Abstract
Cell surface carbohydrates play important roles in a wide variety of biological processes including cell adhesion, fertilization, differentiation, development, and tumor cell metastasis. Lectins are proteins of non-immune origin which recognize and bind to specific carbohydrate structural epitopes. We have recently described the development and use of lectin arrays as tools for the elucidation of the carbohydrate structures expressed on cell surfaces. In the present work this technology is employed for the characterization of differences in carbohydrate expression patterns on normal and tumorigenic human breast cell lines, as well as on sublines differing in their tendency to “home” to different tissues during metastasis. Significant differences were observed, including changes that correlate with metastatic potential as well as with tissue-specific homing of metastatic cells.
Introduction
The mammalian cell surface is decorated with a dense layer of carbohydrates, which are attached to membrane glycoproteins and glycolipids.1 These cell surface carbohydrates are cell-type specific and participate in a wide variety of biological processes including cell adhesion, fertilization, differentiation, development, and tumor-cell metastasis.1–5 Abnormal glycosylation has been associated with many diseases, especially cancers, such as breast cancer, prostate cancer, lung cancer, and colon cancer.6–10 Moreover, tumor-associated alterations of cell surface glycosylation play a crucial role in metastasis of carcinoma cells by altering tumor cell adhesion or motility in a manner that either promotes or inhibits invasion and metastasis.11, 12 The ability to characterize cell surface carbohydrate expression patterns is thus critical both to understanding their role in disease development as well as to provide diagnostic tools to help guide treatment.
Specific carbohydrate-binding ligands such as monoclonal antibodies and lectins have proven to be invaluable tools for the analysis of complex glycoconjugates. The monoclonal antibodies are generally directed towards terminal carbohydrate structures, which limits their utility to the analysis of these terminal components.13 In contrast, lectins, which are proteins of non-immune origin that recognize and bind to specific carbohydrate structural epitopes,14 can detect changes in the carbohydrate core structures.13, 15 Both antibodies and lectins can be tagged with a fluorophore or enzyme and used to stain tissue sections to provide information on the distribution of carbohydrate structures within the sample.16 Although such histochemical approaches are very powerful, they do have some shortcomings. The paraffin-embedding methods commonly used for tissue fixation can render carbohydrates on glycoproteins inaccessible due to protein denaturation, and glycolipids can be lost during the process.16 As a result, methods which work on unfixed, unprocessed material without carbohydrate loss may be advantageous. In addition, the subjective visual analysis of such stained samples is also subject to considerable error, particularly for complex samples with heterogeneous staining patterns,6 and the overall process is fairly lengthy and expensive due to multiple steps and expensive reagents.
We have previously presented an approach to profiling cell surface carbohydrate expression by simple microscopic observation of cell binding.17, 18 Briefly, several different lectins are immobilized on gold substrates in an array format, followed by binding of a cell suspension to the surface. The cells show distinct binding patterns to the lectin array due to differences in their cell surface carbohydrate expression patterns. Both glass and gold substrates are amenable to fabrication of lectin arrays with comparable complexity.17, 19–22 However, gold is more flexible in terms of modes of analysis of cell binding patterns. Both surface plasmon resonance imaging (SPR),23 a label free method of analysis, and MALDI mass spectrometry24, 25 can be performed on gold substrates whereas neither is compatible with glass substrates.
In the present work we extend this approach to evaluate differences in carbohydrate expression on normal and tumorigenic human breast cell lines, as well as on sublines differing in their tendency to “home” to different tissues during metastasis. A quantitative and objective measure of the degree of cell-binding is provided by measuring the fractional surface area coverage of cells within each array feature.
