Abstract
Manganese (Mn) is the second most prevalent transition metal in the Earth's crust but its availability is often limited due to rapid oxidation and low mobility of the oxidized forms. Acclimation to low Mn availability was studied in Arabidopsis seedlings subjected to Mn deficiency. As reported here, Mn deficiency caused a thorough change in the arrangement and characteristics of the root epidermal cells. A proportion of the extra hairs formed upon Mn deficiency were located in atrichoblast positions, indicative of a post-embryonic reprogramming of the cell fate acquired during embryogenesis. When plants were grown under a light intensity of >50 μmol m−2 s−1 in the presence of manganese root hair elongation was substantially inhibited, whereas Mn-deficient seedlings displayed stimulated root hair development. GeneChip analysis revealed several candidate genes with potential roles in the reprogramming of rhizodermal cells. None of the genes that function in epidermal cell fate specification were affected by Mn deficiency, indicating that the patterning mechanism which controls the differentiation of rhizodermal cells during embryogenesis have been bypassed under Mn-deficient conditions. This assumption is supported by the partial rescue of the hairless cpc mutant by Mn deficiency. Inductively coupled plasma-optical emission spectroscopy (ICP-OES) analysis revealed that, besides the anticipated reduction in Mn concentration, Mn deficiency caused an increase in iron concentration. This increase was associated with a decreased transcript level of the iron transporter IRT1, indicative of a more efficient transport of iron in the absence of Mn.
Keywords: Ion homeostasis, iron, light regulation, manganese, root hairs, transcriptional profiling
Introduction
Manganese (Mn) is an essential trace element for metabolism in virtually all living organisms and is required as a cofactor or as an activator for an array of enzymes, such as manganese superoxide dismutase (MnSOD), RNA polymerases, malic enzyme, isocitrate dehydrogenase, and PEP carboxykinase (Marschner, 1995). In plants, Mn is required for the oxygen-evolving photosynthetic machinery, catalysing the water-splitting reaction in PSII. The bio-availability of manganese depends on its oxidation state. Only the divalent cation Mn2+ can be readily taken up by plants, the higher oxidation states Mn(III) and Mn(IV) are not accessible. In sandy soils, soils high in organic matter, and in dry well-aerated soils with high pH, the bio-availability of Mn decreases far below the level that is required for normal plant growth. Mn deficiency is a widespread plant nutritional disorder in agriculture, which is difficult to overcome because Mn2+ is rapidly oxidized when supplemented artificially in the form of fertilizers. Visual symptoms of Mn deficiency include the interveinal chlorosis of younger leaves in dicotyledonous plants and grey specks on the basal leaves in cereals (Marschner, 1995).
Photosynthetic autotrophs have evolved specific routes for the entry of Mn2+ into the cell. A so-called ABC-type permease has been reported to be responsible for the uptake of Mn2+ in cyanobacteria (Bartsevich and Parasi, 1995). In the yeast Saccharomyces cerevisiae, the accumulation of Mn2+ is mediated by the natural resistance-associated macrophage protein (NRAMP) family transporter SMF1 and by the high-affinity phosphate (Pi) transporter PHO84 (Jensen et al., 2003). Similarly, in the alga Chlamydomonas an NRAMP protein was identified as the main component of a Mn2+-selective uptake pathway (Allen et al., 2007). No Mn2+-specific transporter has been identified in plants to date. An explanation for this might be provided by the fact that, in plants, Mn2+ shares the same entry route as iron (Fe). In Arabidopsis, Fe is taken up by IRT1, a member of the ZIP transporter family (Eide et al., 1996; Henriques et al., 2002; Varotto et al., 2002; Vert et al., 2002). IRT1 has a relatively broad substrate spectrum and is reportedly capable of transporting other metal ions including Mn2+ (Korshunova et al., 1999). Recently, a transporter of the cation diffusion facilitator (CDF) family, MTP11, was shown to be crucial for maintaining Mn homeostasis in plants (Peiter et al., 2007; Delhaize et al., 2007). However, MTP11 is not expressed in the root epidermis, nor is its transcript level increased upon Mn starvation, indicating that this protein functions in Mn tolerance rather than in Mn acquisition from the soil (Peiter et al., 2007). This assumption is supported by its localization to prevacuolar compartments (Delhaize et al., 2007). A Golgi-localized P2A-type ATPase, ECA3, was shown to be important for Mn homeostasis under Mn-deplete conditions, probably by mediating the loading of Mn into the Golgi (Mills et al., 2008).
Similar to deficiencies in other immobile mineral nutrients such as Fe and phosphate (Pi), it is the low bio-availability of Mn rather than the concentration in the soil that causes Mn shortage. An efficient means of improving the acquisition of immobile nutrients is to enlarge the soil/root interface. Root epidermal cells can differentiate into root hairs, thereby substantially increasing the absorptive surface of the root. In Arabidopsis, the decision of epidermal cells to enter either the root hair cell fate or the non-hair developmental pathway is controlled by positional information. Epidermal cells that lie over a periclinal cortical cell wall (N position) differentiate into a non-hair cell, whereas epidermal cells that are located over the clefs of two cortical cells (anticlinal) walls (H position) develop into a hair cell (Dolan, 2006). The positional information, derived from the inner cell layer(s), is conveyed by SCRAMBLED (SCM), an LRR-receptor-like kinase, which regulates a subset of transcription factors that ultimately determine the fate of the cell (Kwak et al., 2005). An as yet unidentified signal is perceived by SCM, which in turn causes a bias in the expression of the MYB-type transcription factor WEREWOLF (WER), resulting in a slightly lower expression of the gene in trichoblast files (Bernhardt et al., 2005; Koshino-Kimura et al., 2005). In non-hair cells, WER forms a complex with the R-like bHLH proteins GLABRA3 (GL3), ENHANCER OF GLABRA3 (EGL3), and with the WD40 protein TRANSPARENT TESTA GLABRA1 (TTG1). This complex then promotes the expression of the single-repeat MYB protein CAPRICE (CPC) and of the homeodomain leucine zipper protein GLABRA2 (GL2). GL2 acts as a positive regulator of the non-hair cell fate and blocks the formation of hairs in the N position. The movement of CPC from non-hair cells to hair cells enables the protein to compete with WER to form a complex composed of CPC/GL3/EGL3 and TTG1, which in turn blocks the expression of CPC and GL2 in future hair cells. The cell specification in the root epidermal cells is strengthened by the movement of GL3/EGL3 from hair cells to non-hair cells.
Growth of plants in a medium with low availability of Fe or Pi increases the number of root hairs and alters their characteristics in a manner typical of each growth type (Müller and Schmidt, 2004). Based on pharmacological and genetic evidences, it was shown previously that the signalling pathways, which ultimately lead to the formation of extra root hairs, differ between Pi and Fe deficiency (Schmidt and Schikora, 2001; Müller and Schmidt, 2004). The formation of additional root hairs has also been reported for Mn-deficient plants (Ma et al., 2001; Konno et al., 2006), but no information on how Mn deficiency is perceived and translated into changes in the root epidermal pattern is available. It is reported here that Mn deficiency induces a unique root hair phenotype comprising changes in the length, characteristics, and in the position of root hairs relative to the underlying cortical cells. The Mn deficiency-induced phenotype is dominant over the inhibition of root hair elongation by high light intensities and distinguishable from previously observed changes in root hair patterning in response to other nutrient deficiencies. Based on these findings, we suppose that a Mn-specific signalling pathway is induced below a certain threshold level of external Mn, which triggers post-embryonic developmental processes that acclimatize the plant to low Mn availability. Using GeneChip analysis, potential components of the Mn stress response in Arabidopsis roots were revealed.
