Abstract
Self-illuminating quantum dots, also known as QD-BRET conjugates, are a new class of quantum dots bioconjugates which do not need external light for excitation. Instead, light emission relies on the bioluminescence resonance energy transfer from the attached Renilla luciferase enzyme, which emits light upon the oxidation of its substrate. QD-BRET combines the advantages of the QDs (such as superior brightness & photostability, tunable emission, multiplexing) as well as the high sensitivity of bioluminescence imaging, thus holds the promise for improved deep tissue in vivo imaging. Although studies have demonstrated the superior sensitivity and deep tissue imaging potential, the stability of the QD-BRET conjugates in biological environment needs to be improved for long-term imaging studies such as in vivo cell trafficking. In this study, we seek to improve the stability of QD-BRET probes through polymeric encapsulation with a polyacrylamide gel. Results show that encapsulation caused some activity loss, but significantly improved both the in vitro serum stability and in vivo stability when subcutaneously injected into the animal. Stable QD-BRET probes should further facilitate their applications for both in vitro testing as well as in vivo cell tracking studies.
Keywords: self-illuminating quantum dots, bioluminescence resonance energy transfer (BRET), polymeric encapsulation, molecular imaging, nanotechnology
Semiconductor quantum dots (QDs) have captivated scientists and engineers over the past two decades owing to their fascinating optical and electronic properties, which are not available from either single individual molecules or bulk solids [1]. Compared with organic dyes and fluorescent proteins, semiconductor QDs offer several unique advantages, such as size- and composition-tunable emission from visible to infrared wavelengths, large absorption coefficients across a wide spectral range, and very high levels of brightness and photostability [2]. These properties make them appealing fluorescent probes for bioimaging applications including in situ cell/tissue labeling, live cell imaging and in vivo imaging [3]. However, in vivo imaging using quantum dots still faces challenges such as interferences from tissue autofluorescence [4] and paucity of light available at non-superficial tissue sites [5]. One approach to solve these problems is to use high quality near infrared (NIR) QDs [6], which is an area still under active development. Alternatively, since both problems are related to the need for external excitation light, a “self-illuminating” quantum dot that does not require external light to fluoresce may also help to alleviate these problems.
Recently, work in our lab has demonstrated the feasibility of using QDs as the acceptor in a BRET system [7]. Bioluminescence resonance energy transfer (BRET) is a naturally occurring phenomenon whereby a light-emitting protein (the donor, e.g. R. reniformis luciferase) non-radiatively transfers energy to a fluorescent protein (the acceptor, e.g. green fluorescent protein) in close proximity. In this study, QDs were covalently conjugated by EDC (N-Ethyl-N′-(3-dimethylaminopropyl)carbodiimide hydrochloride) coupling to the donor, Luc8 protein, an eight-mutation variant of the bioluminescent R. reniformis luciferase [8]. Upon the addition of substrate, coelenterazine, the protein emits blue light with a peak at 480 nm and this energy transfers to the QDs in a close proximity, causing QD fluorescence. The advantage of using bioluminescence versus fluorescence lies in the fact that no external excitation is needed for bioluminescence. This “self-illuminating” feature allows imaging in deeper tissue where light sources are limited. Compared with existing QDs, self-illuminating QD conjugates have greatly enhanced sensitivity in small animal imaging, with an in vivo signal-to-background ratio of >1000 for 5 pmole of conjugate subcutaneously injected [7]. However, the stability of the first generation of QD-BRET probes in blood or serum is moderate, thus prohibiting many in vivo applications such as tumor targeting and cell trafficking. Mechanisms for deterioration in bioluminescence activity of QD-BRET probes are not clear; since the protein by itself is relatively stable in serum and the stability of the conjugates in the phosphate buffer is much better, some unfavorable interactions between the QD and protein and inactivation of the protein by serum/blood proteins may both be responsible. In this work, we explored strategies to preserve the stability of the enzyme on the QD in order to achieve longer-lived QD-BRET probes and use them for prolonged animal imaging purposes such as cell trafficking or tumor targeting.