Experimental section
Materials
Lectins used in this study include Concanavalin A (ConA), Sambucus Nigra Lectin (SNA), and Wheat Germ Agglutinin (WGA) from Vector Laboratories (Covington LA); Phaseolus Vulgaris Leucoagglutinin (PHA-L), Maackia Amurensis Agglutinin (MAA), and Soybean Agglutinin (SBA) from Sigma (St. Louis MO), and Helix Pomatia Agglutinin (HPA) from MP Biomedicals (Solon OH). The carbohydrate specificities of these lectins are shown in Table 1. Gold-coated glass substrates were from GenTel BioSciences, Inc. (Madison WI). The thickness of the gold thin films was 30nm in order for the substrates to be sufficiently transparent for cell-binding observations using an inverted optical microscope. 10% Bovine Serum Albumin (BSA) in PBS and Dithiobis[succinimidylpropionate] (DSP) were from Pierce (Rockford IL). Phosphate buffer solution (PBS) pH 7.2, Dulbecco’s phosphate buffer solution (DPBS), fetal bovine serum (FBS), Dulbecco’s modified Eagle’s medium (DMEM), DMEM-F12 Ham medium (DMEM-F12), penicillin/streptomycin, TrypLE™ Express Stable Trypsin Replacement Enzyme were from Invitrogen (Carlsbad CA). Hydrocortisone, insulin, epidermal growth factor (EGF), and 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) were from Sigma (St. Louis MO). MDA-MB-231 and MCF-10A cell lines were from American Type Culture Collection (ATCC) (Manassas VA). 1833, 2287, 4142 and 4175 sublines derived from the MDA-MB-231 parental cell line were a generous gift from Dr. Joan Massagué (Memorial Sloan-Kettering Cancer Center, NY).
Table 1.
Carbohydrate specificities of lectins used in this paper. More detailed specificity information can be found in reference 15.
| Lectin | Binding Specificity | |
|---|---|---|
| α Man | Concanavalin A (ConA) | Terminal α Man, Man α3[Man α6] Man |
| Phaseolus Vulgaris Leucoagglutinin (PHA-L) | β1,6-branched tri and tetra antennary oligosaccharides | |
| GalNAc | Helix Pomatia Agglutinin (HPA) | α-GalNAc > α-GlcNAc~ β-GalNAc> β-GlcNAc33 |
| Soybean Agglutinin (SBA) | α,β-GalNAc33 | |
| Neu5Ac | Maackia Amurensis Agglutinin (MAA) | Neu5Ac α(2,3) Gal |
| Sambucus Nigra Lectin (SNA) | Neu5Ac α(2,6) Gal | |
| βGlcNAc | Wheat Germ Agglutinin (WGA) | (GlcNAc β4GlcNAc) Neu5Ac α(Gal β4GlcNAc) |
GlcNAc: N-acetylglucosamine; GalNAc: N-acetylgalactosamine; NeuAc: N-acetylneuraminic acid (a member of the sialic acid family); Gal: Galactose; Man: Mannose.
Cell Culture
MCF-10A cells were cultured in DMEM-F12 supplemented with 15mM HEPES, 10% (v/v) FBS, 10µg/mL insulin, 0.5 µg/mL hydrocortisone, and 0.02 µg/mL EGF. MDA-MB-231 cells and the four sublines were cultured in DMEM with 10% (v/v) FBS. All media were supplemented with 60U/mL penicillin and 60 µg/mL streptomycin. All cells were grown in standard 60mm×15mm Petri dishes (Fisher Scientific, Hampton NH) at 37°C in a humidified incubator containing 5% CO2.
Array Fabrication
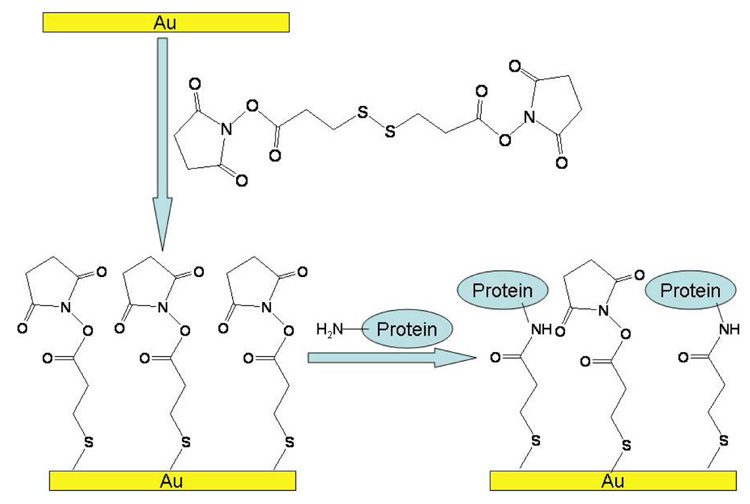
Clean gold substrates were rinsed thoroughly with water and then ethanol and dried gently with nitrogen. The substrates were soaked in saturated DSP/ethanol overnight. After rinsing the surface extensively with ethanol, lectins dissolved in HEPES buffer (10mM HEPES, 1mM CaCl2, pH 8.5) were spotted in triplicate onto the NHS-terminated surface. The volume of each spot was 0.5µL. After 3 hours incubation in a humid chamber, the substrates were soaked in 1% BSA/PBS (diluted from 10% BSA/PBS) to block non-specific binding that may be introduced in the following steps. After 30 minutes incubation, the substrates with covalently immobilized proteins were washed with PBS and ready for use (Figure 1).
