Abstract
Hepatitis C virus (HCV) is a major cause of chronic liver disease including steatosis, cirrhosis and hepatocellular carcinoma. The development of transgenic mice expressing HCV proteins and the successful repopulation of SCID/Alb-uPA mice with human hepatocytes provides an important tool for unraveling virus–host interactions in vivo. Several of these mouse models exhibit aspects of HCV-related liver disease. Thus, these in vivo models play an important role to further understand the pathogenesis of HCV infection and to evaluate the pre-clinical safety and efficacy of new antiviral compounds against HCV. This review summarizes the most important mouse models currently used to study HCV pathogenesis and infection. Finally, the perspective of these models for future HCV research as well as the design of novel small animal models is discussed.
Keywords: Viral infection, Transgenic mice, Antivirals
1. Introduction
More than 170 million individuals worldwide are currently infected with the hepatitis C virus (HCV). Chronic HCV infection frequently results in serious liver disease, including steatosis, cirrhosis and hepatocellular carcinoma [1]. In the United States, hepatitis C is a leading cause for orthotopic liver transplantation. Unfortunately, liver transplantation is not a cure for hepatitis C. Viral recurrence is an invariable problem and a leading cause of graft loss [2]. A vaccine protecting against HCV infection is not available, and current antiviral therapies are characterized by limited efficacy, high cost, and substantial side effects [3].
HCV is a positive strand RNA virus classified in the genus Hepacivirus of the Flaviviridae. Translation of the major open reading frame of the HCV genome results in the production of an approximately 3000 amino acid long polyprotein, which is cleaved co- and post-translationally by the coordinated action of cellular and two viral proteases into its functional subunits Core (C), envelopes 1 and 2 (E1 and E2), p7 and non-structural proteins (NS) 2, NS3, NS4A, NS4B, NS5A and NS5B [4]. HCV replication takes place in the cytoplasm of the host cell, which is primarily the hepatocyte. Until recently, due to the lack of a cell culture system, HCV could not be efficiently propagated in cultured cells to support molecular studies of the virus–host interaction [5]. Robust production of infectious HCV in cell culture has finally been achieved using a unique HCV genome derived from the blood of a Japanese patient with fulminant hepatitis C (JFH-1) [6], [7], [8]. Moreover, virus particles generated from the JFH-1 clone turned out to be infectious in vivo both in chimpanzees and in mice containing human liver xenografts [6], [9]. By introducing multiple adaptive mutations or using DNA expression construct, infectious HCV genotype 1 could also be produced in cell culture [10], [11].
Over the past two decades the chimpanzee (Pan troglodyte) has been commonly used to study mechanisms of acute and chronic HCV infection. These studies have greatly contributed to our current understanding of HCV infection, including immunity and pathogenesis (for review see [12]). However, the chimpanzee model has some important limitations. HCV-infected chimpanzees rarely develop chronic liver disease to the extent seen in HCV-infected humans [13], making the chimpanzee a less than ideal model for studying the mechanisms of HCV pathogenesis. Moreover, chimpanzees are expensive and difficult to handle, and must be housed and cared for in appropriate non-human primate research facilities. Since 1988 the chimpanzee has been listed as an endangered species [14]. These limitations of the chimpanzee model have stimulated progress toward developing alternative animal models for HCV research.
Although HCV infection cannot be propagated in mouse tissues, transgenic technology [15] as well as the availability of mice transplanted with human hepatocytes [16] has made the laboratory mouse an attractive animal model for HCV research. In this article we present an overview of current mouse models for HCV research. The first part of this article focuses on the application of transgenic mouse models in HCV research for the analysis of virus–host interaction. The second part summarizes the emerging role of chimeric transgenic mice populated with human hepatocytes for the study of HCV infection. We conclude with a presentation on future perspectives for these animal models in the HCV research field.
2. Transgenic mouse models
Transgenic animal technology, which allows the germline insertion of exogenous genes or the alteration or disruption of endogenous genes, has emerged as a powerful tool for the in vivo analysis of gene function [17]. By far the most common and well-characterized approach for producing transgenic mice is direct pronuclear microinjection of the “transgene” in one-cell fertilized embryos. Typically 1–200 copies of the exogenous transgene integrates into the host genome at an apparently random site [17].
Several groups have established transgenic mice expressing HCV proteins either individually or together as a polyprotein to study the effect of these proteins on liver pathology. Hepatic steatosis is a common histological feature of chronic hepatitis C. Steatosis is more frequent and severe in patients infected with HCV genotype 3 [18]. The mechanisms underlying this genotype specific steatosis are unknown. However, hepatic steatosis can develop secondary to obesity, diabetes mellitus, alcohol abuse, protein malnutrition, total parenteral nutrition, acute starvation and drug therapy [19]. A number of transgenic mouse model studies indicate that the HCV core protein is sufficient to induce lipid accumulation in hepatocytes (for review see [20]). The core protein is an RNA-binding protein that is a component of the viral nucleocapsid [21]. In infected cells the core protein was found to associate with lipid droplets [22]. Recent studies suggest that the association of core protein with lipid droplets plays an important role in HCV morphogenesis and efficient virus production [23], [24]. Based on the experimental model of the transgenic mouse, the HCV core protein seems to inhibit the microsomal triglyceride transfer protein (MTP) activity [25]. As this is the rate limiting enzyme of hepatic lipoprotein assembly, the direct and likely consequence of its inactivation is accumulation of unsecreted triglycerides, hence steatosis. Moreover, HCV core protein expression in the mouse liver upregulated sterol regulatory element binding protein 1c (SREBP-1c) promoter activity [26]. SREBP-1c is a transcription factor leading to the upregulation of enzymes involved in de novo lipogenesis, an event that can favor intracellular accumulation of triglycerides [27].
