. J., Kunzer, A. R., Letai, A., Li, C., Mitten, M. J., Nettesheim, D. G., Ng, S., Nimmer, P. M., O'Connor, J. M., Oleksijew, A., Petros, A. M., Reed, J. C., Shen, W., Tahir, S. K., Thompson, C. B., Tomaselli, K. J., Wang, B., Wendt, M. D., Zhang, H., Fesik, S. W., and Rosenberg, S. H. (2005) Nature 435 677-681 [DOI] [PubMed] [Google Scholar]
- 26.Konopleva, M., Contractor, R., Tsao, T., Samudio, I., Ruvolo, P. P., Kitada, S., Deng, X., Zhai, D., Shi, Y. X., Sneed, T., Verhaegen, M., Soengas, M., Ruvolo, V. R., McQueen, T., Schober, W. D., Watt, J. C., Jiffar, T., Ling, X., Marini, F. C., Harris, D., Dietrich, M., Estrov, Z., McCubrey, J., May, W. S., Reed, J. C., and Andreeff, M. (2006) Cancer Cell 10 375-388 [DOI] [PubMed] [Google Scholar]
- 27.Chen, S., Dai, Y., Harada, H., Dent, P., and Grant, S. (2007) Cancer Res. 67 782-791 [DOI] [PubMed] [Google Scholar]
- 28.Shoemaker, A. R., Oleksijew, A., Bauch, J., Belli, B. A., Borre, T., Bruncko, M., Deckwirth, T., Frost, D. J., Jarvis, K., Joseph, M. K., Marsh, K., McClellan, W., Nellans, H., Ng, S., Nimmer, P., O'Connor, J. M., Oltersdorf, T., Qing, W., Shen, W., Stavropoulos, J., Tahir, S. K., Wang, B., Warner, R., Zhang, H., Fesik, S. W., Rosenberg, S. H., and Elmore, S. W. (2006) Cancer Res. 66 8731-8739 [DOI] [PubMed] [Google Scholar]
- 29.Chauhan, D., Velankar, M., Brahmandam, M., Hideshima, T., Podar, K., Richardson, P., Schlossman, R., Ghobrial, I., Raje, N., Munshi, N., and Anderson, K. C. (2007) Oncogene 26 2374-2380 [DOI] [PubMed] [Google Scholar]
- 30.Tahir, S. K., Yang, X., Anderson, M. G., Morgan-Lappe, S. E., Sarthy, A. V., Chen, J., Warner, R. B., Ng, S. C., Fesik, S. W., Elmore, S. W., Rosenberg, S. H., and Tse, C. (2007) Cancer Res. 67 1176-1183 [DOI] [PubMed] [Google Scholar]
- 31.Lin, X., Morgan-Lappe, S., Huang, X., Li, L., Zakula, D. M., Vernetti, L. A., Fesik, S. W., and Shen, Y. (2007) Oncogene 26 3972-3979 [DOI] [PubMed] [Google Scholar]
- 32.van Delft, M. F., Wei, A. H., Mason, K. D., Vandenberg, C. J., Chen, L., Czabotar, P. E., Willis, S. N., Scott, C. L., Day, C. L., Cory, S., Adams, J. M., Roberts, A. W., and Huang, D. C. (2006) Cancer Cell 10 389-399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song, J. H., Wang, C. X., Song, D. K., Wang, P., Shuaib, A., and Hao, C. (2005) J. Biol. Chem. 280 12896-12901 [DOI] [PubMed] [Google Scholar]
- 34.Song, J. H., Slot, A. J., Ryan, R. W., and Ross, G. M. (2004) Neuropharmacology 46 984-993 [DOI] [PubMed] [Google Scholar]
- 35.Song, J. H., Tse, M. C., Bellail, A., Phuphanich, S., Khuri, F., Kneteman, N. M., and Hao, C. (2007) Cancer Res. 67 6946-6955 [DOI] [PubMed] [Google Scholar]
- 36.Song, J. H., Song, D. K., Pyrzynska, B., Petruk, K. C., Van Meir, E. G., and Hao, C. (2003) Brain Pathol. 13 539-553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lilly, M., Sandholm, J., Cooper, J. J., Koskinen, P. J., and Kraft, A. (1999) Oncogene 18 4022-4031 [DOI] [PubMed] [Google Scholar]
- 38.Eguchi, H., Suga, K., Saji, H., Toi, M., Nakachi, K., and Hayashi, S. I. (2000) Cell Death Differ. 7 439-446 [DOI] [PubMed] [Google Scholar]
- 39.Yoshida, T., Shiraishi, T., Nakata, S., Horinaka, M., Wakada, M., Mizutani, Y., Miki, T., and Sakai, T. (2005) Cancer Res. 65 5662-5667 [DOI] [PubMed] [Google Scholar]
