Abstract
Gene expression in response to Cu stress in rice leaves was quantified using DNA microarray (Agilent 22K Rice Oligo Microarray) and real-time PCR technology. Rice plants were grown in hydroponic solutions containing 0.3 (control), 10, 45, or 130 μM of CuCl2, and Cu accumulation and photosynthesis inhibition were observed in leaves within 1 d of the start of treatment. Microarray analysis flagged 305 Cu-responsive genes, and their expression profile showed that a large proportion of general and defence stress response genes are up-regulated under excess Cu conditions, whereas photosynthesis and transport-related genes are down-regulated. The Cu sensitivity of each Cu-responsive gene was estimated by the median effective concentration value (EC50) and the range of fold-changes (F) under the highest (130 μM) Cu conditions (|log2F|130). Our results indicate that defence-related genes involved in phytoalexin and lignin biosynthesis were the most sensitive to Cu, and that plant management of abiotic and pathogen stresses has overlapping components, possibly including signal transduction.
Keywords: Copper-sensitivity, DNA microarray, excess copper stress, gene expression, Oryza sativa L
Introduction
Copper is an essential element for plants as a cofactor of enzymes such as plastocyanin, cytochrome c, and Cu/Zn-superoxide dismutase (Cu/Zn-SOD). Cu has a long history in agriculture as an antifungal agent, but in recent years it has been extensively released into the environment by human activities, such as industrial processes, pesticide application, and mining, that often cause environmental pollution. Exposure to excess Cu causes phytotoxicity by inhibiting key cellular processes, including photosynthesis and electron transport, lipid peroxidation, and disruption of protein functions due to Cu-binding to sulphhydryl groups (Sandmann and Böger, 1980; Yruela et al., 1993; Babu et al., 2001). Cu also induces the formation of reactive oxygen species (ROS) based on the Fenton or Haber–Weiss reactions (Halliwell and Gutteridge, 1989; Bartosz, 1997). A positive correlation between Cu exposure and the accumulation of hydroxy radicals has been reported in Arabidopsis (Drążkiewicz et al., 2004). However, plants have ROS scavenging systems that prevent or reduce cellular injury that can be caused by the generation of ROS in response to heavy metal stresses. Some ROS scavenging enzymes (e.g. SOD, CAT, APX) change their activities or transcription levels in response to excess Cu exposure (Luna et al., 1994; Weckx and Clijsters, 1996; Kurepa et al., 1997; Lombardi and Sebastiani, 2005).
Toxic concentrations of heavy metals can, in some cases, be reduced by chelation with metal ligands, or metal ions can be effluxed or sequestered, resulting in lower toxicity (Clemens, 2001; Hall, 2002). Metallothioneins and phytochelatins are well-known metal-binding peptides. Guo et al. (2003) reported that Arabidopsis metallothioneins play a role in Cu tolerance, homeostasis, and long-distance transport for sequestration.
Susceptibility to excess Cu stress varies with plant species. For instance, alfalfa and barley are highly tolerant to Cu stress, but rice and potato are less tolerant (Jones, 1998). In addition, rice is more susceptable to Cu toxicity than to other heavy metals, such as Ni, Co, and Zn (Chino, 1981). Although plant responses to heavy metal exposure have been widely investigated, it is still not completely understood how excess Cu affects the plant, nor how the plant copes with that stress at the gene expression level. Thus, a better understanding of how Cu stress affects gene expression in rice is important for providing an overall understanding of how higher plants adapt to heavy metal stress.
DNA microarrays are one of the most powerful tools for providing an overview of gene expression under various environmental conditions. Weber et al. (2006) examined transcriptome changes upon Cd2+ and Cu2+ exposure in roots of the Cd2+-hypertolerant metallophyte Arabidopsis halleri. Keinänen et al. (2007) identified genes that are up-regulated by CuSO4 exposure in a Cu-tolerant birch clone using macroarrays. The search for genes whose expression is modified by Cu stress has yielded a number of valuable tools that have been used to understand the Cu stress response. Completion of the rice genome sequence has made the comprehensive identification of Cu stress-responsive genes in this model monocot plant possible. The aim of this study is to identify genes which are affected directly or indirectly by toxic levels of Cu, some of which may be involved in ameliorating heavy metal, oxygen radical or other stress damage. Therefore, the effects of CuCl2 doses on rice leaf gene expression were examined using an Agilent 22K Rice Oligo Microarray. Three hundred and five Cu-responsive genes were selected which were either up- or down-regulated depending on CuCl2 dose, and the Cu sensitivity of the genes was analysed to determine what kind of functional genes and pathways might be critically involved in response to excess Cu.
Materials and methods
Plant culture
Rice plants (Oryza sativa L. cv. Nipponbare) were grown hydroponically (Kamachi et al., 1991) in an environment-controlled greenhouse with a photoperiod of 12 h light (25–28 °C) for 6–7 weeks. The basal nutrient solution was prepared as described by Kamachi et al. (1991) and the pH was adjusted to 5.5. Three rice plants were grown in each 500 ml plastic pot containing the nutrient solution, which was renewed once a week. Rice plants whose 8th leaf was fully expanded were used for experimental treatments.
Experimental design
Rice plants which had been grown as described above were treated with hydroponic solutions containing 10 μM, 45 μM, or 130 μM CuCl2. Treatment with the standard rice hydroponic solution containing 0.3 μM Cu was performed simultaneously as a control. Gas exchange measurements were performed using the fully expanded 8th leaf 24–30 h after the start of treatment, after which the leaves were harvested for RNA extraction. In addition, 8th leaf blades, the remainder of the shoot, and roots were separately collected for examining Cu contents.
Gas exchange measurements
Gas exchange was measured using a CIRAS-1 portable system (PP-system, Hitchin, Herts, UK). Measurements were made at a leaf temperature of 28 °C, and a PPFD of 800 μmol quanta m-2 s-1 at the position of the leaf in the chamber. CO2 and H2O partial pressures of the air exiting from the chamber were maintained at 38 Pa and 2.3 kPa, respectively. Irradiance was provided by a halogen lamp attached to an exclusive light unit (PP-system). Gas exchange parameters were calculated according to the equations of von Caemmerer and Farquhar (1981).
Measurement of Cu in rice tissues
For analyses of Cu concentrations in rice tissues, inductively coupled plasma mass spectrometry (ICP-MS) (Elan6100DRC; Perkin Elmer, Norwalk, CT, USA) was used. Rice plant tissues were dried for more than 3 d at 60 °C, followed by wet microwave digestion in 8 ml of concentrated HNO3 using a microwave sample preparation system (MultiWave-3000, Perkin Elmer). The digested samples were brought up to a volume of 50 ml with Milli-Q water and filtered through 5B filter paper (Advantec, Tokyo, Japan). For ICP-MS analysis, a portion of the filtered samples of leaf blade, sheath, and root were diluted 5-, 100-, and 100-fold with Milli-Q water, respectively.
RNA extraction and synthesis of Cy3- and Cy5-labelled cRNA
Total RNA was extracted from three different leaf samples per treatment using an RNeasy® Plant Mini Kit (Qiagen, Hilden, Germany). Cy3- and Cy5-labelled cRNA was prepared from 400 ng of total RNA from rice leaves, using a Low RNA Input Linear Amplification Kit (Agilent Technologies, Inc., Palo Alto, CA, USA) and Cy3- and Cy5-CTP (Perkin Elmer). Labelled cRNA was purified with RNeasy mini spin columns (Qiagen).