Materials and methods
Plant material and mineral nutrients
Plants were grown in a growth chamber on an agar medium as described by Estelle and Somerville (1987). Seeds of Arabidopsis (Arabidopsis thaliana L. Heynh), ecotype Col-0 and cpc were obtained from the Arabidopsis Biological Resource Center (Ohio State University) and surface-sterilized by immersing them in 5% (v/v) NaOCl for 5 min and 96% ethanol for 7 min, followed by four rinses in sterile water. The medium was composed of (mM): KNO3 (5), MgSO4 (2), Ca(NO3)2 (2), KH2PO4 (2.5), (μM): H3BO3 (70), MnCl2 (14), ZnSO4 (1), CuSO4 (0.5), NaCl (10), Na2MoO4 (0.2), and FeEDTA (40), solidified with 0.3% Phytagel (Sigma-Aldrich). Sucrose (43 mM) and 4.7 mM MES were included and the pH was adjusted to 5.5. Seeds were placed onto Petri dishes containing agar medium either with (+Mn plants) or without Mn (–Mn plants) and kept for 1 d at 4 °C in the dark, before being transferred to a growth chamber and grown at 21 °C under continuous illumination (70 μmol m−2 s−1, Phillips TL lamps). Light intensity was varied as indicated by shading with layers of Miracloth (Calbiochem Biosciences, La Jolla, CA), which did not alter the light quality. Mn concentration was varied as indicated. Plants were analysed 6 d after sowing. For gene expression analysis, roots were harvested and immediately frozen in liquid nitrogen.
RNA analysis and real-time RT-PCR
Total RNA was isolated from roots of 100 plants with the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer's instructions. Nucleic acid quantity was evaluated by using a NanoDrop ND-1000 UV-Vis Spectrophotometer (NanoDrop Technologies, Wilmington, USA). One μg of total DNase-treated RNA (Turbo DNase, Ambion) was reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen) with oligo dT primers in a total volume of 20 μl. Real-time quantitative PCR was performed using double-stranded DNA binding dye Sybr® Green PCR Master mix (Applied Biosystems) in an ABI GeneAmp 7000 Sequence Detection System. Each reaction was run in triplicate and the melting curves were constructed using Dissociation Curves Software (Applied Biosystems), to ensure that only a single product is amplified. Validation experiments were performed to test the efficiency of the target amplification and the efficiency of the reference amplification. Duplicate CT values were analysed with Microsoft Excel using the comparative CT(ΔCT) method as described by the manufacturer (Applied Biosystems). The amount of target (2–ΔΔCT) was obtained by normalizing to an endogenous reference (Alpha tubulin, At5g19770) and relative to a calibrator (control tissue). Relative expression (n-fold) of the normalized target gene in the treatment was calculated according to the mathematical model proposed by Pfaffl (2001). Primers were designed by the Primer Express program (Applied Biosystems). Specificity of the primers was ensured through sequence alignment by the BLASTN algorithm (Altschul et al., 1997).
The following primers were used for the validation of the microarray genes. 5′-AACACAAGAAGGTGGAGCAAGTC-3′ and 5′-TTTCCTTCATTGCGCTCTTCA-3′ were used for At1g33890; 5′-CGAGAGGTAACCAAATCGCAAT-3′ and 5′-TGTGGTTGATTCAACCAGCTGT-3′ for At1g56160; 5′-CTTCCGTCGTTCTTGCCTC TT-3′and 5′-GGATCAGACGAAACCAAACGAG-3′ for At1g56430; 5′-CGAACGAGGCTGCTCTTTTG-3′ and 5′-TGGTAGTGGCTCGCAGCATA-3′ for At1g73800; 5′-AGACGACAGAAACAGCCAAGGA-3′ and 5′-TCACGGATTCCACAGACTCCAT-3′ for At1g79840; 5′-TGGTGGATACAGGGAGGAATG-3′ and 5′-TTTGGACCGAACTGAGATAAACC-3′ for At2g24710; 5′-CGATTCTCCAGCTGCTGATGA-3′ and 5′-CCGAGCAACAAAACAATGCA-3′ for At2g20520; 5′-GCGGAGCATAGGGTTCGAA-3′ and 5′-GGGATTTGGTGCCGTGAA-3′ for At3g12900; 5′-GCATGCTT CACCAGCGATG-3′ and 5′-TGCCCAATGGAATGACCACT-3′ for At3g53280; 5′-AATCTAAACCTCAAGGAATATCTGAAAGA-3′ and 5′-ACCCGTGGTCGGTCAAAA-3′ for At4g02900; 5′-CCGATGTTCGAGATGGGATAG-3′ and 5′-CAGCGGCACAAACAACTGTAA-3′ for At4g10510; 5′-TGCTTCGCAGCTGACGAAG-3′ and 5′-CATTCCGGAGGCATAACACCT-3′ for At4g28850; 5′-GCGATGTTGACCGCACAA-3′ and 5′-ACAAAGGGCGGTTGCTGTT-3′ for At4g37050; 5′-AACACGAAGACCGAACGAAT-3′ and 5′-GTGCTGAAGGTGGAGACGAT-3′ for At5g19770; 5′-ACGTGTTTGTGTTCCCCATAGG-3′ and 5′-CAATCGTGATGACAC CAGCATT-3′ for At5g39110.
Microarray experiments
The Affymetrix GeneChip Arabidopsis ATH1 Genome Array was used for cDNA microarray analysis. Total RNA from roots of control and –Mn plants was isolated as described above. All RNA samples were quality assessed by using the Agilent Bioanalyser 2100 (Agilent). cRNA synthesis was performed by use of the GeneChip One-Cycle Target Labeling Kit (Affymetrix). GeneChips were hybridized with 15 μg of fragmented cRNA. Hybridization, washing, staining, and scanning procedures were performed as described in the Affymetrix technical manual.
Microarray data analysis
Data from the mircoarray experiments were imported directly into GeneSpring (version 7.0, Agilent). The software was used to normalize data per chip, to the 50th percentile and per gene to the control samples. The data were then filtered using the following: (i) by removing genes that were flagged as absent in at least three replicates; (ii) by expression level to remove those genes that were deemed to be unchanging between log values 0.8 and 1.2 (>1.5-fold difference); and (iii) by confidence using a one-sample t test and a P-value cut-off of 0.05 so that any gene with a P-value of 0.05 or less when compared to the normalized control was regarded as statistically significant, i.e. up- or down-regulated compared to the expression baseline of 1. To determine those genes that were statistically differentially expressed between groups of samples, GeneSpring’s ANOVA statistical analysis function (a Welch t test) was applied to the filtered data set.