Figure 1.
Reaction of primary amines with N-hydroxy succinimidyl (NHS) ester on gold surfaces. The gold surface is modified with self-assembled NHS-ester monolayer by incubating in a DSP/Ethanol solution. The NHS ester reacts with the free amines of the lysine residues to form a stable amide bond.
Cell binding with lectin arrays
Cells were trypsinized, washed twice with PBS pH 7.2 and resuspended in DPBS containing 1.0mM Ca2+ and 0.5mM Mg2+ at a concentration of ~2×106 cells/mL. Cells were then incubated with gold substrates on ice (0°C) for 30 minutes. After incubation the substrates were washed three times with DPBS to wash off the non-specifically bound cells and imaged with an Olympus IX81 phase contrast microscope (Japan).
Cell binding with different lectin deposition concentrations
Lectins were dissolved in HEPES buffer at different concentrations (from 16nM to 10µM) and then deposited on the NHS-terminated gold substrates. Cell suspensions of ~2×106 cells/mL were incubated with lectin-modified substrates as described above. After washing, substrates were imaged with the microscope.
Cell counting
The densities of the bound cells on lectin spots were counted using the publicly available NIH ImageJ processing software (http://rsb.info.nih.gov/ij/). Briefly, each figure was first converted into a binary image by thresholding. The surface density of cells was calculated as the areas covered by cells divided by the total area of each spot and expressed in percentage of surface area coverage. Surface area coverage was employed as a quantitative measure rather than simply counting cells per unit area in order to avoid biases due to size differences between cells. This analysis provides more quantitative results than are obtained by classical histochemical approaches.
Results and Discussion
Lectin arrays consisting of seven lectins were used to evaluate the cell surface carbohydrate expression of two epithelial cell lines derived from human breast with different invasive potentials: MCF-10A, a non-tumorigenic epithelial breast cell line; and MDA-MB-231, an adenocarcinoma breast cell line with high invasiveness. Four sublines (1833, 2287, 4142, and 4175) derived from parental MDA-MB-231 cell lines and differing in their metastatic behaviors were also tested. The first two sublines (1833 and 2287) metastasize to bone26 and the last two (4142 and 4175) metastasize to lung.27 Cell suspensions of these cell lines at the same concentration (~2×106 cells/mL) were incubated with the lectin arrays and after washing the bound cells were observed by phase contrast microscopy.
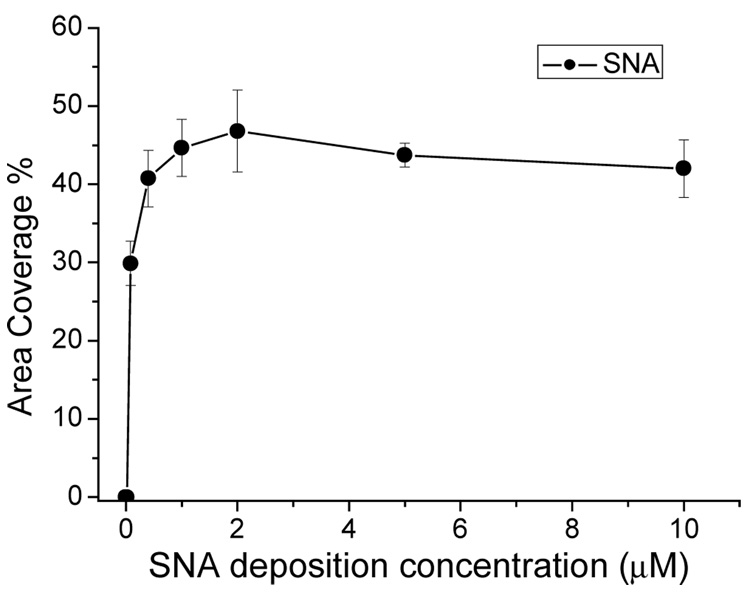
As for any protein array, an important parameter of the lectin arrays is the lectin surface density (molecules of lectin per unit area). At low densities fewer binding sites are present on the surface; whereas at higher densities there is potential for steric interference between the lectins, which could interfere with cell binding. The surface density of the lectins can be modulated by varying the concentration of the lectins in the solution spotted onto the surface (referred to here as the “lectin deposition concentration”). Cell-binding was measured as a function of lectin deposition concentration for each lectin (see Figure 2 for an illustrative example), and in each case the “optimum” lectin deposition concentration was taken as the lowest concentration which yields saturated cell-binding densities (Table 2). These concentrations were employed for all the cell-binding experiments shown.