Enzyme stability improvement or how to preserve its activity under harsh conditions (such as high temperature, organic solvents etc) has been studied for a long time in the industrial biotechnology field. Complementary to genetic modifications such as directed evolution [9] and site mutation [10], chemical modification via conjugation or encapsulation provide an efficient route to preserve enzyme stability under harsh conditions. Different coating materials have been tested including silica [11], polymers [12, 13] and organo-clay. These coating materials, however, may hinder the conformational transition of the enzyme and the transport of substrate, resulting in low biocatalytic activity. A few years ago, Kim and Grate reported an encapsulation scheme that does not cause significant increase in mass transfer resistance [14]. This scheme consists of multiple steps including surface modification, lyophilization, polymerization in organic solvent and shell condensation. More recently, Yan et al reported a new two-step procedure that involves surface acryloylation and in situ aqueous polymerization to encapsulate a single enzyme in a nanogel [15]. The authors applied this technique to both horseradish peroxidase and bovine carbonic anhydrase [15,16], and both showed excellent preservation of their biocatalytic activities (70–80%). Encouraged by these results, in this study, we applied this approach to improve the serum stability of QD-BRET conjugates. We characterized the optical properties of the encapsulated QD-BRET conjugates, and evaluated their in vitro and in vivo stabilities.
Experimental Procedures
Animals and imaging equipments
Nude mice (4–6 weeks old) were from Charles River Breeding Laboratories. Fluorescence and bioluminescence emission spectra were collected with a Fluoro Max-3 (Jobin Yvon Inc., New Jersey); in the case of bioluminescence, the excitation light was blocked, and emission spectra were corrected with a correction file provided by the company. Bioluminescence emission spectra collected with the spectrofluorometer were further corrected for the Luc8 kinetics over the course of data acquisition (typically ~20 s). Enzymatic activity of Luc8 was measured with a 20/20n Luminometer (Turner Biosystems, Inc.). Animal use protocols were reviewed and approved by the Institutional Animal Care Use Committee of Stanford University.
Preparation of QD-Luc8 BRET probe
Quantum dots BRET probes were made according to a procedure published previously by our lab [7]. Brief ly, carboxylated semiconductor quantum dots (QD655, Invitrogen) were mixed with coupling agent EDC (Fluka, 4000 equivalent to QDs) and Luc8 protein (20 equivalent to QDs) in phosphate buffer (pH 7.4) and reacted for 1 hr at room temperature. Afterwards, excessive EDC and protein (37 KDa) were removed by ultrafiltration with a MWCO 100K filter (Millipore) at 5000 rpm for 3 times at 4 °C. The washed conjugates then were measured for bioluminescence and fluorescence using a Fluoro Max3 Fluoremeter (Jobin Yvon). Each quantum dot has approximately 6 protein molecules on the surface.
Conjugates acryloylation & polymeric encapsulation
QD-Luc8 conjugates (100 pmole) were mixed with acryloylating agent N-hydroxysuccinimide ester (8 µg, Sigma) and stabilizer 4-Dimethylaminoantipyrine (2 µg, Sigma) in borate buffer (100 mM, pH 9.3) and incubated at 30 °C. This step converts free amines in the protein to vinyl groups. After 2 hrs’ reaction, excessive reagents (N-hydroxysuccinimide, 4-dimethylaminoantipyrine) were removed by gel filtration (G-25 column) or ultrafiltration filter (MWCO: 100K, Millipore). The acryloylated conjugates were then purged with Argon before adding in 3 µl TEMED (BioRad) and ammonium persulfate (100 µg, Sigma). Subsequently, acrylamide monomer (Invitrogen) and crosslinking agent N, N’-methylene bis-acrylamide (Sigma) dissolved in deoxygenated water were evenly added to the solution over the next hour at a molar ratio of 10 to 1. Total amount of monomer added was approximately 5000 times of protein molecules present on the quantum dots. The mixture was left at room temperature under argon for another hour. At the end, excessive monomer and cross-linking reagents were removed from the conjugates by passing through a G-75 column or ultrafiltration (MWCO: 100K).
TEM and Dynamic light scattering
Sizing of the nanoparticles was achieved by dynamic light scattering (Brookhaven 90 plus nanosizer) and transmission electron microscopy. TEM specimens were prepared by pipetting 5µl of the samples onto ultrathin Carbon Type A 400-mesh copper grids (Tedpella Inc., Redding, CA) that had been glow-discharged. After about five minutes, the grids were rinsed with deionized water to remove any buffer salts that may be present in the samples and wicked to almost dryness using filter paper. Phosphotungstic Acid (PTA) at pH 7.0 was then added onto the grids to negatively stain the samples. After one minute the grids were completely dried using filter paper. All the specimens were analyzed using a Philips CM20 FEG-TEM operated at 200kV at the Stanford Nanocharacterization Laboratory (SNL). The microscope is also equipped with an energy dispersive X-ray spectrometer (EDS) for compositional analysis.
In vitro stability testing
QD-Luc8 conjugates obtained from above were incubated in equal-volume mouse serum (Equitech Bio.) at 37°C. The stability kinetics was checked by sampling at various time points (0, 24, 48, and 120 hr). At each time point, the conjugates were measured for bioluminescence and quantum dots fluorescence (excitation: 480 nm).