Microarray experiment and data analysis
A 22K Rice Oligo Microarray kit (Agilent Technologies) was used for microarray analysis. One microgram of Cy3-labelled cRNA was mixed with the same amount of Cy5-labelled cRNA and used for subsequent hybridization. Hybridization was carried out for 17 h with rotation at 60 °C. After washing, slides were scanned using a GenePix 4000A scanner (Axon Instruments Inc., Foster City, CA, USA) with 550 V and 680 V of PMT voltage for Cy3 and Cy5 detection, respectively, and quantified by Microarray Suite 2.0 (IPLab Spectrum Software, Scanalytics, Fairfax, VA, USA). Subsequent analysis was performed using GeneSpring 7 software (Agilent Technologies).
Genes which were up- or down-regulated with increasing Cu exposure concentration were selected as candidate Cu-responsive genes. Signal intensity, amplitude of expression fluctuation, and standard error of the mean F (F=the ratio of normalized data between experiment and control) were also considered. First, Cu-responsive genes meeting the criteria were selected as follows: the average signal intensity of the control RNA in the nine experiments (10 μM-1, 2, 3; 45 μM-1, 2, 3; 130 μM-1, 2, 3) was within the range 5×103 to 1×107; the F of triplicate samples under the 130 μM (130 μM-1, 2, 3) treatment were all significantly higher or lower than 1 (P < 0.01); and standard errors divided by the mean F in each treatment (10, 45, and 130 μM) were all less than 1. Second, up-regulated Cu-responsive genes were selected which met three additional criteria: the F value in each treatment was 130 μM >45 μM >10 μM; F was >2 in the 130 mM treatment; and F was >1 in both the 10 μM and 45 μM treatments. Third, down-regulated Cu-responsive genes were selected if they met the following additional criteria: F in each condition was 130 μM <45 μM <10 μM; F was <0.5 in the 130 μM treatment, but <1 in both the 10 μM and 45 μM treatments.
For estimating the Cu sensitivity of each Cu-responsive gene, median effective concentrations for F (EC50F), and the amplitude of expression change with the 130 μM treatment (|log2F|130) were determined. EC50Fs were calculated by probit analysis (Finney, 1978).
Descriptions of each Cu-responsive gene were annotated according to the TIGR database (http://www.tigr.org/tdb/e2k1/osa1/). In addition, Cu-responsive genes were classified into rough functional categories based on the Gene Ontology Classification database (http://www.geneontology.org/).
Quantitative real-time PCR
Total RNA was prepared using an RNeasy® plant Mini Kit (Qiagen) with RNase-free DNase I (Qiagen). Primers for each gene were designed using OLIGO Primer Analysis Software (Takara Bio Inc., Otsu, Japan). Primer sequences for the genes examined are summarized in Table 1. Accumulation levels of the target transcripts were analysed by real-time PCR with an ABIPRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) by monitoring amplification with SYBR-Green I dye (Applied Biosystems) as described in Takei et al. (2004).
Table 1.
List of primers used for quantitative real-time PCR
| Genes | Forward primers | Reverse primers |
| AK060724 | 5′-GCCGTTTGGTTTATAGTG-3′ | 5′-CCAAAATACAGTTTAGCGAC-3′ |
| AK062653 | 5′-CAAACTGCTCCTGCGGAAAG-3′ | 5′-CACACCCAGCACGACGG-3′ |
| AK099241 | 5′-CCTCTTCACGTCGGACCAC-3′ | 5′-ACCATGGCCTTCACGAACTT-3′ |
| AK058896 | 5′-CCAGCGTGAACTAATCTG-3′ | 5′-CAAGATACAAAGCGTGAGAC-3′ |
| AK101836 | 5′-TGGCCGTGTTGGAGCAATAC-3′ | 5′-CCAAAGCTTCTCGGAATGGG-3′ |
| AK070467 | 5′-ACAGCGGACGACACCACGAC-3′ | 5′-CGGCAGCCTCACGATGTTG-3′ |
| AK062796 | 5′-ACGAGCTACCAGTACCACTA-3′ | 5′-CGGCAACATGACATACAT-3′ |
| AK058551 | 5′-AGTGGCATTGTTACCGTGAT-3′ | 5′-CGCCTGGTGCTCGTC-3′ |
| AK060904 | 5′-TGCTGGCTTTTGTGGGTTTC-3′ | 5′-CGTGCCAAGCTCAAGGGTAG-3′ |
| AK065381 | 5′-CGATTTGGCGTGACGTGT-3′ | 5′-AATGCGCCACAAGATACCTG-3′ |
| AK067353 | 5′-CTGTTGATCCAGCGTTCTAC-3′ | 5′-TGAACCCGACGATAGCA-3′ |
| AK107472 | 5′-CGGTCGCAGGTGACGCT-3′ | 5′-TGATGAGGAGGGCGAACTTG-3′ |
Statistical analyses
Data were analysed by Dunnett's multiple comparison tests using SPSS software version 14.0J (SPSS Japan Inc., Tokyo, Japan).
Results and discussion
Effect of Cu treatment on Cu accumulation and photosynthesis in leaves
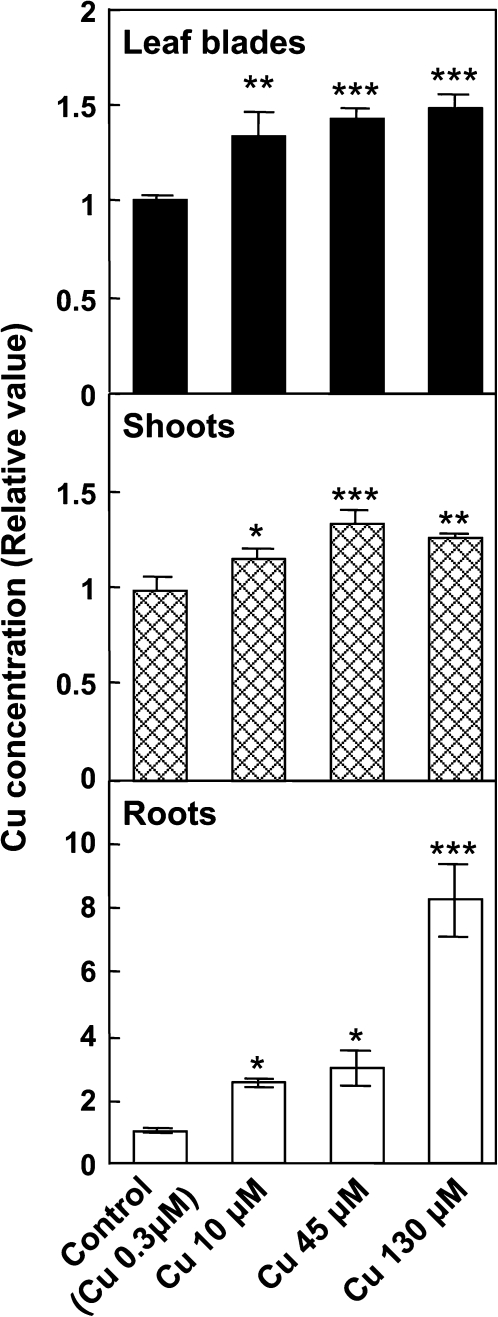
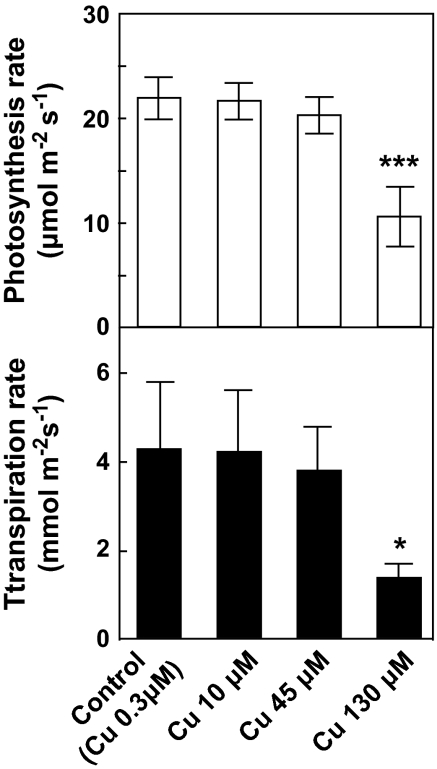
Application of CuCl2 to rice roots caused significant increases in Cu concentrations in the leaf blades, and shoots, as well as in the roots (Fig. 1). These results demonstrated that some of the Cu in hydroponic solution was absorbed by the roots and transported to the leaves. Photosynthetic and transpiration rates were significantly affected at 130 μM of CuCl2 at ambient CO2 levels (Fig. 2). The results confirm that Cu exposure above 45 μM is toxic to rice leaves. The photosynthetic decline at 130 μM (Fig. 2) was accompanied by a decrease in both the intercellular CO2 concentration and stomatal conductance (data not shown), suggesting that intercellular CO2 diffusion was inhibited as a result of stomatal closure. Compared with tissue Cu concentration (Fig. 1), the profile of photosynthetic activity under toxic conditions was consistent with root Cu content (Figs 1, 2). Root-to-shoot stress signalling via chemical components has been widely reported (e.g. ABA, Davies and Gowing, 1999; Sauter et al., 2001). ABA and other compounds may thus provide a mechanism by which root stress induced by excess Cu affects leaf photosynthetic activity by modulating stomatal apertures.