- 40.Yamaguchi, H., and Wang, H. G. (2004) J. Biol. Chem. 279 45495-45502 [DOI] [PubMed] [Google Scholar]
- 41.LeBlanc, H., Lawrence, D., Varfolomeev, E., Totpal, K., Morlan, J., Schow, P., Fong, S., Schwall, R., Sinicropi, D., and Ashkenazi, A. (2002) Nat. Med. 8 274-281 [DOI] [PubMed] [Google Scholar]
- 42.Deng, Y., Lin, Y., and Wu, X. (2002) Genes Dev. 16 33-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goping, I. S., Gross, A., Lavoie, J. N., Nguyen, M., Jemmerson, R., Roth, K., Korsmeyer, S. J., and Shore, G. C. (1998) J. Cell Biol. 143 207-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Desagher, S., Osen-Sand, A., Nichols, A., Eskes, R., Montessuit, S., Lauper, S., Maundrell, K., Antonsson, B., and Martinou, J. C. (1999) J. Cell Biol. 144 891-901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Griffiths, G. J., Dubrez, L., Morgan, C. P., Jones, N. A., Whitehouse, J., Corfe, B. M., Dive, C., and Hickman, J. A. (1999) J. Cell Biol. 144 903-914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ravi, R., Bedi, G. C., Engstrom, L. W., Zeng, Q., Mookerjee, B., Gelinas, C., Fuchs, E. J., and Bedi, A. (2001) Nat. Cell Biol. 3 409-416 [DOI] [PubMed] [Google Scholar]
- 47.Han, J., Goldstein, L. A., Gastman, B. R., Rabinovitz, A., Wang, G. Q., Fang, B., and Rabinowich, H. (2004) Leukemia (Baltimore) 18 1671-1680 [DOI] [PubMed] [Google Scholar]
- 48.Johnson, T. R., Stone, K., Nikrad, M., Yeh, T., Zong, W. X., Thompson, C. B., Nesterov, A., and Kraft, A. S. (2003) Oncogene 22 4953-4963 [DOI] [PubMed] [Google Scholar]
- 49.Huang, S., and Sinicrope, F. A. (2008) Cancer Res. 68 2944-2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Del Gaizo Moore, V., Brown, J. R., Certo, M., Love, T. M., Novina, C. D., and Letai, A. (2007) J. Clin. Investig. 117 112-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shetty, S., Graham, B. A., Brown, J. G., Hu, X., Vegh-Yarema, N., Harding, G., Paul, J. T., and Gibson, S. B. (2005) Mol. Cell. Biol. 25 5404-5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim, H., Kim, E. H., Eom, Y. W., Kim, W. H., Kwon, T. K., Lee, S. J., and Choi, K. S. (2006) Cancer Res. 66 1740-1750 [DOI] [PubMed] [Google Scholar]
- 53.Hussain, A. R., Al-Jomah, N. A., Siraj, A. K., Manogaran, P., Al-Hussein, K., Abubaker, J., Platanias, L. C., Al-Kuraya, K. S., and Uddin, S. (2007) Cancer Res. 67 3888-3897 [DOI] [PubMed] [Google Scholar]
- 54.Kouhara, J., Yoshida, T., Nakata, S., Horinaka, M., Wakada, M., Ueda, Y., Yamagishi, H., and Sakai, T. (2007) Int. J. Oncol. 30 679-687 [PubMed] [Google Scholar]
- 55.Romagnoli, M., Desplanques, G., Maiga, S., Legouill, S., Dreano, M., Bataille, R., and Barille-Nion, S. (2007) Clin. Cancer Res. 13 6010-6018 [DOI] [PubMed] [Google Scholar]
- 56.Braeuer, S. J., Buneker, C., Mohr, A., and Zwacka, R. M. (2006) Mol. Cancer Res. 4 715-728 [DOI] [PubMed] [Google Scholar]
- 57.Oya, M., Ohtsubo, M., Takayanagi, A., Tachibana, M., Shimizu, N., and Murai, M. (2001) Oncogene 20 3888-3896 [DOI] [PubMed] [Google Scholar]
- 58.Khoshnan, A., Tindell, C., Laux, I., Bae, D., Bennett, B., and Nel, A. E. (2000) J. Immunol. 165 1743-1754 [DOI] [PubMed] [Google Scholar]
- 59.Kreuz, S., Siegmund, D., Scheurich, P., and Wajant, H. (2001) Mol. Cell. Biol. 21 3964-3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.