Animal Imaging
Quantum dot conjugates (5 pmole QDs) were injected subcutaneously into 4–6 weeks old nude mice (Charles River Breeding Laboratories). The animals were then anesthetized with isofluorane, and transferred into the light-tight chamber of an IVIS 200 imager (Xenogen). After 2 min, coelenterazine (Nanolight; 10 µg per animal, prepared in 10 µl methanol and 90 µl phosphate buffer) was injected intravenously through the tail vein. Images were acquired with and without filters. Each acquisition took 5 second. To correct for the relatively fast in vivo pharmacokinetics of coelenterazine, we acquired the images sequentially: (i) with filter (5 s); (ii) without filter (5 s); (iii) without filter (5 s); (iv) with filter (5 s). The emission with filter was calculated from the average of 1 and 4, and the emission without filter was the average of 2 and 3.
Results
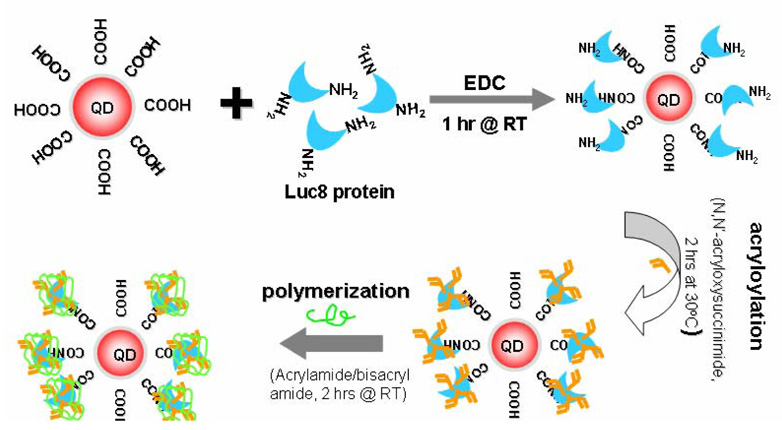
Our modification procedure consists of two major steps: acryloylation of the protein and polymerization afterwards. The detailed procedure is shown schematically in Figure 1. Carboxylated quantum dots and Luc8 proteins were first conjugated by EDC coupling to produce the QD-BRET probes. After purification and removal of excessive reactants (protein and coupling agent), the probes were then incubated with N-acryloxysuccinimide in alkali buffer (pH 9.3) for 2 hrs at 30 °C to generate surface vinyl groups. Encapsulation of each protein molecule was realized by in situ polymerization at room temperature with acrylamide as the monomer, N,N’-methylene bisacrylamide as the cross-linker, and N,N,N,N’-tetramethylethylenediamine/ammonium persulfate as the initiator.
Figure 1.
Schematic of the encapsulation procedure of QD-BRET probes.
Conjugates size change after encapsulation
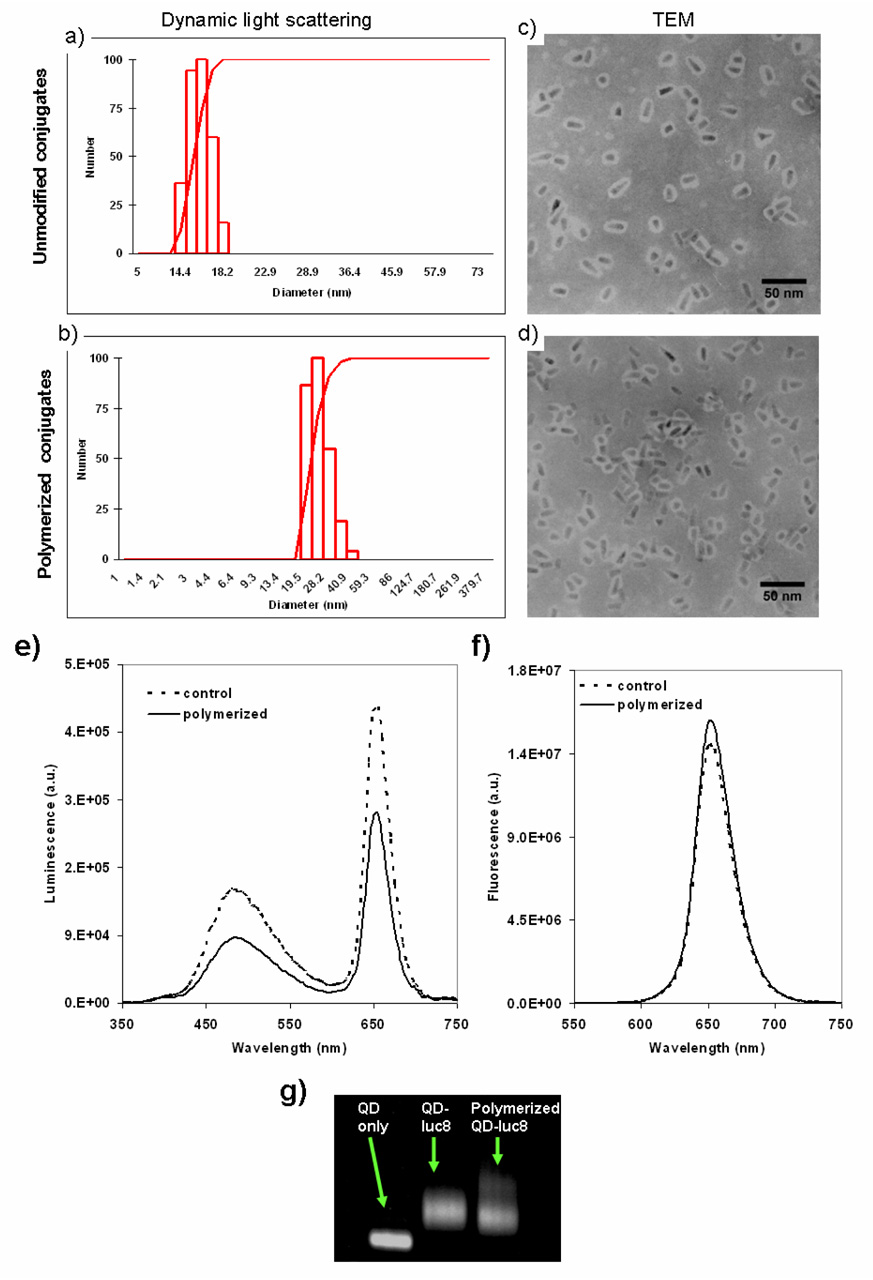