Fig. 1.
The relative concentrations of Cu in the leaf blades, shoots, and roots of rice. Values are means ±SD of three individual samples. Actual Cu concentrations in the control leaf blades, shoots, and roots are 139±3, 165±13, and 2180±90 μg g−1 dry weight, respectively. The statistical significance was determined by Dunnett's multiple comparison tests. Asterisks indicate a significant difference compared with control (*P < 0.05, **P < 0.01, ***P < 0.001).
Fig. 2.
Photosynthetic and transpiration rates after CuCl2 treatment. Values are the means ±SD of three individual leaves. The statistical significance was determined by Dunnett's multiple comparison tests. Asterisks indicate a significant difference compared with control (*P < 0.05, **P < 0.01, ***P < 0.001).
Selection of Cu-responsive genes with DNA microarray analysis
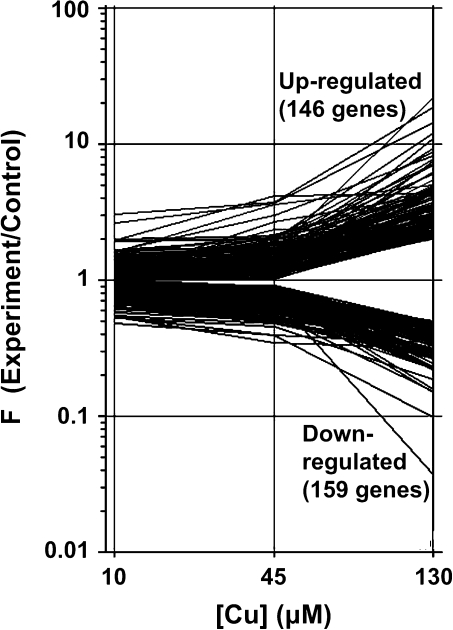
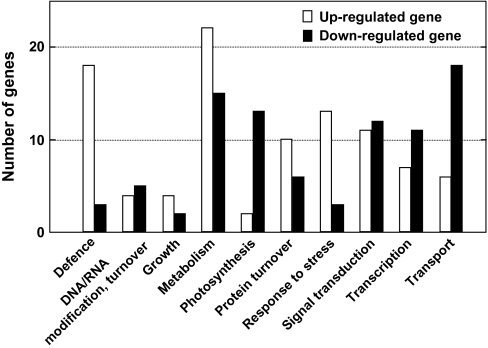
To gain insight into how excess Cu damages cellular processes in rice, a DNA microarray analysis was performed with RNA extracted from CuCl2-treated leaves. 146 genes were up-regulated and 159 were down-regulated in a dose-response manner (Fig. 3).
Fig. 3.
Expression profiles of Cu-responsive genes under excess Cu conditions.
Verification of microarray results by real-time PCR
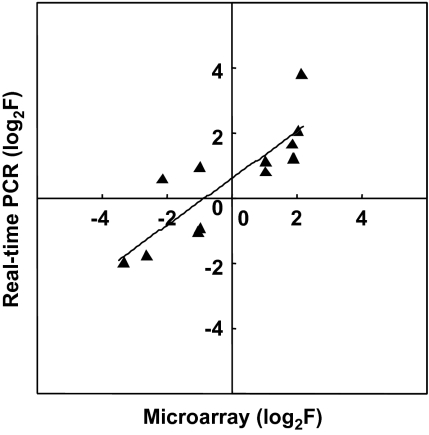
To verify the microarray results, real-time PCR was performed on 12 genes randomly selected from the Cu-responsive genes using the same RNA samples as were used in the microarray hybridization. There was a positive correlation between F from the 130 μM treatment and real-time PCR amplification (r2=0.717, Fig. 4), indicating that the microarray data are valid with respect to Cu dose response.
Fig. 4.
Confirmation of microarray signal ratios by real-time PCR. Real-time PCR analysis of 12 genes selected from Cu-responsive genes was performed with RNA extracted from rice leaves under control or 130 μM Cu treatment: y=0.718x + 0.605, r2=0.717.
Cu-responsive genes
The Cu-responsive genes showed some notable features, and both up- and down-regulated Cu-responsive genes are in each functional category (Fig. 5; a complete list is given in Supplementary Table S1 at JXB online). The number of defence and stress response genes greatly outnumbered the down-regulated genes (Fig. 5). Most of the defence-related genes are involved in the phenylpropanoid pathway for flavonoid, phytoalexin, and lignin biosynthesis (Table 2). Flavonoid accumulation in response to UV-B (Reddy et al., 1994), cold (Christie et al., 1994), and drought stresses (Balakumar et al., 1993) were previously reported. Flavonoids function as scavengers of ROS, and also prevent ROS formation by chelating metals (Scalbert, 1991; Ferrali et al., 1997; Heim et al., 2002). Phytoalexin and lignin biosynthesis are key responses to pathogen attack. CuCl2 treatment increases production of the rice phytoalexins sakuranetin and momilactone A (Rakwal et al., 1996). Our observation of up-regulated defence genes in response to Cu confirms its role as an abiotic elicitor (Graham, 1980).
Fig. 5.
Functional classification of Cu-responsive genes. Up-regulated genes are represented by empty bars and down-regulated genes by filled bars.
Table 2.