Figure 2.
Binding of MCF-10A cells to SNA as a function of SNA deposition concentration. SNA solutions at different concentrations were spotted in triplicate onto NHS-modified gold substrates. The substrates were then incubated in a suspension of MCF-10A cells. Cell binding densities increased with SNA concentration and became saturated at 0.4µM. No significant density change was observed after 0.4µM.
Table 2.
Optimized deposition concentration of lectins
| Lectins | ConA | PHA-L | HPA | SBA | MAA | SNA | WGA |
|---|---|---|---|---|---|---|---|
| Optimized deposition concentration (µM) | 0.4 | 0.08 | 10 | 0.4 | 0.4 | 0.4 | 1.0 |
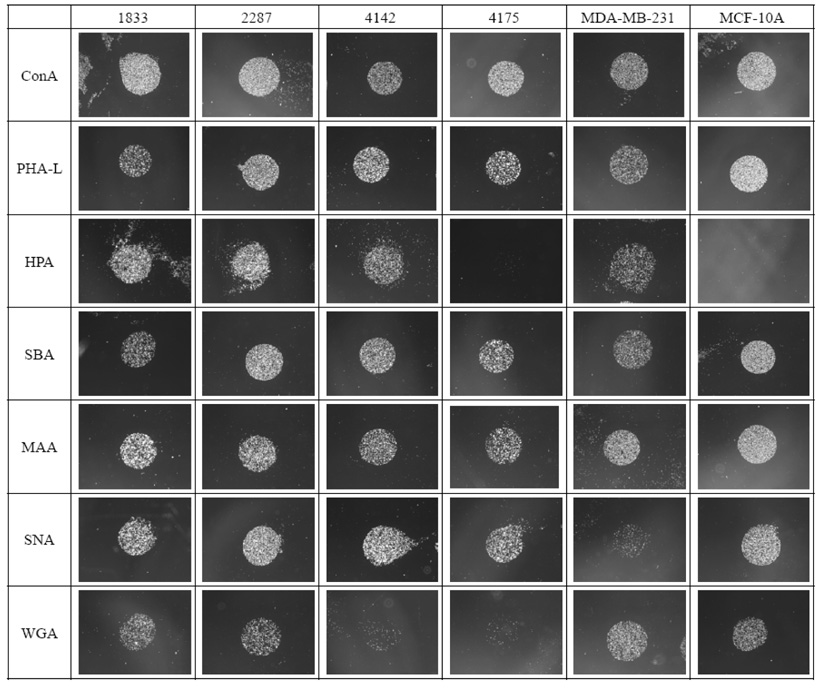
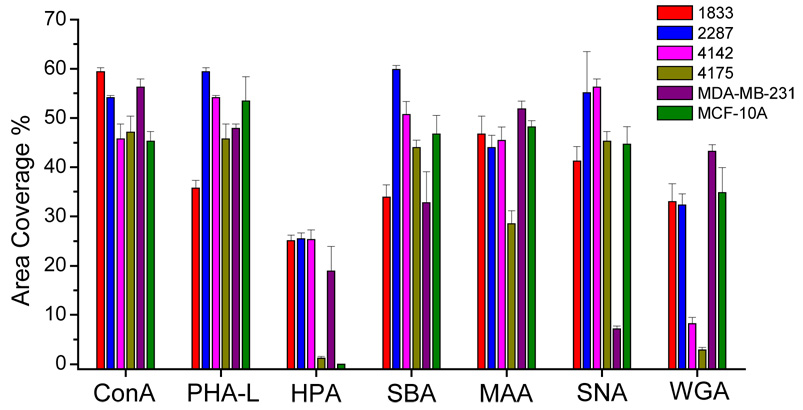
The results of cell-binding assays for each of the six cell lines on the panel of 7 lectins are shown as visual examples in Figure 3, and in graphical form in Figure 4. The lectins ConA and PHA-L bound strongly to all six cell lines. This result was not surprising for ConA, as it is known to bind with high affinity to the core trimannoside moiety, 3,6-di-O-(α-D-mannopyranosyl)- α-D-mannopyranoside, which is present in all asparagine-linked carbohydrates.28 PHA-L binds to β1,6-branched tri- and tetra-antennary oligosaccharides, which are widely observed on tumor cells29 and have been reported to be required for efficient metastatic spread of cancer cells.30–32 Thus it was not surprising to see PHA-L binding for the tumorigenic cell line MDA-MB-231 as well as for the four MDA-MB-231 metastatic sublines, although the observed binding to the non-tumorigenic MCF-10A cell line was less predictable a priori.
Figure 3.
Binding patterns of cell lines to the lectin arrays. MCF-10A, a non-tumorigenic epithelial breast cell line; MDA-MB-231, an adenocarcinoma breast cell line with high invasive potential; 1833 and 2287, cell lines derived from parental MDA-MB-231 with metastatic potential to bone; 4142 and 4175, cell lines derived from parental MDA-MB-231 with metastatic potential to lung. Each lectin was spotted in triplicate onto gold substrates. One representative spot for each lectin and cell line was selected for display in this figure.
Figure 4.
Binding densities represented in area coverage of different cell lines on lectin arrays.