Dynamic light scattering results (Figure 2a & 2c) showed that, after polymerization, the nanopaticles (effective diameter: 51.0 nm) were about twice as big as the unmodified conjugates (effective diameter: 26.0 nm). TEM results revealed that the size increase was mainly due to the cross-linking between the conjugates (Figure 2d). Unmodified conjugates tend to remain as single particles or occasionally cross-linked to another nanoparticle (NP); the encapsulated conjugates are more likely to cross-link, therefore the hydrodynamic size is significantly bigger. This cross-linking in general took place among two to three NPs through the protein molecules, and the solubility & monodispersity of whole solution remains similar to the unmodified conjugates. Agrose gel analysis showed that the encapsulated conjugates moved slightly slower than the unmodified conjugates (Figure 2g).
Figure 2.
Characterization of the QD-luc8 conjugates by dynamic light scattering (a, b), TEM (c, d), spectroscopic analysis (e, f), and electrophoresis (g). Encapsulated QD-BRET conjugates (b, d) are larger than unmodified conjugates (a, c) due to polymeric encapsulation as well as cross-linking of the conjugates, and retain both bioluminescence (e, f) and fluorescence emissions. Dashed lines in e and f represent the spectra of unmodified QD-BRET probes, and solid lines in e and f represent fluorescence of encapsulated QD-BRET conjugates.
BRET effect before & after encapsulation
QD-BRET probes after polymeric encapsulation were tested for fluorescence, bioluminescence and BRET (Figure 2e & 2f). The BRET ratio remained similar (1.33 vs. 1.32) after modification. Biolumiescence signal remained strong although a little attenuated as compared with unmodified conjugates (~ 60%). The decrease in bioluminescence was mainly due to activity loss of Luc8 protein during the acryloylation step. This loss in activity can be controlled by adjusting the amount of acryloylating agent N-hydroxysuccinimide ester. For instance, increasing the molar ratio (acryloylating agent to protein) from low (4.6) to medium (9.3) and to high (93) will cause the remaining luciferase bioactivity to decrease by 20% (low) to 30% (medium) and 84% (high) of the unmodified protein, respectively. The following polymerization step did not cause any significant decline in the bioluminescence activity, regardless of the amount of monomer added (from 800 to 5000 per protein molecule).