Expression profiles of Cu-responsive genes under excess Cu treatment conditions (10, 45, and 130 μM of CuCl2)
| Probe ID | Full length cDNA | Locus_ID | Description |
F (experiment/control) |
||
| Cu-exposure (μM) |
||||||
| 10 | 45 | 130 | ||||
| Defence (up-regulated) | ||||||
| A_71_P105870 | AK060724 | LOC_Os02g41630 | Phenylalanine ammonia-lyase | 1.02 | 1.70 | 2.01 |
| A_71_P105867 | AK068993 | LOC_Os02g41680 | Phenylalanine ammonia-lyase | 1.01 | 1.42 | 5.01 |
| A_71_P105871 | AK102817 | LOC_Os02g41630 | Phenylalanine ammonia-lyase | 1.19 | 1.82 | 2.26 |
| A_71_P113211 | AK067801 | LOC_Os04g43800 | Phenylalanine ammonia-lyase | 1.34 | 1.78 | 4.61 |
| A_71_P126860 | AK099443 | LOC_Os11g02440 | Chalcone-flavonone isomerase | 1.38 | 1.89 | 2.19 |
| A_71_P104485 | AK070746 | LOC_Os02g08420 | Dihydroflavonol-4-reductase | 1.07 | 1.32 | 2.23 |
| A_71_P119630 | AK065515 | LOC_Os08g38910 | Caffeoyl-CoA O-methyltransferase 2 | 1.19 | 2.12 | 3.18 |
| A_71_P115157 | AK104994 | LOC_Os05g25640 | Trans-cinnamate 4-mono-oxygenase | 1.21 | 1.42 | 2.43 |
| A_71_P123533 | AK069308 | LOC_Os10g02880 | O-methyltransferase ZRP4 | 1.17 | 1.19 | 4.27 |
| A_71_P122641 | AK072740 | LOC_Os09g17560 | O-methyltransferase ZRP4 | 1.03 | 1.59 | 21.92 |
| A_71_P111602 | AK065090 | LOC_Os04g59190 | Peroxidase 2 precursor | 1.38 | 1.79 | 7.16 |
| A_71_P113417 | AK106200 | LOC_Os05g04500 | Peroxidase 63 precursor | 1.62 | 2.97 | 8.13 |
| A_71_P117837 | AK072862 | LOC_Os07g47990 | Peroxidase 2 precursor | 1.34 | 1.37 | 3.50 |
| A_71_P103756 | AK099241 | LOC_Os01g22370 | Peroxidase 1 precursor | 1.22 | 1.48 | 4.33 |
| A_71_P120304 | AK069503 | LOC_Os08g02110 | Peroxidase 47 precursor | 1.20 | 1.31 | 3.30 |
| A_71_P117839 | AK073202 | LOC_Os07g48020 | Peroxidase 2 precursor | 1.18 | 1.69 | 9.18 |
| A_71_P103305 | AK107822 | LOC_Os01g72170 | Glutathione S-transferase | 1.21 | 1.24 | 2.07 |
| A_71_P125246 | AK062653 | LOC_Os11g47809 | Metallothionein-like protein 1 | 1.37 | 1.48 | 4.06 |
| Defence (down-regulated) | ||||||
| A_71_P103051 | AK103129 | LOC_Os01g53330 | Anthocyanidin 5,3-O-glucosyltransferase | 0.80 | 0.58 | 0.29 |
| A_71_P119739 | AK067868 | LOC_Os08g07880 | Phosphopantothenate-cysteine ligase | 0.61 | 0.48 | 0.43 |
| A_71_P103162 | AK062796 | LOC_Os01g74300 | Metallothionein-like protein type 2 | 0.92 | 0.82 | 0.16 |
| Response to stress (up-regulated) | ||||||
| A_71_P112980 | AK100788 | LOC_Os04g34600 | ABA/WDS induced protein | 1.47 | 1.88 | 2.17 |
| A_71_P115472 | AK107775 | LOC_Os06g07030 | Dehydration responsive element binding protein | 1.17 | 1.64 | 4.35 |
| A_71_P126985 | AK062422 | LOC_Os09g35010 | Dehydration-responsive element-binding protein 1B | 1.24 | 1.86 | 2.27 |
| A_71_P118699 | AK106022 | LOC_Os07g44250 | Disease resistance response protein 206 | 1.08 | 1.40 | 3.35 |
| A_71_P111503 | AK071013 | LOC_Os04g41680 | Endochitinase A precursor | 1.09 | 1.14 | 2.45 |
| A_71_P114512 | AK060312 | LOC_Os05g42230 | ER6 protein | 1.03 | 1.16 | 2.37 |
| A_71_P124122 | AK065000 | LOC_Os10g22520 | Glucan 1,3-β-glucosidase precursor | 1.14 | 1.40 | 4.57 |
| A_71_P121735 | AK061896 | LOC_Os09g30418 | Heat shock protein 81-3 | 1.37 | 1.78 | 2.39 |
| A_71_P126129 | AK066682 | LOC_Os12g14440 | Jasmonate-induced protein | 1.63 | 1.99 | 11.92 |
| A_71_P103425 | AK062520 | LOC_Os01g24710 | Salt stress-induced protein | 1.16 | 1.28 | 6.20 |
| A_71_P114369 | AK070138 | LOC_Os05g28740 | Universal stress protein | 1.54 | 1.55 | 2.54 |
| A_71_P114262 | AK065866 | LOC_Os05g15770 | Xylanase inhibitor protein 2 precursor | 2.01 | 2.09 | 4.34 |
| A_71_P114261 | AK062114 | LOC_Os05g15770 | Xylanase inhibitor protein 2 precursor | 1.66 | 2.01 | 4.20 |
| Response to stress (down-regulated) | ||||||
| A_71_P117292 | AK099477 | LOC_Os06g47800 | Disease resistance protein RGA3 | 0.77 | 0.61 | 0.27 |
| A_71_P118794 | AK065027 | LOC_Os07g01630 | Disease resistance response protein 206 | 0.95 | 0.77 | 0.49 |
| A_71_P122593 | AK060664 | LOC_Os09g37600 | Erwinia-induced protein 1 | 0.94 | 0.76 | 0.43 |
| Photosynthesis (up-regulated) | ||||||
| A_71_P114297 | AK100910 | LOC_Os05g50380 | Glucose-1-phosphate adenylyltransferase large subunit, chloroplast precursor | 1.39 | 1.47 | 3.93 |
| A_71_P116411 | AK101836 | LOC_Os06g49110 | Δ-Aminolevulinic acid dehydratase, chloroplast precursor | 1.39 | 1.73 | 2.03 |
| Photosynthesis (down-regulated) | ||||||
| A_71_P105099 | AK062994 | LOC_Os02g51470 | ATP synthase delta chain, chloroplast precursor | 0.96 | 0.87 | 0.45 |
| A_71_P115841 | AK060904 | LOC_Os06g21590 | Chlorophyll a-b binding protein 6A, chloroplast precursor | 0.81 | 0.71 | 0.48 |
| A_71_P125058 | AK061295 | LOC_Os11g13890 | Chlorophyll a-b binding protein M9, chloroplast precursor | 0.70 | 0.63 | 0.37 |
| A_71_P118301 | AK109399 | LOC_Os07g37550 | Chlorophyll a-b binding protein of LHCII type III, chloroplast precursor | 0.69 | 0.58 | 0.40 |
| A_71_P121584 | AK109203 | LOC_Os09g32620 | Chloroplastic quinone-oxidoreductase | 0.76 | 0.73 | 0.48 |
| A_71_P101901 | AK066307 | LOC_Os12g10604 | Cytochrome b/b6/petB family protein | 0.61 | 0.54 | 0.32 |
| A_71_P126393 | AK059037 | LOC_Os12g08770 | Photosystem I reaction centre subunit N, chloroplast precursor | 0.73 | 0.67 | 0.40 |
| A_71_P114565 | AK066345 | LOC_Os05g43310 | Photosystem II reaction centre W protein, chloroplast precursor | 0.76 | 0.76 | 0.40 |
| A_71_P108389 | AK058858 | LOC_Os03g55720 | Plastoquinol-plastocyanin reductase | 0.94 | 0.73 | 0.36 |
| A_71_P117917 | AK069170 | LOC_Os07g36080 | Oxygen-evolving enhancer protein 3-1, chloroplast precursor | 0.93 | 0.80 | 0.30 |
| A_71_P117916 | AK058793 | LOC_Os07g36080 | Oxygen-evolving enhancer protein 3-1, chloroplast precursor | 0.70 | 0.61 | 0.29 |
| A_71_P120166 | AK058551 | LOC_Os08g25734 | Glucose-1-phosphate adenylyltransferase small subunit, chloroplast precursor | 0.86 | 0.84 | 0.47 |