Ion content determination
Elemental analysis was carried out by inductively coupled plasma (ICP) atomic absorption with a Perkin Elmer Optima 5300 DV optical emission spectrometer (OES) on 6-d-old seedlings. Two batches of c. 900 plants were grown and treated independently for replicates. For root analysis, two batches were pooled. Samples were digested in nitric acid in a microwave digestion unit (CEM, Matthews, USA). Tomato leaves were used as standard reference material.
Light microscopy
Stereomicroscopy, using a Discovery Z12 (Zeiss, Göttingen, Germany) fitted with an eye piece scale bar, was used to count the number of root hairs per mm along the length of the root. The root remained within the Petri dish in the media during counting. Root hair patterns were analysed in cross-sections for control and –Mn plants. The root samples were fixed, dehydrated, and then embedded in Technovit 7100 (Heraeus Kulzer, Wehrheim) resin in gelatine capsules, in accordance with the manufacturer's instructions. Transverse sections (6 μm) were cut using a RM 2255 Leica microtome (Leica, Nussloch, Germany). Sections were dried and stained with toluidine blue (0.05%) on glass slides and examined using bright-field on an Axioskop 40 (Zeiss, Jena, Germany) microscope. The number of cortical and epidermal cells and the rate of root hairs in the H and N positions root hairs were counted in 60 cross-sections. The bifurcated root hair number was determined by mounting the root in 10% glycerol and examining the root hairs along the length of the root. Thirty roots were used for examining control and –Mn-treated plants.
Confocal microscopy
Seedlings were placed in 10 mg ml−1 propidium iodine solution (PI) for 1 min. The seedling was gently rinsed with water for 2 min. The root was removed and mounted in fresh water. The roots where then observed using a Confocal Laser Scanning Microscope (Zeiss LSM510 Meta). The peak excitation λ and emission λ for PI is 536 nm and 620 nm, respectively.
The cells lengths of trichoblasts and atrichoblasts were measured using ImageJ (http://rsb.info.nih.gov/ij/). The position of each cell was calculated from the cumulative length of all cells between it and the quiescent centre. The data sets were then smoothed and interpolated into 25 mm spaced data points using a kernel-smoothing routine (Beemster and Baskin, 1998); this was done by using it as a macro in Microsoft Excel (version 97), enabling the calculation of the average between replicate roots.
Results
Mn deficiency changes the patterning of root epidermal cells
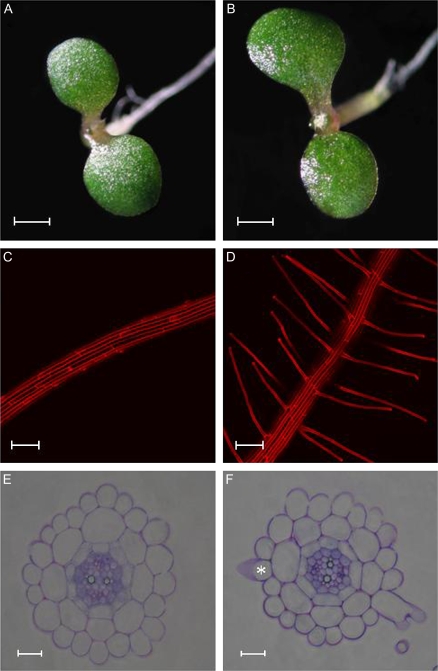
Experiments were conducted to characterize the developmental and molecular responses of Arabidopsis roots to changes in the availability of external Mn. Growing seedlings on Mn-free medium substantially increased the frequency of root hairs and altered their characteristics (Fig. 1C, D; Table 1). The response to Mn deficiency was evident in seedlings 6 d after germination and growth on Mn-free media. No visible symptoms of Mn deficiency were evident in above-ground tissues at this stage (Fig. 1A, B). A substantial percentage of Mn deficiency-induced root hairs were bifurcated with branching starting at the base of the hairs (Fig. 1; Table 1). Besides initiating root hair elongation from cells in the hair (H) position, Mn deficiency also induced the differentiation of epidermal cells in the non-hair (N) position into root hairs (ectopic hairs; Fig. 1F; Table 1), indicative of a change in cell fate. The increase in root hair number in the H position was not the result of an increase in the cortical cell number and, subsequently, in the number of epidermal cells in the hair position (Table 1).
Fig. 1.
Phenotype of shoots and roots and cross sections from 6-d-old control (A, C, E) and Mn-deficient plants (B, D, F). Control plants were grown on media containing 14 μM Mn. Note that root hairs of Mn-deficient plants are frequently branched at their base. The asterisk indicates a developing root hair in an ectopic position. Scale bars (A, B) 1 mm, (C, D) 100 μm, (E, F) 15 μm.
Table 1.
Effects of Mn deficiency on root morphological parameters
| Treatment | Epidermal cell no. | Cortical cell no. | Elongated root hairs in H position | Elongated root hairs in N position | Branched root hairsa |
| Control | 22.2±0.1 | 8.0±0.0 | 0 | 0 | 0 |
| Mn-deficient | 22.5±0.1 | 8.1±0.0 | 1.3±0.1 | 0.4±0.1 | 6.5±0.4 |
Values represent the no. (means ±SE) of the indicated cell type per cell layer. Fifty cross-sections from five roots were scored for each treatment. Control plants were grown on media containing 14 μM Mn. Plants were grown at high light intensity (70 μmol m−2 s−1) and analysed 6 d after sowing.
The number of branched root hairs is given as a percentage of the total root hairs.
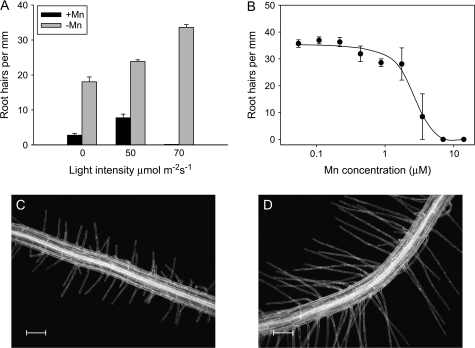
Light intensity affected the elongation of root hairs of both Mn-deficient (–Mn) plants and plants grown in the presence of Mn (+Mn) (Fig. 2). For +Mn seedlings, the highest root hair density observed was at a PAR of 50 μmol m−2 s−1; higher light intensities almost completely inhibited the elongation of the hairs without affecting their initiation (Figs 1, 2). For root hair counting, an epidermal cell was scored as a root hair cell if any protrusion was visible, regardless of its length. The effect of light was due to a direct impact on the roots, since shielding the roots from light by blackening the medium with charcoal prevented the effect (data not shown). In addition to the higher frequency, root hairs formed in –Mn plants were ∼50% longer than those of control plants when grown at light intensities of ≤50 μmol m−2 s−1 (0.52 mm versus 0.25 mm on average, Fig. 2C, D). The difference between +Mn and –Mn plants was less pronounced in older (8-d-old) seedlings. At this age, +Mn plants showed less restricted root hair elongation at a light intensity of 50 μmol m−2 s−1 but developed fewer hairs at higher light intensities (data not shown). In contrast to the control plants, Mn deficiency-induced root hair formation was stimulated by higher light intensity. At 70 μmol m−2 s−1, the difference in root hair elongation between +Mn and –Mn plants was most pronounced, and all subsequent experiments were performed under the latter light regime. Bifurcated or ectopic root hairs were not observed under +Mn conditions regardless of the age or light conditions. The Mn concentration threshold for inducing root hairs under this light intensity was around 3 μM in the growth medium (Fig. 2B). Above this concentration, root hair elongation was inhibited under high light conditions.