Although both HPA and SBA bind terminal N-acetyl-D-galactosamine (GalNAc), HPA binds α-GalNAc preferentially over β-GalNAc, while SBA binds both with equal affinity.33 In the binding experiments both MCF-10A and MDA-MB-231 bound equally strongly to SBA, whereas only MDA-MB-231 bound to HPA. This result shows that although GalNAc is expressed on both cell lines, α-GalNAc is not expressed on the MCF-10A cell surface, consistent with previous reports that HPA binding is an independent prognostic factor of adenocarcinoma (as it is expressed on the tumorigenic cell line MDA-MB-231 but not on the non-tumorigenic cell line MCF-10A).34, 35 HPA binding has also been reported to be associated with metastasis.36, 37 The HPA-binding results obtained for three of the four metastatic MDA-MB-231 sublines are consistent with this observation, with the 4175 subline as an exception.
Sialic acid expressed on cell surfaces plays an important role in many cellular processes including immune recognition, binding of pathogens to host cells, cell adhesion, and apoptosis.38 A number of human diseases, including cancer, are correlated to aberrant expression of sialic acid.39 For example, colon cancer tissues display enhanced expression of α2,6 sialoglycans compared with normal colon tissue;40 the level of α2,6 sialoglycan expression in colorectal tumor cells has been associated with metastatic capacity;41 and enhanced α2,6 sialoglycan and decreased α2,3 sialoglycan expression has been found in cervical carcinoma.42 In the present work the two lectins MAA and SNA, which are respectively specific for α2,3-linked and α2,6-linked sialic acids, were employed for the evaluation of sialic acid expression. All six of the cell lines examined exhibited binding to MAA, consistent with the fact that α2,3-linked sialic acid is the most abundant sialic acid linkage found among mammalian cell surface oligosaccharides.43 In contrast, SNA binding was greatly increased in the four metastatic sublines compared to the MDA-MB-231 parental cell line, suggesting a possible important role for the expression of α2,6-linked sialic acid in metastasis. Further studies, such as gene knock-out experiments on the α2,6 sialytransferase (ST6Gal) in the metastatic cell lines, will be required to confirm or refute this hypothesis.