| A_71_P124217 | AK110705 | LOC_Os06g39730 | Ribulose bisphosphate carboxylase large chain, catalytic domain containing protein | 0.76 | 0.73 | 0.50 |
| Transport (up-regulated) | ||||||
| A_71_P105105 | AK108711 | LOC_Os02g34580 | Ammonium transporter 2 | 1.06 | 1.35 | 2.47 |
| A_71_P119764 | AK065217 | LOC_Os08g03350 | LHT1 | 1.19 | 1.36 | 2.46 |
| A_71_P117869 | AK105311 | LOC_Os07g33780 | PDR-like ABC transporter | 1.09 | 1.17 | 2.61 |
| A_71_P127448 | AK108393 | LOC_Os05g27010 | Peptide transporter PTR2 | 1.17 | 1.33 | 2.22 |
| A_71_P103242 | AK063835 | LOC_Os01g45640 | Tat pathway signal sequence family protein | 1.02 | 1.04 | 7.43 |
| A_71_P100920 | AK103784 | LOC_Os01g31980 | Transparent testa 12 protein | 1.10 | 1.43 | 2.56 |
| Transport (down-regulated) | ||||||
| A_71_P106018 | AK100650 | LOC_Os02g44980 | Amino acid transport protein | 0.99 | 0.76 | 0.45 |
| A_71_P116013 | AK107472 | LOC_Os06g12320 | Amino acid/polyamine transporter II | 0.80 | 0.68 | 0.22 |
| A_71_P115705 | AK072617 | LOC_Os06g03700 | Oligopeptide transporter 9 | 0.87 | 0.84 | 0.24 |
| A_71_P104541 | AK065840 | LOC_Os02g46460 | Peptide transporter PTR2 | 0.74 | 0.57 | 0.29 |
| A_71_P122896 | AK066937 | LOC_Os10g42900 | Peptide transporter PTR2 | 0.77 | 0.73 | 0.49 |
| A_71_P114702 | AK070558 | LOC_Os05g34010 | Peptide transporter PTR2 | 0.90 | 0.77 | 0.48 |
| A_71_P102553 | AK066793 | LOC_Os01g50616 | Phosphatidylinositol transporter/ transporter | 0.75 | 0.68 | 0.44 |
| A_71_P119359 | AK066067 | LOC_Os07g46780 | Tyrosine-specific transport protein | 0.67 | 0.64 | 0.41 |
| A_71_P123937 | AK111957 | LOC_Os10g38910 | ABC-type Co2+ transport system, permease component | 0.91 | 0.73 | 0.44 |
| A_71_P115940 | AK105826 | LOC_Os06g30730 | ATPase, coupled to transmembrane movement of substances | 0.99 | 0.75 | 0.48 |
| A_71_P100064 | AK065048 | LOC_Os01g17214 | Carbohydrate transporter/sugar transporter/transporter | 0.86 | 0.63 | 0.22 |
| A_71_P123327 | AK071193 | LOC_Os10g35140 | Permeases of the drug/metabolite transporter | 0.82 | 0.68 | 0.50 |
| A_71_P104342 | AK071338 | LOC_Os02g56510 | Phosphate transporter 1 | 0.59 | 0.54 | 0.42 |
| A_71_P117558 | AK067110 | LOC_Os06g29790 | Phosphate transporter 1 | 0.55 | 0.48 | 0.27 |
| A_71_P112325 | AK070018 | LOC_Os04g38026 | Sugar transport protein 5 | 0.74 | 0.63 | 0.37 |
| A_71_P108667 | AK067353 | LOC_Os03g09930 | Sulphate transporter 2.1 | 0.53 | 0.39 | 0.10 |
| A_71_P112060 | AK072809 | LOC_Os04g55800 | Sulphate transporter 3.3 | 0.90 | 0.84 | 0.49 |
| A_71_P116372 | AK063490 | LOC_Os06g36450 | Transporter like protein | 0.92 | 0.66 | 0.39 |
Values are means of fold-change (F) calculated from triplicate data of different leaves. The descriptions of each gene were annotated according to the TIGR database (http://www.tigr.org/tdb/e2k1/osa1/), and were classified into rough functional categories based on the Gene Ontology Classification database (http://www.geneontology.org/).
Plants synthesize metal-binding polypeptides, such as metallothionein and phytochelatin, whose apparent function is to maintain cellular metal concentration homeostasis by sequestering and detoxifying excess metal ions. In this study, two genes encoding metallothionein-like proteins were up- and down-regulated by excess Cu (AK062653 and AK062796, respectively; Table 2). At present, the physiological meaning of the differential response of the two genes to excess Cu is not clear. The gene products could be different in their ligand affinity or specificity, and thus functionally specialized to respond to different levels of Cu stress. Cu homeostasis may also be regulated by Cu-containing proteins which act as Cu sinks under excess Cu conditions. Abdel-Ghany et al. (2005) reported that CuSO4 treatment enhanced the production of Cu/Zn-SOD and plastocyanin proteins in Arabidopsis. In this study, the set of Cu-responsive genes contained monocopper oxidase-like protein and L-ascorbate oxidase, which were both up-regulated (see Supplementary Table S1 at JXB online) by excess Cu treatment.
Our results also showed the up-regulation of genes which are known to respond to abiotic stresses such as drought, salt or heat shock (Table 2), suggesting a partial overlap of the signal transduction pathways coping with metal exposure, drought, heat shock or salinity. The dehydration-responsive element (DRE) is involved in response to drought, salt, and cold stresses in Arabidopsis (Yamaguchi-Shinozaki and Shinozaki, 1994), and overexpression of the trans-acting factor DREB confers tolerance to these stresses in transgenic Arabidopsis (Nakashima and Yamaguchi-Shinozaki, 2006). Our results imply that DREB genes may also play a role in Cu tolerance in rice leaves. Because a gene encoding ABA/WDS-induced protein was also up-regulated by excess Cu, metal ions like Cu may also affect the ABA-dependent signal transduction pathway.
The number of photosynthesis and transport-related genes, on the other hand, greatly outnumbered the up-regulated genes (Fig. 5). Generally, photosynthesis-related genes are induced by light and are influenced by circadian rhythms. However, they were often down-regulated under abiotic stresses such as low temperatures (Hahn and Walbot, 1989), heat and/or drought (Rizhsky et al., 2002), salinity (Allakhverdiev et al., 2002; Kore-eda et al., 2004), excess light (Teramoto et al., 2002), or by signal transduction factors, including ROS (Vandenabeele et al., 2003; op den Camp et al., 2003), jasmonate (JA) (Reinbothe et al., 1993) and hexose (Sheen, 1994). These results demonstrate that excess Cu also represses the photosynthetic system at the genetic level (Fig. 5; Table 2) as well as at the physiological level (Fig. 2). Schiavon et al. (2007) reported that excess Cu decreases transcript levels of plastocyanin in Arabidopsis, an observation which is supported by our results.