Fig. 2.
Effect of light intensity (A) and Mn concentration (B) on root hair formation. The effect of Mn concentration on the formation of root hairs was investigated at high light intensity (70 μmol m−2 s−1). Mn-sufficient plants were grown on media containing 14 μM Mn. Plants were analysed 6 d after sowing. Error bars represent standard deviation of means from the measurements. (C) Root hair phenotype of control and Mn-deficient plants (D) grown at a light intensity regime of 50 μmol m−2 s−1.
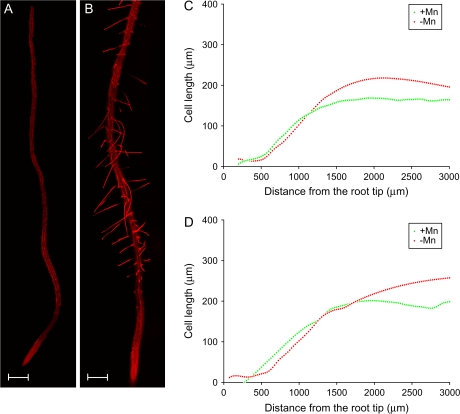
Analysis of longitudinal cell elongation revealed that, relative to the control plants, the length of root cells in the meristem in Mn-deficient seedling were smaller, possibly due to a higher rate of cell division (Fig. 3). No differences were observed in the average root length between +Mn and –Mn plants (data not shown). More distant from the tip, cells from plants grown in the absence of Mn were found to be longer than those of control plants. This effect was more pronounced in atrichoblasts compared to trichoblasts (Fig. 3C, D). The differences in the length of the epidermal cells between –Mn and +Mn is not sufficient to account for the differences in root hair density.
Fig. 3.
Longitudinal cell length of trichoblasts (C) and atrichoblasts (D) of control and Mn-deficient Arabidopsis seedlings. Plants were grown at high light intensity (70 μmol m−2 s−1). Cell length was analysed from compound pictures of roots (A, B). Data in (C) and (D) are averages of five roots per treatment. Control plants were grown on media containing 14 μM Mn. Plants were analysed 6 d after sowing. Scale bars, 200 μm.
Mn deficiency changes the gene expression profile in roots
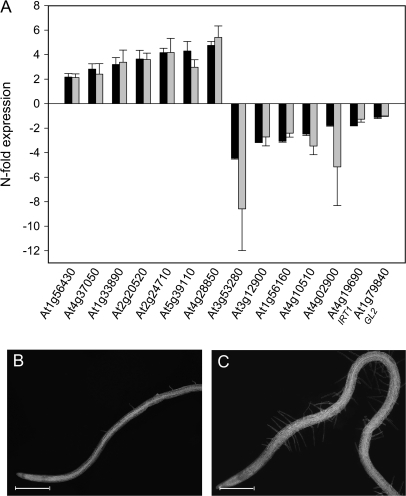
RNA samples were labelled and analysed for expression profiles using the Affymetrix ATH1 GeneChip. Three biological replicates from roots of +Mn and –Mn were examined. The complete datasets are available as supplementary material (see Supplementary Table S1 at JXB online). A total of 71 genes were classified as being differentially expressed in response to Mn deficiency (see Materials and methods for data analysis), the most prominent functional groups being related to the cell-wall, signalling, and transcriptional regulation (Table 2). Nine genes were found to overlap with the root hair transcriptome (Jones et al., 2006). To validate the GeneChip data, the expression of 12 genes have been selected and further analysed by real-time RT-PCR. RNA preparations from four batches of independently grown plants were used for each gene. In addition, two genes that are relevant to this study, namely the iron transporter IRT1 and the homeodomain gene GL2, both of which have not been classified as differentially expressed in Mn-deficient plants, have been included in our analysis. Quantification of the transcript abundance of these genes by RT-PCR revealed a close correlation of message levels with the GeneChip signals. This was also true for the two genes that were not classified as being differentially expressed according to the parameters set for the data analysis (IRT1 and GL2). On average, the variation in total fold-change value between the RT-PCR analysis and the GeneChip signals was 12% (4% for up-regulated and 28% for down-regulated genes). The consistency among the different RNA samples and a comparison to the GeneChip data is shown in Fig. 4A.
Table 2.
Groups of functionally related genes that are differentially regulated in response to Mn deficiency
| Function | Gene locusa | Annotation | Fold change |
| Cell wall-related | At3g60270 | Putative uclacyanin | 5.81 |
| At4g28850* | Xyloglucan endotransglycosylase-like protein | 5.24 | |
| At2g20520* | Fasciclin-like arabinogalactan-protein 6 (FLA6) | 3.55 | |
| At3g01270 | Pectate lyase family protein | 3.07 | |
| At3g20850* | Proline-rich family protein; similar to extensin precursor | 3.02 | |
| At5g39110 | Putative germin-like protein; manganese-ion binding | 2.83 | |
| At1g26250 | Putative proline-rich extensin | 2.46 | |
| At2g24800 | Putative peroxidase | 2.44 | |
| At5g26080* | Proline-rich family protein | 2.25 | |
| At5g57540* | Xyloglucan endotransglycosylase | 2.09 | |
| At2g05520 | Glycine-rich protein (GRP3) | 2.05 | |
| At1g77100 | Putative peroxidase | –2.13 | |
| At1g16160 | WAK-like kinase (WAKL5) | –4.49 | |
| Signalling | At2g24710 | Glutamate receptor 2 (ATGLR2) | 3.85 |
| At1g33890* | Avirulence-responsive protein | 3.13 | |
| At5g58890 | MADS-box family protein | 2.60 | |
| At3g01420 | Pathogen-responsive α-dioxygenase | 2.28 | |
| At1g66500 | Zinc finger (C2H2-type) family protein | 2.11 | |
| At1g29730 | Protein serine/threonine kinase | –2.05 | |
| At4g21210 | Protein kinase | –2.06 | |
| At4g01190 | Phosphatidylinositol phosphate kinase 10 (ATPIPK10) | –2.30 | |
| At1g19960 | Similar to transmembrane receptor | –2.35 | |
| At4g02900 | Early-responsive to dehydration protein-related | –3.51 | |
| Transcriptional regulators and nucleic acid interacting | At1g76420 | No apical meristem (NAM) family protein | 7.35 |
| At2g40350 | ERF/AP2 transcription factor | 2.13 | |
| At5g18090 | Transcriptional factor B3 family protein | 2.11 | |
| At1g34180 | No apical meristem (NAM) family protein | 2.04 | |
| At1g68240 | Transcription factor | 2.02 | |
| At4g18650 | Transcription factor-related | –2.07 | |
| At3g15605 | Nucleic acid binding | –2.30 | |
| At1g56160 | Myb family transcription factor (MYB72) | –2.36 | |
| At1g50350 | Similar to zinc finger (C3HC4-type RING finger) family protein | –2.81 | |
| Protein modification | At4g10550* | Subtilase family protein | 2.00 |
| At3g61930 | Expressed protein, N-terminal myristoylation | –2.25 | |
| At1g79310 | Putative latex-abundant protein | –2.76 | |