Finally, the binding pattern of the lectin WGA revealed further interesting differences between the cell lines. WGA is specific for N-acetylglucosamine oligomers (G1cNAc), although due to configurational similarities between sialic acid and G1cNAc, it can also bind with lower affinity to sialic acid.44 Since the binding results from MAA and SNA, which also bind to sialic acid, showed no significant difference of sialic acid expression among the four metastatic sublines, any differences observed with WGA may be attributed to differences in GlcNAc expression. The results shown in Figure 3 and Figure 4 clearly indicate little or no binding of the lung-metastatic cell lines 4142 and 4175 to WGA, in marked contrast to the binding evident in the bone-metastatic cell lines and the MDA-MB-231 parental cell line. This indicates that whereas GlcNAc is expressed at high levels in the bone-metastatic lines, it is not significantly expressed in the lung-metastatic lines. In previous studies of global gene expression patterns in these and other cell lines,26, 27 the bone-metastatic cell lines were found to be substantially more similar to the parental cell line than were the lung-metastatic cell lines. Interestingly, a similar pattern is evident in the results of the present study, with both the parental and bone-metastatic cell lines showing common high GlcNAc expression levels, while the lung-metastatic cell lines show little or no GlcNAc expression.
The observed differences in WGA-binding may also help to explain conflicting results obtained in previous studies, some of which indicated no significant differences in glycoforms between cell lines differing in metastatic potential,45–47 while others did show differences.48–50 Perhaps differences in carbohydrate expression patterns on metastatic cells are correlated less with general changes in metastatic potential than with tissue-specific homing behavior. As before, further study will be required to investigate this hypothesis.
Conclusion
We present here a study using lectin arrays to evaluate carbohydrate expression patterns on several human breast cell lines. Significant differences were observed in the carbohydrate expression patterns of the different cell types, including changes that correlate with metastatic potential as well as tissue-specific homing. These differences in carbohydrate expression may serve as potential indicators of cancer development and metastasis, and if the differences are shown to be functionally important, could provide useful diagnostic and therapeutic routes to the detection and treatment of cancer metastasis. Although in this study only a small set of several lectins were employed as surface-bound carbohydrate-specific ligands, the approach could readily be extended to include antibody ligands and expanded to a larger panel of lectins, thereby providing additional information on the nature of the cell-surface carbohydrate motifs. The rapid and parallel nature of these measurements makes possible the analysis of many different cell types for binding to a wide variety of ligands, potentially providing a powerful tool for the study of changes occurring during the process of cancer disease progression and other biological processes.
Acknowledgement
We thank Dr. Joan Massagué for providing the metastatic cell lines. This work was supported by NIH grant R01EB000269 and NIH/NHLBI proteomics contract N01-HV-28182.
References