Cu treatment also repressed transport-related genes (Fig. 4). Transport systems are indispensable for keeping metal concentrations in equilibrium in plant species. Metal homeostasis is maintained by chelating, effluxing or sequestering the potentially toxic ions (Clemens, 2001; Hall, 2002). Sancenón et al. (2003) identified a five-member family of Cu transporters (CORT1 to CORT5) in Arabidopsis. In addition, some of the P1B-type heavy metal ATPases (HMAs) have a role in Cu transport in rice (Williams and Mills, 2005; Lee et al., 2007). Excess CuSO4 decreased Arabidopsis transcription of PAA1 and PAA2 (Schiavon et al., 2007), both of which function as Cu transporters for Cu delivery in chloroplasts (Abdel-Ghany et al., 2005). In our results, the genes encoding amino acid and peptide transporters were conspicuously down-regulated (Table 2). Wintz et al. (2003) reported that AtOPT3, a potential oligopeptide transporter of Arabidopsis, is involved in Cu transport. It is, however, still unclear whether the amino acid and peptide transporter genes among the Cu-responsive genes are involved in Cu homeostasis.
Sensitivity of Cu-responsive genes
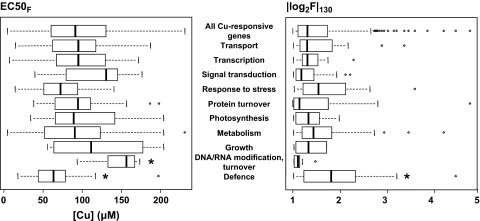
Each of the Cu-responsive genes responds distinctively to Cu concentration, and the fluctuation range of Cu-responsive expression also varied under the 130 μM Cu treatment conditions. These variations can be attributed to ‘Cu-sensitivity’, which can be calculated from the median effective concentration values (EC50F) and fluctuation of expression in the highest Cu concentration (|log2F|130) (see Supplementary Table S1 at JXB online). EC50Fs varied from 4.86 μM to 230 μM with a mean value of 97.9 μM. |log2F|130 ranged from 1.00 to 4.75, with a mean of 1.52 (Fig. 6). Compared with the average value of all Cu-responsive genes, the EC50F and |log2F|130 of defence-related genes are significantly lower and higher than others, respectively, at P < 0.05 (Fig. 6), indicating that the defence-related genes are highly Cu-sensitive to lower concentrations of Cu, and that their expression varies greatly with exposure to Cu.
Fig. 6.
Boxplots of EC50F (left panel) and |log2F|130 (right panel) in each functional category. The empty box indicates the interquartile (25–75%) range. Bars across the boxes represent the median value. Whiskers below and above the box indicate the range of values within 1.5 times the value of the upper or lower edge of the box. Circles represent outliers. The statistical significance of differences was tested by Dunnett's multiple comparison tests. Asterisks indicate significant differences with average values of all Cu-responsive genes (*P < 0.05).
Within the defence-related genes, phytoalexin and lignin biosynthesis pathway genes (phenylalanine ammonia-lyase, caffeoyl-CoA O-methyltransferase, trans-cinnamate 4-mono-oxygenase, O-methyltransferase ZRP4, peroxidase) were particularly sensitive (Table 3). Although one gene encoding a metallothionein-like protein was up-regulated, and the other was down-regulated, their Cu-sensitivities were both higher than many other defence-related genes (Table 3; see Supplementary Table S1 at JXB online). Thus, sequestering mechanisms for heavy metals are also acutely responsive to Cu.
Table 3.
Cu-sensitivity of defence-related genes and some expected genes involved in pathogen resistance mechanisms among the up-regulated Cu-responsive genes
| Probe ID | Full length cDNA | Locus_ID | Description | EC50F | |log2F|130 |
| Genes categorized into ‘defence’ | |||||
| A_71_P105870 | AK060724 | LOC_Os02g41630 | Phenylalanine ammonia-lyase | 96.84 | 1.01 |
| A_71_P105867 | AK068993 | LOC_Os02g41680 | Phenylalanine ammonia-lyase | 66.59 | 2.32 |
| A_71_P105871 | AK102817 | LOC_Os02g41630 | Phenylalanine ammonia-lyase | 79.62 | 1.17 |
| A_71_P113211 | AK067801 | LOC_Os04g43800 | Phenylalanine ammonia-lyase | 39.45 | 2.20 |
| A_71_P126860 | AK099443 | LOC_Os11g02440 | Chalcone-flavonone isomerase | 75.43 | 1.13 |
| A_71_P104485 | AK070746 | LOC_Os02g08420 | Dihydroflavonol-4-reductase | 116.44 | 1.16 |
| A_71_P119630 | AK065515 | LOC_Os08g38910 | Caffeoyl-CoA O-methyltransferase 2 | 50.11 | 1.67 |
| A_71_P115157 | AK104994 | LOC_Os05g25640 | Trans-cinnamate 4-mono-oxygenase | 100.41 | 1.28 |
| A_71_P123533 | AK069308 | LOC_Os10g02880 | O-methyltransferase ZRP4 | 77.19 | 2.09 |
| A_71_P122641 | AK072740 | LOC_Os09g17560 | O-methyltransferase ZRP4 | 44.06 | 4.45 |
| A_71_P111602 | AK065090 | LOC_Os04g59190 | Peroxidase 2 precursor | 32.57 | 2.84 |
| A_71_P113417 | AK106200 | LOC_Os05g04500 | Peroxidase 63 precursor | 18.11 | 3.02 |
| A_71_P117837 | AK072862 | LOC_Os07g47990 | Peroxidase 2 precursor | 65.73 | 1.81 |
| A_71_P103756 | AK099241 | LOC_Os01g22370 | Peroxidase 1 precursor | 54.26 | 2.12 |
| A_71_P120304 | AK069503 | LOC_Os08g02110 | Peroxidase 47 precursor | 78.35 | 1.72 |
| A_71_P117839 | AK073202 | LOC_Os07g48020 | Peroxidase 2 precursor | 39.26 | 3.20 |
| A_71_P103305 | AK107822 | LOC_Os01g72170 | Glutathione S-transferase | 197.47 | 1.05 |
| A_71_P125246 | AK062653 | LOC_Os11g47809 | Metallothionein-like protein 1 | 50.71 | 2.02 |
| Expected genes involved in pathogen resistance mechanism | |||||
| A_71_P126555 | AK066737 | LOC_Os12g37260 | Lipoxygenase 2.1, chloroplast precursor | 4.86 | 4.20 |
| A_71_P107746 | AK061537 | LOC_Os03g57970 | Lipid transfer protein | 17.29 | 2.32 |
| A_71_P125078 | AK061288 | LOC_Os11g24070 | Non-specific lipid-transfer protein 1 precursor | 76.80 | 1.42 |
| A_71_P125472 | AK058896 | LOC_Os11g02369 | Non-specific lipid-transfer protein 2 precursor | 32.98 | 1.84 |
| A_71_P115043 | AK062463 | LOC_Os05g47700 | Non-specific lipid-transfer protein precursor | 25.88 | 2.24 |
| A_71_P101377 | AK067257 | LOC_Os01g03340 | Bowman–Birk-type bran trypsin inhibitor precursor | 38.00 | 2.80 |
| A_71_P101369 | AK070467 | LOC_Os01g03310 | Bowman–Birk-type bran trypsin inhibitor precursor | 62.48 | 1.89 |
| A_71_P111503 | AK071013 | LOC_Os04g41680 | Endochitinase A precursor | 140.75 | 1.29 |
| A_71_P124122 | AK065000 | LOC_Os10g22520 | Glucan 1,3-β-glucosidase precursor | 60.04 | 2.19 |
| A_71_P126129 | AK066682 | LOC_Os12g14440 | Jasmonate-induced protein | 21.62 | 3.58 |
| A_71_P114262 | AK065866 | LOC_Os05g15770 | Xylanase inhibitor protein 2 precursor | 14.42 | 2.12 |
| A_71_P114261 | AK062114 | LOC_Os05g15770 | Xylanase inhibitor protein 2 precursor | 25.71 | 2.07 |
In gene categories other than defence-related, Cu-sensitivity did not differ significantly from the average of all Cu-responsive genes, but DNA, RNA modification, and turnover category genes had relatively lower Cu sensitivity.