Epidemiologic, clinical, and virologic data have shown a close association between chronic HCV infection and the development of hepatocellular carcinoma (HCC). HCC usually arises after 2–4 decades of infection, typically in the context of an underlying cirrhosis [28]. Through the use of transgenic mouse models, it has also become evident that the core protein of HCV has an oncogenic activity in the liver. HCV core protein constitutively expressed in the liver of C57BL/6 mice – a strain which is known to exhibit spontaneous occurrence of HCC only rarely – at levels similar to that found in chronic hepatitis C patients lead to multicentric hepatic adenomas, and developed HCCs in an age-dependent manner [29], [30], [31], [32]. HCC was observed predominantly in males, an observation consistent with the epidemiological data that men chronically infected with HCV are more likely to develop HCC [33]. Transgenic mice expressing the complete HCV polyprotein showed an increased risk of cancer [32] suggesting that other HCV proteins might also play a role in the development of hepatocarcinogenesis. In a diethylnitrosamine (DEN)-based model of hepatocarcinogenesis, transgenic mice expressing core, E1 and E2 structural proteins demonstrated an accelerated tumor growth phenotype suggesting that HCV E1 and/or E2, possibly in conjunction with core protein, can act as tumor enhancer proteins [34]. However, other groups reported transgenic mice expressing the HCV core, E1 and E2 protein did not exhibit any pathological phenotype of the liver [35], [36]. These differences may be related to the expression level of the transgene or different genetic background on which the transgenic models were produced. To address the role of HCV non-structural proteins in liver pathology, transgenic mice expressing HCV NS3, NS4 or NS5A protein were generated. By contrast, expression of HCV non-structural proteins did not cause any spontaneous liver pathology [37], [38], [39].
Based on the experimental model of these transgenic mice, we are beginning to understand the molecular mechanism involved in the development of HCC. Hepatocyte proliferation is influenced by various factors, such as mitogenic chemicals, cytokines, growth factors and transcription factors [40]. One activity of the HCV core protein has been implicated to modify the in vivo expression of cytokines. Indeed, expression of tumor necrosis factor-alpha (TNF-α) and interleukin-1beta (IL-1β) was increased at both protein and mRNA levels in transgenic mice constitutively expressing HCV core protein [41]. Elevated concentrations of TNF-α and IL-1β represent a characteristic feature of chronic liver diseases and liver dysfunction [42].
Pro-inflammatory stimuli, such as TNF-α, induce signal cascades through their cognate receptors to activate IκB kinase (IKK) signalosome and subsequently NF-κB, a major regulator of inflammatory and antiapoptotic genes [43]. Recently, Luedde et al. demonstrated that deletion of the IKK subunit NEMO/IKKγ in liver parenchymal cells caused steatosis and HCC in mice [44] suggesting that NF-κB activity plays an important role in protecting the liver from cancer. Interestingly, in vitro experiments have shown that HCV core protein suppresses IKK signalosome activity in the macrophages [45]. Additional studies are necessary to determine whether HCV core protein mediates in vivo suppression of NF-κB activity in hepatocytes leading to steatosis and HCC.
Experimental data from transgenic mice showed that HCV core protein binds to and activates the DNA-binding domain of the retinoid X receptor alpha (RXRα) [46]. RXRα is abundantly expressed in the liver and plays important roles in regulating cell proliferation and differentiation as well as in lipid metabolism [47]. Furthermore, proteasome activator PA28γ has been identified as an HCV core binding protein in the livers of both HCV core-transgenic mice and a patient with chronic hepatitis C [48]. Interestingly, knockout of the PA28γ gene from PA28γ+/+/HCV core-transgenic mice disrupted the development of both steatosis and HCC [26] suggesting that PA28γ activity may play a crucial role in the development of liver pathology induced by HCV infection. Finally, HCV core-transgenic mice demonstrated an activation of the peroxisome proliferators-activated receptor α (PPARα) [49]. Subsequent studies showed that PPARα is essential for HCV core protein-induced hepatic steatosis and HCC in mice [50]. PPARα regulates the transcription of genes encoding fatty acid-metabolizing enzymes and various cell-cycle regulators and oncogene products such as cyclin D1 and c-Myc are known to be induced in a PPARα-dependent manner [51]. Thus, the modulation of transcriptional activity by the HCV core protein may contribute to the disturbance of cell proliferation and differentiation in the liver, leading to oncogenic potential.
Oxidative stress is a potentially important pathogenic mechanism in chronic liver diseases to initiate and promote multistage carcinogenesis. Oxidants not only are toxic to target cells but also overwhelm cellular antioxidant defenses of neighboring cells, leading to DNA damage [52]. Transgenic mice expressing HCV core alone or in combination with HCV E1 and E2 showed elevated levels of lipid peroxidation and oxidatively damaged DNA [30], [53], [54], [55]. Oxidative stress can trigger signal transducer and activator of transcription 3 (STAT3) tyrosine phosphorylation [56] which is expected to cause significant alteration of the cell growth properties, as STAT3 has been reported as an oncogene [57].
Finally, studies of HCV core protein transgenic mice demonstrated that the expression of HCV core protein in the liver conveys resistance to autoimmune liver injury, induced by the T cell mitogen concanavalin A [58]. Consistent with this observation, the HCV core-transgenic hepatocytes were relatively resistant to death induced by anti-Fas and TNF-α mediated death. This resistance was associated with a shift from STAT1 to STAT3 activation in liver tissue [58]. These findings indicate that HCV core protein may protect infected hepatocytes from destruction by the immune system and promotes their proliferation. Similar observation has been reported in transgenic mice expressing HCV polyprotein [59].
Consistent with the constitutive expression of HCV proteins, conventional transgenic mice are immunotolerant to these proteins. Thus, one desired goal of transgene technology is temporal control of target gene expression in the specific organ. The Cre–loxP recombination system is a useful method of conditional gene expression that allows spatial (cell-type specific) and temporal (inducer-dependent) control. The Cre–loxP system has two components: Cre recombinase and two lox P sites that Cre recognizes. The site-specific recombination is accomplished by Cre-mediated catalysis of reciprocal recombination between the two lox P sites in both tissue-culture cells and mice [60]. Using the Cre–loxP-mediated conditional expression system Wakita and colleagues [61] generated HCV transgenic mice, which express HCV core, E1, E2 and NS2 protein. HCV transgene expression in the liver was induced by intravenous administration of a recombinant adenovirus expressing Cre recombinase [61]. HCV transgenic mice regulated by the Cre–loxP switching system demonstrated an anti-core antibody response and an HCV-specific T cell response after induction of the core transgene expression [61], [62]. Most interestingly, this immune response resulted in hepatitis [61], [62]. Furthermore, Machida and colleagues [63] used the Cre–loxP system to study the effect of HCV proteins on Fas-mediated cell death. Interestingly, transgene expression of HCV transgene mice suppressed Fas-mediated apoptotic cell death suggesting that HCV can evade the innate antiviral mechanism of apoptosis to maintain persistent infection. The Cre–loxP switching system provides a useful “non-immune tolerant” HCV transgene mouse model which allows the study of the host immune response against the HCV proteins. A non-adenoviral gene delivery method for Cre recombinase in future Cre–loxP HCV mouse models may further strengthen the application of this technology for the study of HCV–host interactions by eliminating adenovirus-related effects. A potential strategy could be the production of HCV transgenic mice in which the Cre–loxP switching gene expression system is under the control of a tetracycline-inducible expression system [64].