In vitro stability improvement
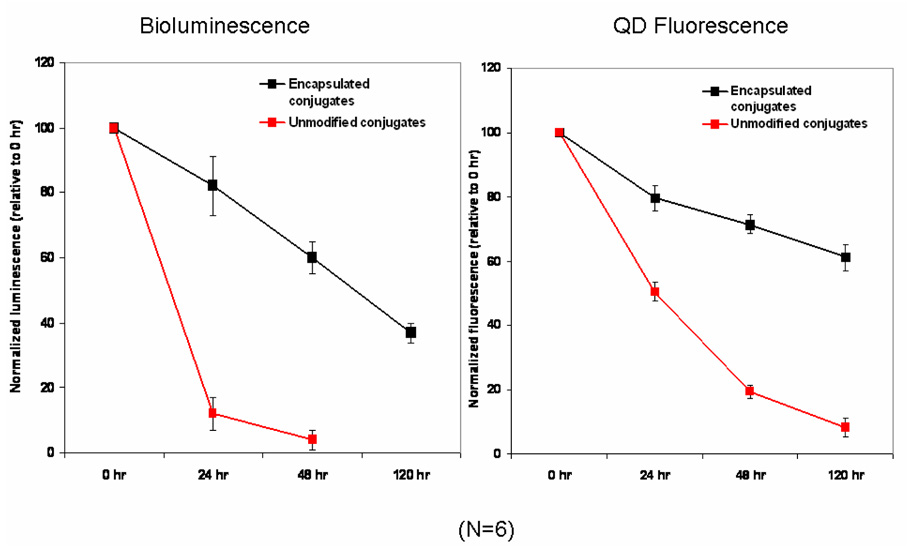
In vitro serum stability results showed that polymeric encapsulation significantly improved the stability of QD-Luc8 conjugates (Figure 3). Unmodified conjugates lost their bioluminescence by 90–95% within 48 hrs in mouse serum at 37 °C, while the encapsulated conjugates still had around 60% bioluminescence remaining during the same period of time. In fact, the encapsulated conjugates can stay in serum for up to 5 days, with still 37% bioluminescence remaining. Stored in phosphate buffer at 4 °C, the encapsulated conjugates can last for over 4 months with significant amount of activity (>30%) remaining. There is also a sharp decline (from 1.33 to 0.07) in BRET ratio for the unmodified conjugates, indicating disruption of the QD-Luc8 assembly in addition to the inactivation of the protein. In contrast, the encapsulated conjugates retained fairly good BRET ratio (0.73 after 40 hrs), suggesting the integrity of the conjugates were largely preserved. Lastly, QD stability (shown as the fluorescence excited by 480 nm) was also significantly improved by polymerization. It seems like the encapsulation not only protected the protein, but also helps to protect the whole assembly.
Figure 3.
In vitro serum stability comparison of unmodified (red line) versus encapsulated QD-BRET conjugates (black line). Left: bioluminescence change over time; Right: change of QD fluorescence intensity over time (0, 16, 24, 48, and 120 hrs). Both are an average of 6 replicates.
In vivo stability improvement
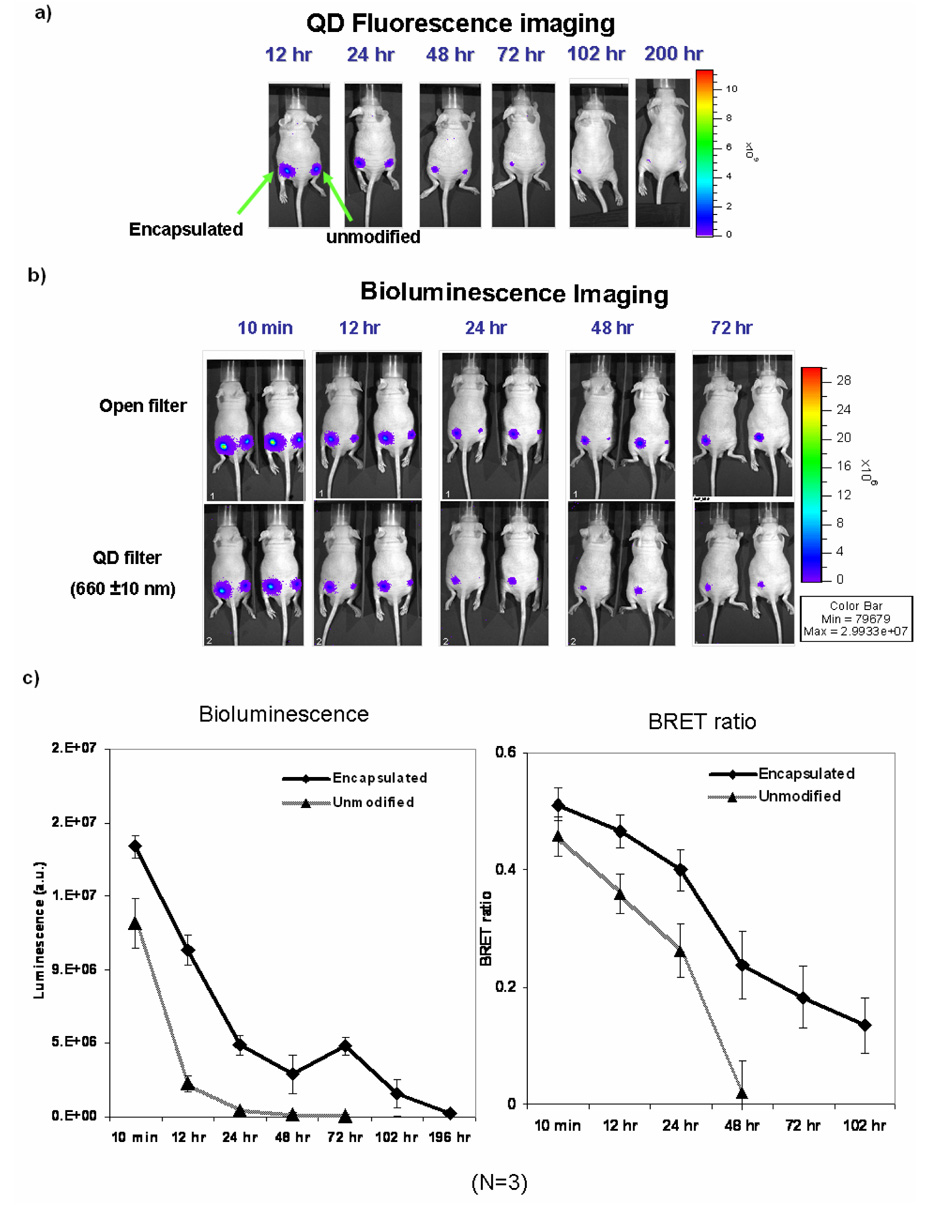
QD-Luc8 conjugates (encapsulated or unmodified, each at 5 pmole QD as determined by the QD fluorescence) were injected subcutaneously in nude mice and bioluminescence and fluorescence images were captured at 6, 12, 24, 48, 72 and 96 hr post injection (Figure 4). Results showed that total bioluminescence signals from the encapsulated conjugates lasted about 200 hrs after injection; and BRET signal remained detectable until 72 hrs. In contrast, bioluminescence signals from the unmodified conjugates became barely detectable at 48 hrs and BRET signal disappeared at 24 hrs post injection. The QD fluorescence inside living animal was also improved by polymeric encapsulation. Signals from the unmodified conjugates disappeared by 72 hrs; while signals from the encapsulated conjugates remained detectable after 200 hrs post injection.
Figure 4.