Sensitivity of defence mechanisms to pathogens and their roles under excess Cu stress
Our results showed that defence-related genes are strikingly up-regulated, with the highest Cu-sensitivity. Considering that Cu is an abiotic elicitor that induces resistance against pathogen attack (Graham, 1980), this result is understandable. According to van Loon and van Strien (1999), there are 14 families of PR proteins (PR-1–14), including β-1,3-glucanase, chitinase, peroxidase, proteinase-inhibitor, and lipid-transfer protein. High Cu sensitivity was also evident in genes encoding glucan β-1,3-glucosidase (β-1,3-glucanase), Bowman–Birk-type bran trypsin inhibitor, lipid-transfer protein, and xylanase inhibitor (Table 3). Furthermore, the sensitivity of JA-induced protein and chloroplast-located lipoxygenase were extraordinarily high (Table 3). Thus, the responses of general defence mechanism genes to Cu treatment suggest either some role in handling Cu stress, or that signal transduction is shared by the stress-response systems. In analysing the Cu-tolerant birch, Keinänen et al. (2007) isolated genes which were suggested to contribute to Cu tolerance mechanisms, including genes encoding HR-induced protein, chitinase, and lipoxygenase. This indicated the involvement of disease defence mechanisms in Cu tolerance.
Concluding remarks
Genome-wide analysis using DNA microarray technology demonstrated the broad response of rice genes to excess Cu. Our results suggest that Cu treatment particularly affected genes involved in defence, various abiotic stresses, photosynthesis, and transport. Further analysis demonstrated the range of defence-related genes for Cu-sensitivity, which suggests one aspect of the Cu-responsive mechanism, and that the defence response has an essential role in the stress response to excess Cu treatment. Defence-related genes could thus be effective targets for increasing tolerance to Cu. Alternatively, the role of Cu as an antifungal agent may act in part by inducing defence-response genes, as well as by inhibiting the pathogen.
Recently, gene expression profiles have been used as indicators of various kinds of stressors, such as environmental pollutants (Lettieri, 2006). The potential use of Cu-responsive genes as an indicator of environmental Cu-pollution was reported previously (Sudo et al., 2006). This study suggests the additional potential of using defence-related genes as biomarkers for very small amounts of Cu-pollution because of their acute sensitivity.
In this study, the focus was on analysing expression profiles in leaves 1 d after inducing Cu stress. Thus, early events, which are indicative of a direct response to some systemic signal that is expressed de novo, or triggered in roots in response to the increase of heavy metal ion concentrations, or to the direct effects of leaf intracellular concentrations, might have been overlooked. Further analysis, including a time-course covering this earlier period, could provide us with information which complements our new understanding of the gene regulatory events that occur in the 1 d timeframe for adaptation to Cu stress.
Supplementary data
Supplementary data for this article are available at JXB online.
Table S1. Expression profiles of all Cu-responsive genes grown with 10, 45, or 130 μM of CuCl2.
Supplementary Material
Acknowledgments
We thank N Makita for her assistance to register our microarray data. This work was supported by a grant for a Leading Project (a project to design sustainable management and recycling systems of biomass, general and industrial wastes) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
References
- Abdel-Ghany SE, Müller-Moulé P, Niyogi KK, Pilon M, Shikanai T. Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. The Plant Cell. 2005;17:1233–1251. doi: 10.1105/tpc.104.030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allakhverdiev SI, Nishiyama Y, Miyairi S, Yamamoto H, Inagaki N, Kanesaki Y, Murata N. Salt stress inhibits the repair of photo-damaged photosystem II by suppressing the transcription and translation of psbA genes in Synechocystis. Plant Physiology. 2002;130:1443–1453. doi: 10.1104/pp.011114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu TS, Marder JB, Tripuranthakam S, Dixon DG, Greenberg BM. Synergistic effects of a photooxidized polycyclic aromatic hydrocarbon and copper on photosynthesis and plant growth: evidence that in vivo formation of reactive oxygen species is a mechanism of copper toxicity. Environmental Toxicology and Chemistry. 2001;20:1351–1358. doi: 10.1897/1551-5028(2001)020<1351:seoapp>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Balakumar T, Hani Babu Vincent V, Paliwal K. On the interaction of UV-B radiation (280–315 nm) with water stress in crop plants. Physiologia Plantarum. 1993;87:217–222. [Google Scholar]
- Bartosz G. Oxidative stress in plants. Acta Physiologiae Plantarum. 1997;19:47–64. [Google Scholar]
- Chino M. Metal stress in rice plants. In: Kitagishi K, Yamane I, editors. Heavy metal pollution in soils of Japan. Tokyo, Japan: Japan Scientific Societies Press; 1981. pp. 65–80. [Google Scholar]
- Christie PJ, Alfenito MR, Walbot V. Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta. 1994;194:541–549. [Google Scholar]
- Clemens S. Molecular mechanisms of plant metal tolerance and homeostasis. Planta. 2001;212:475–486. doi: 10.1007/s004250000458. [DOI] [PubMed] [Google Scholar]
- Davies WJ, Gowing DJG. Plant responses to small perturbations in soil water status. In: Press MC, Scholes JD, Barker MG, editors. Physiological plant ecology. Oxford: Blackwells; 1999. pp. 67–89. [Google Scholar]
- Drążkiewicz M, Skórzyńska-Polit E, Krupa Z. Copper-induced oxidative stress and antioxidant defence in Arabidopsis thaliana. BioMetals. 2004;17:379–387. doi: 10.1023/b:biom.0000029417.18154.22. [DOI] [PubMed] [Google Scholar]
- Ferrali M, Signorini C, Caciotti B, Sugherni L, Ciccoli L, Giachetti D, Comporti M. Protection against oxidative damage of erythrocyte membrane by the flavonoid quercetin and its relation to iron chelating activity. FEBS Letters. 1997;416:123–129. doi: 10.1016/s0014-5793(97)01182-4. [DOI] [PubMed] [Google Scholar]
- Finney DJ, editor. Statistical methods in biological assay. 3rd edn. London: Charles Griffin and Company, Ltd; 1978. [Google Scholar]
- Graham RD. Susceptibility to powdery mildew of wheat plants deficient in copper. Plant and Soil. 1980;56:181–185. [Google Scholar]
- Guo WJ, Bundithya W, Goldsbrough PB. Characterization of the Arabidopsis metallothionein gene family: tissue-specific expression and induction during senescence and in response to copper. New Phytologist. 2003;159:369–381. doi: 10.1046/j.1469-8137.2003.00813.x. [DOI] [PubMed] [Google Scholar]
- Hahn M, Walbot V. Effect of cold-treatment on protein synthesis and mRNA levels in rice leaves. Plant Physiology. 1989;91:930–938. doi: 10.1104/pp.91.3.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JL. Cellular mechanisms for heavy metal detoxification and tolerance. Journal of Experimental Botany. 2002;53:1–11. [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC, editors. Free radicals in biology and medicine. 2nd edn. Oxford: Clarendon Press; 1989. [Google Scholar]