- 1.Fukuda M, Hingsgaul O. Molecular and Cellular Glycobiology. Oxford: Oxford University Press; 2000. [Google Scholar]
- 2.Taylor ME, Drickamer K. Introduction to Glycobiology. Oxford: Oxford University Press; 2003. [Google Scholar]
- 3.Crocker PR, Feizi T. Curr. Opin. Struct. Biol. 1996;6:679–691. doi: 10.1016/s0959-440x(96)80036-4. [DOI] [PubMed] [Google Scholar]
- 4.Feizi T. Immunol. Rev. 2000;173:79–88. doi: 10.1034/j.1600-065x.2000.917310.x. [DOI] [PubMed] [Google Scholar]
- 5.Bertozzi CR, Kiessling LL. Science. 2001;291:2357–2364. doi: 10.1126/science.1059820. [DOI] [PubMed] [Google Scholar]
- 6.Dansey R, Murray J, Ninin D, Bezwoda WR. Oncology. 1988;45:300–302. doi: 10.1159/000226627. [DOI] [PubMed] [Google Scholar]
- 7.Redondo PDG, Nakamura CV, de Souza W, Morgado-Diaz JA. J. Histochem. Cytochem. 2004;52:629–640. doi: 10.1177/002215540405200507. [DOI] [PubMed] [Google Scholar]
- 8.Iwakawa K, Ueda N, Murao S, Kobayashi N. J. Gastroenterol. 1996;31:24–32. doi: 10.1007/BF01211183. [DOI] [PubMed] [Google Scholar]
- 9.Kakari S, Stringou E, Toumbis M, Ferderigos AS, Poulaki E, Chondros K, Dema A, Kotsovoulou V, Pavlidis N. Anticancer Res. 1991;11:2107–2110. [PubMed] [Google Scholar]
- 10.Remani P, Nair RA, Sreelekha TT, Madhavan J, Vijayakumar T, Nair MK. J. Exp. Clin. Cancer Res. 2000;19:519–523. [PubMed] [Google Scholar]
- 11.Lin S, Kemmner W, Grigull S, Schlag PM. Exp. Cell Res. 2002;276:101–110. doi: 10.1006/excr.2002.5521. [DOI] [PubMed] [Google Scholar]
- 12.Hakomori S. Cancer Res. 1996;56:5309–5318. [PubMed] [Google Scholar]
- 13.Dabelsteen E. J. Pathol. 1996;179:358–369. doi: 10.1002/(SICI)1096-9896(199608)179:4<358::AID-PATH564>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 14.Nilsson CL. Anal. Chem. 2003;75:348A–353A. doi: 10.1021/ac031373w. [DOI] [PubMed] [Google Scholar]
- 15.Wearne KA, Winter HC, O'Shea K, Goldstein IJ. Glycobiology. 2006;16:981–990. doi: 10.1093/glycob/cwl019. [DOI] [PubMed] [Google Scholar]
- 16.Leathem A, Atkins N. J. Clin. Pathol. 1983;36:747–750. doi: 10.1136/jcp.36.7.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng T, Peelen D, Smith LM. J. Am. Chem. Soc. 2005;127:9982–9983. doi: 10.1021/ja0505550. [DOI] [PubMed] [Google Scholar]
- 18.Peelen D, Kodoyianni V, Lee J, Zheng T, Shortreed MR, Smith LM. Journal of Proteome Research. 2006;5:1580–1585. doi: 10.1021/pr050467e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pilobello KT, Krishnamoorthy L, Slawek D, Mahal LK. ChemBioChem. 2005;6:985–989. doi: 10.1002/cbic.200400403. [DOI] [PubMed] [Google Scholar]
- 20.Kuno A, Uchiyama N, Koseki-Kuno S, Ebe Y, Takashima S, Yamada M, Hirabayashi J. Nat. Methods. 2005;2:851–856. doi: 10.1038/nmeth803. [DOI] [PubMed] [Google Scholar]
- 21.Hsu KL, Pilobello KT, Mahal LK. Nat. Chem. Biol. 2006;2:153–157. doi: 10.1038/nchembio767. [DOI] [PubMed] [Google Scholar]
- 22.Patwa TH, Zhao J, Anderson MA, Simeone DM, Lubman DM. Anal. Chem. 2006;78:6411–6421. doi: 10.1021/ac060726z. [DOI] [PubMed] [Google Scholar]
- 23.Brockman JM, Nelson BP, Corn RM. Annu. Rev. Phys. Chem. 2000;51:41–63. doi: 10.1146/annurev.physchem.51.1.41. [DOI] [PubMed] [Google Scholar]
- 24.Brockman AH, Orlando R. Anal. Chem. 1995;67:4581–4585. doi: 10.1021/ac00120a024. [DOI] [PubMed] [Google Scholar]
- 25.Bundy JL, Fenselau C. Anal. Chem. 2001;73:751–757. doi: 10.1021/ac0011639. [DOI] [PubMed] [Google Scholar]