| At4g10510 | Subtilase family protein | –3.29 | |
| Transport | At4g12360 | Protease inhibitor/seed storage/lipid transfer family protein | 2.87 |
| At4g25220a | Putative glycerol-3-phosphate permease | 2.79 | |
| At4g13420 | Potassium transporter (HAK5) | 2.60 | |
| At5g49390 | Hydrogen ion transporting ATP synthase | 2.03 | |
| At3g58060 | Cation efflux family protein | –2.05 | |
| At1g04600 | Myosin-like protein XIA ATXIA | –2.49 | |
| At1g08270 | Similar to AAA-type ATPase family protein | –3.41 | |
| Metabolism | At4g37050 | Patatin-like protein 4 (PLA V/PLP4) | 4.78 |
| At2g34490 | Cytochrome P450 (CYP710A2) | 4.42 | |
| At1g34520 | Long-chain-alcohol O-fatty-acyltransferase family protein | 3.57 | |
| At1g78360 | Glutathione transferase (ATGSTU21) | 2.62 | |
| At1g02940 | Glutathione transferase (ATGSTF5) | 2.24 | |
| At1g34540* | Cytochrome P450 (CYP94D) | 2.13 | |
| At1g56430 | Putative nicotianamine synthase | 2.10 | |
| At1g06350 | Fatty acid desaturase family protein | 2.09 | |
| At1g67980 | S-adenosyl-L-methionine: transcaffeoyl | –2.52 | |
| Coenzyme A 3-O-methyltransferase | |||
| At3g12900 | Oxidoreductase, 2OG-Fe(II) oxygenase family protein | –2.53 | |
| At3g53280 | Cytochrome P450 (CYP71B5) | –7.00 |
An asterisk indicates genes that overlap with the root hair transcriptome described by Jones et al. (2006).
Fig. 4.
Validation of the GeneChip signals by real-time RT-PCR (A) and effect of Mn deficiency on the formation of root hairs in the cpc mutant (B, C). Gene expression analysis was performed with four (five for IRT1) RNA preparations from independently grown plants. Black bars indicate means of real-time PCR data, grey bars represent means of normalized GeneChip signals. Error bars shows standard deviation of the means. cpc mutants were grown in either media containing 14 μM Mn or in media deprived of Mn. Plants were analysed 6 d after sowing.
The expression of cell specification genes is not affected by Mn deficiency
Changes in epidermal cell specification are supposed to be associated with differential expression of genes encoding primary determinants of the hair or non-hair fate. Decisions to enter either cell fate are controlled by the WER/GL3/EGL3/TGG gene complex which regulates the expression of CPC and GL2 as positive and negative regulators of hair fate (Bernhardt et al., 2005). None of the cell specification genes were significantly affected in our experiments, suggesting that the patterning mechanism that is active under normal conditions is bypassed under Mn-deficient conditions (see Supplementary Table S2 at JXB online). Overexpression of the receptor kinase gene SCM caused the formation of ectopic root hairs (Kwak and Schiefelbein, 2007). SCM is thus a potential candidate for changing the differentiation of epidermal cells in the N position. A tendency for higher expression of SCM in roots of Mn-deficient plants was noted in all three experiments, but was not statistically significant.
The cpc mutant expresses GL2 in all epidermal cells and forms very few root hairs under control conditions (Wada et al., 1997; Fig. 4B). Under conditions of Mn deficiency, the mutant is partly rescued, further supporting the suggestion that the extra hairs formed upon Mn starvation are not or only partly controlled by the GL2-dependent, basic patterning mechanism (Fig. 4C).
Mn deficiency induces changes in ion homeostasis
Branched root hairs are characteristic of Fe-deficient roots (Müller and Schmidt, 2004). Thus, the bifurcated hairs formed in response to Mn deficiency may be caused by limited Fe availability under Mn-deficient conditions. Mn deficiency was found to induce secondary Fe deficiency in the alga Chlamydomonas (Allen et al., 2007). However, none of the Fe-responsive genes were up-regulated under Mn deficiency, making such a scenario unlikely (see Supplementary Table S2 at JXB online). In fact, genes that were found to be up-regulated in Fe-deficient plants by microarray analysis (Wintz et al., 2003; Colangelo and Guerinot, 2004), were slightly down-regulated under Mn deficiency. A pronounced decrease in transcript abundance under Mn starvation was observed for the iron transporter IRT1. A lower expression of IRT1 in roots of Mn-deficient plants was confirmed by real-time RT-PCR analysis in five independent RNA isolations (Fig. 4A). In addition, the ATFER1 gene, encoding the iron storage protein ferritin also shows higher transcript abundance under Mn-deficient conditions (see Supplementary Table S2 at JXB online), suggesting an alteration in the cellular iron homeostasis.
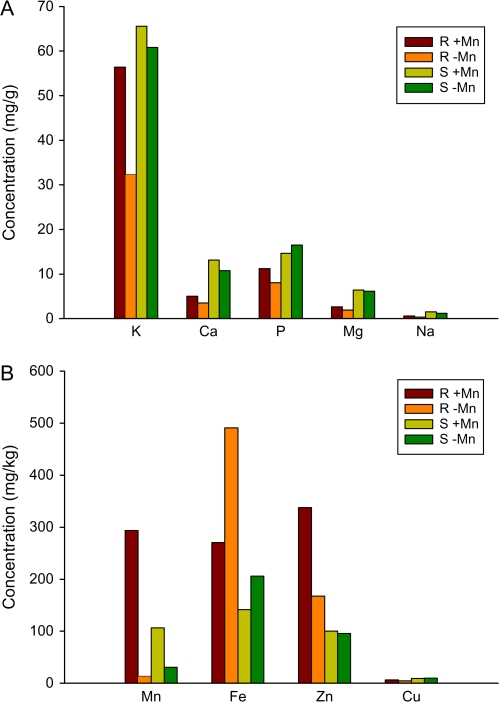
The consequences of Mn deficiency on ion concentration were measured by inductively coupled plasma-optical emission spectroscopy (ICP-OES). Manganese concentrations were dramatically reduced in Mn-deficient plants in both root and leaf tissue (Fig. 5). Concurrent with our GeneChip results, the concentration of Fe was markedly higher in Mn-deficient roots, leaf tissue by comparison, showed a significant but less elevated Fe level. Taking into account of the changes in iron status, some genes that were found to be repressed by Mn deficiency may represent secondary effects of an altered Fe homeostasis rather than changes induced primarily as a result of Mn deficiency. For example, the cytochrome P450-like mono-oxygenase gene At4g31940 was found to be significantly down-regulated under –Mn conditions (see Supplementary Table S2 at JXB online), but dramatically up-regulated in response to Fe deficiency (Colangelo and Guerinot, 2004). Similarly, HMA3, MYB72, and the subtilidase gene At5g03570 were all down-regulated upon Mn starvation, but were found to be highly up-regulated under Fe deficiency (Colangelo and Guerinot, 2004; TJ Buckhout and W Schmidt, unpublished data).