Transgenic mice expressing one or a combination of HCV proteins are unique and irreplaceable tools to elucidate and understand molecular mechanism involved in HCV–host interaction (Table 1 ). They have significantly contributed to our understanding of HCV host interactions in vivo. However, since mice are not permissive to HCV infection, not all results obtained in the transgenic mouse models are directly applicable to pathogenesis of HCV infection in vivo. To overcome this limitation, chimeric transgenic mice repopulated with human hepatocytes have been developed for the study of HCV infection.
Table 1.
Selected transgenic mouse models expressing HCV proteins and liver pathology
| Transgene and promoter | Liver phenotype | Reference | |
|---|---|---|---|
| Core | HBV | Steatosis | Perlemuter et al. (2002) [25] |
| HCC | Moriya et al. (1998) [29] | ||
| Oxidative stress | Moriya et al. (2001) [30] | ||
| Alteration of cytokine expression | Tsutsumi et al. (2002) [41] | ||
| Activation of retinoid X receptor alpha | Tsutsumi et al. (2002) [46] | ||
| Steatosis, HCC | Moriishi et al. (2007) [26] | ||
| Tanaka et al. (2008) [49] | |||
| Tanaka et al. (2008) [50] | |||
| EF-1α | Oxidative stress | Machida et al. (2006) [53] | |
| Core–E1–E2 | Albumin | Steatosis, HCC | Lerat et al. (2002) [32] |
| HCC | Kamegaya et al. (2005) [34] | ||
| Oxidative stress | Okuda et al. (2002) [54] | ||
| Korenaga et al. (2005) [55] | |||
| CMV | Steatosis, HCC | Naas et al. (2005) [92] | |
| Core–E1–E2–NS2 | Cre–loxP system | Hepatitis, cellular immune responses | Wakita et al. (1998) [61], Wakita et al. (2000) [62] |
| Suppression of Fas-mediated cell death | Machida et al. (2001) [63] | ||
| Polyprotein | Alpha1 antitrypsin | Steatosis, intrahepatic T cell recruitment | Alonzi et al. (2004) [93] |
| Albumin | Steatosis, HCC | Lerat et al. (2002) [32] | |
| Impairment of cellular immune response | Disson et al. (2004) [59] | ||
3. Chimeric transgenic mice repopulated with human hepatocytes
The discovery of a hepatocyte-lethal phenotype in mice carrying a urokinase-type plasminogen activator transgene controlled by an albumin promotor (Alb-uPA) and the complete reconstitution of livers of those mice with xenografted rat hepatocytes [65] has laid the foundation for the development of a small animal model of infection with hepatitis B and C viruses using xenografted human hepatocytes.
Petersen and colleagues [66] elegantly applied the uPA-xenograft model for the development of a hepatitis B mouse model by transplanting woodchuck hepatocytes into Alb-uPA mice on an immunodeficient recombinantion activation gene 2 (RAG-2) background. Repopulated woodchuck hepatocytes in Alb-uPA/RAG-2 mice, which lack mature B and T lymphocytes, allowed productive infection with woodchuck hepatitis B virus [66]. Three years later, Mercer et al. demonstrated that the severe combined immunodeficiency disorder (SCID)/Alb-uPA mouse engrafted with primary human hepatocytes can be infected with HCV in vivo [16].
The human albumin level is a reliable marker for the integrity and functional status of the engrafted human hepatocytes. In successfully transplanted mice, the albumin levels reach a plateau of approximately 7 mg/ml around week 7 [67]. Human hepatocytes can occupy up to 87% of liver parenchyma and show no signs of damage or degeneration. However, transplanted liver cells demonstrate an abundant accumulation of glycogen which may be the result of communication failure between mouse ligands or receptors and their human counterparts [67].
Once human hepatocytes are stably engrafted in the SCID/Alb-uPA mouse, these animals can be infected with human hepatotrophic viruses including hepatitis B [67], [68] and C [9], [16], [67], [69], [70]. Inoculation of HCV from serum of HCV patients, infected chimpanzee or HCV produced in cell culture (HCVcc) caused infection with viral titers in the blood of infected mice equal to or higher than those present in patients with chronic HCV infection [9], [16], [67], [69], [70]. Plasma derived from these animals can be used to infect other transplanted naïve mice. The mice displayed the same massive increase of viral load indicating that HCV infection can be serially passaged to naïve animals [9]. The in vivo HCV infection could be maintained for at least 4 months. During this time the HCV infection did not alter the liver function and architecture [67]. However, long-term infection studies with HCV are necessary to study the cytopathic effects of HCV infection in this model.
Although this animal model requires special expertise to isolate and transplant human hepatocytes and the maintenance of a colony of fragile immunodeficient mice with an approximately 35% mortality in newborns [16], it is currently the best available small animal model to study basic biology of HCV. Furthermore, recent studies indicate that this model allows the study of antiviral activities of drugs and neutralizing antibodies in vivo. The successful application of the SCID/Alb-uPA mouse model for drug development has been shown by two recent publications investigating the efficacy of interferon alpha2b (IFN-α) and HCV NS3 protease inhibitor (BILN2061). Treatment with both IFN-α and BILN2061 appeared to produce similar antiviral effect in HCV infection in either humans or this mouse model [69], [70]. For example, after 4 days of therapy with BILN2061, HCV titers decreased by approximately 2 log, similar to the impact seen in clinical trials [69], [70]. These findings indicate that the SCID/Alb-uPA mouse can model the HCV antiviral treatment response in humans with reasonable accuracy and may represent a valuable tool for the study of in vivo drug metabolism during pre-clinical evaluation of candidate therapeutics. In addition, the SCID/Alb-uPA mouse may be a useful animal model to evaluate the pre-clinical toxicity of new antiviral compounds against HCV as shown by the manifestation of the well-characterized clinical and ultrastructural signs of cardiotoxicity in SCID/Alb-uPA mice induced by the antiviral BILN 2061 [70].
In addition, the human SCID/Alb-uPA mouse model has been successfully used to study the efficacy of neutralizing antibodies for control of HCV infection. By using this model, Law and colleagues demonstrated that human monoclonal antibodies are able to neutralize genetically diverse HCV isolates and protect against heterologous HCV quasispecies challenge. Their results provide evidence that broadly neutralizing antibodies to HCV protect against heterologous viral infection and suggest that a prophylactic vaccine against HCV may be achievable [71], [72]. Vanwolleghem and colleagues also addressed the question whether IgG with neutralizing properties from chronically infected patients can prevent de novo HCV infection in vivo. The authors demonstrate evidence that the SCID/Alb-uPA mouse model is useful for passive immunization studies against HCV and that polyclonal IgG from a patient chronically infected with HCV can convey in vivo sterilizing immunity against a homologous and non-mutated ancestral hepatitis C virus [73].