A comparison of in vivo stability of encapsulated vs. unmodified QD-BRET conjugates. The two types of conjugates were injected subcutaneously into the same animal (left: encapsulated; right: unmodified) and monitored for QD fluorescence (a) and bioluminescence (b). QD fluorescence images were taken under excitation with 465 nm, and bioluminescence and BRET images were taken immediately after injection of coelenterazine. Open filter collects light signals across the whole spectrum, and QD filter only collects light emitting between 650–660nm. Bottom graphs (c) show bioluminescence signals and BRET ratios changes over time; note that BRET ratios are smaller than in vitro measurement, because the BRET signals were only collected over 650–660 nm, instead of 600–750 nm as in vitro.
Discussion
Due to their super brightness and photostability, quantum dots have sparkled tremendous interests in the biological sciences community and have rapidly found its way into many applications including in vivo animal imaging [17,18]. Even with their superior optical properties of QDs, the same challenges remain for optical imaging at non-superficial tissue sites, which are the presence of strong background autofluorescence and photon absorbance/scattering by living tissue. One approach to solve these problems is to use QDs emitting in the near-infrared window. However, even in the near-infrared range [19], tissue autofluorescence always remains a major and often overlooked problem that reduces the signal-to-background ratio and, therefore, objects detectability. Self-illuminating quantum dots have great potential as an in vivo imaging and sensitive detection probe. Compared to conventional QDs, it eliminates the need for fluorescent excitation light and takes advantage of QDs blue-increasing extinction coefficient, greatly reduces the high background caused by surface illumination and tissue autofluorescence, efficiently couples chemical energy with light energy, and finally, because energy transfer is resonant, and nonradiative, absorption of excitation photons by hemoglobin and other tissue pigments is virtually eliminated [20]. The first publication on QD-BRET provided convincing evidence that QD-BRET conjugates significantly outperform conventional QDs for in vivo imaging [7]. Since then, the QD-BRET probes already found several applications including live cell imaging [21,22] and protease detection [23]. Despite all these attractive properties, broader & long time in vivo imaging applications have been thus far prohibited because of their relatively low stability inside living body. In this study, we seek to improve the stability of self-illuminating QDs through a polymeric encapsulation method. We adopted the procedure reported by Yan et al [15] for its simplicity (only 2 steps) and compatibility in aqueous media, since treatment with organic solvents such as that reported by Kim & Grate [14] may result in the disruption of the QD-protein assembly and inactivation of the BRET donor--Luc8 protein.