- 26.Kang YB, Siegel PM, Shu WP, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 27.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu WP, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta D, Oscarson S, Raju TS, Stanley P, Toone EJ, Brewer CF. Eur. J. Biochem. 1996;242:320–326. doi: 10.1111/j.1432-1033.1996.0320r.x. [DOI] [PubMed] [Google Scholar]
- 29.Fernandes B, Sagman U, Auger M, Demetrio M, Dennis JW. Cancer Res. 1991;51:718–723. [PubMed] [Google Scholar]
- 30.Dennis JW, Laferte S, Waghorne C, Breitman ML, Kerbel RS. Science. 1987;236:582–585. doi: 10.1126/science.2953071. [DOI] [PubMed] [Google Scholar]
- 31.Dennis JW. Cancer Surv. 1988;7:573–595. [PubMed] [Google Scholar]
- 32.Dennis JW, Laferte S, Vanderelst I. Biochem. Soc. Trans. 1989;17:29–31. doi: 10.1042/bst0170029. [DOI] [PubMed] [Google Scholar]
- 33.Hammarstrom S, Murphy LA, Goldstein IJ, Etzler ME. Biochemistry (Mosc) 1977;16:2750–2755. doi: 10.1021/bi00631a025. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell BS, Schumacher U. Histol. Histopathol. 1999;14:217–226. doi: 10.14670/HH-14.217. [DOI] [PubMed] [Google Scholar]
- 35.Laack E, Nikbakht H, Peters A, Kugler C, Jasiewicz Y, Edler L, Hossfeld DK, Schumacher U. Am. J. Pathol. 2002;160:1001–1008. doi: 10.1016/S0002-9440(10)64921-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fenlon S, Ellis IO, Bell J, Todd JH, Elston CW, Blamey RW. J. Pathol. 1987;152:169–176. doi: 10.1002/path.1711520305. [DOI] [PubMed] [Google Scholar]
- 37.Thies A, Moll I, Berger J, Schumacher U. Br. J. Cancer. 2001;84:819–823. doi: 10.1054/bjoc.2000.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao Y, Merling A, Crocker PR, Keller R, Schwartz-Albiez R. Lab. Invest. 2002;82:1515–1524. doi: 10.1097/01.lab.0000038503.34655.98. [DOI] [PubMed] [Google Scholar]
- 39.Vierbuchen MJ, Fruechtnicht W, Brackrock S, Krause KT, Zienkiewicz TJ. Cancer. 1995;76:727–735. doi: 10.1002/1097-0142(19950901)76:5<727::aid-cncr2820760504>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 40.Dall'Olio F, Chiricolo M, Ceccarelli C, Minni F, Marrano D, Santini D. Int. J. Cancer. 2000;88:58–65. doi: 10.1002/1097-0215(20001001)88:1<58::aid-ijc9>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 41.Harvey BE, Toth CA, Wagner HE, Steele GD, Thomas P. Cancer Res. 1992;52:1775–1779. [PubMed] [Google Scholar]
- 42.Wang P-H, Feng Li Y, Juang C-M, Lee Y-R, Chao H-T, Tsai Y-C, Yuan C-C. Gynecol. Oncol. 2001;83:121–127. doi: 10.1006/gyno.2001.6358. [DOI] [PubMed] [Google Scholar]
- 43.Hennet T, Chui D, Paulson JC, Marth JD. Proc. Natl. Acad. Sci. U. S. A. 1998;95:4504–4509. doi: 10.1073/pnas.95.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monsigny M, Roche AC, Sene C, Magetdana R, Delmotte F. Eur. J. Biochem. 1980;104:147–153. doi: 10.1111/j.1432-1033.1980.tb04410.x. [DOI] [PubMed] [Google Scholar]
- 45.Hagmar B, Erkell LJ, Burns G, Ryd W. Invasion Metastasis. 1990;10:317–328. [PubMed] [Google Scholar]
- 46.Rye PD, Dearing SJ, Walker RA. Cancer J. 1993;6:190–195. [Google Scholar]
- 47.Pacis RA, Pilat MJ, Yamazaki K, Pienta KJ. Int. J. Oncol. 1995;7:1349–1354. doi: 10.3892/ijo.7.6.1349. [DOI] [PubMed] [Google Scholar]
- 48.Penno MB, Demaio A. Exp. Gerontol. 1992;27:493–501. doi: 10.1016/0531-5565(92)90004-j. [DOI] [PubMed] [Google Scholar]
- 49.Zebda N, Bailly M, Brown S, Dore J, Berthiervergnes O. J. Cell. Biochem. 1994;54:161–173. doi: 10.1002/jcb.240540205. [DOI] [PubMed] [Google Scholar]
- 50.Kjonniksen I, Rye PD, Fodstad O. Br. J. Cancer. 1994;69:1021–1024. doi: 10.1038/bjc.1994.200. [DOI] [PMC free article] [PubMed] [Google Scholar]