Chronic HCV infection results in a highly variable course of liver disease, ranging from mild inflammation to rapidly progressive fibrosis, cirrhosis and HCC [1] suggesting that host factors play an important role in both infection outcome and viral pathogenesis. The SCID/Alb-uPA mouse model provides a unique system to analyze host-specific responses to HCV because the animals can be transplanted with hepatocytes from different donors and inoculated with a single source of HCV. Transcriptional profiling of HCV-induced gene expression changes in the SCID/Alb-uPA mouse model demonstrated that animals containing hepatocytes from the same donor showed a more similar response, than animals containing hepatocytes from different donors [74]. For example, all HCV-infected animals showed activation of the host innate antiviral signaling pathways, but the response was highly variable in the number and intensity of interferon (IFN)-stimulated genes indicating that host factors may influence the effectiveness of the innate immune response [74].
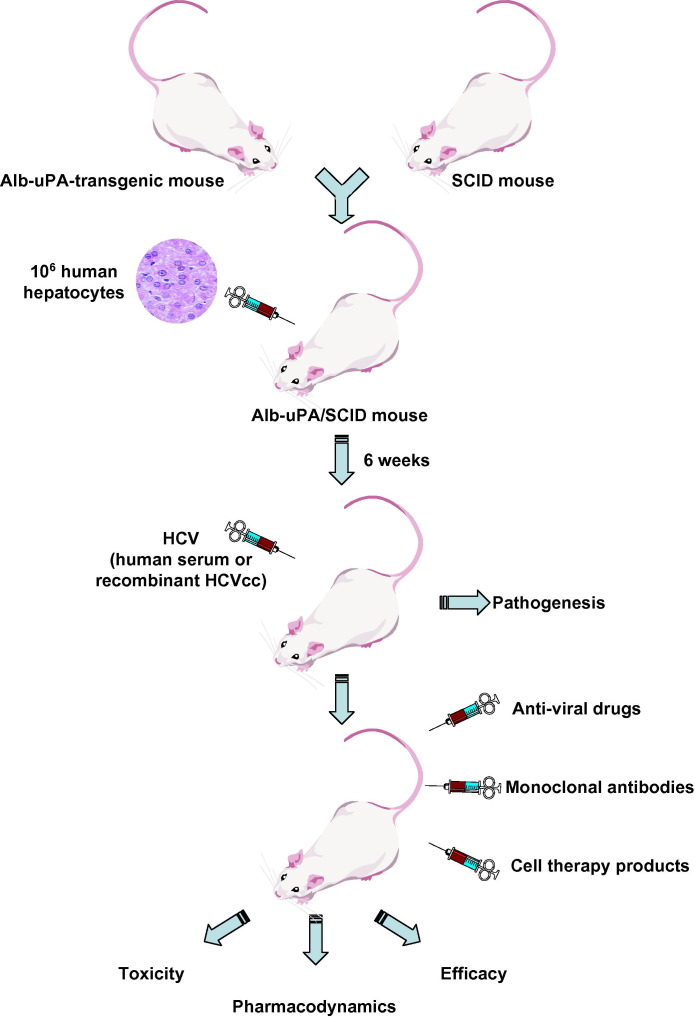
The lack of an adaptive immune response in these animals makes it possible to distinguish viral-mediated from immune-mediated effects on host gene expression. Thus, the SCID/Alb-uPA mouse model is a valuable tool to investigate the role of host factors in the development of liver disease and can be used to study aspects of the innate antiviral immune response that may play a fundamental role in limiting HCV replication. The potential infection of chimeric mice with recombinant tissue-culture derived HCV further broadens the scope of its application [9], [75] (Fig. 1 ).
Fig. 1.
Chimeric transgenic mice repopulated with human hepatocytes for the study of HCV infection. Hemizygous Alb-uPA mice were crossed to homozygosity with homozygous SCID/bg mice. The resulting Alb-uPA/SCID mice can be transplanted with human hepatocytes and support HCV infection from human infected-serum or recombinant cell culture-derived HCV (HCVcc). This model has been successfully used to study virus–host interactions in infected hepatocytes as well as the evaluation of antiviral strategies including antiviral drugs and monoclonal antibodies. The evaluation of cell therapy products may represent another application of this model in the future.
Most recently, liver repopulation by human hepatocytes was demonstrated for the severely immunodeficient fumarylacetoacetate hydrolase (Fah)-deficient mouse [76]. In this model engrafted human hepatocytes can be serially transplanted from primary donors allowing the expansion of human hepatocytes of the same donor through several generations of recipient mice [76]. Further studies are needed to determine whether this model can be used to study HCV infection and is superior to uPA-based models.
4. Perspectives
Appropriate animal models are essential for studying human diseases, developing therapeutic strategies and testing drug safety. Transgenic mice expressing HCV proteins have been shown to play an important role in elucidating the molecular mechanisms of HCV–host interaction and HCV-related liver pathology. Since steatosis and HCC are relevant clinical manifestations seen in HCV-infected individuals [18], transgenic mouse models more metabolically similar to human are needed. First, unlike in humans, mouse’s primary circulating lipoprotein is HDL. Second, whereas human liver produces only apoB100, mouse liver produce both apoB100 and its truncated form, apoB48. Also, CETP, a plasma glycoprotein that facilitates transfer of cholesteryl esters from HDL to apoB-containing lipoproteins such as VLDL and LDL, is present in humans but not in mice (for review see [77]). Developing HCV transgenic mice with more human-like lipid metabolism may help to better understand the impact of viral proteins in the pathogenesis of liver disease as well as the development and characterization of antivirals.