- Heim KE, Tagliaferro AR, Bobilya DJ. Flavonoid antioxidants: chemistry, metabolism and structure-activity relationships. Journal of Nutritional Biochemistry. 2002;13:572–584. doi: 10.1016/s0955-2863(02)00208-5. [DOI] [PubMed] [Google Scholar]
- Jones JB., Jr . The micronutrients. In: Jones JB Jr, editor. Plant nutrition manual. Boca Raton, Florida USA: CRC Press; 1998. pp. 55–76. [Google Scholar]
- Kamachi K, Yamaya T, Mae T, Ojima K. A role for glutamine synthetase in the remobilization of leaf nitrogen during natural senescence in rice leaves. Plant Physiology. 1991;96:411–417. doi: 10.1104/pp.96.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinänen SI, Hassinen VH, Kärenlampi SO, Tervahauta AI. Isolation of genes up-regulated by copper in a copper-tolerant birch (Betula pendula) clone. Tree Physiology. 2007;27:1243–1252. doi: 10.1093/treephys/27.9.1243. [DOI] [PubMed] [Google Scholar]
- Kore-eda S, Cushman MA, Akselrod I, Bufford D, Fredrickson M, Clark E, Cushman JC. Transcript profiling of salinity stress responses by large-scale expressed sequence tag analysis in Mesembryanthemum crystallinum. Gene. 2004;341:83–92. doi: 10.1016/j.gene.2004.06.037. [DOI] [PubMed] [Google Scholar]
- Kurepa J, van Montagu M, Inzé D. Expression of sodCp and sodB genes in Nicotiana tabacum: effects of light and copper excess. Journal of Experimental Botany. 1997;48:2007–2014. [Google Scholar]
- Lee S, Kim Y-Y, Lee Y, An G. Rice P1B-type heavy metal ATPase, OsHMA9, is a metal efflux protein. Plant Physiology. 2007;145:831–842. doi: 10.1104/pp.107.102236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettieri T. Recent applications of DNA microarray technology to toxicology and ecotoxicology. Environmental Health Perspectives. 2006;114:4–9. doi: 10.1289/ehp.8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi L, Sebastiani L. Copper toxicity in Prunus cerasifera: growth and antioxidant enzymes reponses of in vitro-grown plants. Plant Science. 2005;168:797–802. [Google Scholar]
- Luna CM, González CA, Trippi VS. Oxidative damage caused by an excess of copper in oat leaves. Plant and Cell Physiology. 1994;35:11–15. [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K. Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plant. Physiologia Plantarum. 2006;126:62–71. [Google Scholar]
- op den Camp RGL, Przybyla D, Ochsenbein C, et al. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. The Plant Cell. 2003;15:2320–2332. doi: 10.1105/tpc.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakwal R, Tamogami S, Kodama O. Role of jasmonic acid as a signal molecule in copper chloride-elicited rice phytoalexin production. Bioscience, Biotechnology and Biochemistry. 1996;60:1046–1048. [Google Scholar]
- Reddy VS, Goud KV, Sharma R, Reddy AR. Ultraviolet-B-responsive anthocyanin production in rice cultivar is associated with a specific phase of phenylalanine ammonia lyase biosynthesis. Plant Physiology. 1994;105:1059–1066. doi: 10.1104/pp.105.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S, Reinbothe C, Parthier B. Methyl jasmonate-regulated translation of nuclear-encoded chloroplast proteins in barley (Hordeum vulgare L. cv. Salome) Journal of Biological Chemistry. 1993;268:10606–10611. [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiology. 2002;130:1143–1151. doi: 10.1104/pp.006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancenón V, Puig S, Mira H, Thiele DJ, Peñarrubia L. Identification of a copper transporter family in Arabidopsis thaliana. Plant Molecular Biology. 2003;51:577–587. doi: 10.1023/a:1022345507112. [DOI] [PubMed] [Google Scholar]
- Sandmann G, Böger P. Copper-mediated lipid peroxidation processes in photosynthetic membranes. Plant Physiology. 1980;66:797–800. doi: 10.1104/pp.66.5.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter A, Davies WJ, Hartung W. The long-distance abscisic acid signal in the droughted plant: the fate of the hormone on its way from root to shoot. Journal of Experimental Botany. 2001;52:1991–1997. doi: 10.1093/jexbot/52.363.1991. [DOI] [PubMed] [Google Scholar]
- Scalbert A. Antimicrobial properties of tannins. Phytochemistry. 1991;30:3875–3883. [Google Scholar]
- Schiavon M, Zhang L, Abdel-Ghany SE, Pilon M, Malagoli M, Pilon-Smits EAH. Variation in copper tolerance in Arabidopsis thaliana accessions Columbia, Landsberg erecta and Wassilewskija. Physiologia Plantrum. 2007;129:342–350. [Google Scholar]
- Sheen J. Feedback control of gene expression. Photosynthesis Research. 1994;39:427–438. doi: 10.1007/BF00014596. [DOI] [PubMed] [Google Scholar]
- Sudo E, Itouga M, Yoshida K, Ono Y, Sakakibara H. Mitigation of Cu-toxicity through ‘bryo-filtration’: an evaluation using rice leaf photosynthesis and gene expression profile. Hikobia. 2006;14:419–429. [Google Scholar]
- Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H. AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant and Cell Physiology. 2004;45:1053–1062. doi: 10.1093/pcp/pch119. [DOI] [PubMed] [Google Scholar]
- Teramoto H, Nakamori A, Minagawa J, Ono T. Light-intensity-dependent expression of Lhc gene family encoding light-harvesting chlorophyll a/b proteins of photosystem II in Chlamydomonas reinhardtii. Plant Physiology. 2002;130:325–333. doi: 10.1104/pp.004622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S, Van Der Kelen K, Dat J, et al. A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proceedings of the National Academy of Sciences, USA. 2003;100:16113–16118. doi: 10.1073/pnas.2136610100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, van Strien EA. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiological and Molecular Plant Pathology. 1999;55:85–97. [Google Scholar]
- von Caemmerer S, Farquhar GD. Some relationships between the biochemistry of photosynthesis and the gas exchange of leaves. Planta. 1981;153:376–387. doi: 10.1007/BF00384257. [DOI] [PubMed] [Google Scholar]
- Weber M, Trampczynska A, Clemens S. Comparative transcriptome analysis of toxic metal reponses in Arabodopsis thaliana and the Cd2+-hypertolerant facultative metallophyte Arabidopsis halleri. Plant, Cell and Environment. 2006;29:950–963. doi: 10.1111/j.1365-3040.2005.01479.x. [DOI] [PubMed] [Google Scholar]
- Weckx JEJ, Clijsters HMM. Oxidative damage and defence mechanisms in primary leaves of Phaseolus vulgaris as a result of root assimilation of toxic amounts of copper. Physiologia Plantarum. 1996;96:506–512. [Google Scholar]
- Williams LE, Mills RF. P1B-ATPase: an ancient family of transition metal pumps with diverse functions in plants. Trends in Plant Science. 2005;10:491–502. doi: 10.1016/j.tplants.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Wintz H, Fox T, Wu Y-Y, Feng V, Chen W, Chang H-S, Zhu T, Vulpe C. Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. Journal of Biological Chemistry. 2003;278:47644–47653. doi: 10.1074/jbc.M309338200. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yruela I, Alfonso M, Oritz de Zaerte I, Montoya G, Picorel R. Precise location of the Cu(II)-inhibitory binding site in higher plant and bacterial photosynthetic reaction centers as probed by light-induced absorption change. Journal of Biological Chemistry. 1993;268:1684–1689. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.