Surface modification of the protein, Luc8, resulted in activity loss proportional to the degree of acryloylation; likely due to the fact that the lysine (which contain the free amines to be modified by) residues are important for Luc8 function [8]. On the other hand, if there is too little acryloylation, polymerization wouldn’t happen and the stability improvement is minimal. Therefore, we have optimized the amount of acryloylating reagents to retain reasonable activity of the enzyme but still achieve the stability improvement. Our experiments have shown that the polymerization step did not cause significant additional activity loss in the conjugates, indicating the polymer coating did not present much barrier for the substrate transport (data not shown). Our studies also showed that the amount of monomer needed (monomer: protein ratio of 5000) for enhanced stability is much higher than that reported by Yan et al (monomer: protein ratio of 400 to 800). Below this level, we did not observe any significant stability improvements. One possible reason is that proteins anchored on the surface of a nanoparticle may behave very differently from free proteins in solution. For instance, free Luc8 protein remained active for a much longer time than those immobilized onto the QD surface. Additionally, our TEM data suggested cross-linking took place between the nanoparticles. Based on this, we expect cross-linking between protein molecules took place on the same nanoparticle as well and we believe this inter- and inner- nanoparticle cross-linking are responsible for the observed stability improvement.
There are at least two possible mechanisms that might account for improved stability of the polymerized conjugates. First, the polymer coating acts as a physical barrier to large biomolecules such as proteases, thus keeping them from attacking the Luc8 protein on the QD surface. The second reason for improved stability is likely due to flexibility change of the protein molecules on the QD surface. Inter- and inner-nanoparticle cross-linking by the acrylamide/bisacrylamide might have altered the flexibility of the protein and thus prevented them from conformational changes that may be detrimental to their activity.
Although the encapsulated conjugates outperformed the unmodified conjugates in both in vitro and in vivo (subcutaneous injection) studies, the advantages were not significant when both conjugates were systematically injected into the animals. Both types of conjugates were rapidly taken up by the reticuloendothelial system (RES) and degraded soon afterwards. RES uptake is a challenging issue with almost all kinds of nanostructures and it has been reported that surface chemistry of the nanostructure plays a critical role. Further engineering of the surface chemistry, for instance, attaching poly-ethleneglycol, may help alleviate this problem.
Acknowledgements
We thank Dr Samira Guccione the access to their DLS instrument and Dr Yi-shan Yang for technical assistance with the instrument. We also want to acknowledge support from the small animal imaging facility at MIPS. This work was supported by the Department of Defense Breast Cancer Research Program Concept Award and the National Cancer Institute Centers of Cancer Nanotechnology Excellence (1U54CA119367-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gao X, Yang L, Petros JA, Marshall FF, Simons JW, Nie S. In vivo molecular and cellular imaging with quantum dots. Curr. Opin. Biotechnol. 2005;16:63–72. doi: 10.1016/j.copbio.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Pinaud F, Michalet X, Bentolila LA, Tsay JM, Doose S, Li JJ, Iyer G, Weiss S. Advances in fluorescence imaging with quantum dot bio-probes. Biomaterials. 2006;27:1679-16. doi: 10.1016/j.biomaterials.2005.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alivisatos AP, Gu W, Larabell C. Quantum dots as cellular probes. Annu. Rev. Biomed. Eng. 2005;7:55–76. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- 4.Troy T, Jekic-McMullen D, Sambucetti L, Rice B. Quantitative comparison of the sensitivity of detection of fluorescent and bioluminescent reporters in animal models. Mol. Imaging. 2004;3:9–23. doi: 10.1162/15353500200403196. [DOI] [PubMed] [Google Scholar]