In the future, the ideal mouse model would be a mouse permissive for HCV infection. Host cell surface receptors are recognized as important determinants of virus–host range and tissue tropism [78]. The HCV-receptor interaction is a multistep process itself and the virus uses several binding and entry receptors simultaneously or sequentially to enter the host cell (for review see [5]). It is of interest to note that although HCV cannot infect mouse hepatocytes, replication of the HCV prototype JFH-1 strain in mouse hepatoma cell lines has been reported [79]. One future strategy to overcome species barriers in studying HCV infection would be to establish a transgenic mouse line expressing human HCV-receptor molecules. In severe acute respiratory syndrome (SARS) coronavirus infection, the development of a transgenic mouse line expressing the human angiotensin-converting enzyme 2 (hACE2) has been demonstrated to be a suitable model to study SARS infection in mice [80]. The tetraspanin CD81 has been proposed to play a role in HCV entry (for review see [5]). However, transgenic mice expressing human CD81 in the liver did not confer susceptibility to HCV infection [81] suggesting that human CD81 is not the only cellular factor required for HCV infection in the mouse. Thus, the development of a permissive HCV mouse model may require the co-expression of several human HCV entry molecules (such as human liver specific heparan sulfate [82], [83] and scavenger receptor class B type I [84], and claudin [85] as well as other not yet identified host factors restricting HCV tropism to the human hepatocyte.
Finally, a HCV permissive mouse model reconstituted with a human immune system would allow us to study mechanisms of HCV immunopathogenesis. In Rag2-/-gamma(c)-/-mice double knockout mice [86], [87] and non-obese diabetic/SCID/interleukin-2 receptor gamma chain-knocked-out mice [88], [89], the development of a functional human immune system with human CD34+ hematopoietic stem cells has been demonstrated. These mouse models are a valuable tool to study HIV infection and pathogenesis [90], [91] and could conceivably be adopted to study the immunopathogenesis of HCV infection in a mouse model.
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft, Germany (Ba1417/11-2; T.F.B.), the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, NIH (H.B. and T.J.L.), the European Union, Germany (LSHM-CT-2004-503359 “VIRGIL” T.F.B.), the ANR chair of excellence program, France (ANR-05-CEXC-008, T.F.B.), ANRS, France (Grant 06221, T.F.B.; Young scientist career award, E.R.), and Université Louis Pasteur, France (CONECTUS program, T.F.B.).
Associate Editor: M.U. Mondelli
Footnotes
The authors declare that they do not have anything to disclose regarding funding from industries or conflict of interest with respect to this manuscript. NIH funded study.
Contributor Information
Heidi Barth, Email: barthh@niddk.nih.gov.
Thomas F. Baumert, Email: Thomas.Baumert@viro-ulp.u-strasbg.fr.
References
- 1.Chisari F.V. Unscrambling hepatitis C virus–host interactions. Nature. 2005;436:930–932. doi: 10.1038/nature04076. [DOI] [PubMed] [Google Scholar]
- 2.Brown R.S. Hepatitis C and liver transplantation. Nature. 2005;436:973–978. doi: 10.1038/nature04083. [DOI] [PubMed] [Google Scholar]
- 3.Feld J.J., Hoofnagle J.H. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967–972. doi: 10.1038/nature04082. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager R., Sparacio S. Hepatitis C virus molecular clones and their replication capacity in vivo and in cell culture. Virus Res. 2007;127:195–207. doi: 10.1016/j.virusres.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 5.Barth H., Liang T.J., Baumert T.F. Hepatitis C virus entry: molecular biology and clinical implications. Hepatology. 2006;44:527–535. doi: 10.1002/hep.21321. [DOI] [PubMed] [Google Scholar]
- 6.Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindenbach B.D., Evans M.J., Syder A.J., Wolk B., Tellinghuisen T.L., Liu C.C. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 8.Zhong J., Gastaminza P., Cheng G., Kapadia S., Kato T., Burton D.R. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindenbach B.D., Meuleman P., Ploss A., Vanwolleghem T., Syder A.J., McKeating J.A. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc Natl Acad Sci USA. 2006;103:3805–3809. doi: 10.1073/pnas.0511218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi M., Villanueva R.A., Thomas D.L., Wakita T., Lemon S.M. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci USA. 2006;103:2310–2315. doi: 10.1073/pnas.0510727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato T., Matsumura T., Heller T., Saito S., Sapp R.K., Murthy K. Production of infectious hepatitis C virus of various genotypes in cell cultures. J Virol. 2007;81:4405–4411. doi: 10.1128/JVI.02334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bukh J. A critical role for the chimpanzee model in the study of hepatitis C. Hepatology. 2004;39:1469–1475. doi: 10.1002/hep.20268. [DOI] [PubMed] [Google Scholar]
- 13.Bassett S.E., Brasky K.M., Lanford R.E. Analysis of hepatitis C virus-inoculated chimpanzees reveals unexpected clinical profiles. J Virol. 1998;72:2589–2599. doi: 10.1128/jvi.72.4.2589-2599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oates J.F. Is the chimpanzee, Pan troglodytes, an endangered species? It depends on what “endangered” means. Primates. 2006;47:102–112. doi: 10.1007/s10329-005-0149-5. [DOI] [PubMed] [Google Scholar]
- 15.Palmiter R.D., Brinster R.L. Transgenic mice. Cell. 1985;41:343–345. doi: 10.1016/s0092-8674(85)80004-0. [DOI] [PubMed] [Google Scholar]
- 16.Mercer D.F., Schiller D.E., Elliott J.F., Douglas D.N., Hao C., Rinfret A. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 17.Bockamp E., Maringer M., Spangenberg C., Fees S., Fraser S., Eshkind L. Of mice and models: improved animal models for biomedical research. Physiol Genomics. 2002;11:115–132. doi: 10.1152/physiolgenomics.00067.2002. [DOI] [PubMed] [Google Scholar]