Fig. 5.
Analysis of mineral ion concentration of roots and shoots from control and Mn-deficient plants. (A) Macronutrients, (B) micronutrients. Control plants were grown on media containing 14 μM Mn. Plants were analysed 6 d after sowing. R, roots, S, shoots.
Consistent with the assumption that Mn deficiency causes a down-regulation of genes involved in Fe acquisition, in particular IRT1, the root concentration of Zn2+ was also reduced by approximately 50% in Mn-deficient plants. IRT1 transports both Mn2+ and Zn2+ (Korshunova et al., 1999) and the reduced Zn levels may be caused by the down-regulation of the IRT1 gene. Apparently, the lower concentration of Zn cannot be balanced by increasing the expression of transporters that are able to complement a yeast strain defective in Zn2+ uptake, such as ZIP1, ZIP2, ZIP3, and ZIP4 (Grotz and Guerinot, 2006). Among these genes only the GeneChip signals for ZIP2 were consistently higher in –Mn plants, suggesting that ZIP2 is Zn-responsive and mediates Zn2+ transport under these conditions. The weak response of Zn2+ transporters may be due to the fact that the Zn status of the leaves was not affected by Mn deficiency (Fig. 5B).
Mn deficiency has been shown to induce secondary P deficiency in Chlamydomonas (Allen et al., 2007) and P-deficient conditions triggers a root hair phenotype resembling that of Mn-deficient plants in some respects (i.e. the formation of long and ectopic root hairs), it was therefore investigated whether Mn deficiency caused changes in the expression of genes that are responsive to P-deficiency. In line with the ICP analysis, no changes in transcript abundance were observed for genes that were reported to be affected by P-deficient conditions (Misson et al., 2005; Bari et al., 2006), suggesting that P homeostasis is not affected by Mn deficiency (see Supplementary Table S2 at JXB online).
Discussion
Mn deficiency may act late in cell specification
Plants have developed sophisticated mechanisms to improve the acquisition of immobile nutrients, comprising of biochemical, physiological, and morphological responses. Whereas substantial progress has been made towards the understanding of how the uptake of Fe and Pi is balanced in plants, the mechanisms of Mn acquisition and the control of Mn homeostasis are poorly understood. A profound result from this study is the unique root hair phenotype that is induced specifically by Mn deficiency, which differs from the root hair phenotypes observed in response to a lack of Pi or Fe (see Schmidt, 2008, for a review). The Mn deficiency phenotype is evident at the seedling stage and the signal appears to overrule the inhibition of root hair elongation by high light intensity, indicating a critical importance of developmental responses for maintaining a sufficient Mn supply. The Mn deficiency-induced changes include the re-differentiation of atrichoblasts into root hair-forming cells as shown by the formation of root hairs in ectopic positions. This indicates that Mn deficiency does not simply promote the elongation of hairs from trichoblasts, but alters the developmental programme of rhizodermal cells.
Under ordinary conditions, root epidermal cell fate is controlled by the combined action of patterning genes, the expression of which is biased by a cortical signal. Surprisingly, none of the genes with functions in cell fate specification were markedly affected by Mn deficiency. Although the possibility cannot be ruled out that minor differences in spatial expression may account for the changes in cell fate and might potentially be below the detection limit in an analysis of the whole root, the GeneChip signals for GL2 were almost identical in control and Mn-deficient roots, a finding which was confirmed by real-time RT-PCR. This does not support the hypothesis that the root hair phenotype is a result of a change in GL2 transcript abundance (see Supplementary Table S2 at JXB online). It can thus be considered that the patterning mechanism active under control condition is bypassed under conditions of Mn deficiency. A similar scenario has been assumed for root hairs induced by phosphate deficiency. The cpc mutant, in which root hair development is restricted as a result of a GL2 being expressed in all epidermal cells rather than in atrichoblasts only (Wada et al., 1997), can be rescued by growing cpc mutant plants in –P media (Müller and Schmidt, 2004). This suggests that the –P signal is perceived downstream of the cell specification cascade. This scenario is supported by a mathematical model congruent with experimental data, assuming a phosphate-responsive activator–inhibitor mechanism that causes a position-independent increase in root hair density that is not directly affected by GL2 and the genes controlling its expression (Savage and Schmidt, 2008). Similar to P deficiency, Mn deficiency partly rescued the cpc phenotype, further supporting the assumption that the Mn-starvation-induced root hairs are not entirely controlled by GL2 (Fig. 4B, C). Superimposition of environmental signals over the cell specification mechanism that is active during embryogenesis has previously been demonstrated. For example, hormones such as ethylene can alter the cell fate without changing the expression of key determinants of the epidermal cell fate (Masucci and Schiefelbein, 1996). It thus appears that plasticity of the root hair pattern is conferred by a downstream mechanism that adapts the number and characteristics of root hairs to the prevailing conditions.
Cross-talk of light signalling and root hair development
An unexpected finding was the effect of light intensity on the root hair phenotype. A small increase in light intensity from 50 μmol m−2 s−1 to 70 μmol m−2 s−1 caused a marked decrease in root hair elongation under control conditions, but had no effect when plants were grown on media deprived of Mn. It should be noted that even the higher light intensity can be considered as being sub-optimal for Arabidopsis. The lack of light-induced restriction of root hair elongation was also observed in phosphate-deficient plants (data not shown), suggesting that nutrient starvation-induced root hair development interacts with the components of the light signalling pathway. The light effect was dependent on the developmental stage and more pronounced in younger seedlings. A connection between light signalling pathways and signalling cascades involved in root development has been reported in a number of studies. For example, in seedlings carrying a mutation in the bZIP gene HY5, a positive regulator of photomorphogenic responses, the number and length of lateral roots, and the length of root hairs were found to be increased (Oyama et al., 1997). CONSTANS-LIKE3 (COL3), a protein interacting with COP1 (CONSTITUTIVE PHOTOMORPHOGENIC1) that mediates the ubiquitin-dependent degradation of HY5 and other transcription factors, regulates light-dependent developmental processes and affects the development of lateral roots (Gyula et al., 2003; Datta et al., 2005). A promotive effect of light intensity on root hair formation in Arabidopsis has been reported by De Simone et al. (2000), and was shown to be reduced in phyA and phyB mutants. AtMYC2 interacts with light-responsive elements and thereby negatively regulates the expression of light-regulated genes. AtMYC2 has been show to act as a positive regulator of lateral root formation (Yadav et al., 2005), and as a negative regulator of the formation of root hairs (Ryosuke et al., 2003), suggesting that AtMYC2 has multiple roles in root development. Interestingly, red light was shown to induce genes involved in root hair differentiation and elongation, such as CPC and RHD3 (Molas et al., 2006). Together with the data in the present study, it appears that many players in light signalling also play an important role in the environmentally induced changes in root developmental programmes. More work is required to elucidate the cross-talk of light and other environmental signals.