- 5.Tuchin V. Tissue optics. Bellingham, Washington: SPIE Press; 2000. [Google Scholar]
- 6.Kim S, Lim YT, Soltesz EG, De Grand AM, Lee J, Nakayama A, Parker JA, Mihaljevic T, Laurence RG, Dor DM, Cohn LH, Bawendi MG, Frangioni JV. Near-infrared fluorescent type II quantum dots for sentinel lymph node mapping. Nat Biotechnol. 2004;22:93–97. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.So MK, Xu C, Loening AM, Gambhir SS, Rao J. Self-illuminating quantum dot conjugates for in vivo imaging. Nat Biotechnol. 2006;24:339–343. doi: 10.1038/nbt1188. [DOI] [PubMed] [Google Scholar]
- 8.Loening AM, Fenn TD, Wu AM, Gambhir SS. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng. Des. Sel. 2006;19:391–400. doi: 10.1093/protein/gzl023. [DOI] [PubMed] [Google Scholar]
- 9.Arnold FH. Combinatorial and computational challenges for biocatalyst design. Nature. 2001;409:253–258. doi: 10.1038/35051731. [DOI] [PubMed] [Google Scholar]
- 10.Korkegian A, Black ME, Baker D, Stoddard BL. Computational thermostabilization of an enzyme. Science. 2005;308:857–860. doi: 10.1126/science.1107387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frenkel-Mullerad H, Avnir D. Sol-Gel materials as efficient enzyme protectors: preserving the activity of phosphatases under extreme pH conditions. J. Am. Chem. Soc. 2005;127:8077–8081. doi: 10.1021/ja0507719. [DOI] [PubMed] [Google Scholar]
- 12.Dziubla TD, Shuvaev VV, Hong NK, Hawkins BJ, Madesh M, Takano H, Simone E, Nakada MT, Fisher A, Albelda SM, Muzykantov VR. Endothelial targeting of semi-permeable polymer nanocarriers for enzyme therapies. Biomaterials. 2005;29:215–227. doi: 10.1016/j.biomaterials.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupeyrón D, González M, Sáez V, Ramón J, Rieumont J. Nano-encapsulation of protein using an enteric polymer as carrier. IEE Proc. Nanobiotechnol. 2005;152:165–168. doi: 10.1049/ip-nbt:20050005. [DOI] [PubMed] [Google Scholar]
- 14.Kim J J, Grate JW. Single-Enzyme nanoparticles armored by a nanometer-scale organic/inorganic network. Nano Lett. 2003;3:1219–1222. [Google Scholar]
- 15.Yan M, Ge J, Lu Z, Ouyang P. Encapsulation of single enzyme in nanogel with enhanced biocatalytic activity and stability. J. Am. Chem. Soc. 2008;128:11008–11009. doi: 10.1021/ja064126t. [DOI] [PubMed] [Google Scholar]
- 16.Yan M, Liu ZX, Lu DN, Liu Z. Fabrication of single carbonic anhydrase nanogel against denaturation and aggregation at high temperature. Biomacomolecules. 2007;8:560–565. doi: 10.1021/bm060746a. [DOI] [PubMed] [Google Scholar]
- 17.Gao X, Cui Y, Levenson RM, Chung LW, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat. Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 18.Cai W, Shin DW, Chen K, Gheysens O, Cao Q, Wang SX, Gambhir SS, Chen X. Peptide-labeled near-infrared quantum dots for imaging tumor vasculature in living subjects. Nano Lett. 2006;6:669–676. doi: 10.1021/nl052405t. [DOI] [PubMed] [Google Scholar]
- 19.De Grand AM, Lomnes SJ, Lee DS, Pietrzykowski M, Ohnishi S, Morgan TG, Gogbashian A, Laurence RG, Frangioni JV. Tissue-like phantoms for near-infrared fluorescence imaging system assessment and the training of surgeons. J. Biomed. Opt. 2006;11:014007. doi: 10.1117/1.2170579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frangioni JV. Self-illuminating quantum dots light the way. Nat. Biotechnol. 2006;24:326–328. doi: 10.1038/nbt0306-326. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, So MK, Loening AM, Yao H, Gambhir SS, Rao J. HaloTag protein-mediated site-specific conjugation of bioluminescent proteins to quantum dots. Angew. Chem. Int. Ed. Engl. 2006;45:4936–4940. doi: 10.1002/anie.200601197. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, So MK, Rao J. Protease-modulated cellular uptake of quantum dots. Nano Lett. 2006;6:1988–1992. doi: 10.1021/nl0611586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao H, Zhang Y, Xiao F, Xia Z, Rao J. Quantum dot/bioluminescence resonance energy transfer based highly sensitive detection of proteases. Angew. Chem. Int. Ed. Engl. 2007;46:4346–4349. doi: 10.1002/anie.200700280. [DOI] [PubMed] [Google Scholar]