- 18.Negro F. Mechanisms and significance of liver steatosis in hepatitis C virus infection. World J Gastroenterol. 2006;12:6756–6765. doi: 10.3748/wjg.v12.i42.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark J.M., Brancati F.L., Diehl A.M. Nonalcoholic fatty liver disease. Gastroenterology. 2002;122:1649–1657. doi: 10.1053/gast.2002.33573. [DOI] [PubMed] [Google Scholar]
- 20.Kremsdorf D., Brezillon N. New animal models for hepatitis C viral infection and pathogenesis studies. World J Gastroenterol. 2007;13:2427–2435. doi: 10.3748/wjg.v13.i17.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubuisson J. Hepatitis C virus proteins. World J Gastroenterol. 2007;13:2406–2415. doi: 10.3748/wjg.v13.i17.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rouille Y., Helle F., Delgrange D., Roingeard P., Voisset C., Blanchard E. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J Virol. 2006;80:2832–2841. doi: 10.1128/JVI.80.6.2832-2841.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shavinskaya A., Boulant S., Penin F., McLauchlan J., Bartenschlager R. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J Biol Chem. 2007;282:37158–37169. doi: 10.1074/jbc.M707329200. [DOI] [PubMed] [Google Scholar]
- 24.Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 25.Perlemuter G., Sabile A., Letteron P., Vona G., Topilco A., Chretien Y. Hepatitis C virus core protein inhibits microsomal triglyceride transfer protein activity and very low density lipoprotein secretion: a model of viral-related steatosis. FASEB J. 2002;16:185–194. doi: 10.1096/fj.01-0396com. [DOI] [PubMed] [Google Scholar]
- 26.Moriishi K., Mochizuki R., Moriya K., Miyamoto H., Mori Y., Abe T. Critical role of PA28gamma in hepatitis C virus-associated steatogenesis and hepatocarcinogenesis. Proc Natl Acad Sci USA. 2007;104:1661–1666. doi: 10.1073/pnas.0607312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimano H., Horton J.D., Shimomura I., Hammer R.E., Brown M.S., Goldstein J.L. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang T.J., Heller T. Pathogenesis of hepatitis C-associated hepatocellular carcinoma. Gastroenterology. 2004;127:S62–S71. doi: 10.1053/j.gastro.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 29.Moriya K., Fujie H., Shintani Y., Yotsuyanagi H., Tsutsumi T., Ishibashi K. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998;4:1065–1067. doi: 10.1038/2053. [DOI] [PubMed] [Google Scholar]
- 30.Moriya K., Nakagawa K., Santa T., Shintani Y., Fujie H., Miyoshi H. Oxidative stress in the absence of inflammation in a mouse model for hepatitis C virus-associated hepatocarcinogenesis. Cancer Res. 2001;61:4365–4370. [PubMed] [Google Scholar]
- 31.Koike K., Moriya K., Kimura S. Role of hepatitis C virus in the development of hepatocellular carcinoma: transgenic approach to viral hepatocarcinogenesis. J Gastroenterol Hepatol. 2002;17:394–400. doi: 10.1046/j.1440-1746.2002.02763.x. [DOI] [PubMed] [Google Scholar]
- 32.Lerat H., Honda M., Beard M.R., Loesch K., Sun J., Yang Y. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology. 2002;122:352–365. doi: 10.1053/gast.2002.31001. [DOI] [PubMed] [Google Scholar]
- 33.Fattovich G., Stroffolini T., Zagni I., Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127:S35–S50. doi: 10.1053/j.gastro.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Kamegaya Y., Hiasa Y., Zukerberg L., Fowler N., Blackard J.T., Lin W. Hepatitis C virus acts as a tumor accelerator by blocking apoptosis in a mouse model of hepatocarcinogenesis. Hepatology. 2005;41:660–667. doi: 10.1002/hep.20621. [DOI] [PubMed] [Google Scholar]
- 35.Kawamura T., Furusaka A., Koziel M.J., Chung R.T., Wang T.C., Schmidt E.V. Transgenic expression of hepatitis C virus structural proteins in the mouse. Hepatology. 1997;25:1014–1021. doi: 10.1002/hep.510250437. [DOI] [PubMed] [Google Scholar]
- 36.Pasquinelli C., Shoenberger J.M., Chung J., Chang K.M., Guidotti L.G., Selby M. Hepatitis C virus core and E2 protein expression in transgenic mice. Hepatology. 1997;25:719–727. doi: 10.1002/hep.510250338. [DOI] [PubMed] [Google Scholar]
- 37.Frelin L., Brenndorfer E.D., Ahlen G., Weiland M., Hultgren C., Alheim M. The hepatitis C virus and immune evasion: non-structural 3/4A transgenic mice are resistant to lethal tumour necrosis factor alpha mediated liver disease. Gut. 2006;55:1475–1483. doi: 10.1136/gut.2005.085050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Majumder M., Steele R., Ghosh A.K., Zhou X.Y., Thornburg L., Ray R. Expression of hepatitis C virus non-structural 5A protein in the liver of transgenic mice. FEBS Lett. 2003;555:528–532. doi: 10.1016/s0014-5793(03)01337-1. [DOI] [PubMed] [Google Scholar]
- 39.Majumder M., Ghosh A.K., Steele R., Zhou X.Y., Phillips N.J., Ray R. Hepatitis C virus NS5A protein impairs TNF-mediated hepatic apoptosis, but not by an anti-FAS antibody, in transgenic mice. Virology. 2002;294:94–105. doi: 10.1006/viro.2001.1309. [DOI] [PubMed] [Google Scholar]
- 40.Tanimizu N., Miyajima A. Molecular mechanism of liver development and regeneration. Int Rev Cytol. 2007;259:1–48. doi: 10.1016/S0074-7696(06)59001-1. [DOI] [PubMed] [Google Scholar]
- 41.Tsutsumi T., Suzuki T., Moriya K., Yotsuyanagi H., Shintani Y., Fujie H. Alteration of intrahepatic cytokine expression and AP-1 activation in transgenic mice expressing hepatitis C virus core protein. Virology. 2002;304:415–424. doi: 10.1006/viro.2002.1702. [DOI] [PubMed] [Google Scholar]
- 42.Tilg H., Wilmer A., Vogel W., Herold M., Nolchen B., Judmaier G. Serum levels of cytokines in chronic liver diseases. Gastroenterology. 1992;103:264–274. doi: 10.1016/0016-5085(92)91122-k. [DOI] [PubMed] [Google Scholar]
- 43.Hoffmann A., Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 44.Luedde T., Beraza N., Kotsikoris V., van Loo G., Nenci A., De Vos R. Deletion of NEMO/IKKgamma in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell. 2007;11:119–132. doi: 10.1016/j.ccr.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 45.Joo M., Hahn Y.S., Kwon M., Sadikot R.T., Blackwell T.S., Christman J.W. Hepatitis C virus core protein suppresses NF-kappaB activation and cyclooxygenase-2 expression by direct interaction with IkappaB kinase beta. J Virol. 2005;79:7648–7657. doi: 10.1128/JVI.79.12.7648-7657.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsutsumi T., Suzuki T., Shimoike T., Suzuki R., Moriya K., Shintani Y. Interaction of hepatitis C virus core protein with retinoid X receptor alpha modulates its transcriptional activity. Hepatology. 2002;35:937–946. doi: 10.1053/jhep.2002.32470. [DOI] [PubMed] [Google Scholar]