The Mn deficiency phenotype is not caused by secondary Fe deficiency
In contrast to the model alga Chlamydomonas, in which transporters such as NRAMP1 and a member of the cation efflux family were found to be responsive to Mn deficiency (Allen et al., 2007), none of the genes encoding candidate Mn2+ transporters such as ZIPs, OPTs or NRAMPs were induced in the absence of Mn in Arabidopsis roots. NRAMPs are a class of integral membrane proteins that can transport a broad range of metals (Cellier et al., 1995). The NRAMPs 1, 3, and 4 can complement yeast mutants defective in Mn2+ uptake (Curie et al., 2000; Thomine et al., 2000), and are therefore likely candidates for a Mn2+ transporter in Arabidopsis roots. However, none of the NRAMPs was up-regulated in roots of –Mn plants (see Supplementary Table S2 at JXB online). Thus, increased uptake of Mn2+ under conditions of low Mn availability could either be achieved by post-transcriptional activation of transition metal transporters, or Mn2+ uptake might be mediated by a transporter yet to be identified. The iron transporter IRT1 has relatively low substrate specificity and transports Mn2+ as well, which may represent an alternative route of entry for Mn2+ into the root. In any case, the Mn-deficiency-induced formation of root hairs is not due to secondary Fe deficiency; IRT1 was significantly down-regulated in Mn-deficient roots, most likely as a consequence of the high Fe concentration under these conditions (Figs 4A, 5B). Iron overload is further indicated by higher signals on the GeneChips for FER1 (see Supplementary Table S2 at JXB online). In Chlamydomonas on the other hand, Mn deficiency induces secondary Fe deficiency, probably as a consequence of reduced Fe uptake in order to avoid oxidative stress due to reduced activity of MnSODs (Allen et al., 2007).
An alternative route for the uptake of Mn2+ could be represented by OPT3, a gene that has been found to be dramatically up-regulated by Fe and Mn deficiency (Wintz et al., 2003). Although in the present study only a relatively small increase in transcript abundance was noted, a role for OPT3 in Mn homeostasis could be inferred from the fact that OPT3 behaves differently from IRT1 under Mn deficiency. However, OPT3 was shown to be preferentially expressed in the vasculature and to be absent in root hairs and root tips (Stacey et al., 2006). Thus a function of OPT3 in the uptake of Mn2+ from the soil solution appears unlikely.
The formation of root hairs may be crucial for Mn uptake efficiency
Only relatively few genes are induced in response to Mn deficiency, indicating that the plant's innate response to counteract the nutritional imbalance under these conditions is limited. This suggests that the evolutionary force to develop a Mn deficiency response syndrome was low and the situation is not very common in natural habitats. Apparently, the capacity for Mn2+ uptake under most conditions appears to exceed the demand for the metal by far (Clarkson, 1988). The lack of a specific physiological response to Mn deficiency, which aids in the acquisition of Mn2+ and the robust root hair phenotype of Mn-deficient plants, implies that developmental acclimations are more important for the acquisition of Mn. A lack of physiological reactions such as increased expression of a gene coding for a transporter protein may be due to the fact that, in most situations, suboptimal availability of Mn is associated with Fe-deficient conditions. Similar to Fe, the availability of Mn decreases with increasing pH (Marschner, 1995). Induction of the complex Fe starvation syndrome of strategy I plants will mobilize Mn in addition to Fe and the uptake of Mn2+ is then mediated by IRT1. The formation of extra root hairs may represent a ‘rescue’ back-up, which is induced when the Fe deficiency response is not triggered by low availability of Fe. The situation is apparently different from that of Chlamydomonas. Therefore, sophisticated mechanisms to balance Fe and Mn homeostasis, to avoid oxidative stress in the absence of Mn, are probably less important in plants due to the presence of Fe-dependent SODs in addition to MnSOD. It remains to be elucidated how the absence of Mn is sensed and translated into the changes in root epidermal patterning.
Supplementary data
The following materials are available at JXB online in the online version of this article.
Supplementary Table S1. GeneChip data from roots of control and Mn-deficient plants
Supplementary Table S2. Groups of functionally related genes that are differentially regulated in response to Mn deficiency
Acknowledgements
Affymetrix GeneChip assays were performed by the Affymetrix Gene Expression Service Laboratory (http://ipmb.sinica.edu.tw/affy/), supported by Academia Sinica. We thank TJ Buckhout (Humboldt University Berlin) for helpful comments on microarray data analysis, and KC Yeh (ABRC, Taipei) for his help with ICP-OES analysis. This work was supported by a grant to WS from the National Science Council
Note Added in Proof
While this article was in press, Pedas et al. (2008) reported that a gene of the ZIP family with high similarity to OsIRT1, designated HvIRT1, is controlling manganese uptake in barley roots.