- 47.Mangelsdorf D.J., Evans R.M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 48.Moriishi K., Okabayashi T., Nakai K., Moriya K., Koike K., Murata S. Proteasome activator PA28gamma-dependent nuclear retention and degradation of hepatitis C virus core protein. J Virol. 2003;77:10237–10249. doi: 10.1128/JVI.77.19.10237-10249.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka N., Moriya K., Kiyosawa K., Koike K., Aoyama T. Hepatitis C virus core protein induces spontaneous and persistent activation of peroxisome proliferator-activated receptor alpha in transgenic mice: implications for HCV-associated hepatocarcinogenesis. Int J Cancer. 2008;122:124–131. doi: 10.1002/ijc.23056. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka N., Moriya K., Kiyosawa K., Koike K., Gonzalez F.J., Aoyama T. PPARalpha activation is essential for HCV core protein-induced hepatic steatosis and hepatocellular carcinoma in mice. J Clin Invest. 2008;118:683–694. doi: 10.1172/JCI33594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mandard S., Muller M., Kersten S. Peroxisome proliferator-activated receptor alpha target genes. Cell Mol Life Sci. 2004;61:393–416. doi: 10.1007/s00018-003-3216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lieber C.S. Role of oxidative stress and antioxidant therapy in alcoholic and nonalcoholic liver diseases. Adv Pharmacol. 1997;38:601–628. doi: 10.1016/s1054-3589(08)61001-7. [DOI] [PubMed] [Google Scholar]
- 53.Machida K., Cheng K.T., Lai C.K., Jeng K.S., Sung V.M., Lai M.M. Hepatitis C virus triggers mitochondrial permeability transition with production of reactive oxygen species, leading to DNA damage and STAT3 activation. J Virol. 2006;80:7199–7207. doi: 10.1128/JVI.00321-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okuda M., Li K., Beard M.R., Showalter L.A., Scholle F., Lemon S.M. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122:366–375. doi: 10.1053/gast.2002.30983. [DOI] [PubMed] [Google Scholar]
- 55.Korenaga M., Wang T., Li Y., Showalter L.A., Chan T., Sun J. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. J Biol Chem. 2005;280:37481–37488. doi: 10.1074/jbc.M506412200. [DOI] [PubMed] [Google Scholar]
- 56.Carballo M., Conde M., El Bekay R., Martin-Nieto J., Camacho M.J., Monteseirin J. Oxidative stress triggers STAT3 tyrosine phosphorylation and nuclear translocation in human lymphocytes. J Biol Chem. 1999;274:17580–17586. doi: 10.1074/jbc.274.25.17580. [DOI] [PubMed] [Google Scholar]
- 57.Bromberg J.F., Wrzeszczynska M.H., Devgan G., Zhao Y., Pestell R.G., Albanese C. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 58.Kawamura H., Govindarajan S., Aswad F., Machida K., Lai M.M., Sung V.M. HCV core expression in hepatocytes protects against autoimmune liver injury and promotes liver regeneration in mice. Hepatology. 2006;44:936–944. doi: 10.1002/hep.21360. [DOI] [PubMed] [Google Scholar]
- 59.Disson O., Haouzi D., Desagher S., Loesch K., Hahne M., Kremer E.J. Impaired clearance of virus-infected hepatocytes in transgenic mice expressing the hepatitis C virus polyprotein. Gastroenterology. 2004;126:859–872. doi: 10.1053/j.gastro.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 60.Brault V., Besson V., Magnol L., Duchon A., Herault Y. Cre/loxP-mediated chromosome engineering of the mouse genome. Handb Exp Pharmacol. 2007:29–48. doi: 10.1007/978-3-540-35109-2_2. [DOI] [PubMed] [Google Scholar]
- 61.Wakita T., Taya C., Katsume A., Kato J., Yonekawa H., Kanegae Y. Efficient conditional transgene expression in hepatitis C virus cDNA transgenic mice mediated by the Cre/loxP system. J Biol Chem. 1998;273:9001–9006. doi: 10.1074/jbc.273.15.9001. [DOI] [PubMed] [Google Scholar]
- 62.Wakita T., Katsume A., Kato J., Taya C., Yonekawa H., Kanegae Y. Possible role of cytotoxic T cells in acute liver injury in hepatitis C virus cDNA transgenic mice mediated by Cre/loxP system. J Med Virol. 2000;62:308–317. doi: 10.1002/1096-9071(200011)62:3<308::aid-jmv2>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 63.Machida K., Tsukiyama-Kohara K., Seike E., Tone S., Shibasaki F., Shimizu M. Inhibition of cytochrome c release in Fas-mediated signaling pathway in transgenic mice induced to express hepatitis C viral proteins. J Biol Chem. 2001;276:12140–12146. doi: 10.1074/jbc.M010137200. [DOI] [PubMed] [Google Scholar]
- 64.Manickan E., Satoi J., Wang T.C., Liang T.J. Conditional liver-specific expression of simian virus 40 T antigen leads to regulatable development of hepatic neoplasm in transgenic mice. J Biol Chem. 2001;276:13989–13994. doi: 10.1074/jbc.M009770200. [DOI] [PubMed] [Google Scholar]
- 65.Sandgren E.P., Palmiter R.D., Heckel J.L., Daugherty C.C., Brinster R.L., Degen J.L. Complete hepatic regeneration after somatic deletion of an albumin-plasminogen activator transgene. Cell. 1991;66:245–256. doi: 10.1016/0092-8674(91)90615-6. [DOI] [PubMed] [Google Scholar]
- 66.Petersen J., Dandri M., Gupta S., Rogler C.E. Liver repopulation with xenogenic hepatocytes in B and T cell-deficient mice leads to chronic hepadnavirus infection and clonal growth of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1998;95:310–315. doi: 10.1073/pnas.95.1.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meuleman P., Libbrecht L., De Vos R., de Hemptinne B., Gevaert K., Vandekerckhove J. Morphological and biochemical characterization of a human liver in a uPA-SCID mouse chimera. Hepatology. 2005;41:847–856. doi: 10.1002/hep.20657. [DOI] [PubMed] [Google Scholar]
- 68.Meuleman P., Libbrecht L., Wieland S., De Vos R., Habib N., Kramvis A. Immune suppression uncovers endogenous cytopathic effects of the hepatitis B virus. J Virol. 2006;80:2797–2807. doi: 10.1128/JVI.80.6.2797-2807.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kneteman N.M., Weiner A.J., O’Connell J., Collett M., Gao T., Aukerman L. Anti-HCV therapies in chimeric scid-Alb/uPA mice parallel outcomes in human clinical application. Hepatology. 2006;43:1346–1353. doi: 10.1002/hep.21209. [DOI] [PubMed] [Google Scholar]
- 70.Vanwolleghem T., Meuleman P., Libbrecht L., Roskams T., De Vos R., Leroux-Roels G. Ultra-rapid cardiotoxicity of the hepatitis C virus protease inhibitor BILN 2061 in the urokinase-type plasminogen activator mouse. Gastroenterology. 2007;133:1144–1155. doi: 10.1053/j.gastro.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 71.Law M., Maruyama T., Lewis J., Giang E., Tarr A.W., Stamataki Z. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med. 2007;14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 72.Zeisel MB, Cosset FL, Baumert TF. Host neutralizing responses and pathogenesis of hepatitis C virus infection. Hepatology 2008, in press. [DOI] [PubMed]
- 73.Vanwolleghem T, Bukh J, Meuleman P, Desombere I, Meunier JC, Alter H, et al. Polyclonal immunoglobulins from a chronic HCV patient protect human liver-chimeric mice from infection with a homologous HCV strain. Hepatology 2008, in press. [DOI] [PubMed]
- 74.Walters K.A., Joyce M.A., Thompson J.C., Smith M.W., Yeh M.M., Proll S. Host-specific response to HCV infection in the chimeric SCID-beige/Alb-uPA mouse model: role of the innate antiviral immune response. PLoS Pathog. 2006;2:e59. doi: 10.1371/journal.ppat.0020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hiraga N., Imamura M., Tsuge M., Noguchi C., Takahashi S., Iwao E. Infection of human hepatocyte chimeric mouse with genetically engineered hepatitis C virus and its susceptibility to interferon. FEBS Lett. 2007;581:1983–1987. doi: 10.1016/j.febslet.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 76.Azuma H., Paulk N., Ranade A., Dorrell C., Al-Dhalimy M., Ellis E. Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Il2rg−/− mice. Nat Biotechnol. 2007;25:903–910. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zadelaar S., Kleemann R., Verschuren L., de Vries-Van der Weij J., van der Hoorn J., Princen H.M. Mouse models for atherosclerosis and pharmaceutical modifiers. Arterioscler Thromb Vasc Biol. 2007;27:1706–1721. doi: 10.1161/ATVBAHA.107.142570. [DOI] [PubMed] [Google Scholar]
- 78.Marsh M., Helenius A. Virus entry: open sesame. Cell. 2006;124:729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Uprichard S.L., Chung J., Chisari F.V., Wakita T. Replication of a hepatitis C virus replicon clone in mouse cells. Virol J. 2006;3:89. doi: 10.1186/1743-422X-3-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tseng C.T., Huang C., Newman P., Wang N., Narayanan K., Watts D.M. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human angiotensin-converting enzyme 2 virus receptor. J Virol. 2007;81:1162–1173. doi: 10.1128/JVI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Masciopinto F., Freer G., Burgio V.L., Levy S., Galli-Stampino L., Bendinelli M. Expression of human CD81 in transgenic mice does not confer susceptibility to hepatitis C virus infection. Virology. 2002;304:187–196. doi: 10.1006/viro.2002.1631. [DOI] [PubMed] [Google Scholar]
- 82.Barth H., Schafer C., Adah M.I., Zhang F., Linhardt R.J., Toyoda H. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J Biol Chem. 2003;278:41003–41012. doi: 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- 83.Barth H., Schnober E.K., Zhang F., Linhardt R.J., Depla E., Boson B. Viral and cellular determinants of the hepatitis C virus envelope–heparan sulfate interaction. J Virol. 2006;80:10579–10590. doi: 10.1128/JVI.00941-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zeisel M.B., Koutsoudakis G., Schnober E.K., Haberstroh A., Blum H.E., Cosset F.L. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology. 2007;46:1722–1731. doi: 10.1002/hep.21994. [DOI] [PubMed] [Google Scholar]
- 85.Evans M.J., von Hahn T., Tscherne D.M., Syder A.J., Panis M., Wolk B. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 86.Traggiai E., Chicha L., Mazzucchelli L., Bronz L., Piffaretti J.C., Lanzavecchia A. Development of a human adaptive immune system in cord blood cell-transplanted mice. Science. 2004;304:104–107. doi: 10.1126/science.1093933. [DOI] [PubMed] [Google Scholar]
- 87.Gimeno R., Weijer K., Voordouw A., Uittenbogaart C.H., Legrand N., Alves N.L. Monitoring the effect of gene silencing by RNA interference in human CD34+ cells injected into newborn RAG2−/− gammac−/− mice: functional inactivation of p53 in developing T cells. Blood. 2004;104:3886–3893. doi: 10.1182/blood-2004-02-0656. [DOI] [PubMed] [Google Scholar]
- 88.Watanabe S., Ohta S., Yajima M., Terashima K., Ito M., Mugishima H. Humanized NOD/SCID/IL2Rgamma(null) mice transplanted with hematopoietic stem cells under nonmyeloablative conditions show prolonged life spans and allow detailed analysis of human immunodeficiency virus type 1 pathogenesis. J Virol. 2007;81:13259–13264. doi: 10.1128/JVI.01353-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Watanabe S., Terashima K., Ohta S., Horibata S., Yajima M., Shiozawa Y. Hematopoietic stem cell-engrafted NOD/SCID/IL2Rgamma null mice develop human lymphoid systems and induce long-lasting HIV-1 infection with specific humoral immune responses. Blood. 2007;109:212–218. doi: 10.1182/blood-2006-04-017681. [DOI] [PubMed] [Google Scholar]
- 90.Baenziger S., Tussiwand R., Schlaepfer E., Mazzucchelli L., Heikenwalder M., Kurrer M.O. Disseminated and sustained HIV infection in CD34+ cord blood cell-transplanted Rag2−/−gamma c−/− mice. Proc Natl Acad Sci USA. 2006;103:15951–15956. doi: 10.1073/pnas.0604493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang L., Kovalev G.I., Su L. HIV-1 infection and pathogenesis in a novel humanized mouse model. Blood. 2007;109:2978–2981. doi: 10.1182/blood-2006-07-033159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Naas T., Ghorbani M., Alvarez-Maya I., Lapner M., Kothary R., De Repentigny Y. Characterization of liver histopathology in a transgenic mouse model expressing genotype 1a hepatitis C virus core and envelope proteins 1 and 2. J Gen Virol. 2005;86:2185–2196. doi: 10.1099/vir.0.80969-0. [DOI] [PubMed] [Google Scholar]
- 93.Alonzi T., Agrati C., Costabile B., Cicchini C., Amicone L., Cavallari C. Steatosis and intrahepatic lymphocyte recruitment in hepatitis C virus transgenic mice. J Gen Virol. 2004;85:1509–1520. doi: 10.1099/vir.0.19724-0. [DOI] [PubMed] [Google Scholar]