Pedas P, Ytting CK, Fuglsang AT, Jahn TP, Schjoerring JK, Husted S. 2008. Manganese efficiency in barley: identification and characterization of the metal ion transporter HvIRT1. Plant Physiology (published online 9 July 2008; doi:10.1104/pp.108.118851)
Supplementary Material
References
- Allen MD, Kropat J, Tottey S, Del Campo JA, Merchant SS. Manganese deficiency in Chlamydomonas results in loss of photosystem II and MnSOD function, sensitivity to peroxides, and secondary phosphorous and iron deficiency. Plant Physiology. 2007;143:263–277. doi: 10.1104/pp.106.088609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Pant BD, Stitt M, Scheible WR. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiology. 2006;141:988–999. doi: 10.1104/pp.106.079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsevich VV, Parasi HB. Molecular identification of an ABC transporter complex for manganese: analysis of a cyanobacterial mutant strain impaired in the photosynthetic oxygen evolution process. EMBO Journal. 1995;14:1845–1853. doi: 10.1002/j.1460-2075.1995.tb07176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GTS, Baskin TI. Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiology. 1998;116:1515–1526. doi: 10.1104/pp.116.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt C, Zhao MZ, Gonzalez A, Lloyd A, Schiefelbein J. The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development. 2005;132:291–298. doi: 10.1242/dev.01565. [DOI] [PubMed] [Google Scholar]
- Cellier M, Privé G, Belouchi A, Kwan T, Rodrigues V, Chia W, Gros P. Nramp defines a family of membrane proteins. Proceedings of the National Academy of Sciences, USA. 1995;92:10089–10093. doi: 10.1073/pnas.92.22.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson DT. Solute transport in plant cells. New York, NY, USA: Longman; 1988. [Google Scholar]
- Colangelo EP, Guerinot ML. The essential basic helix–loop–helix protein FIT1 is required for the iron deficiency response. The Plant Cell. 2004;16:3400–3412. doi: 10.1105/tpc.104.024315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Alonso JM, Le Jean M, Ecker JR, Briat JF. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochemical Journal. 2000;347:749–755. [PMC free article] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi GHCM, Deng XD, Holm M. Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. The Plant Cell. 2005;18:70–84. doi: 10.1105/tpc.105.038182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone S, Oka Y, Inoue Y. Effect of light on root hair formation in Arabidopsis thaliana phytochrome-deficient mutants. Journal of Plant Research. 2000;113:63–69. [Google Scholar]
- Delhaize E, Gruber BD, Pittman JK, White RG, Leung H, Miao Y, Jiang L, Ryan PR, Richardon AE. A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. The Plant Journal. 2007;51:198–210. doi: 10.1111/j.1365-313X.2007.03138.x. [DOI] [PubMed] [Google Scholar]
- Dolan L. Positional information and mobile transcriptional regulators determine cell pattern in the Arabidopsis root epidermis. Journal of Experimental Botany. 2006;57:51–54. doi: 10.1093/jxb/erj037. [DOI] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proceedings of the National Academy of Sciences, USA. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle MA, Somerville C. Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Molecular and General Genetics. 1987;206:200–206. [Google Scholar]
- Grotz N, Guerinot ML. Molecular aspects of Cu, Fe, and Zn homeostasis in plants. Biochimica et Biophysica Acta. 2006;1763:595–608. doi: 10.1016/j.bbamcr.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Gyula P, Schäfer E, Nagy F. Light perception and signalling in higher plants. 2003;6:446–452. doi: 10.1016/s1369-5266(03)00082-7. [DOI] [PubMed] [Google Scholar]
- Henriques R, Jasik J, Klein M, Martinoia E, Feller U, Schell J, Pais MS, Koncz C. Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defect. Plant Molecular Biology. 2002;50:587–597. doi: 10.1023/a:1019942200164. [DOI] [PubMed] [Google Scholar]
- Jensen LT, Ajua-Alemanji M, Cizewski Culatta V. The Saccharomyces cerevisiae high affinity phosphate transporter encoded by PHO84 also functions in manganese homeostasis. Journal of Biological Chemistry. 2003;278:42036–42040. doi: 10.1074/jbc.M307413200. [DOI] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Smirnoff N. Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. The Plant Journal. 2006;45:83–1000. doi: 10.1111/j.1365-313X.2005.02609.x. [DOI] [PubMed] [Google Scholar]
- Konno M, Ooishi M, Inoue Y. Temporal and positional relationships between Mn uptake and low-pH-induced root hair formation in Lactuca sativa cv. Grand Rapids seedlings. 2006;119:439–447. doi: 10.1007/s10265-006-0006-7. [DOI] [PubMed] [Google Scholar]
- Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB. The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Molecular Biology. 1999;40:37–44. doi: 10.1023/a:1026438615520. [DOI] [PubMed] [Google Scholar]
- Koshino-Kimura Y, Wada T, Tachibana T, Tsugeki R, Ishiguro S, Okada K. Regulation of CAPRICE transcription by MYB proteins for root epidermis differentiation in Arabidopsis. Plant and Cell Physiology. 2005;46:817–826. doi: 10.1093/pcp/pci096. [DOI] [PubMed] [Google Scholar]
- Kwak SH, Schiefelbein J. The role of the SCRAMBLED receptor-like kinase in patterning the Arabidopsis root epidermis. Developmental Biology. 2007;302:118–131. doi: 10.1016/j.ydbio.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Kwak SH, Shen R, Schiefelbein J. Positional signaling mediated by receptor-like kinase in Arabidopsis. Science. 2005;307:1111–1113. doi: 10.1126/science.1105373. [DOI] [PubMed] [Google Scholar]
- Ma Z, Bielenberg DG, Brown KM, Lynch JP. Regulation of root hair density by phosphorous availability in Arabidopsis thaliana. Plant, Cell and Environment. 2001;24:459–467. [Google Scholar]
- Marschner H. Mineral nutrition of higher plants. 2nd edn. Cambridge, UK: Academic Press; 1995. [Google Scholar]
- Masucci JD, Schiefelbein JW. Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. The Plant Cell. 1996;8:1505–1517. doi: 10.1105/tpc.8.9.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RF, Doherty ML, López-Marqués RL, Weimar T, Dupree P, Palmgren MG, Pittman JK, Williams LE. ECA3, a Golgi-localised P2A-type-ATPase, plays a crucial role in manganese nutrition in Arabidopsis. Plant Physiology. 2008;146:116–128. doi: 10.1104/pp.107.110817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N. A genome-wide transcriptional analysis using Arabidopsis thaliana affymetrix gene chips determined plant responses to phosphate deprivation. Proceedings of the National Academy of Sciences, USA. 2005;102:11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molas ML, Kiss JZ, Correll MJ. Gene profiling of the red light signaling pathway in roots. Journal of Experimental Botany. 2006;57:3217–3229. doi: 10.1093/jxb/erl086. [DOI] [PubMed] [Google Scholar]
- Müller M, Schmidt W. Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiology. 2004;134:409–419. doi: 10.1104/pp.103.029066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes and Development. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiter E, Montanini B, Gobert A, Pedas P, Husted S, Maathuis FJM, Blaudez D, Chalot M, Sanders D. A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Plant Biology. 2007;104:8532–8537. doi: 10.1073/pnas.0609507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryosuke S, Ryoko N, Kayoko I, et al. Analysis of bHLH (MYC) genes involved in root hair and trichome differentiation. 14th International Conference on Arabidopsis Research. 2003 [Google Scholar]
- Savage NS, Schmidt W. From priming to plasticity: the changing fate of rhizodermal cells. Bioessays. 2008;30:75–81. doi: 10.1002/bies.20693. [DOI] [PubMed] [Google Scholar]
- Schmidt W. Inner voices meet outer signals: the plasticity of rhizodermal cells. Plant Science. 2008;174:239–245. [Google Scholar]
- Schmidt W, Schikora A. Different pathways are involved in phosphate and iron stress-induced alterations of root epidermal cell development. Plant Physiology. 2001;125:2078–2084. doi: 10.1104/pp.125.4.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, Osawa H, Patel A, Gassmann W, Stacey G. Expression analyses of Arabidopsis oligopeptide transporters during seed germination, vegetative growth and reproduction. Planta. 2006;223:291–305. doi: 10.1007/s00425-005-0087-x. [DOI] [PubMed] [Google Scholar]
- Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to NRAMP gene. Proceedings of the National Academy of Sciences, USA. 2000;97:4991–4996. doi: 10.1073/pnas.97.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F, Leister D. The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. The Plant Journal. 2002;31:589–599. doi: 10.1046/j.1365-313x.2002.01381.x. [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. The Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog. CPC. Science. 1997;277:1113–1116. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- Wintz H, Fox T, Wu YY, Feng V, Chen W, Chang HS, Zhu T, Vulpe C. Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. Journal of Biological Chemistry. 2003;278:47644–47653. doi: 10.1074/jbc.M309338200. [DOI] [PubMed] [Google Scholar]
- Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay S. A basic helix-loop transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. The Plant Cell. 2005;17:1953–1966. doi: 10.1105/tpc.105.032060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.